Significance
Recombination and chromosome remodeling are tightly regulated to ensure accurate chromosome segregation during meiosis. However, how these important steps are regulated remains poorly understood. Here, we show that RAD-51 foci, which form at the sites of meiotic DNA double-strand breaks (DSBs), exhibit a biased distribution toward off-centered positions along Caenorhabditis elegans chromosomes, and we identify two meiotic roles for chromatin-associated protein HIM-17. During early prophase I, HIM-17 regulates the distribution of DSB-dependent RAD-51 foci and crossovers on chromosomes. During late prophase I, HIM-17 promotes the normal localization of protein phosphatases GSP-1/GSP-2 and potentially GSP-1 phosphorylation, thereby antagonizing Aurora B kinase/AIR-2. Therefore, HIM-17 plays distinct roles during meiotic progression that converge to promote normal chromosome remodeling and accurate chromosome segregation.
Keywords: HIM-17, DNA double-strand breaks, crossovers, late prophase I remodeling, meiosis
Abstract
The position of recombination events established along chromosomes in early prophase I and the chromosome remodeling that takes place in late prophase I are intrinsically linked steps of meiosis that need to be tightly regulated to ensure accurate chromosome segregation and haploid gamete formation. Here, we show that RAD-51 foci, which form at the sites of programmed meiotic DNA double-strand breaks (DSBs), exhibit a biased distribution toward off-centered positions along the chromosomes in wild-type Caenorhabditis elegans, and we identify two meiotic roles for chromatin-associated protein HIM-17 that ensure normal chromosome remodeling in late prophase I. During early prophase I, HIM-17 regulates the distribution of DSB-dependent RAD-51 foci and crossovers on chromosomes, which is critical for the formation of distinct chromosome subdomains (short and long arms of the bivalents) later during chromosome remodeling. During late prophase I, HIM-17 promotes the normal expression and localization of protein phosphatases GSP-1/2 to the surface of the bivalent chromosomes and may promote GSP-1 phosphorylation, thereby antagonizing Aurora B kinase AIR-2 loading on the long arms and preventing premature loss of sister chromatid cohesion. We propose that HIM-17 plays distinct roles at different stages during meiotic progression that converge to promote normal chromosome remodeling and accurate chromosome segregation.
Sexual reproduction depends on the cell division program of meiosis to generate haploid gametes from diploid germ cells, which will combine during fertilization to form a diploid zygote. During meiosis I, several coordinated events take place, including homologous chromosome pairing, synapsis, recombination, late chromosome remodeling, and stepwise loss of sister chromatid cohesion (SCC), which are critical for accurate segregation of homologs at meiosis I and sister chromatids at meiosis II. Meiotic recombination is initiated by the formation of programmed DNA double-strand breaks (DSBs) by the topoisomerase-like protein Spo11 and its cofactors (1–6). Programmed DSBs are made in excess to ensure that at least one crossover (CO) is formed between each pair of homologous chromosomes via homologous recombination (7). In Caenorhabditis elegans, due to absolute CO interference, a single CO is formed per homologous chromosome pair (8). This single CO is made at an off-center position on the holocentric chromosome to generate a cruciform-shaped asymmetric bivalent with distinct short and long arms (9–12). Establishment of these two distinct domains during late prophase I is critical for the relocalization of a specific set of proteins, which is required for the regulated stepwise loss of SCC (11, 13–16). The mechanisms promoting the off-centered distribution of COs remain unknown.
Upon CO formation in C. elegans, a wave of chromosome remodeling is initiated, which involves the asymmetric disassembly of the synaptonemal complex (SC) along the longest distance from the site of the CO to a chromosome end (long arm of the bivalent) and retention of SC proteins on the short arm of the bivalent (14). In contrast, axis-associated proteins HTP-1, HTP-2, and LAB-1 are lost from the short arm of the bivalent and relocalize to the long arm of the bivalent (13, 16). At late diakinesis, SC proteins on the short arm are then replaced by the highly conserved Aurora B kinase, AIR-2. The localization of AIR-2 to the short arm facilitates the phosphorylation of the meiotic cohesin subunit REC-8 licensing its cleavage by separase at anaphase I and subsequent segregation of homologous chromosomes to opposite spindle poles (17, 18). Immunofluorescence analysis has revealed the characteristic ring-like localization of AIR-2 to the short arms of the bivalents in the last two oocytes at diakinesis in C. elegans (-2 and -1 oocytes, which are positioned most proximally to the spermatheca; Fig. 1A), with the strongest signal detected in the -1 oocyte (16). AIR-2 localization is restricted to the short arm by the PP1/Glc7 protein phosphatases GSP-1 and GSP-2, which have been proposed to be targeted to the long arm by LAB-1 to antagonize AIR-2 loading along that chromosome subdomain (16, 18, 19). This function of LAB-1 is akin to Shugoshin, which protects SCC near the pericentromeric regions of mammalian chromosomes (19, 20). However, it remained unclear whether additional factors play a role in regulating arm identity and chromosome remodeling in late prophase I.
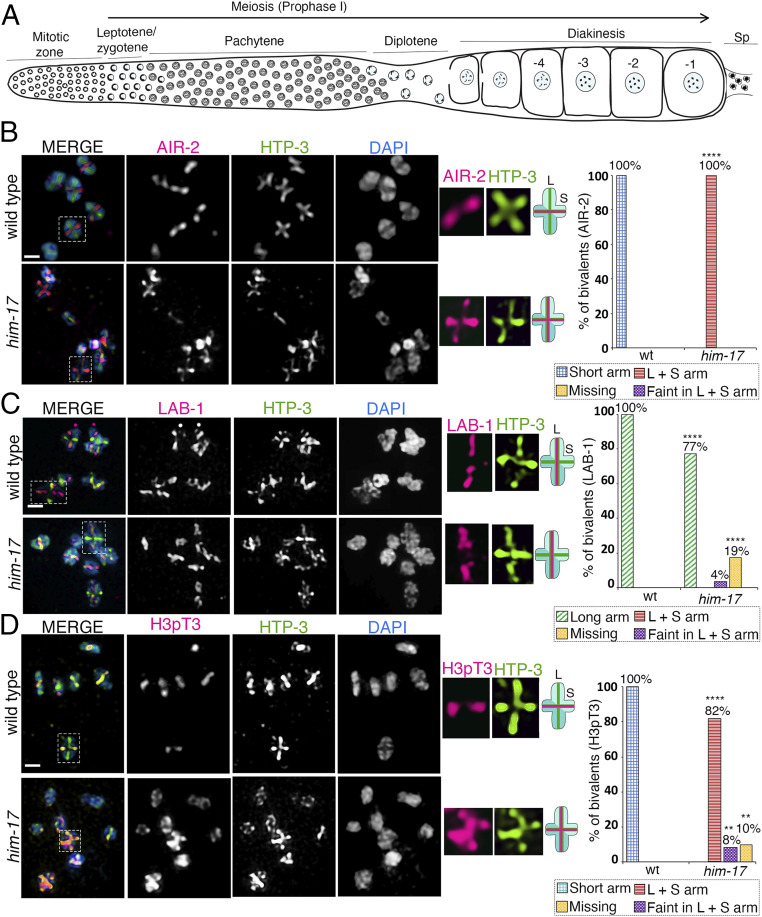
Fig. 1.
HIM-17 regulates AIR-2 and H3pT3 localization on the short arm of the bivalents. High-magnification images of -1 oocytes at diakinesis in wild type and him-17 mutant are pictured. (A) Schematic representation of an adult C. elegans hermaphrodite gonad arm indicating nuclei in different stages of meiotic progression. The most proximal oocyte next to the spermatheca (Sp) is referred to as the -1 oocyte. (B) Costaining with anti–AIR-2 (magenta), anti–HTP-3 (green), and DAPI (blue) indicates that AIR-2 is not restricted to the short arm of the bivalents in him-17 mutants compared to wild type. Dashed boxes indicate the bivalents for which AIR-2 and HTP-3 localization are shown at higher magnification. (Middle) The chromosome configuration observed at this stage showing distinct short (S) and long (L) arms, and the localization of AIR-2 (magenta) and HTP-3 (green) for each genotype. (Right) Histogram indicates the percentage of bivalents at diakinesis with AIR-2 restricted to the short arm, mislocalized to both long and short arm, faint but in both long and short arms, or missing. A total of 102 and 97 bivalents were analyzed from the gonad arms of 18 and 112 animals for wild type and him-17, respectively, from three biological replicates. (C) Costaining with anti–LAB-1 (magenta), anti–HTP-3 (green), and DAPI (blue) indicates that a large number of bivalents in him-17 mutants exhibit LAB-1 localization restricted to the long arm as observed in wild type. (Right) Histogram indicates the percentage of bivalents at diakinesis with LAB-1 restricted to the long arm, long and short arm, faint but in both long and short arms, or missing on the chromosomes. A total of 90 and 96 bivalents from the gonad arms of 15 and 103 animals were analyzed for wild type and him-17, respectively, from three biological replicates. (D) Costaining with anti-H3pT3 (magenta), anti–HTP-3 (green), and DAPI (blue) indicates that H3pT3 signal is frequently observed on both long and short arms of the bivalents in him-17 mutants in contrast to the restricted localization only to the short arms observed in wild type. (Right) Histogram indicates the percentage of bivalents at diakinesis with H3pT3 localization restricted to the short arm, mislocalized to both long and short arms, faint but on both long and short arms, or missing on the chromosomes. A total of 99 and 72 bivalents from the gonad arms of 17 and 69 animals were analyzed for wild type and him-17, respectively, from three biological replicates. ****P < 0.0001, **P < 0.005 by the two-tailed Fisher’s exact test. (Scale bars, 2 μm.) For complete/additional statistical analysis, see Dataset S1.
