Significance
Why biological complexity evolves is a major question in the life sciences, but the specific selection pressures favoring simple or complex traits remain unclear. Using high-resolution measurements of venom complexity in North American pitvipers, we link changes in complexity to natural history via phylogenetic diversity of snake diets. The results indicate that venom complexity evolves in response to phylogenetic diversity in a community of species, likely reflecting divergence in the physiological targets of venom. The nature of a species community, rather than their richness alone, is an important feature in the evolution of complex traits.
Keywords: diversity, predator, toxin, transcriptomics, diet breadth
Abstract
The role of natural selection in the evolution of trait complexity can be characterized by testing hypothesized links between complex forms and their functions across species. Predatory venoms are composed of multiple proteins that collectively function to incapacitate prey. Venom complexity fluctuates over evolutionary timescales, with apparent increases and decreases in complexity, and yet the causes of this variation are unclear. We tested alternative hypotheses linking venom complexity and ecological sources of selection from diet in the largest clade of front-fanged venomous snakes in North America: the rattlesnakes, copperheads, cantils, and cottonmouths. We generated independent transcriptomic and proteomic measures of venom complexity and collated several natural history studies to quantify dietary variation. We then constructed genome-scale phylogenies for these snakes for comparative analyses. Strikingly, prey phylogenetic diversity was more strongly correlated to venom complexity than was overall prey species diversity, specifically implicating prey species’ divergence, rather than the number of lineages alone, in the evolution of complexity. Prey phylogenetic diversity further predicted transcriptomic complexity of three of the four largest gene families in viper venom, showing that complexity evolution is a concerted response among many independent gene families. We suggest that the phylogenetic diversity of prey measures functionally relevant divergence in the targets of venom, a claim supported by sequence diversity in the coagulation cascade targets of venom. Our results support the general concept that the diversity of species in an ecological community is more important than their overall number in determining evolutionary patterns in predator trait complexity.
Darwin evoked an entangled riverbank when hypothesizing how natural selection could promote the evolution and maintenance of complexity in nature (1), placing an early focus on the link between trait evolution and species interactions. Diverse species interactions give rise to varied forms of natural selection that are expected to favor complexity in functional traits (2, 3). The specific pressures a trait responds to can have critical impacts on the trait’s evolutionary trajectory. For example, a host defending against many parasites might evolve unique defenses for each parasite species (a response to overall community richness or species diversity) (4–7), or, alternatively, modular defensive units reserved for phylogenetically distant parasite guilds with divergent infection strategies (8–10). Whether the number or nature of interacting species is most relevant for understanding the evolution of trait complexity remains unresolved (11, 12).
Complex traits are defined by multiple components contributing to the final functional phenotype (13, 14) and often by the amount of information in the genome required to produce them (15). Molecular traits involved in antagonistic interactions are emerging as models for linking trait complexity to the diversity of ecological communities because their complexity can be precisely quantified by the number and abundance of unique components (16–19). This allows the use of diversity metrics such as Shannon’s Index (16, 20) to summarize trait complexity. For example, Shannon’s was used to link major histocompatibility complex diversity with the richness of local parasite species (21, 22), as well as phytochemical defenses in plant leaves to the -diversity of herbivorous insects (16, 17).
Animal venoms function by disrupting homeostatic physiological processes to rapidly incapacitate prey (23) and provide the opportunity to understand the forces mediating trait complexity in predator–prey interactions. As protein mixtures, venom phenotype complexity can be quantified through chromatographic separation. Additionally, the venom-gland transcriptome directly links the venom proteome to the genotype (24–26). Transcriptomes, therefore, provide a second independent measure of venom complexity via expressed genomic sequence complexity (14). Recent studies across several venomous lineages have shown that the transcript (24), protein (25), and enzymatic activities (26) of venom are more complex when more prey classes are consumed. However, these studies do not directly test alternative hypotheses for how prey diversity may select for either reductions or increases in venom complexity. Disentangling the roles of species richness and phylogenetic diversity of diets in the evolution of venom complexity requires taxon-wide diet and venom characterization in a system known to exhibit adaptive evolution of prey-specific venoms.
