Abstract
Cryptococcus neoformans is a dimorphic fungus that causes lethal meningoencephalitis mainly in immunocompromised individuals. Different morphotypes enable this environmental fungus and opportunistic pathogen to adapt to different natural niches and exhibit different levels of pathogenicity in various hosts. It is well-recognized that C. neoformans undergoes bisexual or unisexual reproduction in vitro to generate genotypic, morphotypic, and phenotypic diversity, which augments its ability for adaptation. However, if and how sexual reproduction and the meiotic machinery exert any direct impact on the infection process is unclear. This review summarizes recent discoveries on the regulation of cryptococcal life cycle and morphogenesis, and how they impact cryptococcal pathogenicity. The potential role of the meiotic machinery on ploidy regulation during cryptococcal infection is also discussed. This review aims to stimulate further investigation on links between fungal morphogenesis, sexual reproduction, and virulence.
Keywords: Cryptococcus neoformans, Sexual cycles, Ploidy, Meiosis, DNA damage response, Morphogenesis, Dimorphism, Pathogenesis, Host-pathogen interactions, Vaccination
1. Introduction
Cryptococcus neoformans has been recognized as an environmental fungus and an opportunistic pathogen since its description in 1894 and 1895 when C. neoformans was isolated from a bone infection and fermented fruit juice (Otto, 1894; Sanfelice, 1895). This basidiomycete is ubiquitous in the environment and is commonly isolated from avian excreta, soil, and trees. Consequently, asymptomatic exposure through inhalation of spores or desiccated yeast cells is common in the general population, but it could lead to pulmonary and systemic cryptococcosis in individuals with compromised immune systems. Systemic cryptococcosis, with the most common clinical manifestation as cryptococcal meningoencephalitis, causes 15% of AIDS-related deaths globally (Dromer et al., 2011; Perfect, 2015). In the C. neoformans pathogenic species complex, C. neoformans generally causes systemic cryptococcosis in immunocompromised patients. A sibling species in this complex, Cryptococcus gattii, affects mostly immunocompetent individuals, although it also causes infections in immunocompromised patients (Dromer et al., 2011; Perfect, 2015). Compared to C. neoformans, C. gattii is more commonly isolated from tropical and subtropical regions, and it is responsible for the ongoing cryptococcosis outbreak in otherwise healthy individuals in North America and Canada (Kwon-Chung et al., 2002).
C. neoformans usually propagates mitotically in the unicellular yeast form through budding. Under conditions that induce sexual reproduction, such as dehydration or nitrogen starvation, the fungus undergoes morphological transition from yeast to hyphae. Hyphal formation enables nutrient scavenging from surroundings and the production of stress tolerant infectious spores. Because Cryptococcus yeast-to-hypha transition is uniquely and tightly associated with its bisexual and unisexual reproduction, it has not been considered a classic dimorphic pathogen, which typically encompasses thermally dimorphic species like Histoplasma capsulatum, Blastomyces dermatitidis, and Talaromyces marneffei (Sil and Andrianopoulos, 2015). In this review, we will focus on recent discoveries on Cryptococcus sexual cycles, the regulation of morphogenesis, and their impact on cryptococcal pathogenicity. We will also discuss the potential impact of its meiotic machinery on ploidy regulation during cryptococcal infection.
2. Cryptococcus sexual cycles
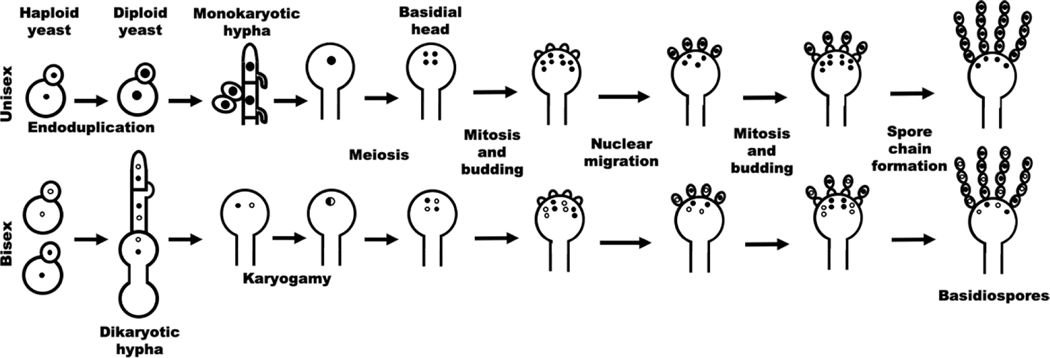
The bipolar heterothallic sexual cycle of C. neoformans was discovered in the 1970s (Kwon-Chung, 1975). Under mating-inducing conditions (e.g. low nitrogen and dehydration), cells of the opposite mating type (α and a) conjugate in response to pheromone and fuse to form a dikaryon in which two parental nuclei congress but do not fuse (Figure 1). The resulting zygote then extends filamentous growth through the a parental cell in the form of dikaryotic hyphae. Clamp connections, a characteristic of heterothallic mating in basidiomycetes, ensure the inheritance of two parental nuclei in each newly generated hyphal compartment. The tips of aerial hyphae develop into club-shaped basidium heads where karyogamy and meiosis occur. The following repeated mitotic divisions in parallel with sporogenesis give rise to four chains of sexual spores (Figure 1). In addition to recombinant meiotic basidiospores formed on the fruiting structure of basidial heads, mitotic blastospores (yeast-like cells) bud off from hyphae or occasionally from the clamp connections. The mitotic blastospores inherit one of the parental nuclear genotypes. Although the dikaryotic hyphae, the blastospores, and the meiotic basidiospores inherit different nuclear genomes, all predominantly inherit mitochondrial DNA from the a parental strain (Matha and Lin, 2020). Thus, there is an amazing variety of cell types and genotypes generated during the sexual reproduction process.
Figure 1. Cryptococcus sexual cycles.
Cryptococcal cells of a single mating type can undergo unisexual reproduction mainly through endoduplication to generate monokaryotic hyphae. The clamp connections formed during unisexual reproduction are unfused. Sporogenesis occurs in basidia through repeated mitosis and budding following one round meiosis. During bisexual reproduction, a and α cells fuse, undergo morphological switch to dikaryotic hyphae, generate fruiting bodies, and produce basidiospores.
