Abstract
Background
COVID-19 prediction models based on clinical characteristics, routine biochemistry and imaging, have been developed, but little is known on proteomic markers reflecting the molecular pathophysiology of disease progression.
Methods
The multicentre (six European study sites) Prospective Validation of a Proteomic Urine Test for Early and Accurate Prognosis of Critical Course Complications in Patients with SARS-CoV-2 Infection Study (Crit-COV-U) is recruiting consecutive patients (≥ 18 years) with PCR-confirmed SARS-CoV-2 infection. A urinary proteomic biomarker (COV50) developed by capillary-electrophoresis-mass spectrometry (CE-MS) technology, comprising 50 sequenced peptides and identifying the parental proteins, was evaluated in 228 patients (derivation cohort) with replication in 99 patients (validation cohort). Death and progression along the World Health Organization (WHO) Clinical Progression Scale were assessed up to 21 days after the initial PCR test. Statistical methods included logistic regression, receiver operating curve (ROC) analysis and comparison of the area under the curve (AUC).
Findings
In the derivation cohort, 23 patients died, and 48 developed worse WHO scores. The odds ratios (OR) for death per 1 standard deviation (SD) increment in COV50 were 3·52 (95% CI, 2·02–6·13, p <0·0001) unadjusted and 2·73 (1·25–5·95, p = 0·012) adjusted for sex, age, baseline WHO score, body mass index (BMI) and comorbidities. For WHO scale progression, the corresponding OR were 2·63 (1·80–3·85, p<0·0001) and 3·38 (1·85–6·17, p<0·0001), respectively. The area under the curve (AUC) for COV50 as a continuously distributed variable was 0·80 (0·72–0·88) for mortality and 0·74 (0·66–0·81) for worsening WHO score. The optimised COV50 thresholds for mortality and worsening WHO score were 0·47 and 0·04 with sensitivity/specificity of 87·0 (74·6%) and 77·1 (63·9%), respectively. On top of covariates, COV50 improved the AUC, albeit borderline for death, from 0·78 to 0·82 (p = 0·11) and 0·84 (p = 0·052) for mortality and from 0·68 to 0·78 (p = 0·0097) and 0·75 (p = 0·021) for worsening WHO score. The validation cohort findings were confirmatory.
Interpretation
This first CRIT-COV-U report proves the concept that urinary proteomic profiling generates biomarkers indicating adverse COVID-19 outcomes, even at an early disease stage, including WHO stages 1–3. These findings need to be consolidated in an upcoming final dataset.
Funding
The German Federal Ministry of Health funded the study.
Keywords: COVID-19, Disease severity, Risk score, SARS-CoV-2, Urinary proteomics
Research in context.
Evidence before this study
A PubMed search without limitations on publication date or language using the terms COVID-19 AND risk prediction produced 1734 or 11 results, resp., if proteomics was added. Studies focused on the molecular basis of COVID-19 manifestations, one study focused on blood metabolomics and risk assessement. One preliminary report ahead of peer review described a proteomic profile in the serum of 49 COVID-19 patients predicting critical illness and death. Other studies addressed inflammatory, immunological, and T-cell responses to infection or were literature reviews.
Added value of this study
This study is the first to include a specific urinary proteomic biomarker (COV50) in a model predicting death and worsening WHO scores up to 21 days from PCR-confirmed SARS-CoV-2 infection. The COV50-associated AUC was 0·80 (95% CI, 0·72–0·88) for mortality and 0·74 (0·66-0·81) for worsening WHO severity score.
Implications of all the available evidence
This first report of the Prospective Validation of a Proteomic Urine Test for Early and Accurate Prognosis of Critical Course Complications in Patients with SARS-CoV-2 Infection Study (CRIT-COV-U) proves the concept that UPP generates biomarkers indicative of disease outcome even at the preclinical stage of SARS-CoV-2 infection. These findings need to be consolidated in an upcoming final dataset.
Alt-text: Unlabelled box
1. Introduction
Since the outbreak of the COVID-19 pandemic in Wuhan, People's Republic of China, an exponentially growing literature has described the clinical characteristics of infected patients at high risk of severe disease and death [[1], [2], [3]]. The burden on health care has led to the development of models that predict progression to adverse outcomes with the principal objective of supporting clinical decision-making. A systematic review of the literature published in April 2020 and updated thereafter summarised over 50 prognostic models, which commonly include sex, age, comorbidities, C-reactive protein, lymphocyte count, body temperature, serum creatinine, and imaging features; however, the review cautioned that the models are vulnerable to bias and not clinically applicable [4]. More recently published models, including the Coronavirus Clinical Characterisation Consortium score (4C), have been properly calibrated and have improved in accuracy [[5], [6], [7]], but none has considered the molecular pathophysiology of the progression from silent infection to critical disease.