Functional studies of snake venom have revealed several examples of variable complexity and evolved prey specificity. For example, feeding solely on fish eggs has relaxed selection in the sea snake Aipysurus eydouxii, resulting in a simplified venom arsenal and frequent null and deleterious mutations in venom genes (27, 28). Additionally, adaptive specialization of snake venom function occurs toward congeneric prey species (29) and populations (30–32). Finally, highly expressed venom gene paralogs from the same snake can display taxonomic specificity. Paralogous three-finger toxins in the Amazon puffing snake (Spilotes sulphureus) are alternatively lethal to mammals or lizards (33), while snake venom metalloproteinase (SVMP) paralogs in the Brazilian lancehead (Bothrops neuwiedi) differentially enact procoagulant function in blood of either mammals or birds (34). Despite deep conservation of the individual metabolic and homeostatic targets of venom (35, 36), the sequence divergence that has accumulated in these systems appears to influence venom protein function and may exert selection on venom complexity.
Unlike other venomous groups (24, 25), dietary ecology in viperid snakes has been researched extensively (Dataset S1 A and B), allowing detailed quantification of dietary variation. Most vipers feed mostly on small vertebrates, with the ancestral diet of the clade likely being mammals, lizards, and frogs (37). Rattlesnakes (Crotalus and Sistrurus), copperheads, cottonmouths, and cantils (Agkistrodon) comprise the largest clade of the front-fanged venomous snake species in North America, with between 45 and 64 recognized species (38). Species range widely in dietary ecology. For example, the Timber Rattlesnake (Crotalus horridus) is a mammal specialist (39), whereas the exceptionally diverse diet of the Florida Cottonmouth (Agkistrodon piscivorus conanti) (40) includes fish, frogs, mammals, snakes, lizards, birds, and even turtles. The venoms of these three genera consist of between 10 and 70 proteins from 15 to 25 distinct gene families (41, 42). Their venom cocktails underlie diverse functionality, with venoms enacting neurotoxic, coagulopathic, hemorrhagic, and myotoxic effects in accordance with their composition (41). Functional variation also exists within gene families, where neofunctionalization has generated phospholipase A2 (PLA2) paralogs that produce both cytotoxic and neurotoxic functions (43, 44) and SVMP paralogs with hemorrhagic and coagulopathic functions.
To test alternative hypotheses for the link between ecological communities and complexity, we have combined analysis of snake diets and independent, standardized venom proteomic and venom-gland transcriptomic complexity measures in a comparative framework. We collected venom and venom-gland samples from Agkistrodon, Crotalus, and Sistrurus across North America to generate the largest dataset of proteomes and venom-gland transcriptomes for this group to date (68 lineages). We generated a phylogeny by harvesting 1,525 nonvenom genetic loci from transcriptomes and collated diet studies on these snakes. Specifically, we tested three primary hypotheses for the evolution of venom complexity levels (shown in SI Appendix, Fig. S1). First, if intense pairwise coevolution favored trait complexity, we expected to observe a negative relationship between venom complexity and prey species diversity, as the intensity of coevolutionary selection is expected to be highest when only a few key prey species are consumed (45, 46). In contrast, if diffuse coevolution with multiple species exerted the strongest selection on trait complexity, we predicted venom complexity to be positively associated with prey species diversity, as diffuse coevolution is expected to be strongest when several prey species are important (17, 47). If prey divergence, rather than the simple number of prey, exerted the strongest selection on trait complexity, we expected venom complexity to be correlated with phylogenetic diversity of prey consumed and for prey phylogenetic diversity to explain more variation in complexity than prey species diversity alone (48, 49). Finally, a lack of any relationship between dietary ecology and venom complexity accompanied by strong phylogenetic signal in venom complexity would support neutral evolution of complexity levels.
Results
Phylogenetic Relationships.
We inferred phylogenies using both species-tree and concatenation approaches (Fig. 1 and SI Appendix, Figs. S2–S4) using sequences from 169 individuals representing 46 species of Agkistrodon, Crotalus, and Sistrurus (71.8% of species-level diversity) (38). To capture maximum diversity within our focal clade, we expanded our sampling to include phylogenetically discrete subspecies and lineages with described phylogenetic diversity, resulting in a final dataset of 9 Agkistrodon lineages, 5 Sistrurus lineages, and 54 Crotalus lineages. See SI Appendix, Results for an abbreviated discussion of rattlesnake relationships based on our topologies. We used these trees for phylogenetic comparative analyses of the relationship between diet diversity and venom complexity.