C. neoformans produces equal numbers of a and α meiotic progeny from heterothallic sexual reproduction. However, natural cryptococcal populations are sharply distorted favoring the α mating type (99.9% α for serotype A, 95% α for serotype B, C, and D) (Cogliati, 2013; Kwon-Chung and Bennett, 1978; Yan et al., 2002), which spurs the view that the a-α bisexual reproduction may be rare or even absent in nature except in regions where MATa isolates are more commonly present (e.g. C. neoformans serotype A VNB isolates in Africa or C. gattii isolates in Brazil) (Chen et al., 2015; Hagen et al., 2013). The discovery of unisexual reproduction in Cryptococcus provides a plausible explanation for the dominance of the α mating type. Monokaryotic fruiting or haploid fruiting, a process that resembles the a-α bisexual cycle with yeast-to-hypha transition and basidiospore production, involves cells of only a single mating type (Wickes et al., 1996). Since the opposite mating partner is absent, monokaryotic fruiting was previously considered asexual and mitotic. In 2005, Lin et al. demonstrated that meiosis and high levels of recombination occur during monokaryotic fruiting, indicating its sexual nature (Lin et al., 2005). Haploid cells of a single mating type can diploidize either through nuclear fusion or through endoreplication (Fu and Heitman, 2017; Lin et al., 2005; Lin et al., 2009b). Diploidization could take place either prior to or after the yeast-to-hypha transition. Meiosis and sporulation happen in the fruiting body basidial heads as observed in bisexual reproduction. Quantitative analyses of recombination during cryptococcal sexual reproduction (Roth et al., 2018; Sun et al., 2014) provide further evidence that monokaryotic fruiting is a meiotic process, the hallmark of sexual reproduction. The identification of natural homozygous and heterozygous α-α diploids, including αAAα, αADα and αBDα hybrids, further supports the occurrence of same-sex mating in nature (Lin et al., 2007; Lin et al., 2009b; Rhodes et al., 2017). Yadav et al. recently reported the occurrence of uniparental reproduction in Cryptococcus, known as hybridogenesis where both parents are physically required but only one parent contributes its genetic material to the hemi-clonal progeny (Yadav et al., 2020). Hybridogenesis might also contribute to the markedly skewed mating-type distribution in C. neoformans.
3. Cryptococcus yeast-to-hypha transition
3.1. Cryptococcus yeast-to-hypha transition and pheromone signaling
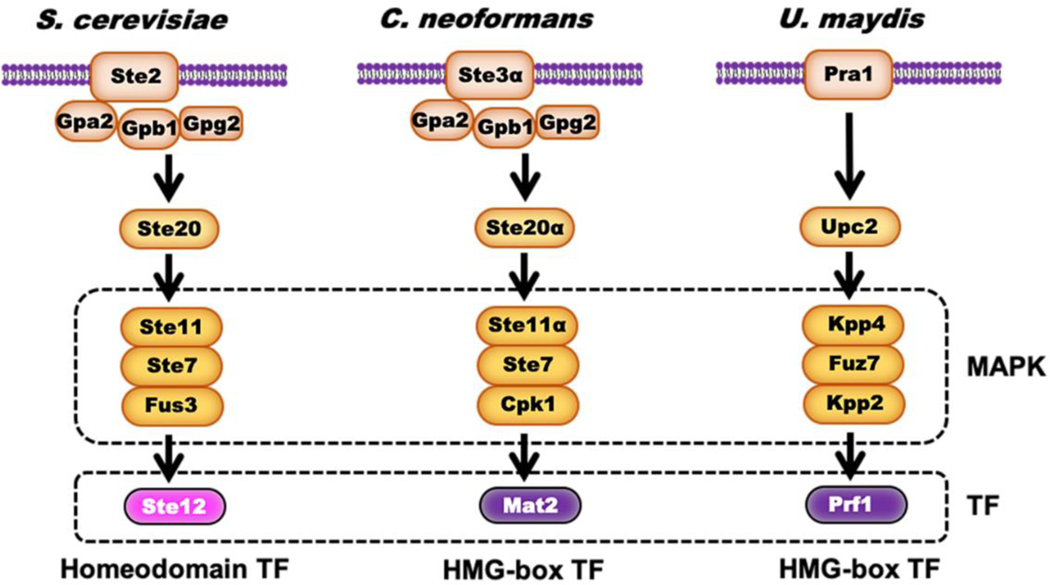
Yeast-to-hypha transition is tightly associated with sexual cycles in C. neoformans (Wang et al., 2014). Pheromone signaling triggers non-self-recognition and cell fusion during a-α bisexual mating, and the resulting dikaryotic zygote initiates hyphal growth (Figure 1). Therefore, yeast-to-hypha transition in Cryptococcus has historically been considered a pheromone-dependent process. Secreted pheromones are recognized by the G protein-coupled receptors (GPCRs), which reside on the plasma membrane of the recipient cells (Xue et al., 2008). Pheromone activation of the GPCR initiates a signal transduction cascade that includes a mitogen-activated protein kinase (MAPK) pathway (Figure 2) (Davidson et al., 2003). The central elements of the MAPK signaling are conserved among diverged fungal species, yet the downstream transcription factors that regulate mating usually differ between species (Figure 2) (Jones and Bennett, 2011). In S. cerevisiae, Ste12 is the transcription factor required for mating gene expression activated by the pheromone signaling (Roberts and Fink, 1994). A STE12 homolog resides in the mating type locus in C. neoformans, but it is dispensable for mating (Wickes et al., 1997): deletion of STE12 in C. neoformans reduces but does not abolish bilateral mating (Chang et al., 2000). In Ustilago maydis, a dimorphic basidiomycete that infects plants, it is the high mobility group (HMG)-box transcription factor Prf1 that recognizes the pheromone response elements (PREs) and activates mating genes (Hartmann et al., 1999). C. neoformans is evolutionarily close to U. maydis, yet deletion of the PRF1 homolog does not affect cryptococcal mating (Lin et al., 2010). The transcription factor Mat2 in C. neoformans was eventually identified through a forward genetics screening for mutants unable to initiate filamentation under mating-inducing conditions (Lin et al., 2010). Mat2, as an HMG-box transcription factor, functions downstream of the Cpk1 MAPK pathway, recognizes PREs, and is essential for pheromone sensing and response (Kruzel et al., 2012; Lin et al., 2010). Overexpression of MAT2 is sufficient to increase the transcript levels of pheromone genes, evoke shmoo cell formation, and promote cell fusion (Wang et al., 2012), as expected for roles of the transcription factor in the pheromone sensing and response pathway.