SARS-CoV-2 preferentially infects the respiratory tract cells but also penetrates the heart, liver, brain, kidneys, and blood vessels [8] The infection is therefore a systemic disease, leading to potential multiorgan failure [9]. Urine contains an array of over 20,000 endogenous peptides, which are partly generated along the nephron or from the circulation passing through the glomerular barrier. The urinary peptidome profile (UPP) therefore provides a system-wide molecular signature of progressing SARS-CoV-2 infection. Sequencing of urinary peptides allows identification of the parental proteins [10,11]. Multidimensional urinary peptide profiles can already provide a specific molecular signature in the preclinical phase of heart failure [12]. chronic kidney disease (CKD) [13], and diabetic nephropathy [14]. A proof-of-concept study suggested that the UPP at the initial SARS-CoV-2 infection stage may feasibly predict outcome [15]. The Prospective Validation of a Proteomic Urine Test for Early and Accurate Prognosis of Critical Course Complications in Patients with SARS-CoV-2 Infection Study (CRIT-COV-U) was therefore designed to develop and validate a UPP biomarker for prediction of the outcome of SARS-CoV-2–infected patients [[5], [6], [7]].
2. Methods
2.1. Study design and participants
The CRIT-COV-U project complies with the Helsinki declaration [16]. The Ethics Committee of the German-Saxonian Board of Physicians, Dresden, Germany (number EK-BR-88/20.1) and the Institutional Review Boards of the recruiting sites provided ethical clearance. The protocol was registered with the German Register for Clinical Studies (www.drks.de), number DRKS00022495, which is interconnected with the WHO International Clinical Trial Registry Platform (www.who.int/clinical-trials-registry-platform).
CRIT-COV-U is a prospective multicentre cohort study that aims to identify UPP biomarkers predictive of the clinical course in adults with PCR-confirmed SARS-CoV-2 infection [17]. To be eligible, patients had to be ≥ 18 years old, not anuric and able to give informed written consent. Six European study sites, two located in Germany (161/6 pts.) and one each in France (50 pts.), the Republic of North Macedonia (69 pts.), Poland (19 pts.) and Sweden (22 pts.), enrolled consecutive patients. The patients were diagnosed while in ambulatory care or on the first day of hospital care and followed up for at least 21 days or until hospital discharge or death, whichever occurred first. Study procedures were explained and patients were asked to provide written informed consent after positive SARS-CoV-2 test. At each of three timepoints (timepoint 1: days 0–2, timepoint 2: days 4–7, and timepoint 3: days 10–21 after initial positive PCR-confirmed diagnosis, the patients were staged according to the WHO Clinical Progression Scale in eight categories [18]: (1) ambulatory without limitation of activity; (2) ambulatory with limited activity; (3) hospitalised without oxygen therapy; (4) hospitalised on oxygen therapy by mask or nasal prongs; (5) hospitalised receiving non-invasive ventilation or high-flow oxygen therapy; (6) hospitalised with intubation and mechanical ventilation; (7) hospitalised with mechanical ventilation and additional organ support, such as vasopressors, renal replacement therapy, or extracorporeal membrane oxygenation; and (8) death. The information collected via electronic case report forms (MARVIN EDC, XClinical GmbH, Munich, Germany) included demographic and clinical characteristics, such as ethnicity, sex, age, body mass index, and blood pressure and routine biochemical measurements, such as glomerular filtration derived from serum creatinine [19].
The sample size calculations informed by the proof-of-concept study [15], proposed a derivation phase sample size of 212 patients with critical COVID-19 (WHO stage ≥6) to be contrasted with 271 patients with mild symptoms to identify an UPP, yielding a sensitivity and specificity of 75% and 80%, respectively. Following a request from the German regulators faced with the extraordinary burden placed on health care, the CRIT-COV-U database was frozen on 17 December 2020 for an interim analysis with 228 and 99 patients enrolled in the derivation and validation cohorts, respectively.
2.2. Urinary proteome analysis, biomarker identification and classifier generation
For UPP, 8 ml urine samples were collected in borated test tubes (ExactoBac-U®, Sarstedt, Nümbrecht, Germany) at each clinical staging timepoint (days 0–2, 4–7, and 10–21). The samples were kept at -20 °C until assayed. The methods for capillary electrophoresis coupled with mass spectrometry, peptide sequencing, and, for the evaluation, the calibration and quality control of the mass spectrometric data have been published [11,20,21] and are outlined in the Appendix. For identification of the urinary biomarker, 186 urine samples were randomly selected from those available in the derivation dataset at timepoints 2 and 3, excluding samples from the patients at COVID-19 stages 4 and 5 according to the WHO classification, allowing contrast of the UPP profiles at stages 1–3 (n = 116) and stages 6–8 (n = 88). After transformation of the mass spectrometric spectra, the levels of peptides with known amino acid sequences were compared between the patients with mild disease and those with critical disease, using the Wilcoxon rank sum test with adjustment of the significance for multiple comparisons by the Benjamini–Hochberg method [22]. The disease-specific classifier was developed using support vector machine modelling and cross-validated by a leave-one-out procedure with significance adjusted for the false-discovery rate set at 0·05.