A functional genomics approach to identify regulators of AIR-2 localization to the short arm of the bivalents revealed the chromatin-associated protein HIM-17 (this study). HIM-17 is required for meiotic DSB formation and histone H3 dimethylation on lysine 9 (H3K9me2) on germline chromatin (21). HIM-17 is a nematode-specific protein that carries six THAP DNA-binding domains and is implicated in chromatin regulation through genetic interaction with the retinoblastoma-like protein LIN-35/Rb. Moreover, THAP domain-containing proteins present in other organisms, including humans, play important roles in cell proliferation, apoptosis, chromosome segregation, cell cycle, chromatin modification, and transcriptional regulation (21–23). him-17 null mutants exhibit a reduction/absence of CO formation which is rescued by introduction of exogenous DSBs by gamma-irradiation, indicating that all other factors required for DNA damage repair and CO formation are present in him-17 mutants (21).
Here, we found two roles for HIM-17 in regulating the distribution of DSB-dependent RAD-51 foci and COs along chromosomes in early meiotic prophase I and in the establishment of short arm identity on bivalents during late prophase I chromosome remodeling. In him-17 mutants, AIR-2 and Haspin-mediated phosphorylation of H3T3 (H3pT3), which is required for the recruitment of AIR-2 to the short arm of the bivalents, are mislocalized to both long and short arms. In contrast, the restricted localization of LAB-1 to the long arm is not affected. Consistent with previous studies, him-17 mutants exhibit fewer DSBs and lower levels of CO formation. In addition, analysis of RAD-51 foci along computationally straightened wild-type chromosomes revealed higher levels of RAD-51 foci at off-center positions compared to the center of the chromosomes. This enrichment for DSB-dependent RAD-51 foci at off-center positions may partly explain the off-center bias in CO distribution observed in wild-type worms. We found that the distribution of DSB-dependent RAD-51 foci and COs is altered, resulting in a significant number of these events being made on the center of the chromosomes in him-17 mutants compared to wild type. Further, we found that GSP-1 and GSP-2 expression and localization on the surface of the bivalents were decreased in oocytes at diakinesis in him-17 mutants. Interestingly, the phosphorylation state of GSP-1 partially rescues the late prophase I chromosome remodeling defect observed in him-17 mutants. We propose a model whereby HIM-17 ensures normal chromosome remodeling in late prophase I, and therefore subsequent stepwise loss of SCC and accurate chromosome segregation, by two mechanisms: 1) regulating DSB and CO position along the chromosome, which is critical for establishment of short arm identity, and 2) preventing the loading of AIR-2 on the long arm of the bivalent by regulating the expression, localization, and possibly phosphorylation of GSP-1 and GSP-2.
Results
HIM-17 Regulates Short Arm Identity during Late Prophase I Chromosome Remodeling.
To identify novel regulators of chromosome remodeling and the short/long arm identity of bivalents, we conducted a targeted RNA interference (RNAi) screen in transgenic animals expressing GFP-tagged AIR-2. In wild type, AIR-2 localization is restricted to the short arm of the bivalents in the diakinesis stage oocyte (14, 17). Using high-resolution microscopy, we scored for the mislocalization of AIR-2::GFP upon RNAi-mediated depletion of candidate genes. We screened 381 germline-enriched genes defined as such based on microarray analysis (24) and identified 44 potential candidates that affect AIR-2 localization. Details of the screen will be described elsewhere. him-17 was one of the first candidate genes identified as a potential regulator of AIR-2 localization from this screen, given that upon RNAi-mediated depletion of HIM-17, AIR-2::GFP is mislocalized to both short and long arms of the bivalents instead of being restricted to the short arms. Of note, both bivalents and univalents were observed in oocytes at diakinesis, since HIM-17 has been implicated in promoting normal numbers of meiotic DSBs (21). To verify the RNAi phenotype, we analyzed a him-17 null mutant for altered AIR-2 localization. Similar to RNAi depletion, we observed both bivalents and univalents in diakinesis oocytes, with 67% of the oocytes (n = 78) carrying two or more bivalents and 15% exhibiting only univalents (SI Appendix, Fig. S1A). These are higher levels of bivalents than reported by Reddy and Villeneuve (21) and consistent with our previous findings (25) (see Materials and Methods). We found that AIR-2 is mislocalized to both long and short arms in 100% (n = 97) of the bivalents detected in him-17 mutant worms (Fig. 1B). However, LAB-1 localization remained restricted to the long arm of the bivalents, and therefore was indistinguishable from wild type, in 77% (74/96) of the bivalents in -1 oocytes in him-17 mutants and was either detected as a faint signal on both long and short arms in 4% (4/96) or missing in 19% (18/96) of the bivalents (Fig. 1C). The latter category might be explained by the limited number of DSBs/COs made in him-17 mutants, given that a subthreshold level of CO designation within the nucleus (less than four) results in markers that are normally restricted to the short arm localizing instead along both long and short arms on those bivalents with a CO-designated site, which could be accompanied by LAB-1 depletion along the entire bivalent (26, 27). Since LAB-1 localization is not altered in the majority of bivalents in him-17 mutants, we set out to determine whether overall short arm identity or only AIR-2 localization is altered in him-17 mutants. H3pT3 has been shown to promote recruitment of AIR-2 onto the short arm of the bivalent in diakinesis stage oocytes (18). In wild type, H3pT3 is restricted to the short arm of the bivalents (18), whereas in him-17 mutants, we found that H3pT3 signal intensity is increased and no longer restricted to the short arms (Fig. 1D). H3pT3 was mislocalized to both short and long arms in 82% (59/72) of bivalents in -1 oocytes, and H3pT3 signal was either faint on both long and short arms in 8% (6/72) or missing in 10% (7/72) of bivalents (Fig. 1D). Altogether, our data suggest that the short arm identity of the bivalents in diakinesis stage oocytes is altered in him-17 mutants although LAB-1 localization to the long arm is not affected in the majority of the bivalents.
Exogenous DSBs Partially Rescue the Short Arm Identity Defect in him-17 Mutants.