Figure 2. Pheromone signaling.
Pheromone binds to a cell surface receptor that in turn activates a downstream signaling cascade through the protein kinase Ste20 homologs in S. cerevisiae and C. neoformans, or Upc2 in U. maydis. The signal is relayed through a highly conserved MAPK cascade, in which the MAP kinase phosphorylates and activates the downstream transcription factor, resulting in gene transcription. The transcription factors diverge in different fungi. Ste12 in S. cerevisiae is a homeodomain transcription factor, while the Prf1 in U. maydis is an HMG-box transcription factor. In C. neoformans, the Prf1 homolog is dispensable for mating, but another HMG-box transcription factor Mat2 regulates pheromone signaling. Some components of this pathway are omitted for clarity.
For sexual reproduction, diploidization is necessary for meiosis, a reductive division. Diploidization is achieved through a-α nuclear fusion during bisexual reproduction, while it is more likely attained through endoreplication during unisexual reproduction as unisexual reproduction is largely independent from cell-cell fusion (Figure 1) (Fu and Heitman, 2017). Thus, the pheromone signaling pathway, which is required for non-self-recognition between a and α cells, should not be essential for unisexual development. Indeed, deletion of some major components of the pheromone pathway, including the pheromone receptor gene CPRα (STE3α), the pheromone transporter gene STE6, the G-protein α subunit genes GPA2 and GPA3, or the pheromone MF genes abolishes bisexual mating but not self-filamentation (Chang et al., 2003; Hsueh and Shen, 2005; Hsueh et al., 2007; Lin et al., 2005). In addition, despite the essentiality of the cell identity homeodomain Sxi1α-Sxi2a complex for bisexual development, it is dispensable for unisexual development. However, deletion of the pheromone-responsive transcription factor gene MAT2 abolished self-filamentation under all laboratory conditions that had been previously tested (Lin et al., 2010). The observation that Mat2 was necessary for both bisexual and unisexual development was contradictory to the notion that the pheromone pathway should not be essential for unisexual development. This conundrum was resolved recently when mat2Δ mutants were found to undergo robust self-filamentation in the presence of excessive copper, after heat-shock and exposure to high concentrations of calcium, or in the presence of glucosamine as the only carbon source (Gyawali et al., 2017; Xu et al., 2017). These discoveries strongly suggest that cryptococcal filamentation is regulated through complex pathways including the pheromone signaling pathway and other unidentified pathways. The natural or laboratory conditions and the unique signaling pathways that promote bisexual versus unisexual reproduction remain to be defined.
3.2. The regulation of Cryptococcus yeast-to-hypha transition
In many heterothallic fungi, the mating-type-specific homeodomain transcription factors form a heterodimer after cell-cell fusion, which regulates the subsequent sexual development. In U. maydis, the bE-bW heterodimer initiates the switch from budding yeast growth to polarized filamentous growth by targeting RBF1, which is necessary and sufficient for b-dependent filamentation (Heimel et al., 2010). In C. neoformans, Sxi1α and Sxi2a form a heterodimer that regulates post-zygotic bisexual development without any apparent pre-zygotic role in pheromone sensing or cell fusion (Hull et al., 2005; Hull et al., 2002). However, no homolog of Ustilago Rbf1 exists in Cryptococcus based on the analysis of the Sxi1α-Sxi2a binding sites (Mead et al., 2015b). Thus, a Sxi heterodimer-independent transcription factor must govern the yeast-to-hypha transition in C. neoformans.
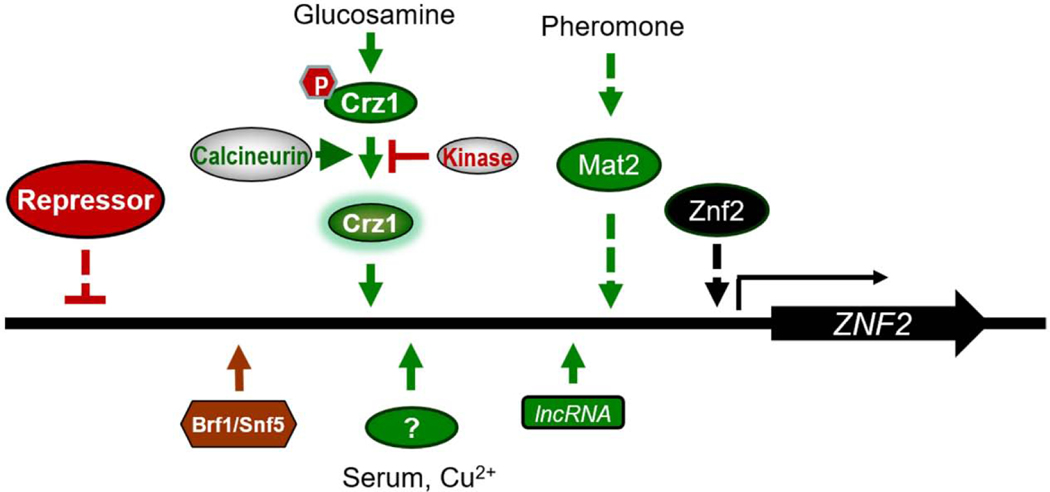
Transcriptome profiling of non-filamentous versus filamentous strains coupled with genetic studies revealed that Znf2 functions as the master regulator of yeast-to-hypha transition in C. neoformans (Lin et al., 2010). Deletion of this zinc finger transcriptional factor gene does not abolish cell fusion or pheromone response, similar to deletion of SXI1α or SXI2a. However, Znf2 is required for cryptococcal hyphal growth during both bisexual and unisexual reproduction (Lin et al., 2010; Wang et al., 2012), in contrast to the Sxi1α-Sxi2a complex. Deletion of ZNF2 locks Cryptococcus cells in the yeast form, and overexpression of ZNF2 drives robust hyphal growth regardless of growth conditions (Wang et al., 2012). It is not surprising that ZNF2 is not regulated by the Sxi1α/Sxi2a heterodimer (Mead et al., 2015a). By contrast, the pheromone transcription factor Mat2 is one of the upstream regulators of Znf2 (Figure 3) (Lin et al., 2010). A forward genetics approach to identify mutations that recapitulate the deletion of ZNF2 revealed a long non-coding RNA (lncRNA) RZE1 that regulates the total number of the ZNF2 transcripts and the export of ZNF2 transcripts from the nucleus to the cytosol for translation (Figure 3) (Chacko et al., 2015; Fu et al., 2018; Xu et al., 2017). Other upstream factors of Znf2 are yet to be identified (Figure 3).