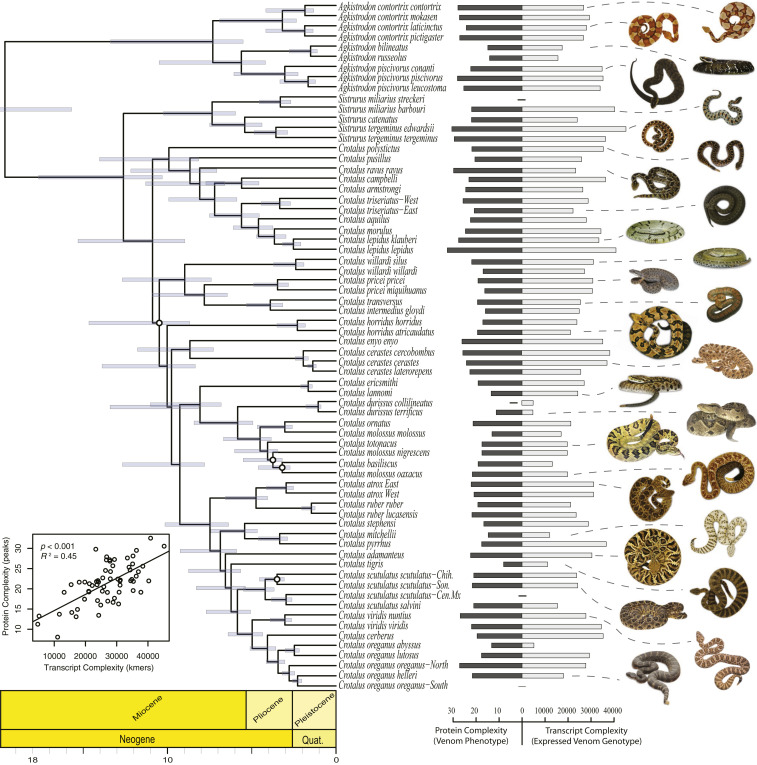
Fig. 1.
Dated phylogeny of Agkistrodon, Crotalus, and Sistrurus produced using Astral-III and dated with TreePL and fossil calibration. Nodes include 95% CIs (blue bars) for divergence date estimates, and open circles on nodes indicate posterior support values less than 70%. Bars next to the tips of phylogeny indicate mean protein complexity of venom samples from each species (dark gray) and mean transcriptome complexity of reads aligning to venom gene transcript coding regions (light gray). Dashes in place of bars indicate taxa for which venom complexity measures were not available. The scatterplot (Inset) shows the significant positive relationship between transcriptomic complexity and protein complexity of the venom across species. Crotalus pricei pricei, Crotalus viridus nuntius, Crotalus cerastes cerastes, and Crotalus scutulatus scutulatus images credit: Travis Fisher (Howard College, San Angelo, TX).
Complex Genotypes Underlie Complex Phenotypes.
Venom transcriptomic and proteomic complexities were positively related (R2 = 0.45, = 0.0004, T2,62 = 7.1, P < 0.001; Fig. 1, Inset [scatterplot]), affirming a link between the expressed genotype and the phenotype complexity in snake venoms. The lack of a stronger relationship between these metrics likely underscores the nature of our sequence data as a richer source of information. In high-performance liquid chromatography (HPLC), similar proteins may elute together as a single peak in chromatograms, potentially underrepresenting phenotype complexity at the level of individual components. Our transcriptomic complexity measure will instead quantify such differences, while the equal weighting of all unique sequences regardless of transcript length provides a complimentary focus on total sequence diversity, rather than organization into transcripts. There was phylogenetic signal in both transcriptomic complexity (Pagel’s = 0.79 0.014, P = 0.002) and proteomic complexity ( = 0.96 0.009, P < 0.0001; Fig. 1), as well as marked variation among lineages. The transcriptomic complexity underlying venoms varied by a factor of 10 among lineages, with the most complex, the Desert Massasauga Rattlesnake (Sistrurus tergeminus edwardsii), expressing 45,191 effective -mers and the simplest, the South American Rattlesnake (Crotalus durissus terrificus), expressing only 4,723 effective -mers. Venom proteomic complexity also varied considerably, with the most complex being the Mottled Rock Rattlesnake (Crotalus lepidus lepidus) with a mean of 32 effective protein peaks and the simplest being the Tiger Rattlesnake (Crotalus tigris), having only 8 effective peaks on average. Ancestral state reconstructions using the ace function in ape version 5 (54) indicate intermediate protein (22.3 peaks) and transcriptomic complexity (28,751 effective k-mers) in the common ancestor of Agkistrodon and rattlesnakes, suggesting that evolution toward both simpler and more complex venoms has occurred over time (SI Appendix, Fig. S5).