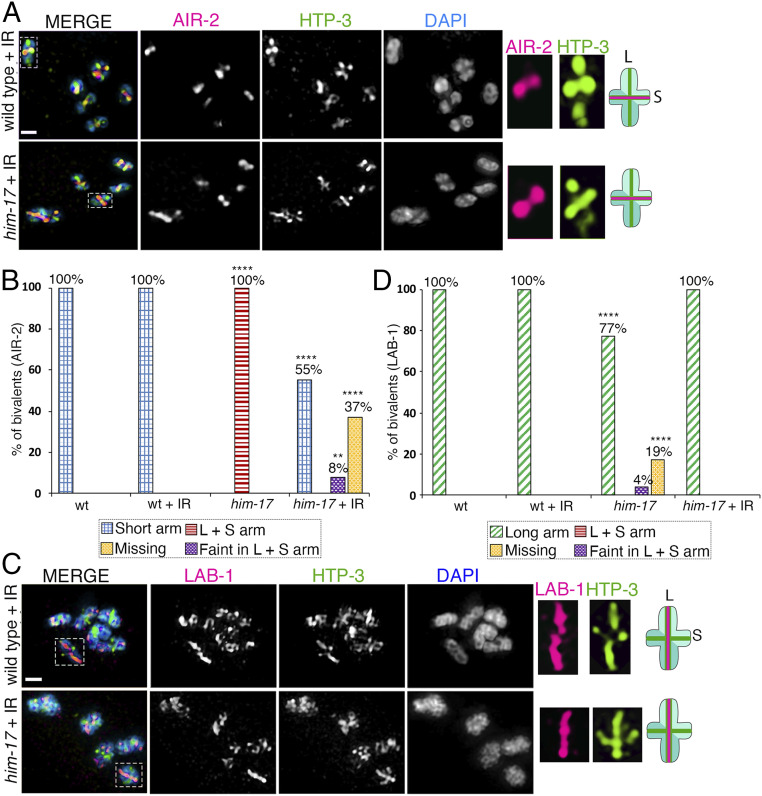
HIM-17 has been shown to be required for DSB formation, and fewer DSB-dependent RAD-51 foci are detected in a him-17 null mutant (21). To test whether lower levels of DSB formation may be the cause of short arm identity problems in him-17 mutants, we exposed ∼18 h post-L4 larval stage wild type and him-17 mutant animals to 60 Gy of gamma-irradiation (γ-IR), thereby introducing exogenous DSBs. Their germlines were dissected ∼24 h post IR and immunostained with anti–AIR-2. This dose of γ-IR fully rescued bivalent formation (six bivalents) in 97% of the -1 oocytes in the him-17 mutant (SI Appendix, Fig. S1A). Furthermore, it resulted in 39.1 and 39.8 DSB-dependent RAD-51 foci per nucleus (∼6.5 and ∼6.6 RAD-51 foci per chromosome) in wild type and him-17 mutant worms, respectively (SI Appendix, Fig. S1B). We found that AIR-2 localization is rescued, being once again restricted to the short arms upon γ-IR in him-17 mutants compared to unexposed mutants in 55% (48/88) of bivalents in -1 oocytes at diakinesis. AIR-2 signal is either faint on both long and short arms in 8% (7/88) of bivalents or missing in 37% (33/88) of bivalents (Fig. 2 A and B). In addition, we analyzed LAB-1 localization after γ-IR in wild type and him-17 mutant animals. We found that LAB-1 is localized to the long arms in 100% (103/103) of bivalents after γ-IR in him-17 mutants (Fig. 2 C and D). Altogether, our data suggest that the AIR-2 mislocalization observed in him-17 mutants is partially rescued by introduction of exogenous DSBs with γ-IR.
Fig. 2.
AIR-2 and LAB-1 localization on bivalents in oocytes at diakinesis in wild type and him-17 mutants after exogenous DSB formation. High-magnification images of diakinesis stage oocytes in wild type and him-17 mutant after exogenous DSB formation by γ-IR (60 Gy). (A) Immunolocalization of AIR-2 (magenta), HTP-3 (green), and DAPI (blue) in -1 oocytes at diakinesis indicates that AIR-2 is restricted to the short arm of the bivalents in him-17 mutants, similar to wild type, after IR treatment. A total of 106 and 88 bivalents from the gonad arms of 20 and 16 animals were analyzed for wild type and him-17, respectively, from three biological replicates. Dashed boxes indicate the bivalents for which AIR-2 and HTP-3 localization are shown at higher magnification. (Middle) The chromosome configuration observed at this stage showing distinct short (S) and long (L) arms, and the localization of AIR-2 (magenta) and HTP-3 (green) for each genotype following IR treatment. (B) Histogram indicates the percentage of bivalents at diakinesis with AIR-2 restricted to the short arm, mislocalized to both long and short arms, faint but on both long and short arms, or missing from the chromosomes in wild type and him-17 mutants with and without IR treatment. (C) Immunolocalization of LAB-1 (magenta), HTP-3 (green), and DAPI (blue) in -1 oocytes at diakinesis. A total of 91 and 103 bivalents from the gonad arms of 16 and 18 animals were analyzed for wild type and him-17, respectively, from three biological replicates. (D) Histogram indicates the percentage of bivalents at diakinesis with LAB-1 restricted to the long arms, mislocalized to both long and short arms, faint but on both long and short arms, or missing from the chromosomes in wild type and him-17 mutants with and without IR. ****P < 0.0001, **P < 0.005 by the two-tailed Fisher’s exact test. (Scale bars, 2 μm.) For complete/additional statistical analysis, see Dataset S1.
Most DSB-Dependent RAD-51 Foci Occur along the Arms of the Chromosomes in Wild Type, whereas Increased Numbers of RAD-51 Foci and COs Are Made at the Center of the Chromosomes in him-17 Mutants.
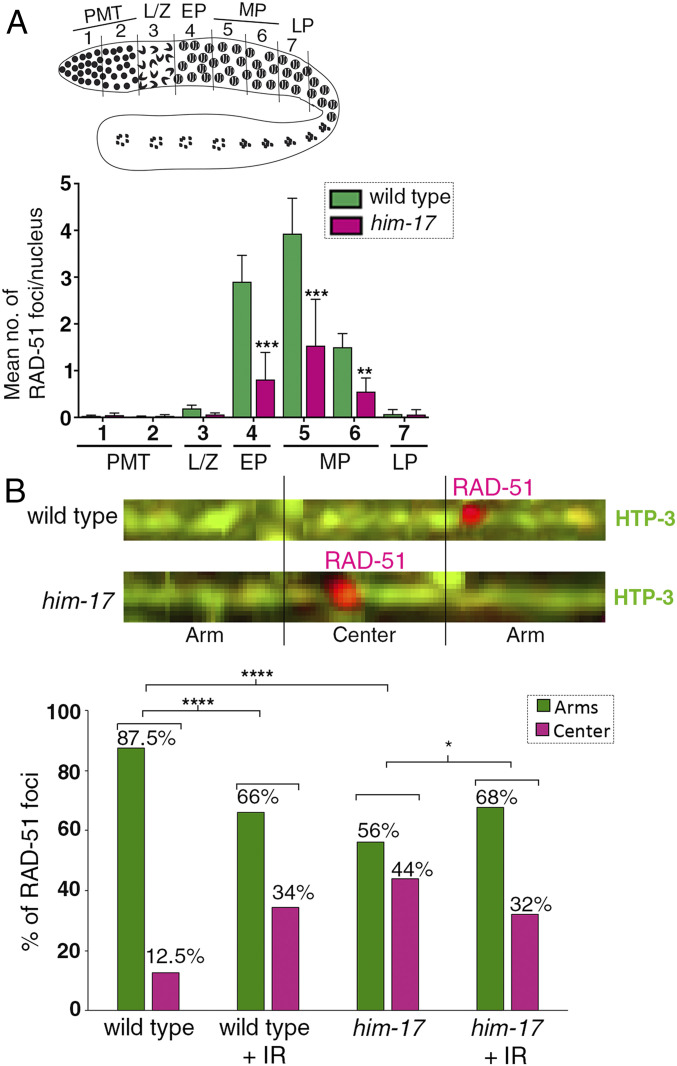
We previously proposed that, in C. elegans, meiotic programmed DSBs are made in excess to ensure that at least one CO is made at the preferred off-centered position to promote normal chromosome remodeling, which in turn is critical for accurate chromosome segregation (28). Since introduction of exogenous DSBs by γ-IR can rescue chromosome remodeling in him-17 mutants, we hypothesized there might be an alteration in the distribution of DSBs in him-17 mutants. To test this, we coimmunostained whole-mounted germlines from wild type and him-17 mutants with RAD-51 and HTP-3 antibodies to mark DSB repair sites and chromosome axes, respectively. Consistent with previous findings, we observed a significant decrease in the levels of RAD-51 foci in him-17 null mutants compared to wild type (Fig. 3A). Computational straightening of chromosomes and analysis of the distribution of RAD-51 foci along chromosomes divided into three equal parts (two “arm” regions flanking a middle region referred to as the “center”), each corresponding to 1/3 of the total length of the chromosomes, revealed that in wild-type animals, around 87.5% of DSB-dependent RAD-51 foci are located on the arms, and 12.5% are located on the center region of the chromosomes (Fig. 3B). This enrichment at off-centered regions is in contrast to the even distribution of RAD-51 foci previously detected along chromosomes in a rad-54 mutant background where DSBs are formed, ends are resected, and RAD-51 associates with the 3′ single-strand overhangs but is not removed due to lack of RAD-54 (29). In him-17 mutants, 56% of DSB-dependent RAD-51 foci are detected on the arms of the chromosomes and 44% on the center of the chromosomes. Interestingly, in him-17 mutants, despite a 37% decrease in the total number of DSB-dependent RAD-51 foci formed compared to wild type, we observed a 32% increase in RAD-51 foci on the center and a 31% decrease on the arms of the chromosomes relative to wild type (SI Appendix, Fig. S2). Therefore, in wild-type animals, DSB-dependent RAD-51 foci are enriched along the arms of the chromosomes, a biased distribution that had not been previously defined, and there is a shift in the position of RAD-51 foci to the less-favored center region of the chromosomes in him-17 mutants.