2.3. Statistical analysis
For database management and statistical analysis, SPSS (version 22·0) and SAS (version 9·4) software were used. Significance was a two-tailed significance of 0·05 or less. Means and proportions were compared using the large-sample z-test or analysis of variance and Fisher's exact test, respectively. The predefined endpoints were mortality and progression across the WHO COVID-19 severity scale. In the derivation dataset, the incidence of endpoints was related to the proteomic classifier using single and multiple logistic regression analysis, taking into account previously reported risk factors such as sex, age, the WHO scale at timepoint 1, and comorbidities, including hypertension, diabetes, heart failure, and cancer. The performance of COV50 in risk stratification was assessed by the area under the receiver operating curve (AUROC) and the Delong approach to compare the areas under the ROCs between the nested models. Internal validation was performed by calculating the leave-one-out cross-validated AUC. Prior to computing the sensitivity and specificity, the COV50 thresholds were optimised using the Youden index. The AUC, sensitivity, and specificity in the validation dataset were calculated from the logistic model and the thresholds derived from the derivation cohort. For further external validation, the distribution of the COV50 classifier was evaluated in 981 controls randomly selected from the human CE-MS proteome database available at Mosaiques-Diagnostics, Hanover, Germany. The controls sampled before the end of 2019 were therefore free of COVID-19, and they were matched for sex, age (± 5 years), and body mass index (± 1 kg/m2) in a 3:1 proportion with the 327 patients enrolled in the current analysis. Finally, we compared the performance of COV50 with the 4C score [6] to predict mortality.
2.4. Role of the funding source
The funder of the study had no role in the study design, data collection, data analysis, data interpretation, or the writing of the report. All the authors had full access to all the data in the study and had final responsibility for the decision to submit the manuscript.
3. Results
Comparing the UPP at stages 1–3 with the UPP at stages 6–8 of COVID disease at timepoints 2 and 3 identified 1132 significantly deregulated peptides. To generate the COV50 classifier, 100 peptides in the top tail of the significance distribution were combined by support vector modelling and reduced to 50 by applying leave-one-out cross-validation. The 50 sequenced peptides making up the UPP biomarker and the parental proteins from which the peptide fragments were derived are listed in the Appendix. Using the urine samples collected at timepoint 1, the association of the severity of infection during follow-up was prospectively studied in relation to the 50-peptide urinary biomarker (COV50) and potential confounders, first in the 228 patients included in the derivation dataset and next in the 99 patients enrolled in the validation dataset. The 228 participants enrolled in the derivation cohort (Table 1) were on average 63·1 years old and included 94 (41·2%) women and 152 (66·7%) patients with comorbidities (Appendix Fig. 1), including hypertension (n = 137), heart failure (n = 30), diabetes mellitus (n = 65), and cancer (n = 13) and 119 (52·2%) patients being treated with inhibitors of the renin-angiotensin system, either angiotensin-converting enzyme inhibitors (n = 67) or angiotensin-receptor blockers (n = 58). The WHO score at enrolment was 1–3 in 90 (39·5%) patients, 4–5 in 107 (46·9%) patients, and 6 in 31 (13·6%) participants.
Table 1.
Baseline (time point 1) characteristics.
| Characteristic | Derivation cohort | Validation cohort | p value |
|---|---|---|---|
| Number in cohort | 228 | 99 | |
| Main study variables | |||
| Number (%) in WHO class | |||
| 1–3 | 90 (39.5) | 37 (37.4) | <0.0001 |
| 4–5 | 107 (46.9) | 60 (60.6) | |
| 6 | 31 (13.6) | 2 (2.0) | |
| Mean (SD) of COV50 level | -0.19 (1.52) | -0.17 (1.23) | 0.92 |
| Number with characteristic (%) | |||
| White ethnicity | 205 (89.9) | 91 (91.9) | 0.68 |
| Women | 94 (41.2) | 43 (43.4) | 0.72 |
| Non-smoker | 109 (47.8) | 58 (58.6) | 0.031 |
| Hypertension | 137 (60.1) | 66 (66.7) | 0.27 |
| Heart failure | 30 (13.6) | 27 (27.8) | 0.0039 |
| Body mass index ≥30 kg/m2 | 68 (29.8) | 26 (26.3) | 0.60 |
| Diabetes mellitus | 65 (28.5) | 41 (41.4) | 0.028 |
| Cancer | 13 (5.7) | 7 (7.1) | 0.62 |
| Use of RAS blockers, | 119 (52.2) | 55 (55.6) | 0.27 |
| Mean (SD) of characteristic | |||
| Age, yr | 63.1 (17.1) | 66.8 (16.1) | 0.68 |
| Systolic blood pressure, mm Hg | 130.0 (23.4) | 127.5 (20.2) | 0.35 |
| Diastolic blood pressure, mm Hg | 79.9 (55.0) | 75.6 (12.2) | 0.45 |
| Heart rate, beats per minute | 83.4 (15.0) | 82.8 (17.9) | 0.75 |
| Body mass index, kg/m2 | 28.1 (6.0) | 27.4 (4.6) | 0.24 |
| Glomerular filtration rate, ml/min/1.73 m2 | 76.7 (30.9) | 78.7 (30.4) | 0.63 |
RAS blockers indicate blocker of the renin-angiotensin system, including angiotensin-converting enzyme inhibitors and angiotensin-receptor blockers. The glomerular filtration rate was derived from serum creatinine, using the Chronic Kidney Disease Epidemiology Collaboration. The p value indicates the difference between the timepoint 1 characteristics of the derivation and validation cohort.