Phylogenetic Diet Diversity Predicts Venom Complexity.
Snake species ranged widely in both the simple species diversity (measured as effective number of species) and MPD of prey consumed. Prey species diversity ranged from 2.9 to 15 (mean = 6.7) effective species, while prey MPD ranged from 118 to 731 (mean = 365) million years of divergence (Fig. 2 and SI Appendix, Fig. S6).
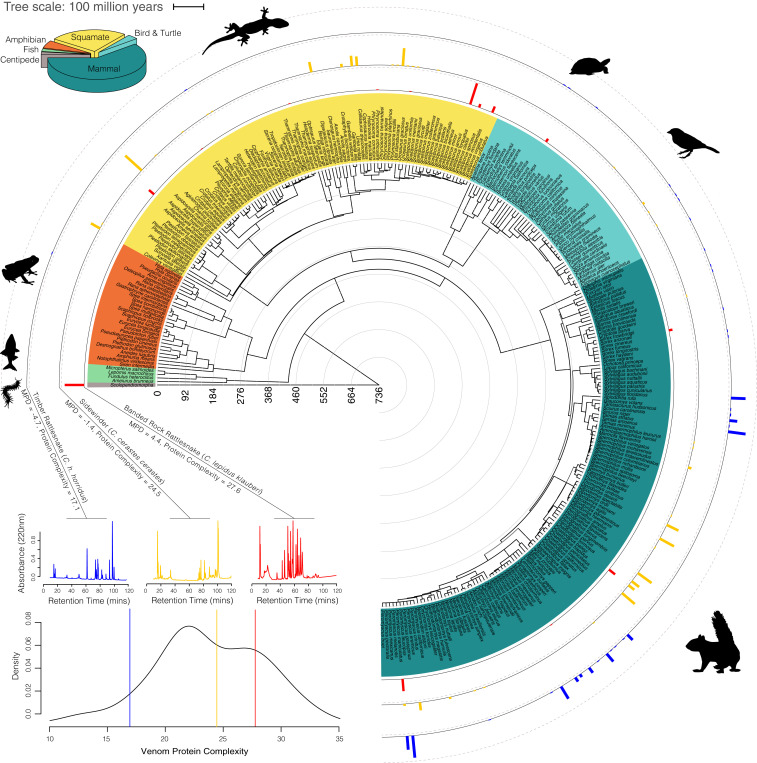
Fig. 2.
Phylogenetic diversity of all prey species used to calculate diet diversity. Topology was produced by joining published supertrees (50–53) using divergence dates from TimeTree.org (52). The pie chart (Top Left) shows the overall percentage of recorded snake stomach contents belonging to each prey taxa, and the pie chart colors correspond to colors on the prey phylogeny. Tracks outside the tree represent the diet composition of species with the lowest (blue), nearest to average (orange), and highest (red) phylogenetic diet diversity, as measured by standardized MPD. Gray dashed lines on track mark 50% of the diet. Insets show three representative HPLC chromatograms of a venom sample from these snake lineages with absorbance values standardized to the tallest peak in each sample and a density plot of all species’ mean HPLC complexity values for which diet data were also tabulated, with values for species featured in chromatograms marked with vertical lines and color-coded as for diet tracks.