Fig. 3.
Quantification of RAD-51 foci and distribution on the chromosomes in wild type and him-17 mutants. (A) Schematic representation of a C. elegans germline indicating the different zones scored for the number of RAD-51 foci/nucleus. (Bottom) Histogram depicts the mean number of RAD-51 foci/nucleus observed in different zones of him-17 mutant germlines compared to wild type. X-axis shows the position along the germline. PMT: premeiotic tip (germ cells in mitosis), L/Z: meiotic nuclei in leptotene/zygotene stage, EP: meiotic nuclei in early pachytene, MP: meiotic nuclei in midpachytene, and LP: meiotic nuclei in late pachytene. The number of nuclei scored per zone is indicated in Dataset S1. ***P < 0.001 and **P < 0.02 by the two-tailed Mann–Whitney test, 95% CI. (B, Top) Representative images of linearized chromosomes costained with anti–HTP-3 (green) to trace chromosome axes and anti–RAD-51 (magenta) to mark DSB repair sites. Linearized chromosomes from pachytene nuclei were divided into three equal portions referred to as arms and center. Only chromosomes with clear start and end points were scored using PRIISM software as in (25). (Bottom) Histogram indicates the distribution of RAD-51 foci on the center versus the arm regions of the chromosomes in wild type, wild type + IR, him-17, and him-17 + IR. A total of 115, 33, 113, and 36 chromosomes from pachytene stage nuclei from the gonad arms of 35, 6, 33, and 6 animals were analyzed for wild type, wild type + IR, him-17, and him-17 + IR, respectively. ****P < 0.0001, *P < 0.024 by the two-tailed Fisher’s exact test.
To test whether there is any bias in the pattern of RAD-51 foci in him-17 mutants compared to wild type upon introduction of exogenous DSBs, we irradiated 22 to 24 h post-L4 worms with 60 Gy of γ-IR and scored the number of RAD-51 foci on computationally straightened chromosomes from whole-mounted germlines 1 h post-IR. There is a significant difference between wild type with and without IR given that with IR, RAD-51 foci decrease from 87.5 to 66% on the arms and increase from 12.5 to 34% on the center of the chromosomes. However, there is no difference in the pattern of DSB-dependent RAD-51 foci formation between wild type and him-17 mutants after IR with both exhibiting an enrichment for RAD-51 foci on the arm regions (66 and 68%, respectively) compared to the center of the chromosomes (34 and 32%, respectively) (Fig. 3B).
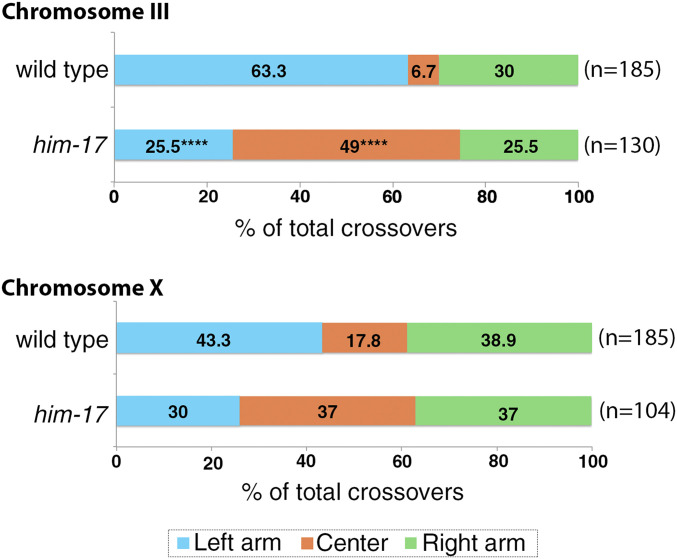
A failure to form COs was previously reported for him-17 null mutants as measured by genetic recombination in an interval spanning 80% of the X chromosome (21). Our analysis of CO distribution using single-nucleotide polymorphism mapping (snip-SNP method examining the segregation of codominant SNP markers) for both chromosomes III and X performed on eggs laid, and therefore unbiased by bypassing assessment restricted to viable progeny, revealed that CO distribution is shifted from the arms to the center for both autosomes and sex chromosomes in him-17 null mutants compared to wild type (Fig. 4). While this shift is significant for chromosome III, a similar, albeit not significant trend is detected for the X chromosome. Altogether, our data support a role for HIM-17 in regulating the distribution of DSB-dependent RAD-51 foci and COs along the chromosomes during meiosis.
Fig. 4.
HIM-17 regulates CO distribution. An analysis of CO distribution on chromosomes III and X in wild type and him-17 mutants is shown. The positions on the chromosomes are indicated as left (blue), center (orange), and right (green). ****P < 0.0001 by two-tailed Fisher's exact test, 95% CI n = number of embryos scored.
GSP-1/2 Expression and Localization Are Affected in him-17 Mutants.
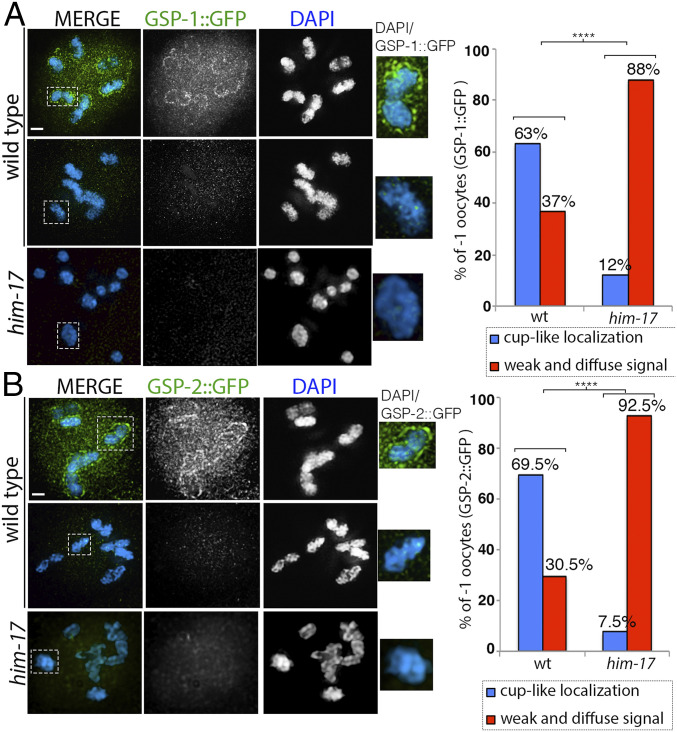
Protein phosphatase 1 (PP1) is conserved from yeast to humans and well characterized for its diverse roles in meiotic division, cell division, centriole duplication, and germ cell immortality (30–32). In C. elegans, the homologs of catalytic subunits PP1β and PP1α, respectively GSP-1 and GSP-2, are broadly expressed, including expression in the germline (33). During meiotic prophase I, LAB-1 has been proposed to antagonize the loading of AIR-2 on the long arm of the chromosomes by recruiting PP1 (19). PP1 antagonizes phosphorylation of H3T3 in mammalian cells (34). Similarly, in C. elegans, GSP-1/2 are required to antagonize phosphorylation of H3T3 on the long arm of the bivalents in oocytes at diakinesis (18). In him-17 mutants, although LAB-1 is properly localized to the long arms of most bivalents, AIR-2 and H3pT3 are still present on the long arms of the chromosomes. Using a transgenic line expressing HIM-17::GFP and GFP antibodies for mass spectrometry analysis, we identified GSP-1 among the proteins that were more enriched in coimmunoprecipitations from whole worm lysates with HIM-17::GFP compared to wild type (SI Appendix, Table S1). Therefore, we hypothesized that the protein phosphatases GSP-1/2 that act downstream of LAB-1 and upstream of H3T3 and AIR-2 might be affected in him-17 mutants. To test whether GSP-1/2 protein expression or localization is affected in him-17 mutants, we analyzed the localization patterns of GSP-1 and GSP-2 in wild-type animals. We found that GSP-1 and GSP-2 protein expression is enriched in -1 oocytes at diakinesis. GSP-1::GFP and GSP-2::GFP exhibited similar cup-like localizations around the bivalents in 63% (19/30) and 69.5% (16/23), respectively, of -1 oocytes, whereas in the remaining 37% (11/30) and 30.5% (7/23) of -1 oocytes, weak and diffuse signals (not on the chromosomes) were observed inside the nuclei (Fig. 5 A and B). In contrast, in him-17 mutants, we saw decreased expression of GSP-1::GFP and GSP-2::GFP in -1 oocytes and a significant decrease in the number of -1 oocytes with a cup-like localization around the bivalents (Fig. 5 A and B). Our data suggest that GSP-1/2 protein expression and localization depend on HIM-17.