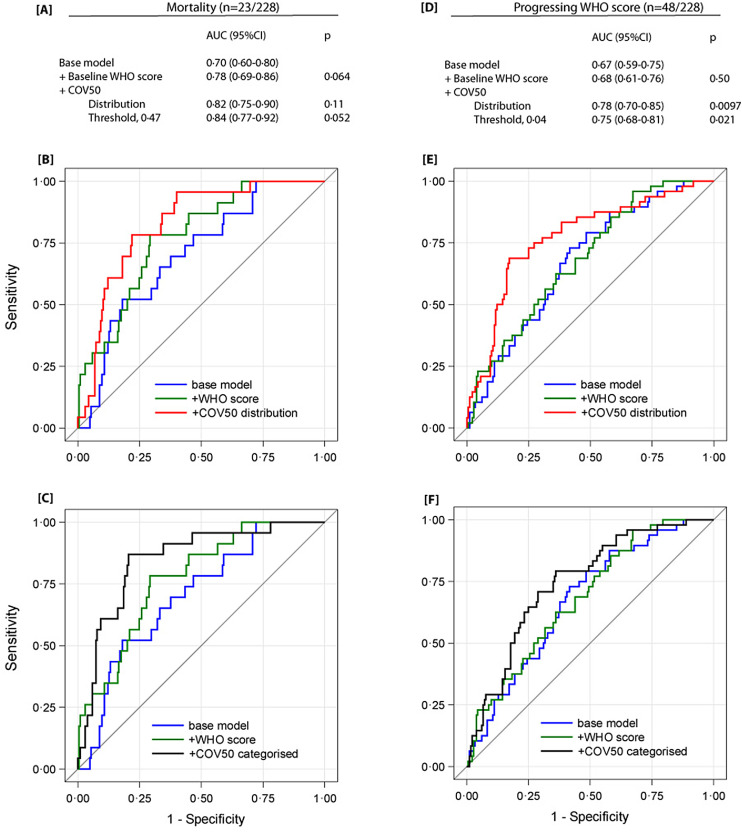
Fig. 1.
Performance of COV50 on top of other baseline risk factors in the derivation cohort to discriminate death from survival (panels A-C) and progression from non-progression in the time point 1 WHO score during follow-up (panels D-F) in the derivation cohort
The base model included sex, age, body mass index and the presence of comorbidities: hypertension, heart failure, diabetes or cancer. In subsequent steps, the time point 1 WHO score was added and next COV50 as a continuously distributed variable (panels B and E) or as a categorised variable based on an optimised threshold of 0.47 for mortality (panel C) or 0.04 for a worsening WHO score (panel F).
Across increasing fourths of the COV50 distribution (Table 2), the proportion of women decreased and age and the prevalence of hypertension, heart failure, and diabetes increased. During follow-up, 23 patients died and 48 progressed along the WHO scores. The COV50 distribution at timepoint 1 shifted upward (p < 0·0001) when plotted against the worst WHO score attained during follow-up (Appendix Fig. 2). For death (Table 3), the relative risk expressed per 1 SD increment in COV50 was 3·52 (95% CI, 2·02–6·13, p < 0·0001) unadjusted and 2·73 (1·25–5·95, p = 0·012) when fully adjusted for sex, age, timepoint 1 WHO score, body mass index, and the presence of comorbidities. For progression along WHO scores (Table 3), the corresponding OR were 2·63 (1·80–3·85, p < 0·0001) and 3·38 (1·85–6·17, p < 0·0001), respectively. The AUC for COV50 analysed as a continuously distributed variable was 0·82 (95% confidence interval, 0·74–0·89) for total mortality and 0·75 (0·67–0·82) for progressing WHO score (Table 4). The optimised COV50 thresholds for total mortality and progressing WHO score were 0·47 and 0·04 and resulted in estimates of sensitivity/specificity of 87·0 (74·6%) and 77·1 (63·9%), respectively (Table 4).
Table 2.
Baseline (timepoint 1) characteristics by fourths of the baseline COV50 distribution in the derivation cohort.