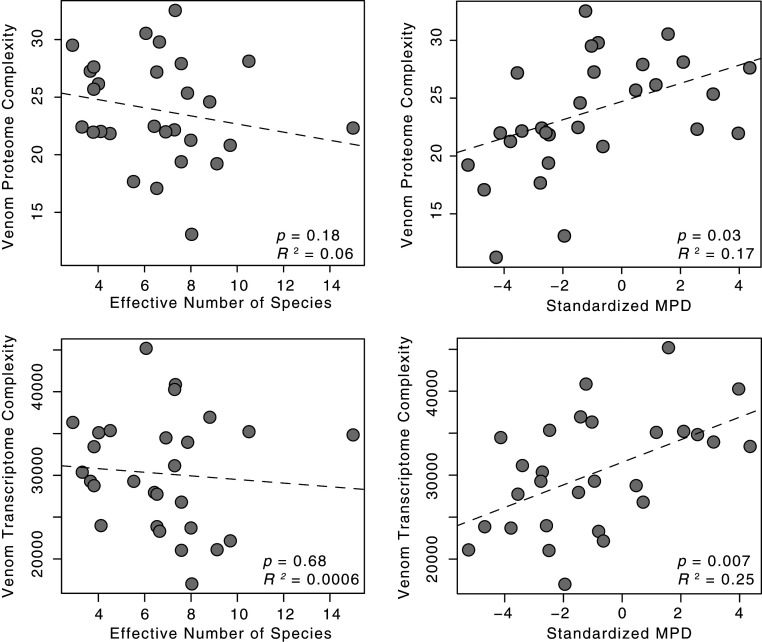
The observed variation in both venom proteomic and transcriptomic complexity was modeled best by prey MPD across all alternative phylogenetic trees used to represent this relationship (change in sample-size corrected Akaike Information Criterion [AICc] > 2 for all MPD models compared to complementary species diversity models; Table 1). Furthermore, the MPD models were significantly better fits to the data compared with intercept-only null phylogenetic generalized least squares (pgls) models (AICc > 2), suggesting neutral evolution alone does not explain variation in venom complexity. Together with positive slopes for the relationship between venom complexity and MPD (Fig. 3), model selection supports the phylogenetic diversity hypothesis for the evolution of more complex venom traits across these predators. For brevity, we discuss pgls analyses using our Astral-III species tree here, with replicate analyses using other phylogenetic hypotheses reported in Table 1. Venom proteomic complexity and MPD were positively correlated (, , T2,26 = 2.3, P = 0.028; Figs. 2 and 3), but we did not detect a significant association between species diversity and venom protein complexity ( = −0.43, R2 = 0.07, T2,26 = −1.3, P = 0.18). MPD also predicted venom transcriptomic complexity (R2 = 0.25, T2,26 = 2.9, P = 0.007; Fig. 3). We also did not detect a significant relationship between raw species diversity and venom transcriptomic complexity ( = −216.2, R2 = 0.006, T2,26 = −0.42, P = 0.68).
Table 1.
Model comparison via AICc of prey species diversity and prey MPD as predictors of venom transcript complexity
| Null | Sp. Div. | MPD | |||
| Analysis/tree | AICc | AICc | P | AICc | P |
| Protein complexity | |||||
| 1. Astral-III | 164.0 | 164.4 | 0.18 | 161.9 | 0.031 |
| 2. RAxML | 164.3 | 164.7 | 0.18 | 162.0 | 0.029 |
| 3. Alencar et al. (55) | 101.5 | 102.7 | 0.28 | 98.7 | 0.020 |
| 4. Blair et al. (56) | 101.7 | 102.4 | 0.21 | 98.7 | 0.019 |
| Transcript complexity | |||||
| 1. Astral-III | 575.5 | 577.6 | 0.68 | 569.7 | 0.009 |
| 2. RAxML | 575.2 | 577.6 | 0.68 | 569.7 | 0.007 |
| 3. Alencar et al. (55) | 389.6 | 390.8 | 0.29 | 382.4 | 0.004 |
| 4. Blair et al. (56) | 389.5 | 390.7 | 0.28 | 382.4 | 0.003 |
Fig. 3.
Relationship between species diversity (effective number of species) (Left) or MPD (Right) and proteomic complexity (Top) or transcriptomic complexity (Bottom) of all sequencing reads mapping to coding regions of venom gene transcripts.
To assess the robustness of our results to other phylogenetic hypotheses, we used the phylogenies of Blair et al. (56) and Alencar et al. (55). This required that we recalculate our estimates of venom complexity and diet diversity using their lineage assignments. Prey MPD again remained the best model of both proteomic and transcriptomic complexity in these tests as well (Table 1), demonstrating robustness of our result using alternative summations of both diet and trait data.
Diet Diversity Predicts Expressed Complexity in Three Venom Gene Families.