Fig. 5.
GSP-1::GFP and GSP-2::GFP expression in wild type and him-17 mutant. (A) High-magnification images of diakinesis stage -1 oocytes from animals expressing GSP-1::GFP stained with anti-GFP (green) and DAPI (blue) in wild type and him-17 mutant backgrounds. Dashed boxes indicate the bivalents shown at higher magnification to the right for clearer visualization of either the presence or absence of GSP-1::GFP cup-like localization. (Right) Histogram indicates the percentage of -1 oocytes with cup-like localization of GSP-1::GFP on the chromosomes (blue) or a weak and diffuse signal not on the chromosomes (red) in wild type and him-17 mutant. A total of 30 and 33 -1 oocytes from 30 and 33 animals were scored for wild type and him-17, respectively, from three biological repeats. (B) High-magnification images of diakinesis stage -1 oocytes from animals expressing GSP-2::GFP stained with anti-GFP (green) and DAPI (blue) in wild type and him-17 mutants. Dashed boxes indicate the bivalents shown at higher magnification to the right for clearer visualization of either the presence or absence of GSP-2::GFP cup-like localization. (Right) Histogram indicates the percentage of -1 oocytes with cup-like localization of GSP-2::GFP on the chromosomes (blue) or weak and diffuse signal not on the chromosomes (red) in wild type and him-17 mutants. A total of 23 and 27 -1 oocytes from 23 and 27 animals were scored for wild type and him-17, respectively. ****P < 0.0001 two-tailed Fisher’s exact test. (Scale bars, 2 μm.)
Phosphorylation State of GSP-1 Partially Rescues Late Prophase I Remodeling Defect in him-17 Mutant.
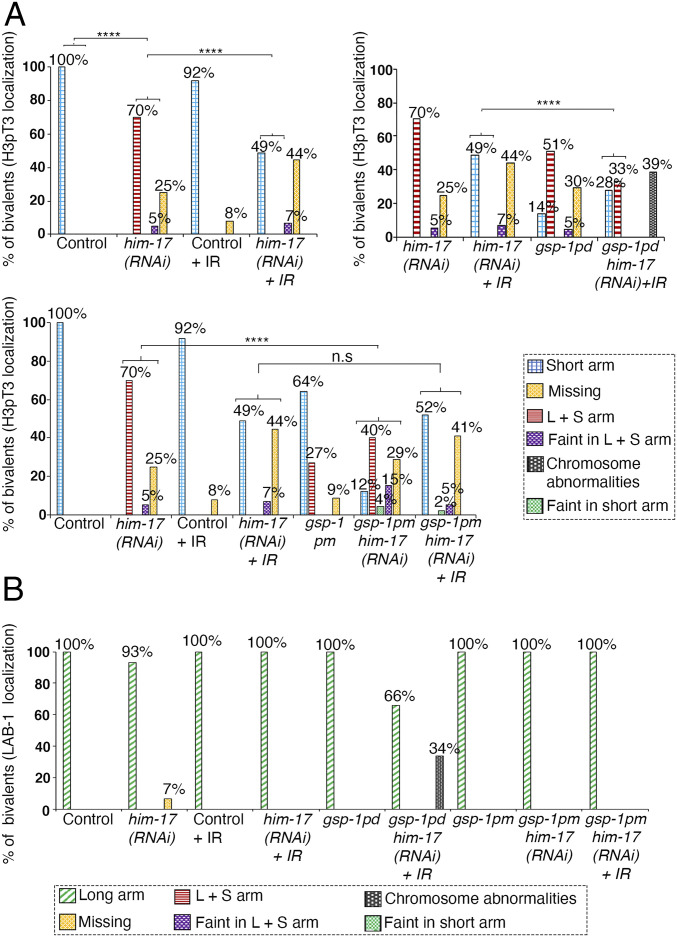
Reversible phosphorylation of protein phosphatase 1 is important for its intracellular localization and activity during meiosis in mouse oocytes (35). To determine whether GSP-1 and GSP-2 are phosphorylated, we used the PHOsphorylation SIte DAtabase (PHOSIDA; 36, 37), which identified S2 in GSP-1 as a phosphorylation site by mass spectrometry. To test whether GSP-1 phosphorylation at the S2 site affects its function in chromosome remodeling during late prophase I, we mutated this site from a serine to an alanine to generate a gsp-1(S2A) phospho-dead mutant (referred to herein as gsp-1pd) using the CRISPR/Cas9 system. Interestingly, analysis of the gsp-1pd mutant revealed defects in restriction of H3pT3 localization to the short arm of the bivalents, similar to him-17 null mutants (Fig. 6A). In 51% (54/105) of the bivalents, H3pT3 is mislocalized to both short and long arms, and in 5% (5/105) and 30% (31/105) of the bivalents, H3pT3 signal is either faint on both long and short arms or undetectable (missing), respectively. Also, similar to him-17 mutants, LAB-1 localization is not affected in the gsp-1pd mutant (Fig. 6B). This data suggests that phosphorylation of GSP-1 at the S2 site plays a role in specifying short arm identity during late prophase I.
Fig. 6.
Phosphorylation of GSP-1 at S2 partially rescues the mislocalization of H3pT3 observed in him-17 mutants. (A) Histograms indicate the percentage of bivalents at diakinesis with H3pT3 localization restricted to the short arm, mislocalized to both long and short arms, faint but on both long and short arms, faint but only on short arms, or missing on the chromosomes in the indicated genotypes. Chromosome abnormalities, consisting of aggregates and presence of DNA fragments, were observed in gsp-1pd him-17(RNAi) + IR. At least 100 bivalents were scored for each genotype from between 12 to 65 animals from three biological replicates. (B) Histogram indicates the percentage of bivalents at diakinesis with LAB-1 localization observed restricted to the long arm, mislocalized to both long and short arms, faint on both long and short arms, faint only on the short arm, or missing from chromosomes. Chromosome abnormalities, consisting of aggregates and DNA fragments, were observed in gsp-1pd him-17(RNAi) + IR. At least 72 bivalents were scored for each genotype from three biological replicates. ****P < 0.0001 two-tailed Fisher’s exact test. For complete/additional statistical analysis, see Dataset S1.
In him-17 mutants, H3pT3 and AIR-2 are mislocalized to both short and long arms of the bivalents, which could be partially rescued by introduction of exogenous DSBs by γ-IR. We hypothesized that if HIM-17 regulates late prophase I chromosome remodeling via phosphorylation of GSP-1, then introduction of exogenous DSBs in gsp-1pd him-17 double mutants should not be able to rescue the short arm marker mislocalization phenotype. To test this hypothesis, we depleted him-17 by RNAi in the gsp-1pd mutant background. We opted for depletion of him-17 by RNAi instead of generating a double mutant with gsp-1 because him-17 and gsp-1 are only two map units apart on the same chromosome, and him-17 mutants produce few and sterile progeny. First, upon depletion of him-17 by RNAi, H3pT3 was mislocalized to both long and short arms in 70% (65/93) of bivalents and either observed as a faint signal on both long and short arms in 5% (5/93) or missing in 25% (23/93) of bivalents in -1 oocytes at diakinesis as expected based on the localization observed in him-17 null mutants (Fig. 6A). Second, we could partially rescue the H3pT3 mislocalization phenotype in him-17 (RNAi) worms by introducing exogenous DSBs with γ-IR (Fig. 6A). H3pT3 localization is restricted to the short arms in 49% (57/117) of bivalents, observed as a faint signal on both short and long arms in 7% (8/117) and missing in 44% (52/117) of bivalents. Third, we analyzed H3pT3 localization in gsp-1pd him-17 double mutants after IR. H3pT3 localization is restricted to the short arm in 28% (30/109) of bivalents and observed on both long and short arms in 33% (36/109) of bivalents, indicating a significant decrease in the ability of exogenous DSBs to rescue the localization defect. Moreover, 39% of animals showed chromosome abnormalities, such as aggregates and fragments, suggesting that phosphorylation of GSP-1 at the S2 site could be playing a role in DNA damage repair in addition to its role in late prophase I chromosome remodeling.