| Characteristic | Low | Medium-low | Medium-high | High | p value for trend |
|---|---|---|---|---|---|
| COV50 limits | -1.23 | [-1.23, -0.20[ | [-0.20, 0.90[ | ≥0.90 | |
| Number in group | 57 | 57 | 57 | 57 | |
| Main study variables | |||||
| Number (%) in WHO class | |||||
| 1-3 | 50 (87.7) | 20 (35.1) | 19 (33.3) | 1 (1.8) | <0.0001 |
| 4-5 | 7 (12.3) | 35 (61.4) | 37 (64.9) | 28 (49.1) | |
| 6-8 | 0 | 2 (3.5) | 1 (1.8) | 28 (49.9) | |
| Mean (SD) of COV50 level | -2.13 (0.50) | -0.77 (0.30) | 0.29 (0.28) | 1.85 (0.59) | <0.0001 |
| Number with characteristic (%) | |||||
| Women | 28 (49.1) | 27 (47.4) | 25 (43.9) | 14 (24.6) | 0.0087 |
| Non-smoker | 32 (56.1) | 22 (38.6) | 22 (38.6) | 33 (57.9) | 0.36 |
| Hypertension | 23 (40.4) | 35 (61.4) | 42 (73.7) | 37 (64.9) | 0.0034 |
| Heart failure | 1 (1.8) | 7 (12.5) | 14 (25.5) | 8 (14.5) | 0.016 |
| Body mass index ≥30 kg/m2 | 14 (24.6) | 18 (31.6) | 16 (28.1) | 20 (35.1) | 0.30 |
| Diabetes mellitus | 6 (10.5) | 9 (15.8) | 20 (35.1) | 30 (52.6) | <0.0001 |
| Cancer | 2 (3.5) | 6 (10.5) | 2 (3.5) | 3 (5.3) | 0.62 |
| Use of RAS blockers, | 16 (28.1) | 30 (52.6) | 39 (68.4) | 34 (59.4) | 0.0024 |
| Mean (SD)of characteristic | |||||
| Age | 49.5 (16.8) | 63.9 (17.2) | 71.0 (13.8) | 67.8 (12.1) | <0.0001 |
| Systolic blood pressure, mm Hg | 128.9 (23.9) | 130.1 (23.1) | 134.8 (21.6) | 126.3 (24.5) | 0.83 |
| Diastolic blood pressure, mm Hg | 79.8 (13.0) | 77.9 (13.0) | 77.1 (11.8) | 70.6 (20.0) | 0.0014 |
| Heart rate, beats per minute | 81.6 (12.1) | 81.3 (13.5) | 81.6 (14.1) | 89.0 (18.5) | 0.011 |
| Body mass index, kg/m2 | 27.2 (5.2) | 28.2 (6.3) | 28.2 (5.3) | 29.0 (7.0) | 0.12 |
| Glomerular filtration rate, ml/min/1.73 m2 | 86.3 (23.1) | 81.8 (26.7) | 74.7 (31.9) | 69.6 (35.3) | 0.0083 |
RAS blockers indicate blocker of the renin-angiotensin system, including angiotensin-converting enzyme inhibitors and angiotensin-receptor blockers.
The glomerular filtration rate was derived from serum creatinine, using the Chronic Kidney Disease Epidemiology Collaboration.
Table 3.
Odds ratios relating outcome to COV50.
| Outcome | Number E/R | Odds ratio (95% confidence interval) | p value |
|---|---|---|---|
| Mortality | 23/228 | ||
| Unadjusted | 3.52 (2.02–6.13) | <0.0001 | |
| Adjusted | |||
| Sex and age | 3.23 (1.81–5.74) | <0.0001 | |
| + timepoint 1 WHO score | 2.63 (1.21–5.69) | 0.014 | |
| + body mass index and comorbidities | 2.73 (1.25–5.95) | 0.012 | |
| Progressing WHO score | 48/228 | ||
| Unadjusted | 2.63 (1.80–3.85) | <0.0001 | |
| Adjusted | |||
| Sex and age | 2.37 (1.58–3.54) | <0.0001 | |
| + timepoint 1 WHO score | 3.34 (1.83–6.07) | <0.0001 | |
| + body mass index and comorbidities | 3.38 (1.85–6.17) | <0.0001 |
Number E/R indicates the number of events/number at risk. Odds ratios express the risk for 1-SD increment in COV50. Comorbidities include hypertension, heart failure, diabetes and cancer.
Table 4.
Discriminative performance of COV50.
| Outcome | Derivation cohort | Validation cohort |
|---|---|---|
| Mortality | ||
| Number events/at risk | 23/228 | 10/99 |
| Continuously distributed COV50 | ||
| AUC (95% confidence interval) | 0.82 (0.74–0.89) | 0.83 (0.71–0.94) |
| Categorised COV50 | ||
| Youden cut-off threshold | 0.47 | 0.47 |
| Sensitivity | 87.0 (73.2–1.00) | 80.0 (55.0–1.00) |
| Specificity | 74.6 (68.7–80.6) | 70.8 (61.3–80.2) |
| Progressing WHO score | ||
| Number events/at risk | 48/228 | 23/99 |
| Continuously distributed COV50 | ||
| AUC (95% confidence interval) | 0.75 (0.67–0.82) | 0.70 (0.58–0.88) |
| Categorised COV50 | ||
| Youden cut-off threshold | 0.04 | 0.04 |
| Sensitivity (95% confidence interval) | 77.1 (65.2–89.0) | 73.9 (56.0–91.9) |
| Specificity (95% confidence interval) | 63.9 (56.9–70.9) | 63.2 (52.3–74.0) |
AUC indicates area under the curve. The AUC in the validation cohort was derived from the probabilities as predicted by the logistic model in the derivation cohort. Sensitivity and specificity in the validation cohort were based on the thresholds obtained in the derivation cohort. NA indicates not applicable.
The AUCs of the timepoint 1 risk factors were 0·57 (0·46–0·68) for age, 0·65 (0·54–0·75) for the WHO score, 0·80 (0·72–0·82) for COV50 in relation to mortality (Appendix Table 2), and 0·59 (0·51–0·68) for age, 0·52 (0·43–0·61) for the WHO score, and 0·74 (0·66–0·81) for COV50, respectively, in relation to worsening WHO score (Appendix). In the derivation cohort, on top of sex, age, body mass index, comorbidities, and the timepoint 1 WHO score, COV50 was analysed as a continuously distributed variable slightly enlarged the AUC. When assessed per threshold as categorized variable (Fig. 1) COV50 significantly improved the AUC. For mortality in relation to the continuously distributed COV50 marker, the AUC increased from 0·78 to 0·82 (p = 0·11). For worsening WHO score, the AUC increased from 0·68 to 0·78 (p = 0·0097). When applying the threshold and investigating as categorized variable, adding COV50 increased the AUC from 0.78 to 0·84 (p = 0·052) for mortality prediction, and from 0.68 to 0·75 (p = 0·021) for worsening WHO score.