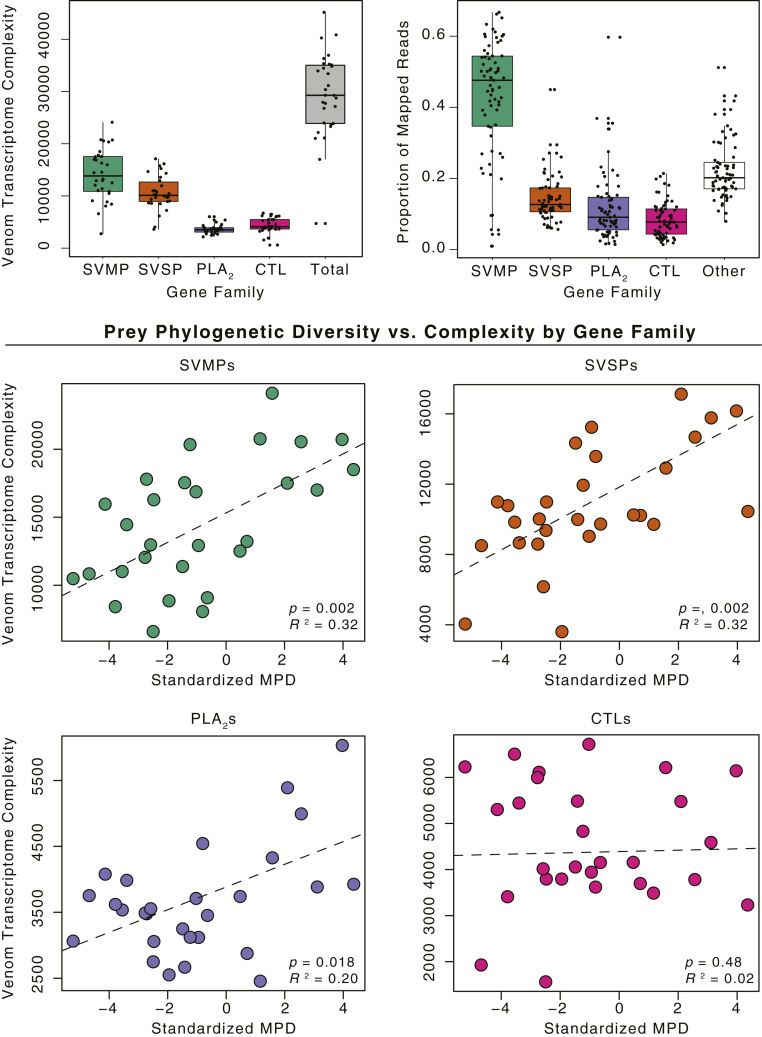
We next assessed the relationship between venom transcriptomic complexity and phylogenetic diversity of prey at the gene family level for the four largest venom gene families in viper venoms (SVMPs: mean sequence length = 1,641 base pairs [bp]; snake venom serine proteases [SVSPs]: mean sequence length = 783 bp; : mean sequence length = 418 bp; and C-type lectins [CTLs]: mean sequence length = 463 bp). These analyses permitted an assessment of levels of heterogeneity in the relationships between transcriptomic complexity and MPD among gene families, while controlling for average transcript length based on similar lengths of transcripts from a single gene family. Significant positive relationships between transcriptomic complexity and MPD were found for the SVMP gene family (R2 = 0.32, T2,26 = 3.5, P = 0.002; Fig. 4), the SVSP gene family (R2 = 0.32, T2,26 = 3.5, P = 0.002), and the gene family (R2 = 0.20, T2,26 = 2.5, P = 0.018). The CTL gene family, however, did not show a significant association between MPD and transcriptomic complexity (T2,26 = −0.71, P = 0.48). Parsing transcriptomic complexity by gene family in this way demonstrates an evolutionary response to selection from more phylogenetically diverse prey occurs across several gene families.
Fig. 4.
Boxplots of transcriptomic complexity estimates (Top Left) and proportion of venom reads mapped for each gene family (Top Right). Scatterplots show relationships between transcriptomic complexity and MPD for the four most prevalent gene families in the venoms of viperid snakes.
Prey Phylogenetic Diversity Predicts Amino Acid Divergence in Key Venom Targets.
Lastly, we performed a direct test of whether prey phylogenetic diversity better reflects functional diversity in prey than does raw species diversity using amino acid sequence variation in homologous venom targets of different prey. We compiled available amino acid sequences for eight different coagulation cascade proteins targeted by SVSPs (57): FV, FVIII, FXIII, prothrombin, kininogen, fibrinogen, plasminogen, and protein C (Dataset S8). We generated gene trees for each protein and used these to recalculate mean phylogenetic distances for each snake as a mean coagulation cascade distance (MCCD) in amino acid substitutions per site (SI Appendix, Methods). Across the snake species, MPD was strongly correlated with MCCD (R2 = 70%, P < 0.001; SI Appendix, Fig. S7A), whereas prey species diversity was significantly but considerably less strongly associated with MCCD (R2 = 30%, P = 0.002). Prey MCCD was also a significant predictor of SVSP transcriptome complexity (R2 = 26%, P = 0.006; SI Appendix, Fig. S7B), as would be predicted under the hypothesis that MCCD is the source of selection maintaining complexity in SVSPs. Thus, prey MPD relates more strongly to protein sequence diversity in functionally important venom targets than does prey species diversity, and amino acid sequence diversity in specific venom targets has power to predict venom complexity.