Our data supports a model where HIM-17 may regulate the phosphorylation of GSP-1 at S2 to prevent phosphorylation of H3T3 at the long arm, thereby impeding loading of AIR-2 on the long arm. Consistent with this, an analysis of a gsp-1(S2D) phosphomimetic mutant (gsp-1pm) in a him-17(RNAi) background revealed a partial rescue of the H3pT3 mislocalization phenotype observed in him-17(RNAi) worms with 12% (14/118) of bivalents exhibiting H3pT3 signal restricted to the short arms, 40% (47/118) on both short and long arms, 15% (18/118) faint on long and short arms, 4% (5/118) faint only on the short arms, and 29% (34/118) missing. Whereas in gsp-1pm single mutants, H3pT3 signal is restricted to the short arms in 64% (89/138) of bivalents, is present on both short and long arms in 27% (37/138) of bivalents, and is missing in 9% (12/138) of bivalents (Fig. 6A). In addition, we found that the introduction of exogenous DSBs by γ-IR further rescues the phenotype of gsp-1pm him-17(RNAi) worms with 52% (47/90) of bivalents exhibiting H3pT3 signal restricted to the short arms, 2% (2/90) faint only on the short arm, 5% (4/90) faint on both long and short arms, and 41% (37/90) missing. However, there was no significant difference between him-17(RNAi) and gsp-1pm him-17(RNAi) after IR (Fig. 6A). It is possible that constitutive phosphorylation of GSP-1 affects its expression, folding, localization, or binding to GSP-2, thereby hindering further rescue. Finally, LAB-1 localization is not altered in either gsp-1 phospho-dead or phosphomimetic mutants (Fig. 6B).
Taken together, our data suggests at least two mechanisms through which HIM-17 regulates short arm identity of the bivalents: one by regulating DSB-dependent RAD-51 foci/CO distribution such that they occur in an off-centered position and another via expression, localization, and possibly phosphorylation of GSP-1/2 to restrict H3pT3 and AIR-2 localization.
Discussion
CO formation is a critical step during meiosis that generates genetic diversity and forms physical linkages (chiasmata) which hold homologous chromosomes together until their separation at meiosis I. To ensure CO formation, an excess number of programmed meiotic DSBs are made during prophase I (10, 38). DSBs are not uniformly distributed throughout chromosomes, as evident by the presence of DSB hotspots, regions where DSBs are preferentially made, and regions that are devoid of DSBs, to avoid a potential deleterious effect on achieving accurate chromosome segregation (39–41). HIM-17 had been previously shown to be required for normal levels and timing of DSB formation (21). We found that HIM-17 also plays a role in regulating the distribution of DSB-dependent RAD-51 foci along the chromosomes. In this study, we detected fewer RAD-51 foci at the center of the chromosomes compared to the arms in wild type, indicating that DSB-dependent RAD-51 foci are suppressed in the center of the chromosomes. However, we had previously detected a uniform distribution of RAD-51 foci on chromosomes (25), although they were scored in a rad-54 mutant background in order to measure the total number and distribution of DSBs. There are at least three possible reasons for the differences observed in the distribution of RAD-51 foci between these two studies: 1) The rad-54 mutant background in which DSBs are made and RAD-51 associates at resected 3′ ends, but is not ejected from those ends, might affect distribution of RAD-51 foci on the chromosomes. 2) DSBs are continuously made, and only when a DSB is designated to be repaired as a CO, a negative feedback loop is activated, thereby turning off additional DSB formation on that chromosome (42). When CO designation or formation is blocked by interrupting the DNA repair pathway, such as in the rad-54 mutant background, the DSB machinery might end up making more breaks even at sites that are usually not preferred in wild type, possibly giving rise to a more uniform DSB distribution. 3) In this current study, we are capturing only a subset of events; although if these were normally evenly distributed, it is unclear why scoring RAD-51 foci in an unbiased manner, without any other mutation in the background, would fail to detect this. Therefore, we favor the model that HIM-17 function results in suppression of DSBs along the center of the chromosomes.
While an excess number of DSBs are made in wild type, only some of these breaks are processed into COs. Moreover, CO frequency and distribution are not random. DSBs at preferred positions along the chromosomes are repaired into COs, which contributes to distinct CO distribution patterns as observed, for example, between human males in which COs are enriched at the distal region and females in which they are more interstitially placed (43). In C. elegans, COs are always made at an off-center position, resulting in asymmetric bivalents with distinct short and long arms (7), which is critical for the accurate segregation of homologous chromosomes during meiosis I and sister chromatids during meiosis II. Off-center position CO triggers disassembly of LAB-1 from the short arm, and its localization is restricted to the long arm, whereas AIR-2 loading on the short arm allows the separation of homologous chromosomes during meiosis I.
In mouse, plants, and yeast, when limited numbers of DSBs are made, CO homeostasis ensures that at least one CO is made per homolog (5, 44, 45). Similarly, in C. elegans, if only one DSB is made, it will be converted into a CO even if it is positioned at the center of the chromosome and will therefore result in the formation of a symmetric bivalent which fails to undergo proper chromosome remodeling (28). As a consequence, AIR-2 loads on both the long and short arms of the chromosome, resulting in premature loss of SCC during meiosis I and chromosome segregation defects (28). Errors in achieving accurate chromosome segregation lead to a reduced brood size and sterility. In him-17 mutants, only a limited number of DSBs are formed, and these are shifted toward the center of the chromosomes, resulting in an equivalent change in CO position toward the center as well. However, this shift in CO position may not be the sole contributor to the late prophase I remodeling defects observed in him-17 mutants. This is supported by our observation that while AIR-2 is mislocalized in 100% of bivalents in him-17 mutants, only between 37 to 49% of COs are made at the center of the chromosomes. HIM-17 has been proposed to link chromatin state and DSB formation during meiosis in C. elegans (21). him-17 mutants exhibit greatly reduced H3K9me2 signal in germline nuclei, which can be rescued by introduction of exogenous DSBs with γ-IR (SI Appendix, Fig. S3). HIM-17 therefore provides an entry point to analyze a potential link between H3K9 methylation, CO distribution, and regulation of chromosome remodeling. A potential additional mechanism involved in exerting the formation of chromosome subdomains could involve the spreading of histone modifications along chromosomes. Especially repressive histone modifications such as H3K9 methylation are enriched on the arms compared to the center of the chromosomes, whereas other modifications such as H3K4me3, H3K36me3, and H3K79me3 exhibit the opposite localization pattern (46). Since this histone methylation pattern coincides with the recombination pattern, at least at a broad scale, it is tempting to argue that they influence each other. Furthermore, the study of proteins involved in mitotic DNA repair supports the speculation that histone modifications could serve as a platform for the loading of specific resolvases during meiosis, thereby promoting distinct repair pathways (47, 48).
There is also substantial evidence that the occurrence of certain histone modifications correlates with the site of DSB formation in various organisms. For example, modifications associated with an open chromatin state, such as H3K4me3 or H3K9ac, have been shown to influence recombination (49). However, in C. elegans, which is holocentric, recombination seems to follow marks associated with heterochromatin that are enriched on chromosome arms, at least on a broad scale (9, 50). As to whether DSB sites also correlate with these marks and how the recombination and histone landscapes correlate on a finer scale remain unclear. The fact that the H3K9me2 pattern is not changed in spo-11 and mre-11 mutants, which both lack endogenous DSB formation (21, 51), argues that the altered histone landscape in him-17 mutants is independent of SPO-11–induced DSBs. Furthermore, mutations in histone H3 lysine 9 methyltransferases met-2 and set-25 do not result in meiotic defects such as chromosome mis-segregation. Even met-2;set-25 double mutants that exhibit increased apoptosis and a reduction in brood size show elevated RAD-51 levels, arguing that H3K9me2 might not be necessary for DSB formation (52). In general, the defects observed in met-2;set-25 double mutants were linked to R-loop formation due to stalled replication forks (53). Interestingly, the loss of H3K9me2 does not necessarily lead to a derepression of heterochromatin genes, indicating that additional factors or mechanisms are involved in the regulation of gene silencing and expression. Since the regulation of CO position affects chromosome remodeling, and therefore the production of viable progeny, an interaction of multiple regulatory factors and mechanisms seems to be plausible in order to tightly regulate CO position even in the event of failure of one involved factor or mechanism.
Regulation of Late Prophase I via GSP-1 and GSP-2.
In wild type, similar to its Shugoshin counterpart, LAB-1 has been proposed to recruit GSP-1 and GSP-2 to the long arm of the bivalents and prevent H3T3 phosphorylation by Haspin, thereby preventing AIR-2 loading on the long arm of the bivalents during late diakinesis in oogenesis (16, 18). Whereas in him-17 mutants, although LAB-1 is localized properly only on the long arm of the bivalents, H3T3 located on the long arms is still phosphorylated and recruits AIR-2 to the long arms. Moreover, we see an increase in the intensity of H3pT3 signal on bivalents in the him-17 mutant background, which might be due to a positive feedback loop. This is not unprecedented, as several studies on mitotic cell division have shown the existence of a Haspin-H3pT3-Aurora B kinase positive feedback loop regulating the accumulation of the chromosome passenger complex at centromeres (54, 55). We found that HIM-17 protects SCC on the long arm in several ways. First, HIM-17 regulates the expression of GSP-1 and GSP-2 in the -1 oocyte since in him-17 mutants, we see a decrease in GSP-1::GFP and GSP-2::GFP signal. This reduction in GSP-1/2 expression may not be due to a role for HIM-17 in transcription since defects in late prophase I chromosome remodeling can be partially rescued by the introduction of exogenous DSBs by γ-IR. Second, HIM-17 regulates the localization of GSP-1/2 on the bivalents. Finally, we propose that HIM-17 may regulate the role of GSP-1 in restricting AIR-2 localization via phosphorylation of the S2 site on GSP-1.