Compared to the derivation cohort (Table 1), the baseline (timepoint 1) characteristics of the validation cohort, including the distribution of COV50 and comorbidities (Appendix Figs. 1 and 3), were broadly similar. Using the predicted probabilities and the optimised thresholds derived from the derivation cohort, the results in the validation cohort confirmed the discriminatory performance of the COV50 biomarker, irrespective of whether it was analysed as a continuously distributed variable or as a categorised risk factor (Fig. 1). Compared with the 327 patients included in the current analyses, the 981 matched COVID-19-free controls had comparable characteristics (Appendix; 0.084≤p≤0.87). When applying 0·47 and 0·04 as COV50 thresholds, only two and seven controls scored positive, yielding specificities of 99·8% and 99·3%, respectively. Finally, the 4C mortality score consisting of eight variables to grade was applicable only to 257 hospitalised CRIT-CoV-U patients without missing data, of whom 31 died. In these 257 patients, a 4C score of ≥15, indicating critical disease and COV50 as a stand-alone biomarker had a similar AUC in relation to mortality (0·77 versus 0·76; p = 0·79; Appendix Fig. 5).
4. Discussion
COV50 is a novel multidimensional urinary biomarker (Appendix Table 1) consisting of 50 deregulated urinary peptides mainly derived from collagen alpha 1(1) but also from other proteins previously recognised to be involved in COVID-19 pathogenesis. On its own and adjusted for clinical risk factors, COV50 predicted the incidence of death and progression across WHO stages. This association was robust and withstood internal validation in the derivation cohort by the leave-one-out AUC approach and by correction for overfitting. External validation in the validation cohort produced confirmatory results. Moreover, on top of the established clinical risk factors commonly used in predictive models, COV50 analysed as a continuously distributed variable and per threshold (Fig. 1) improved the AUC, albeit the data were stronger for worsening WHO score than for mortality, given the number of study endpoints.
COV50 is registered in Germany and immediately applicable for clinical and research purposes. A scheme to stimulate early anti-replicative therapies by combined usage of the 4C score and the COV50 test in patients with non-discriminative 4C results is under development. The UPP does not undergo significant changes when urine is stored for 5 days at room temperature in borated test tubes [23,24], thereby providing a wide time window for handling urine samples, for instance as collected at the homes of patients with PCR-confirmed SARS-CoV-2 infection. Furthermore, urine can be stored for years at -20 °C without UPP alteration, opening opportunities for research [25]. From a clinical perspective, COV50 might contribute to the personalised management of COVID-19 patients, which could range from observation and follow-up at home to non-invasive and invasive hospital care, such as treatment with remdesivir, corticosteroids, monoclonal antibodies, convalescent plasma, or mechanical ventilation combined with other life-supporting interventions. Patients with a COV50 level of less than -1 could be managed at home; those with a level ranging from 1 to 0·40 might require in-hospital management with intermediate care, such as intensified oxygen supply; and those with a level of ≥ 0·40 are likely to require intensive care and invasive life-support measures. The discriminatory performance of COV50 as a stand-alone test is comparable with the 4C score but has the advantage of not including any clinical or biochemical variable that is already indicative of evolving respiratory insufficiency. From this perspective, UPP followed by the identification of the parental proteins by sequencing the urinary peptides is a powerful instrument in generating multidimensional biomarkers that reflect the molecular processes underlying various illnesses. Disease-specific peptidomic signatures have become evident in the subclinical run-on to critical illness, as demonstrated for diastolic left ventricular dysfunction (HF1)[12] and CKD or diabetic nephropathy (CKD273) [13,14]. The number of peptide fragments making up HF1 is 85 and 273 for CKD273. These UPP are mutually exclusive, highlighting their specificity for the target disease. COV50 shares 13 urinary peptides with CKD273 and only one with HF1. Only two fragments are common to COV50, HF1 and CKD273 (Appendix Fig. 4). Along similar lines, COV50 levels 0·47 and 0·04 scored seven or fewer of the 981 matched controls as at risk for critical COVID-19, thereby confirming the >99·0% specificity of the marker.
The most prominent characteristic of the COV50 signature (Appendix Table 1) is the shift in collagen fragments, in particular collagen alpha 1(1). Deregulation of collagen homeostasis is a hallmark of SARS-CoV-2 infection [26] and has also been observed in CKD [13,27]. Several studies have reported that CKD and biomarkers indicative of renal impairment predict critical COVID-19, while survivors remain at high risk of CKD [28]. The COV50 urinary signature showed upregulation of α1-antitrypsin degradation products, which is in line with reports that α1-antitrypsin deficiency is a major risk factor for life-threatening COVID-19 [29]. No information in the context of COVID-19 is currently available on CD99, which is involved in cell recruitment, leukocyte trans-endothelial migration, and maintaining the integrity of the endothelial barrier [30,31]. Reduction of CD99 might interfere with appropriate immune responses and indicate endothelial damage. The polymeric immunoglobulin receptor (pIgR), highly expressed in the trachea and the lung and responsible for transcytosis, especially IgA, has not yet been investigated in COVID-19. It is downregulated in chronic obstructive pulmonary disease and is associated with disease severity [32]. In our study, the reduction in urinary pIgR fragments was associated with COVID-19 severity. In line with the reduction of urinary gelsolin fragments, patients with an unfavourable COVID-19 outcome have lower plasma levels of gelsolin [33]. The sodium/potassium-transporting ATPase subunit gamma (FYXD2) is highly expressed in the kidney. Reduced abundance of a peptide from FYXD2 in our current study was associated with severe COVID-19, in keeping with the same observation in IgA nephropathy [34].