Figure 3.
The established and predicted regulatory pathways upstream of the master regulator of filamentation Znf2.
In addition to the genetic factors, the role of epigenetic factors in regulating cryptococcal yeast-to-hypha transition has also been investigated. Feretzaki et al. found that RNAi is hyperactive during sexual reproduction. Znf3, a zinc finger protein that regulates pheromone production and cell fusion, is required for this sex-induced silencing (Feretzaki et al., 2016; Feretzaki and Heitman, 2013). Factors that modify chromatin structures, including histone modifications and chromatin remodeling complexes, also regulate cryptococcal filamentation. For instance, the histone deacetylase and acetyltransferase regulate mating and/or virulence (Brandao et al., 2018; O’Meara et al., 2010). Five out of 15 examined plant homeodomain (PHD) finger proteins, which are potential readers of histone modifications, regulate hyphal growth either as suppressors or as activators in C. neoformans (Meng et al., 2018). A PAS (Per-Arnt-Sim) domain-containing protein Pas3 regulates Cryptococcus hyphal growth through interacting with the E3 ubiquitin ligase Bre1, which mediates the ubiquitination of histone H2B (Zhao et al., 2018). Given the role of H2B mono-ubiquitination in regulating transcription activation (Deng et al., 2020), it is reasoned that the regulation of cryptococcal filamentation is a combination of genetic and epigenetic regulation. The SWI/SNF chromatin remodeling complex that slides or evicts nucleosomes is essential for cryptococcal filamentation (Lin et al., 2019; Walton et al., 2005). The SWI/SNF complex facilitates effective binding of Znf2 to the promoters of its many downstream targets and is required to open the chromatin for the transcription of these genes (Lin et al., 2019). Collectively, these studies suggest that epigenetic regulation intertwines with genetic regulation to control sexual development in C. neoformans (Figure 3).
4. Cryptococcus morphogenesis and virulence
Canonical Cryptococcus virulence traits, such as the production of capsule and melanin, and tolerance to the host temperature and CO2 levels, have been previously reviewed (Nosanchuk and Casadevall, 2006; Perfect, 2006; Zaragoza et al., 2009b) or recently published (Krysan et al., 2019). Here, we focus on recently emerging concepts in the relationship between Cryptococcus morphogenesis and virulence.
4.1. The effect of morphotypes on host-Cryptococcus interactions
Heterogeneity in morphotype provides a hedge-betting strategy for C. neoformans to adapt to different niches or stresses. Basidiospores produced during sexual reproduction are resistant to various environmental stresses tested, including high temperature, desiccation, and oxidative stress (Botts et al., 2009). Their small size (~1–2 μm) allows basidiospores to get deep into lung alveoli and prevents effective clearance by the airway ciliary movement (Botts and Hull, 2010). Previous studies showed that spores of serotype D are more infectious than yeast cells in a murine inhalation model (Sukroongreung et al., 1998), while disease progression by serotype A spores is modestly delayed compared to that of yeast cells (Velagapudi et al., 2009). These findings suggest that the difference in virulence between spores and yeast cells may be serotype-dependent, and the molecular bases for such difference are unclear. A recent study showed that parental yeasts that are not virulent by themselves produce meiotic basidiospores that cause fatal meningitis in mice (Walsh et al., 2019). This difference could be attributed to the association of spores, but not yeast cells, to the lung-draining lymph nodes. In addition, mice infected intranasally with a mixture of JEC20 and JEC21 yeasts (1:1) showed a lower fungal burden in lungs than those infected with equal inoculum of spores derived from a JEC20 × JEC21 cross (Walsh et al., 2019), consistent with the previous conclusion that spores of serotype D are more infectious than yeast cells (Sukroongreung et al., 1998).
Filamentation is critical for Cryptococcus to produce spores or defend itself against predation by soil amoeba in the environment (Casadevall, 2012; Lin et al., 2015; Steenbergen et al., 2001). However, during infection within a mammalian host, C. neoformans cells grow almost exclusively in the yeast form. This is opposite from the basidiomycete plant pathogen U. maydis or the ascomycete human pathogen C. albicans, of which the filamentous form is the characteristic morphotype associated with host invasion (Madhani and Fink, 1998). Intravenous or intracranial infection with purified filaments from a self-filamentous C. neoformans strain causes cryptococcosis at lower rates compared to yeast cells of the same strain (Shadomy and Utz, 1966; Zimmer et al., 1983). Furthermore, the RAM mutants that propagate as pseudohyphae are drastically attenuated in virulence in a murine model of cryptococcosis (Magditch et al., 2012).
The molecular link between morphogenesis and virulence in C. neoformans was further strengthened with the characterization of Znf2 (Lin et al., 2010; Wang et al., 2012). Overexpression of ZNF2 drives cryptococcal hyphal formation and greatly attenuates virulence. Collectively, these studies using natural isolates and isogenic morphological mutants support an inverse relationship between Cryptococcus filamentation and virulence in a mammalian host. The difference in virulence of these morphotypes (yeast and filament) in mammalian hosts could be caused by the apparent physiological differences, chemical differences in molecular patterns at the cell surface, or the combination of all these factors. For instance, the surface of yeast and hyphal cells display different sets of molecules that elicit different host immune responses. Previous studies showed that wild-type yeast cells induce Th2 response, while hyphae induce Th1 response (Zhai et al., 2015). In addition, the polysaccharide capsule, which masks pathogen-associated molecular patterns (PAMPs) and is antiphagocytic, is thinner in hyphae comparing to that in yeast cells (Zhao et al., 2019). The polysaccharide capsule helps Cryptococcus avoid and survive the attack from phagocytic cells such as macrophages, dendritic cells, and neutrophils (Zaragoza et al., 2009a). The ability of capsulated yeast cells to proliferate both intracellularly and extracellularly in host cells facilitates cryptococcal dissemination to other organs including into the central nervous system (Alvarez and Casadevall, 2006; Charlier et al., 2009; Ma et al., 2006; Tucker and Casadevall, 2002). Therefore, the yeast form of Cryptococcus is advantageous to establish infection, evade host immune response, disseminate, and to cause fatal disease.