The NetPhos 3.1 program (56) predicts that the S2 site in GSP-1 can be phosphorylated by a number of kinases among which the highest ranking are casein kinase 1, GSK3, and p38 MAP kinase. Interestingly, we identified CDK-1, KIN-18, KIN-20, MEK-1, PLK-1, F52B5.2, and AIR-1 as interactors of HIM-17 by coimmunoprecipitation of HIM-17 tagged with GFP and mass spectrometry (SI Appendix, Table S1). One of the kinases we identified as a HIM-17 interactor could be a potential kinase that phosphorylates and regulates GSP-1 activity. CDK-1 is a serine/threonine kinase previously shown to control the timing of AIR-2 loading on the chromosomes (18). However, depletion of CDK-1 does not affect phosphorylation of H3pT3. Instead, CDK-1 functions between H3pT3 phosphorylation and AIR-2 loading onto chromosomes, therefore possibly ruling out CDK-1 as the kinase that phosphorylates and regulates GSP-1 expression and activity. KIN-18 is a TAO kinase with MAP kinase activity that is expressed in the C. elegans germline and is found to coordinate meiotic recombination with meiotic progression (57). KIN-18 is also required for the proper timing of MAP kinase MPK-1 activity in the germline (57). Interestingly, short arm identity is altered in kin-18 mutants (57). In wild-type worms, phosphorylated histone H3 (pH3) is observed localized to the short arm of the bivalent chromosome in late diakinesis stage oocytes. Whereas in kin-18 mutants, this restriction is lost and pH3 is frequently seen on both long and short arms of the bivalents. KIN-18 could be playing a role in phosphorylating GSP-1 at the S2 site directly or through a kinase cascade. We also found KIN-20, which is an ortholog of human casein kinase I delta (CSNK1D) and is expressed in the C. elegans germline (modENCODE data in ref. 58). GSP-1 could be phosphorylated by casein kinase either directly or indirectly. In addition, we also found several kinases that are enriched in the germline (59). PLK-1 (polo-like kinase 1) plays several roles during meiosis along with PLK-2 including the regulation of chromosome movements, which is important for the stabilization of pairing and the initiation of SC assembly between homologs during the leptotene and zygotene stages (60, 61). Later in pachytene, PLK-1/2 phosphorylate SYP-4 in response to CO designation and prevent further DSB formation on chromosomes (42). In late prophase I, PLK-1/2 play an important role in stepwise loss of SCC via phosphorylation of HTP-1 (18). F52B5.2 is an uncharacterized protein with predicted serine/threonine kinase activity. Its homolog Cdc28 is important for meiotic progression in yeast (62). MEK-1 is a MAPK kinase shown to play a role in heavy metal-induced germ cell apoptosis in C. elegans (63, 64). Finally, AIR-1 encodes for Aurora A kinase, which is required for germline development and the formation of meiotic spindles in C. elegans (65, 66). Any of these kinases identified as an interactor of HIM-17 could be potentially involved in the phosphorylation of GSP-1 either directly or indirectly. However, we cannot rule out the possibility that yet another kinase that we did not identify in our pull-downs could play a role in regulating GSP-1 phosphorylation and expression since the interactions between kinases and their substrates are transient, and therefore the kinase acting on GSP-1 may not be among the interactors we identified for HIM-17.
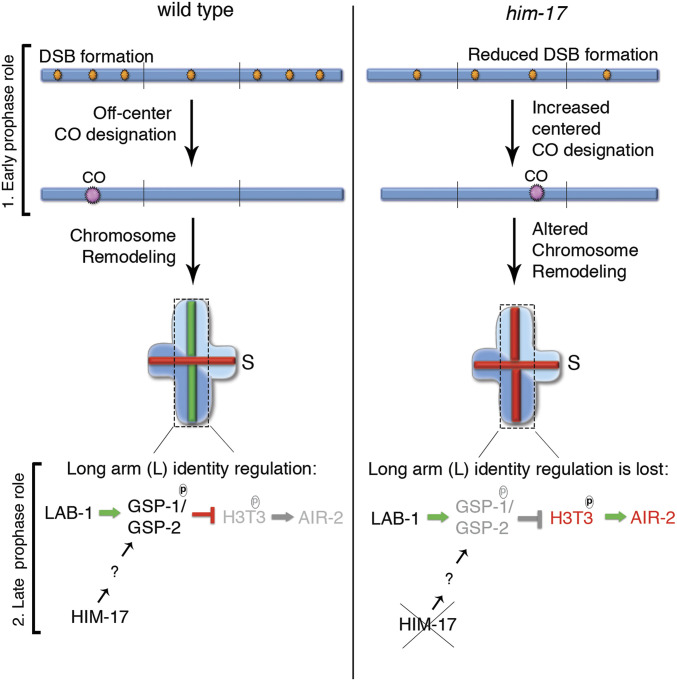
HIM-17 contains evolutionarily conserved THAP domains, which are motifs present in proteins involved in various cellular functions including cell proliferation, apoptosis, cell cycle, chromosome segregation, chromatin modification, and transcriptional regulation (21–23). Similar to HIM-17, the THAP domain-containing protein THAP7 in humans is found to function in histone post-translational modification (67). The divergent THAP domains present in HIM-17 are predicted to have lost their DNA-binding activity and instead function as protein–protein interaction modules (21, 22, 68). Akin to what is observed in C. elegans, during mouse meiosis, the intracellular localization of PP1 to the nucleus is important for its function to achieve meiotic competence. In addition, PP1 activity is regulated by phosphorylation (35). In conclusion, our findings suggest that HIM-17 plays two potentially conserved roles that converge in promoting normal late prophase I chromosome remodeling: 1) HIM-17 regulates DSB-dependent RAD-51 foci and CO distribution to achieve formation of bivalents with distinct long and short arm identities, and 2) HIM-17 protects SCC via the regulation of intracellular expression, localization, and potential phosphorylation of GSP-1/2 (Fig. 7).
Fig. 7.
Model for how HIM-17 regulates DSB/CO distribution and late prophase I chromosome remodeling. HIM-17 acts in early prophase I by regulating DSB levels and DSB-dependent RAD-51 foci/CO distribution. In wild type, an excess number of DSBs are made, and a higher number of DSB-dependent RAD-51 foci is observed on the arms compared to the center region of the chromosomes in C. elegans. One DSB on the arm regions of the chromosomes is processed into a CO. This off-center CO position results in the production of an asymmetric bivalent with long (L) and short (S) arms. During late prophase I, recruitment of GSP-1/2 by LAB-1 and phosphorylation of GSP-1 at the S2 site, which requires HIM-17 function, prevents phosphorylation of H3T3 on the long arm, thereby preventing AIR-2 loading on the long arm. In contrast, in him-17 mutants, a reduced number of DSBs are made, and their distribution is altered such that now higher levels of DSB-dependent RAD-51 foci are detected at the center of the chromosomes. An increase in the levels of COs on the center of the chromosomes is observed, which potentially contributes to the short arm identity defect. In addition, expression, localization, and probably phosphorylation of GSP-1/2 is impaired, leading to phosphorylation of H3pT3 on the long arms even when LAB-1 is properly localized to the long arm of the bivalents. Therefore, HIM-17 functions at early and late prophase I are required to impede altered CO position and impaired GSP-1/2 activity to promote normal chromosome remodeling and subsequent regulated stepwise loss of SCC, leading to accurate chromosome segregation.
Materials and Methods
C. elegans Strains.
C. elegans strains were cultured at 20 °C under standard conditions, and the N2 Bristol strain was used as the wild-type background (69). All strains were outcrossed at least six times. In addition, we routinely outcrossed the him-17(ok424)/nT1 strain with N2 and rebalanced it since we observed that otherwise, the levels of univalents in diakinesis oocytes progressively increased through multiple generations. The following mutations and chromosome rearrangements were used:
LG III: gsp-2(lt27[GFP::gsp-2), unc-119(ed3), unc-119(e2498); LG IV: ltIs37[pAA64; pie-1/mCherry::his-58; unc-119 (+)], nT1 [unc-?(n754) let-? qIs50] (IV;V); LG V: him-17(ok424), gsp-1(rj59[S2A]), gsp-1(rj60[S2D]), gsp-1(lt94 [GFP::gsp-1]), meIs5[unc-119(+) + him-17::GFP] (unmapped), ojIs50[pie-1p::GFP::air-2 + unc-119(+)] (unmapped).