Discussion
Snake venom complexity evolves in association with the phylogenetic diversity of snake diets, with the evolution of both simpler and more complex venoms over time. Our work builds upon previous findings linking categorically defined taxonomic diet breadth and venom trait complexity in several important ways (25, 26). First, our study system allowed testing of multiple alternative hypotheses for the link between dietary ecology and venom complexity. By revealing a continuous, linear relationship between venom complexity and MPD, our results argue that the amount of divergence among prey species matters for the evolution of targeted venoms, rather than only differences among differences among prey species or major taxonomic groups, regardless of their phylogenetic relatedness (24, 26). Also, densely sampling this relatively young clade of snakes showed how the effective complexity of venom has doubled (or been halved) largely within the past million years, with several stark examples of closely related species exhibiting marked variation in venom complexity. These insights were prevented by the sporadic sampling of more ancient families (Conid snails) (24) or orders (Spiders or Snakes) (25, 26). Second, we show that the relationship between diet diversity and venom complexity involves the parallel evolution of complexity levels within multiple independent venom gene families.
We argue that prey phylogenetic diversity likely predicts the evolution of venom complexity because of the subtle variation in conserved toxin targets that has arisen during prey species’ divergence. The targets of venom mediate fundamental physiological processes, such as members of the coagulation cascade, ion channels, or receptors, to confer lethality (23, 58). Since the evolution of extant tetrapods, only one duplication and one loss of a coagulation factor has occurred (35). Although their presence is conserved, we showed that a snake species’ prey MPD predicts prey MCCD based on homologs of coagulation factors. There are well-documented functional consequences of coagulation factor sequence variation for their interaction with heterologous proteins (59) that may extend to SVSPs in snake predators. While SVSP specificity for divergent coagulation factor homologs is largely unexplored in vivo, sequence variation in other conserved proteins, such as ion channels (60) and nicotinic acetylcholine receptors (61), carries functional relevance for taxonomic specificity of some venoms.
Consistent with the hypothesis that prey phylogenetic divergence is associated with the accrual of functionally relevant changes in venom targets, snake venom potency decreases with increasing phylogenetic distance from natural prey (62, 63) and taxonomic breadth in a snakes’ diet predicts venom that is highly lethal to a more diverse set of prey (64). These trends in toxicity combined with the relationships between venom complexity and MPD/MCCD shown here suggest phylogenetic generalism in dietary preference selects for venom proteins with taxonomic specificity. The result is a continuum from simple venoms in phylogenetic specialists and complex venoms in phylogenetic generalists (65, 66). The taxonomic specificity of procoagulant SVMP paralogs from B. neuwiedi (34) and the differential lethality of three-finger toxin paralogs in S. sulphureus (33) provide concrete examples where evolving more venom complexity facilitates venom function to disrupt physiological targets in phylogenetically distinct prey (34). We hypothesize that such functional specialization explains much of the results shown in the present paper.
The evolutionary outcomes of species interactions are dependent on several contextual factors (67), where communities of varying diversity represent some, but not all, of that context. While unmeasured processes likely play a role in the evolution of trait complexity in this system, based on , MPD explains a substantial portion of the variation in venom complexity. One caveat of our approach is that both diet and venom composition can vary among populations of snakes (68–70). Diet studies used here varied in geographic scale from single locations to whole species ranges. Similarly, while we controlled for ontogenetic variation in venoms by sampling adult animals, population-level variation is undoubtedly undersampled. It is possible that matched localities between diet and venom sampling would measure an even stronger relationship between prey MPD and venom complexity, assuming intraspecific venom local adaptation (30, 31). Future work estimating geographic and temporal variation in prey availability could address if long-term stable trends in prey diversity are reflected in evolved venom complexity. Other key processes include coevolution with resistant prey (30, 31), which can accelerate the evolution of traits (45), and defense against a snake’s predators (58, 71). We suggest that the phylogenetic diversity of snake prey communities exerts selection on venom complexity levels broadly to form patterns that are modulated by other evolutionary processes (72–75).