4.2. Protective immunity against cryptococcal infection
Most human fungal pathogens reside in the environment and are opportunistic pathogens. Hosts recognize their presence by detecting fungal antigens or conserved PAMPs through pattern recognition receptors (PRRs) expressed on host immune cells. Early recognition and inflammation could clear fungal pathogens and/or stimulate the adaptive antifungal immunity, supporting the benefit of vaccination against fungal infections. In B. dermatitidis, the adhesin Bad1 is specifically expressed in the virulent yeast-form and it blocks T cell activation (Finkel-Jimenez et al., 2002). The bad1Δ mutant evokes a protective immune response and serves as a live-attenuated vaccine to protect the host from a subsequent lethal infection by a wild-type strain (McBride et al., 2018). The C. albicans hypha-specific surface adhesin Als3 mediates fungal attachment and invasion into host cells (Liu and Filler, 2011). An anti-Candida vaccine (NDV-3A) designed based on Als3 is now in clinical trials (Edwards et al., 2018). This is the first vaccine composed of a recombinant fungal protein antigen tested in humans. Those pioneering studies affirm that vaccination can be an effective strategy to prevent and/or treat fungal infections.
Animals inoculated intraperitoneally with live cryptococcal cells in the pseudohyphal form develop immunity against cryptococcosis. These animals became resistant to a subsequent challenge by virulent C. neoformans yeast cells (Fromtling et al., 1979). Interestingly, immunocompromised mice immunized with a live C. neoformans strain expressing interferon-γ are completely protected from a challenge by the wild-type C. neoformans H99 strain (Wormley et al., 2007). This suggests that cryptococcal vaccination could work even in immunocompromised hosts (Caballero Van Dyke and Wormley, 2018). We showed that either live or heat-killed ZNF2oe filamentous cells can offer complete protection to mice from the subsequent infection with an otherwise highly aggressive isolate (Zhai et al., 2015). Recent discoveries of immune-protection from different cryptococcal mutants, including the live attenuated sterylglucosidase mutant (sgl1Δ) (Rella et al., 2015), the inactivated chitosan mutant (cda1–3Δ) (Lam et al., 2019), and the inactivated fbp1Δ mutant (Wang et al., 2019) further support the potential of inactivated whole cell vaccines or protein subunit vaccines to prevent the deadly cryptococcosis. Given that Znf2’s regulon is enriched with extracellular proteins, the dimorphism-associated factors likely profoundly shape host-pathogen interactions, providing a fertile ground to identify vaccine candidates and guide the development of immunotherapies. As most research is focused on molecules specific to the virulent morphological form, research on the virulence-attenuated morphological form is underexplored in Cryptococcus and other environmental dimorphic fungi.
4.3. Meiotic machinery-involved ploidy variation during infection
As an opportunistic fungal pathogen, Cryptococcus cells can stay dormant without causing any symptoms for decades in the lungs of immunocompetent individuals. Reactivation of latent pulmonary infections occur primarily in immunocompromised individuals. It is of great significance to unveil what promotes cryptococcal latency and how this fungus reactivates from dormancy.
Cryptococcus typically exists in a haploid state in the yeast form of 3–10 μm in diameter in vitro. However, heterogeneity in cryptococcal cell size in animal/human lungs has been documented for many decades. In a murine model of cryptococcosis, about 10 to 20 percent of cryptococcal H99 cells in the lungs form titan cells of 10–100 μm in diameter with polyploid DNA content (Okagaki et al., 2010; Zaragoza et al., 2010). The capsule layer of titan cells are highly cross-linked and tightly attached to a thicker cell wall compared to normal haploid cells (Zaragoza et al., 2010). These cells are resistant to phagocytosis, oxidative and nitrosative stresses, and antifungals (Crabtree et al., 2012; Gerstein et al., 2015; Okagaki and Nielsen, 2012). Interestingly, polyploid titan cells generate offspring with reduced-ploidy, primarily haploid daughter cells (Gerstein et al., 2015). Under stressful conditions (e.g. in the presence of antifungals), titan cells can produce diploid or aneuploid progeny with better adaptation to the stresses compared to progeny produced by normal yeast cells (Gerstein et al., 2015). It is proposed that titan cell formation may contribute to cryptococcal dormancy and regeneration of haploid cells may contribute to cryptococcal re-activation. What triggers polyploidization and what are the routes of depolyploidization remain unsolved.
During sexual reproduction, ploidy increase and reduction occur naturally. Meiosis is a form of reductive division that occurs following an increase in nuclear DNA resulting from karyogamy or endoduplication and pre-meiotic replication. In C. neoformans, meiosis occurs in the club-shaped basidial head developed at the tip of hyphae. Blocking meiosis does not affect hyphal growth or the development of basidial heads in vitro, but it does abolish or severely impair sporulation (Figure 1) (Feretzaki and Heitman, 2013; Liu et al., 2018). Given that yeast-to-hypha morphological differentiation associated with sexual reproduction in vitro is typically absent in vivo, and that disruption of the pheromone pathway specifically has no or minimal impact on cryptococcal virulence, meiosis had not been considered for any direct role during infection (Lin et al., 2009a). Meiosis is initiated intrinsically with programmed DNA double stranded breaks and might have been evolved prior to the emergence of the non-self-recognition system. Non-self-recognition chiefly primes cryptococcal cells for cell-cell fusion designed for bisexual reproduction (Gyawali et al., 2017). As diploidization during unisexual reproduction can be achieved through endoreplication, it is possible that meiosis can be uncoupled from non-self-recognition or morphogenesis under certain settings where bisexual reproduction is inhibited. We recently found that extreme genotoxic stresses, such as gamma radiation that causes DNA double stranded breaks, can efficiently trigger cryptococcal cell enlargement and polyploidization in vitro (Zhao et al., 2020). Cryptococcus likely experiences similar stress in the lungs of immunocompetent hosts. When these polyploid cells are released to a stress-free condition, some cells form multiple smaller nuclei, mimicking the meiotic features. Interestingly, blocking meiosis increases the proportion of titan cells in the lungs, likely by impairing ploidy reduction rather than polyploidization (Zhao et al., 2020). Consistently, cryptococcal meiosis-specific genes are activated in a subset of the population during infection. Surprisingly, cryptococcal isolates with meiosis-specific genes activated in vivo displayed phenotypic diversity in terms of resistance to specific genotoxic stress that is not observed in sibling isolates recovered from the same tissue in the same host (Zhao et al., 2020). The findings strongly suggest that Cryptococcus uses the sexual program (gametogenesis) for direct adaptation to promote its individual survival in the host. So far, meiotic events in fungi occurring during invasive infection in a mammalian host have only been documented in the obligate pathogen Pneumocystis (Aliouat el et al., 1999; Almeida et al., 2015; Cushion et al., 2007; Kutty et al., 2010; Peters et al., 2001). That said, multiple eukaryotic pathogens, particularly parasites, undergo sexual differentiation during infection. Cancer cells, which in essence act like eukaryotic pathogens, are known to use the sexual program to promote their own cellular propagation at the cost of the whole organism (Erenpreisa et al., 2015). Thus, understanding sexual reproduction, even in a relatively ‘simple’ eukaryotic microbe like C. neoformans, will yield important novel insights into a conserved mechanism for adaptation and rejuvenation in diverse eukaryotic species.