The urgency associated with the COVID-19 pandemic in Germany, Europe, and beyond justified the generation of this interim report, as other investigators working in this field have also done [35]. Nevertheless, we have complied with all the quality criteria as outlined in a recent commentary on the COVID-19 literature [36]. In accordance with the scientific rigour required in this research field, the CRIT-COV-U consortium is preparing a protocol amendment describing the statistical analysis plan and significance levels required for a second look at the CRIT-CoV-U data in the final analysis. As can be expected for an interim report, the current study has potential limitations. First, the sample size of the validation cohort was small compared with the derivation cohort. Second, the models were not adjusted for glomerular filtration rate because intravenous fluid administration confounds this renal function measurement. Presumably, this limitation is also applicable to other scoring algorithms. Third, the power of the study was reduced in comparison to the initial planning, which apparently resulted in the inability to demonstrate statistical significance for all aspects. Finally, at the current stage of data collection, calibrating the predictive models was not yet possible, limiting the generalisability of the COV50 biomarker. However, the data also support the power calculations performed when planning the study, and we expect that in the full cohort of 1000 subjects all the above limitations will be eliminated due to the larger power.
In conclusion, this first CRIT-COV-U report supports the concept that UPP generates biomarkers indicative of adverse COVID-19 outcomes, even at WHO stages 1–3. The current findings obviously need consolidation in the full dataset of 1000 patients, but open up potential for patient management, health policy planning, and for providing an intermediate UPP endpoint in randomised clinical trials of novel COVID-19 treatment modalities. COV50 is licensed in Germany and available for clinical use.
Contributors
RW, CL, HM, JM, HvdL, and JB conceptualised the study. HM, JS, JR, and JM performed the proteomic urine analyses. LT, JAS, and JB did the statistical analysis and wrote the first draft of the manuscript. JAS was the principal writer of the final draft. RW, SK, AM, ED, GS, MM, ACT, BC, AW, BP, ÅN, MS, KR, CL, JB and the Crit-CoV investigators were study investigators or participated in the conduct of the study, including the recruitment and follow-up of the patients. BN and HvdL conducted and supervised the eCRF database construction, the data retrieval, the data management, and the data quality control. All the authors interpreted the results, commented on successive drafts of the manuscript, and approved the final version.
Data sharing
The study protocol is available at the German Register for Clinical Studies (www.drks.de), number DRKS00022495. Anonymised participant data will be made available when the study is complete, upon request directed to the corresponding author. Proposals will be reviewed and approved by the funder, investigators, and collaborators based on scientific merit. After approval of a proposal, data can be shared through a secure online platform after signing a data access and confidentiality agreement. All data will be made available for a minimum of five years from the end of the study.
Declaration of Competing Interest
AW, BC, BN, HvdL, AN, JB, AM, MM, ACT, BP, KR, CL, RW, SK, ED, JS report payment for study inclusion from the Federal Ministry of Health (Germany) during the conduct of the study; MS reports Scientific Research Activity from Robert Bosch Stiftung during the conduct of the study, grants for clinical trials from Green Cross Wellbeing Co. Ltd. And Gilead Sciences Inc. and grants for research activity from Robert Bosch GmbH, consulting fees as reviewer for Research Impact Fund Hongkong and EU Horizon 2020, honoraria for lectures from CED Service GmbH and ALL Akademie, supporting for meetings from CED Service GmbH and ALL Akademie, participation on advisory board (European PGx Advisory Board) from Agena Bioscience GmbH and other financial interests (Editor) for Phrmacogenetics and Genomics, Drug Research and Genome Medicine; JM, JS and JR are past (JM) or current employees of Mosaiques-Diagnostics, Hanover, Germany, HM is a co-funder and co-owner of Mosaiques-Diagnostics.
Acknowledgments
Mrs Vera De Leebeeck and Mrs Renilde Wolfs provided expert clerical assistance.
Footnotes
CRIT-COV-U investigators and key investigators are listed in the appendix (pp 06-07).
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2021.100883.