Additional procedures are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to the Caenorhabditis Genetics Center for providing strains. We thank Arshad Desai for providing animals expressing GSP-1::GFP and GSP-2::GFP and Monique Zetka and Yumi Kim for the HTP-3 antibody. We thank the members of the M.P.C. laboratory for the critical reading of this manuscript and for providing helpful suggestions. This work was supported by NIH Grant R01GM072551 to M.P.C. and a fellowship from the Lalor Foundation to S.N.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016363118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Keeney S., Lange J., Mohibullah N., Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu. Rev. Genet. 48, 187–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergerat A., et al., An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Dernburg A. F., et al., Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Gray S., Cohen P. E., Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Copenhaver G. P., Meiotic recombination: Mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Perez E., Colaiácovo M. P., Distribution of meiotic recombination events: Talking to your neighbors. Curr. Opin. Genet. Dev. 19, 105–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meneely P. M., Farago A. F., Kauffman T. M., Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162, 1169–1177 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes T. M., Kohara Y., Coulson A., Hekimi S., Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141, 159–179 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones G. H., The control of chiasma distribution. Symp. Soc. Exp. Biol. 38, 293–320 (1984). [PubMed] [Google Scholar]
- 11.Schvarzstein M., Wignall S. M., Villeneuve A. M., Coordinating cohesion, co-orientation, and congression during meiosis: Lessons from holocentric chromosomes. Genes Dev. 24, 219–228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lui D. Y., Colaiácovo M. P., Meiotic development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757, 133–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Perez E., et al., Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22, 2886–2901 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nabeshima K., Villeneuve A. M., Colaiácovo M. P., Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 168, 683–689 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riddle D. L., et al., Chromosome Organization, Mitosis, and Meiosis (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 2, 1997). [PubMed] [Google Scholar]
- 16.de Carvalho C. E., et al., LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22, 2869–2885 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogers E., Bishop J. D., Waddle J. A., Schumacher J. M., Lin R., The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157, 219–229 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrandiz N., et al., Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 9, 834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzur Y. B., et al., LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion. PLoS Biol. 10, e1001378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitajima T. S., et al., Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441, 46–52 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Reddy K. C., Villeneuve A. M., C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118, 439–452 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Clouaire T., et al., The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. U.S.A. 102, 6907–6912 (2005).15863623 [Google Scholar]
- 23.Gervais V., Campagne S., Durand J., Muller I., Milon A., NMR studies of a new family of DNA binding proteins: The THAP proteins. J. Biomol. NMR 56, 3–15 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Reinke V., Gil I. S., Ward S., Kazmer K., Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Saito T. T., Mohideen F., Meyer K., Harper J. W., Colaiácovo M. P., SLX-1 is required for maintaining genomic integrity and promoting meiotic noncrossovers in the Caenorhabditis elegans germline. PLoS Genet. 8, e1002888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandt J. N., Hussey K. A., Kim Y., Spatial and temporal control of targeting Polo-like kinase during meiotic prophase. J. Cell Biol. 219, e202006094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato-Carlton A., et al., Phosphoregulation of HORMA domain protein HIM-3 promotes asymmetric synaptonemal complex disassembly in meiotic prophase in Caenorhabditis elegans. PLoS Genet. 16, e1008968 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altendorfer E., Láscarez-Lagunas L. I., Nadarajan S., Mathieson I., Colaiácovo M. P., Crossover position drives chromosome remodeling for accurate meiotic chromosome segregation. Curr. Biol. 30, 1329–1338.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mets D. G., Meyer B. J., Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139, 73–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceulemans H., Bollen M., Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84, 1–39 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Billmyre K. K., et al., The meiotic phosphatase GSP-2/PP1 promotes germline immortality and small RNA-mediated genome silencing. PLoS Genet. 15, e1008004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peel N., et al., Protein phosphatase 1 down regulates ZYG-1 levels to limit centriole duplication. PLoS Genet. 13, e1006543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sassa T., Ueda-Ohba H., Kitamura K., Harada S., Hosono R., Role of Caenorhabditis elegans protein phosphatase type 1, CeGLC-7 beta, in metaphase to anaphase transition during embryonic development. Exp. Cell Res. 287, 350–360 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Qian J., Lesage B., Beullens M., Van Eynde A., Bollen M., PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 21, 766–773 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Wang X., et al., Endogenous regulators of protein phosphatase-1 during mouse oocyte development and meiosis. Reproduction 128, 493–502 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Gnad F., et al., PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 8, R250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gnad F., Gunawardena J., Mann M., PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 39, D253–D260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokoo R., et al., COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tock A. J., Henderson I. R., Hotspots for initiation of meiotic recombination. Front. Genet. 9, 521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukaszewicz A., Lange J., Keeney S., Jasin M., Control of meiotic double-strand-break formation by ATM: Local and global views. Cell Cycle 17, 1155–1172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan J., Keeney S., Molecular cartography: Mapping the landscape of meiotic recombination. PLoS Biol. 5, e333 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadarajan S., et al., Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife 6, 265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruhn J. R., Rubio C., Broman K. W., Hunt P. A., Hassold T., Cytological studies of human meiosis: Sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One 8, e85075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole F., et al., Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue M., et al., The number of meiotic double-strand breaks influences crossover distribution in Arabidopsis. Plant Cell 30, 2628–2638 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu T., et al., Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21, 227–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.House N. C. M., Koch M. R., Freudenreich C. H., Chromatin modifications and DNA repair: Beyond double-strand breaks. Front. Genet. 5, 296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L., Oberdoerffer P., Chromatin dynamics in DNA double-strand break repair. Biochim. Biophys. Acta 1819, 811–819 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Székvölgyi L., Ohta K., Nicolas A., Initiation of meiotic homologous recombination: Flexibility, impact of histone modifications, and chromatin remodeling. Cold Spring Harb. Perspect. Biol. 7, a016527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockman M. V., Kruglyak L., Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5, e1000419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bessler J. B., Reddy K. C., Hayashi M., Hodgkin J., Villeneuve A. M., A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics 175, 2029–2037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeller P., et al., Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48, 1385–1395 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Ahringer J., Gasser S. M., Repressive chromatin in Caenorhabditis elegans: Establishment, composition, and function. Genetics 208, 491–511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang F., et al., A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21, 1061–1069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu F., et al., Aurora-A promotes the establishment of spindle assembly checkpoint by priming the Haspin-Aurora-B feedback loop in late G2 phase. Cell Discov. 3, 16049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blom N., Gammeltoft S., Brunak S., Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Yin Y., Donlevy S., Smolikove S., Coordination of recombination with meiotic progression in the Caenorhabditis elegans germline by KIN-18, a TAO kinase that regulates the timing of MPK-1 signaling. Genetics 202, 45–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Celniker S. E.et al.; modENCODE Consortium , Unlocking the secrets of the genome. Nature 459, 927–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzur Y. B., et al., Spatiotemporal gene expression analysis of the Caenorhabditis elegans germline uncovers a syncytial expression switch. Genetics 210, 587–605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper N. C., et al., Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell 21, 934–947 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labella S., Woglar A., Jantsch V., Zetka M., Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell 21, 948–958 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17, 1524–1539 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pei B., et al., Arsenite-induced germline apoptosis through a MAPK-dependent, p53-independent pathway in Caenorhabditis elegans. Chem. Res. Toxicol. 21, 1530–1535 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Villanueva A., et al., jkk-1 and mek-1 regulate body movement coordination and response to heavy metals through jnk-1 in Caenorhabditis elegans. EMBO J. 20, 5114–5128 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sumiyoshi E., Fukata Y., Namai S., Sugimoto A., Caenorhabditis elegans Aurora A kinase is required for the formation of spindle microtubules in female meiosis. Mol. Biol. Cell 26, 4187–4196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furuta T., Baillie D. L., Schumacher J. M., Caenorhabditis elegans Aurora A kinase AIR-1 is required for postembryonic cell divisions and germline development. Genesis 34, 244–250 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Macfarlan T., Parker J. B., Nagata K., Chakravarti D., Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol. Endocrinol. 20, 335–347 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Sun L., Liu A., Georgopoulos K., Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 15, 5358–5369 (1996). [PMC free article] [PubMed] [Google Scholar]
- 69.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.