5. Summary of key points
Cryptococcal sexual reproduction, the pheromone pathway for non-self-recognition, and morphogenesis are intimately associated. During unisexual reproduction, however, the sexual program could be uncoupled from the latter processes under certain conditions.
Znf2 is the master regulator that governs the ultimate yeast-to-hypha transition in C. neoformans. Multiple signaling pathways converge at Znf2 to promote or inhibit cryptococcal filamentation. The internal and external stimuli and their cognate receiving pathways that convey the information to Znf2 remain largely unknown.
Cell surface antigens likely elicit the immune-protection observed in filamentous strains (e.g. ZNF2oe). Identifying the key immune-protective antigens would be the key to guide the development of subunit vaccines or to monitor the quality of inactivated whole cell vaccines.
The Cryptococcus sexual machinery likely contributes to its disease progression. Future challenges include identifying the host factors that activate sexual machinery and dissecting the molecular connection between sexual development and cryptococcal latency and reactivation during infection.
Highlights.
1) Cryptococcal sexual reproduction, the pheromone pathway for non-self-recognition, and morphogenesis are intimately associated.
2) Znf2 is the master regulator that governs the ultimate yeast-to-hypha transition in C. neoformans.
3) Cell surface antigens likely elicit the immune-protection observed in filamentous strains (e.g. ZNF2oe).
4) The Cryptococcus sexual machinery likely contributes to its disease progression.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliouat el M, Dujardin L, Martinez A, Duriez T, Ricard I, Dei-Cas E, 1999. Pneumocystis carinii growth kinetics in culture systems and in hosts: involvement of each life cycle parasite stage. J Eukaryot Microbiol 46, 116S–117S. [PubMed] [Google Scholar]
- Almeida JM, Cisse OH, Fonseca A, Pagni M, Hauser PM, 2015. Comparative genomics suggests primary homothallism of Pneumocystis species. mBio 6, e02250–02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A, 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16, 2161–2165. [DOI] [PubMed] [Google Scholar]
- Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM, 2009. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell 8, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botts MR, Hull CM, 2010. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol 13, 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao F, Esher SK, Ost KS, Pianalto K, Nichols CB, Fernandes L, Bocca AL, Pocas-Fonseca MJ, Alspaugh JA, 2018. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci Rep 8, 5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero Van Dyke MC, Wormley FL Jr., 2018. A call to arms: quest for a cryptococcal vaccine. Trends Microbiol 26, 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, 2012. Amoeba provide insight into the origin of virulence in pathogenic fungi. Adv Exp Med Biol 710, 1–10. [DOI] [PubMed] [Google Scholar]
- Chacko N, Zhao Y, Yang E, Wang L, Cai JJ, Lin X, 2015. The lncRNA RZE1 controls cryptococcal morphological transition. PLoS Genet 11, e1005692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ, 2003. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect Immun 71, 4953–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Wickes BL, Miller GF, Penoyer LA, Kwon-Chung KJ, 2000. Cryptococcus neoformans STE12a regulates virulence but is not essential for mating. J Exp Med 191, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F, 2009. Evidence of a role for monocytes in dissemination and brain Invasion by Cryptococcus neoformans. Infect Immun 77, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Litvintseva AP, Frazzitta AE, Haverkamp MR, Wang LY, Fang C, Muthoga C, Mitchell TG, Perfect JR, 2015. Comparative analyses of clinical and environmental populations of Cryptococcus neoformans in Botswana. Mol Ecol 24, 3559–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, 2013. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an atlas of the molecular types. Scientifica, 675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, Nielsen K, 2012. Titan cell production enhances the virulence of Cryptococcus neoformans. Infect Immun 80, 3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J, 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One 2, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Nicholls CB, Cox GM, Perfect JR, Heitman J, 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol 49, 469–485. [DOI] [PubMed] [Google Scholar]
- Deng ZH, Ai HS, Lu CP, Li JB, 2020. The Bre1/Rad6 machinery: writing the central histone ubiquitin mark on H2B and beyond. Chromosome Res 28(3–4):247–258. [DOI] [PubMed] [Google Scholar]
- Dromer F, Casadevall A, Perfect J, Sorrell T, 2011. Cryptococcus neoformans: Latency and disease. ASM Press, Washington, DC.. [Google Scholar]
- Edwards JE Jr., Schwartz MM, Schmidt CS, Sobel JD, Nyirjesy P, Schodel F, Marchus E, Lizakowski M, DeMontigny EA, Hoeg J, Holmberg T, Cooke MT, Hoover K, Edwards L, Jacobs M, Sussman S, Augenbraun M, Drusano M, Yeaman MR, Ibrahim AS, Filler SG, Hennessey JP Jr., 2018. A fungal immunotherapeutic vaccine (NDV-3A) for treatment of recurrent vulvovaginal candidiasis-a phase 2 randomized, double-Blind, placebo-controlled Trial. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 66, 1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erenpreisa J, Salmina K, Huna A, Jackson TR, Vazquez-Martin A, Cragg MS, 2015. The “virgin birth”, polyploidy, and the origin of cancer. Oncoscience 2, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Billmyre RB, Clancey SA, Wang XY, Heitman J, 2016. Gene network polymorphism illuminates loss and retention of novel RNAi silencing components in the Cryptococcus pathogenic species complex. PLoS Genet 12, e1005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Heitman J, 2013. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet 9, e1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel-Jimenez B, Wüthrich M, Klein BS, 2002. BAD1, an essential virulence factor of Blastomyces dermatitidis, suppresses host TNF-α production through TGF-β-dependent and -independent mechanisms. J Immunol 168, 5746–5755. [DOI] [PubMed] [Google Scholar]
- Fromtling RA, Blackstock R, Hall NK, Bulmer GS, 1979. Immunization of mice with an avirulent pseudohyphal form of Cryptococcus neoformans. Mycopathologia 68, 179–181. [DOI] [PubMed] [Google Scholar]