Appendix. Supplementary materials
References
- 1.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W., Ni Z., Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson E.J., Walker A.J., Bhaskaran K. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wynants L., Van Calster B., Collins G.S. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. Br Med J. 2020;369:1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montero-Odasso M., Hogan D.B., Lam R. Age alone is not adequate to determine health-care resource allocation during the COVID-19 pandemic. Can Geriatr J. 2020;23:152–154. doi: 10.5770/cgj.23.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight S.R., Ho A., Pius R. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: development and validation of the 4C mortality score. Br Med J. 2020;370:3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myrstad M., Ihle-Hansen H., Tveita A.A. National Early Warning Score 2 (NEWS2) on admission predicts severe disease and in-hospital mortality from COVID-19 - a prospective cohort study. Scan J Trauma Resusc Emerg Med. 2021;28:66. doi: 10.1186/s13049-020-00764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimhofer T., Lodge S., Whiley L. Integrative modeling of quantitative plasma lipoprotein, metabolic, and amino acid data reveals a multiorgan pathological signature of SARS-CoV=2 infection. J Proteome Res. 2020;19:4442–4454. doi: 10.1021/acs.jproteome.0c00519. [DOI] [PubMed] [Google Scholar]
- 10.Zürbig P., Renfrow M.B., Schiffer E. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis. 2006;27:2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
- 11.Klein J., Papadopoulos T., Mischak H., Mullen W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis. 2014;35:1060–1064. doi: 10.1002/elps.201300327. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z.Y., Nkuipou-Kenfack E., Staessen JA. Urinary peptidomic biomarker for personalized prevention and treatment of diastolic left ventricular dysfunction. Proteom Clin Appl. 2019;13 doi: 10.1002/prca.201800174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pontillo C., Jacobs L., Staessen J.A. A urinary proteome-based classifier for the early detection of decline in glomerular filtration. Nephrol Dial Transplant. 2017;32:1510–1516. doi: 10.1093/ndt/gfw239. [DOI] [PubMed] [Google Scholar]
- 14.Tofte N., Lindhardt M., Adamova K. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinol. 2020;8:301–312. doi: 10.1016/S2213-8587(20)30026-7. [DOI] [PubMed] [Google Scholar]
- 15.Wendt R., Kalbitz S., Lübbert C. Urinary proteomics associate with COVID-19 severity: pilot proof-of-principle data and design of multicentric diagnostic study. Proteomics. 2020;20 doi: 10.1002/pmic.202000202. [DOI] [PubMed] [Google Scholar]
- 16.Association W.M. World Medical Association declaration of Helsinki. Ethical principles for medical research involving human subjects. J Am Med Assoc. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 17.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization . WHO R&D Blueprint Novel Coronavirus; 2020. Ordinal scale for clinical improvement. [Google Scholar]
- 19.Levey A.S., Bosch J.P., Lewis J.B. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 20.Mischak H., Vlahou A., Ioannidis JPA. Technical aspects and inter-laboratory variability in native peptide profiling: the CE-MS experience. Clin Biochem. 2013;46:432–443. doi: 10.1016/j.clinbiochem.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Latosinska A., Siwy J., Mischak H., Frantzi M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: the past, the present, and the future. Electrophoresis. 2019;40:2294–2308. doi: 10.1002/elps.201900091. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]
- 23.Schaub S., Wilkins J., Weiler T., Sangster K., Rush D., Nickerson P. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Intern. 2004;2004:323–332. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- 24.Theodorescu D., Wittke S., Ross M.M. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 25.Fliser D., Novak J., Thongboonkerd V. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 26.Nunnari G., Sanfilippo C., Castrogiovanni P. Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection. Exp Cell Res. 2020;395 doi: 10.1016/j.yexcr.2020.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schanstra J.P., Zürbig P., Alkhalaf A. Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol. 2015;26:1999–2010. doi: 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ERA-EDTA Council and the ERACODA Working Group Chronic kidney disease is a risk factor for severe COVID-19: a call to action by ERA-EDTA. Nephrol Dial Transplant. 2021;36:314. doi: 10.1093/ndt/gfaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang C., Chapman K.R., Wong A., Liu M. A1-antitrypsin deficiency and the risk of COVID-19: an urgent call to action. Lancet Respir Med. 2021;9 doi: 10.1016/S2213-2600(21)00018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winger R.C., Koblinski J.E., Kanda T., Ransohoff R.M., Muller W. Rapid remodeling of tight junctions during oaracellular diapedesis in a human model of the bloodpbrain barrier. J Immunol. 2014;193:2427–2437. doi: 10.4049/jimmunol.1400700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller W.A. Transendothelial migration: unifying principles from the endothelial perspective. Immunol Rev. 2016;273:61–75. doi: 10.1111/imr.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gohy S.T., Detry B.R., Lecocq M. Polymeric immunoglobulin receptor down-regulation in chronic obstructive pulmonary disease. Persistence in the cultured epithelium and role of transforming growth factor-β. Am J Respir Crit Care Med. 2014;190:509–521. doi: 10.1164/rccm.201311-1971OC. #x03B2. [DOI] [PubMed] [Google Scholar]
- 33.Abers M.S., Delmonte O.M., Ricotta E.E. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6 doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudnicki M., Siwy J., Wendt R. Urine proteomics for prediction of disease progression in patients with IgA nephropathy. Nephrol Dial Transplant. 2020;35:9faa307. doi: 10.1093/ndt/gfaa307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P., Nirula A., Heller B. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with COVID-19. N Engl J Med. 2020;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira J.P., Epstein M., Zannad F. The decline of the experimental paradigm during the COVID-19 pandemic: a template for the future. Am J Med. 2021;134:166–175. doi: 10.1016/j.amjmed.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.