- Fu C, Donadio N, Cardenas ME, Heitman J, 2018. Dissecting the roles of the calcineurin pathway in unisexual reproduction, stress responses, and virulence in Cryptococcus deneoformans. Genetics 208, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Heitman J, 2017. PRM1 and KAR5 function in cell-cell fusion and karyogamy to drive distinct bisexual and unisexual cycles in the Cryptococcus pathogenic species complex. PLoS Genet 13, e1007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Fu MS, Mukaremera L, Li ZM, Ormerod KL, Fraser JA, Berman J, Nielsen K, 2015. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 6, e01340–01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali R, Zhao Y, Lin J, Fan Y, Xu X, Upadhyay S, Lin X, 2017. Pheromone independent unisexual development in Cryptococcus neoformans. PLoS Genet 13, e1006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Ceresini PC, Polacheck I, Ma H, van Nieuwerburgh F, Gabaldón T, Kagan S, Pursall ER, Hoogveld HL, van Iersel LJJ, Klau GW, Kelk SM, Stougie L, Bartlett KH, Voelz K, Pryszcz LP, Castañeda E, Lazera M, Meyer W, Deforce D, Meis JF, May RC, Klaassen CHW, Boekhout T, 2013. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One 8, e71148-e71148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann HA, Kruger J, Lottspeich F, Kahmann R, 1999. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell 11, 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimel K, Scherer M, Vranes M, Wahl R, Pothiratana C, Schuler D, Vincon V, Finkernagel F, Flor-Parra I, Kamper J, 2010. The transcription factor Rbf1 Is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog 6, e1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Shen WC, 2005. A homolog of Ste6, the a-factor transporter in Saccharomyces cerevisiae, is required for mating but not for monokaryotic fruiting in Cryptococcus neoformans. Eukaryot Cell 4, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue CY, Heitman J, 2007. G protein signaling governing cell fate decisions involves opposing G alpha subunits in Cryptococcus neoformans. Mol Biol Cell 18, 3237–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily MJ, Heitman J, 2005. Sex-specific homeodomain proteins Sxi1 alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell 4, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, Heitman J, 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1 alpha. Gene Dev 16, 3046–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SK, Bennett RJ, 2011. Fungal mating pheromones: choreographing the dating game. Fungal Genet Biol 48, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruzel EK, Giles SS, Hull CM, 2012. Analysis of Cryptococcus neoformans sexual development reveals rewiring of the pheromone-response network by a change in transcription factor identity. Genetics 191, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan DJ, Zhai B, Beattie SR, Misel KM, Wellington M, Lin XR, 2019. Host carbon dioxide concentration is an independent stress for Cryptococcus neoformans that affects virulence and antifungal susceptibility. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty G, Achaz G, Maldarelli F, Varma A, Shroff R, Becker S, Fantoni G, Kovacs JA, 2010. Characterization of the meiosis-specific recombinase Dmc1 of Pneumocystis. J Infect Dis 202, 1920–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67, 1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE, 1978. Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol 108, 337–340. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M, 2002. (1557) Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). TAXON 51, 804–806. [Google Scholar]
- Lam WC, Upadhya R, Specht CA, Ragsdale AE, Hole CR, Levitz SM, Lodge JK, 2019. Chitosan biosynthesis and virulence in the human fungal pathogen Cryptococcus gattii. mSphere 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Idnurm A, Lin X, 2015. Morphology and its underlying genetic regulation impact the interaction between Cryptococcus neoformans and its hosts. Med Mycol 53, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JF, Zhao YB, Ferraro AR, Yang EC, Lewis ZA, Lin XR, 2019. Transcription factor Znf2 coordinates with the chromatin remodeling SWI/SNF complex to regulate cryptococcal cellular differentiation. Commun Biol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J, 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434, 1017–1021. [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J, 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 6, e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J, 2007. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet 3, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J, 2009a. Diploids in the Cryptococcus neoformans serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog 5, e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XR, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J, 2009b. Diploids in the Cryptococcus neoformans Serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathogens 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LX, He GJ, Chen L, Zheng J, Chen YY, Shen L, Tian XY, Li EW, Yang EC, Liao GJ, Wang LQ, 2018. Genetic basis for coordination of meiosis and sexual structure maturation in Cryptococcus neoformans. eLife 7, e38683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Filler SG, 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10, 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HS, Croudace JE, Lammas DA, May RC, 2006. Expulsion of live pathogenic yeast by macrophages. Curr Biol 16, 2156–2160. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR, 1998. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol 8, 348–353. [DOI] [PubMed] [Google Scholar]
- Magditch DA, Liu T-B, Xue C, Idnurm A, 2012. DNA Mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog 8, e1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matha AR, Lin XR, 2020. Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans. Pathogens 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JA, Gauthier GM, Klein BS, 2018. Turning on virulence: mechanisms that underpin the morphologic transition and pathogenicity of Blastomyces. Virulence, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead ME, Stanton BC, Kruzel EK, Hull CM, 2015a. Targets of the Sex Inducer homeodomain proteins are required for fungal development and virulence in Cryptococcus neoformans. Mol Microbiol 95, 804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead ME, Stanton BC, Kruzel EK, Hull CM, 2015b. Targets of the sex inducer homeodomain proteins are required for fungal development and virulence in Cryptococcus neoformans. Mol Microbiol 95, 804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Fan Y, Liao W, Lin X, 2018. Plant homeodomain genes play important roles in cryptococcal yeast-hypha transition. Appl Environ Microb 84, e01732–01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk JD, Casadevall A, 2006. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Chemother 50, 3519–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Hay C, Price MS, Giles S, Alspaugh JA, 2010. Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot Cell 9, 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Nielsen K, 2012. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell 11, 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K, 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 6, e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto B, 1894. Ueber parasitäre Zelleinschlüsse und ihre Züchtung. Centralblatt für Bakteriologie und Parasitenkunde 16, 175–180. [Google Scholar]
- Perfect JR, 2006. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Res 6, 463–468. [DOI] [PubMed] [Google Scholar]
- Perfect JR, 2015. Cryptococcosis (Cryptococcus neoformans and Cryptococcus gattii), in: Bennett JE, Dolin R, Blaser MJ (Eds.), Mandell, Douglas, and Bennett’s principles and practice of infectious diseases (Eighth Edition), Philadelphia, pp. 2934–2948. [Google Scholar]
- Peters SE, English K, Rana A, Akter S, Malik S, Warburton NC, Duckett JG, 2001. Synaptonemal complexes in the pre-cyst of Pneumocystis carinii. J Eukaryot Microbiol Suppl, 134S. [DOI] [PubMed] [Google Scholar]
- Rella A, Mor V, Farnoud AM, Singh A, Shamseddine AA, Ivanova E, Carpino N, Montagna MT, Luberto C, Del Poeta M, 2015. Role of Sterylglucosidase 1 (Sgl1) on the pathogenicity of Cryptococcus neoformans: potential applications for vaccine development. Front Microbiol 6, 836–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Desjardins CA, Sykes SM, Beale MA, Vanhove M, Sakthikumar S, Chen Y, Gujja S, Saif S, Chowdhary A, Lawson DJ, Ponzio V, Colombo AL, Meyer W, Engelthaler DM, Hagen F, Illnait-Zaragozi MT, Alanio A, Vreulink JM, Heitman J, Perfect JR, Litvintseva AP, Bicanic T, Harrison TS, Fisher MC, Cuomo CA, 2017. Tracing genetic exchange and biogeography of Cryptococcus neoformans var. grubii at the global population level. Genetics 207, 327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Fink GR, 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Gene Dev 8, 2974–2985. [DOI] [PubMed] [Google Scholar]
- Roth C, Sun S, Billmyre RB, Heitman J, Magwene PM, 2018. A high-resolution map of meiotic recombination in Cryptococcus deneoformans demonstrates decreased recombination in unisexual reproduction. Genetics 209, 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfelice F, 1895. Sull’azione patogena dei blastomiceti. Annali d’Igiene Sperimentale 5, 239–262. [Google Scholar]
- Shadomy HJ, Utz JP, 1966. Preliminary studies on a hyphaforming mutant of Cryptococcus neoformans. Mycologia 58, 383–390. [PubMed] [Google Scholar]
- Sil A, Andrianopoulos A, 2015. Thermally dimorphic human fungal pathogens-polyphyletic pathogens with a convergent pathogenicity trait. CSH Perspect Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Shuman HA, Casadevall A, 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A 98, 15245–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukroongreung S, Kitiniyom K, Nilakul C, Tantimavanich S, 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med Mycol 36, 419–424. [PubMed] [Google Scholar]
- Sun S, Billmyre RB, Mieczkowski PA, Heitman J, 2014. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet 10, e1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SC, Casadevall A, 2002. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc Natl Acad Sci U S A 99, 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J, 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun 77, 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NM, Botts MR, McDermott AJ, Ortiz SC, Wuthrich M, Klein B, Hull CM, 2019. Infectious particle identity determines dissemination and disease outcome for the inhaled human fungal pathogen Cryptococcus. PLoS Pathog 15, e1007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FJ, Idnurm A, Heitman J, 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol 57, 1381–1396. [DOI] [PubMed] [Google Scholar]
- Wang L, Tian X, Gyawali R, Upadhyay S, Foyle D, Wang G, Cai JJ, Lin X, 2014. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog 10, e1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhai B, Lin X, 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog 8, e1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Wang KY, Masso-Silva JA, Rivera A, Xue CY, 2019. A heat-killed Cryptococcus mutant strain induces host protection against multiple invasive mycoses in a murine vaccine model. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Edman U, Edman JC, 1997. The Cryptococcus neoformans STE12a gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol 26, 951–960. [DOI] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC, 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc Natl Acad Sci U S A 93, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormley FL, Perfect JR, Steele C, Cox GM, 2007. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect Immun 75, 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Li JF, Zhao YB, Kirkman E, So YS, Bahn YS, Lin XR, 2017. Glucosamine stimulates pheromone-independent dimorphic transition in Cryptococcus neoformans by promoting Crz1 nuclear translocation. PLoS Genetics 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue CY, Hsueh YP, Heitman J, 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev 32, 1010–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Sun S, Heitman J, 2020. Uniparental sexual reproduction following cell-cell fusion of opposite mating-type partners. 10.1101/2020.12.03.410415 [DOI]
- Yan Z, Li XG, Xu JP, 2002. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol 40, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A, 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog 6, e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A, 2009a. The capsule of the fungal pathogen Cryptococcus neoformans, Adv Appl Microbiol. Academic Press, pp. 133–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A, 2009b. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68, 133–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Wozniak KL, Masso-Silva J, Upadhyay S, Hole C, Rivera A, Wormley FL Jr., Lin X, 2015. Development of protective inflammation and cell-mediated immunity against Cryptococcus neoformans after exposure to hyphal mutants. mBio 6, e01433–01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Upadhyay S, Lin X, 2018. PAS domain protein Pas3 interacts with the chromatin modifier Bre1 in regulating cryptococcal morphogenesis. mBio 9, e02135–02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YB, Lin JF, Fan YM, Lin XR, 2019. Life cycle of Cryptococcus neoformans. Annu Rev Microbiol 73, 17–42. [DOI] [PubMed] [Google Scholar]
- Zhao YB, Wang YN, Upadhyay S, Xue CY, Lin XR, 2020. Activation of meiotic genes mediates ploidy reduction during cryptococcal Infection. Curr Biol 30, 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer BL, Hempel HO, Goodman NL, 1983. Pathogenicity of the hyphae of Filobasidiella neoformans. Mycopathologia 81, 107–110. [DOI] [PubMed] [Google Scholar]