The SVMP, SVSP, , and CTL venom gene families evolved before the divergence of vipers from other snakes (76). Hence, our study evaluates the evolution of expressed complexity since divergence from a common ancestor where all these proteins were present. Giorgianni et al. (77) recently showed the rapidity of SVMP gene family expansion and contraction in Crotalus, where counts of distinct SVMP paralogs in a tandem array can range between 5 and 30 (77), and hypothesized a role for positive selection in expansion. Similarly, rattlesnake venom s have been previously hypothesized to have evolved from an ancestral set of seven genes in a tandem array (43). Dowell et al. (43) hypothesized that lineage-specific gene loss in rattlesnakes could be due to selective screening of genes that remain functional during niche differentiation. Our results support this notion, suggesting that expression of complex sets of SVMPs and s, as well as SVSPs, are perhaps best maintained by the balancing selection of an evolutionarily diverse diet. Thus, selection for more complex venoms, rather than merely increasing expression to produce more of the same functional toxin (77), is a likely key for the maintenance of large venom gene families.
The lack of a relationship between CTL expressed transcriptomic complexity and MPD suggests heterogeneity in the response to selection from diet across gene families. Two major differences between CTLs and the other venom gene families standout: They function only as multimeric heterodimers and act not as enzymes but instead bind to platelet receptors such as von Willebrand factor (vWF) or GP1b (78, 79). Forming quaternary structures and ligand-binding sites composed of multiple protein chains can constrain rates of protein evolution (80, 81), but levels of extant variation are likely not an issue given similar levels of variation in sequence complexity between s and CTLs. The CTLs may also be influenced by their function in defense against coevolving predators. Opossums (Didelphidae) are known to feed on pitvipers, including rattlesnakes, and these mammals have coevolved resistance to CTLs by rapid evolution of the binding sites on the vWF (71). Different evolutionary dynamics among toxin classes are not unprecedented, occurring among phytochemical toxin classes in parsnip plants coevolving with webworms (20). We provide an example of multigene protein families with such class-specific evolutionary dynamics.
Broadly, the extension of our results to three of the four gene families tested highlights a variable response to ecologically mediated selection across multiple unlinked genomic regions, as these gene families occur on different chromosomes (82). There is mounting evidence that venom varies in a modular fashion, with separate functional units composed of proteins from multiple families (83–85). Under the simplest scenario, a module of SVMP, SVSP, and genes might exist for phylogenetically clustered prey guilds, such as reptiles or mammals, and simplified diets may lead to the underexpression or genomic deletion of entire sets of genes as balancing selection wanes. The -mer approach measured expressed gene sequence as unique strings of 60 bases and their relative abundance, with the benefit of quantifying sequence complexity free of a reference. However, this approach is limited by its inability to parse the contributions of coding sequence length, counts of paralogs, and expression variation to the complexity scores generated. The increasing availability of whole-genome sequence data combined with functional characterization of venom paralogs will be crucial for testing, in a comparative framework, the relative roles of gene copy number variation, allelic variation, and expression variation (31, 86, 87) in forming modular responses to diverse prey species.
The primary contribution of our study is showing that in the context of species interactions, it is the nature (in the form of phylogenetic diversity) rather than the number (species diversity alone) of prey species that predicts molecular trait complexity. The evolutionary trends we have quantified suggest that molecular trait complexity evolves and is maintained by ecological generalism (16, 17), specifically elevating phylogenetic generalism as a selection pressure. The phylogenetic context for dietary generalism, where functional and/or physiological variation accumulates with distance between taxa, can help explain important global trends in biological diversity. For example, the negative relationship between dietary species diversity of herbivorous insects and the species richness of plant families (88) can be explained by the existence of speciose but chemically similar plant clades. Our results lead us to predict areas of higher community phylogenetic diversity will produce more complex defense traits on average within a community. Indeed, taxa ranging from invertebrates to plants tend to be better chemically defended against multiple enemies in the tropics (89), where phylogenetic diversity also happens to be highest in many taxa (90, 91). Overall, our results suggest the following general principle governing trait evolution in the planet’s entangled bank: In a given ecological community, species’ phylogenetic diversity plays a large role in shaping the evolutionary trajectory of complex traits of community members.
Materials and Methods
Sample Collection and Transcriptome Analyses.
Snakes were collected from the field in numerous localities across the United States, Mexico, and Brazil (Dataset S2 A and B). Venom was collected from each snake for proteomic analysis and venom glands were dissected 4 d post-venom collection for RNA sequencing. Transcriptomes were assembled following Holding et al. (92). See SI Appendix, Methods for full details.
Phylotranscriptomic Analysis of Nontoxin Genes.
We extracted nontoxin coding regions from the Trinity assembly of a subsample of up to five individuals of each known lineage within our sampled transcriptomes (169 individuals from the 68 focal taxa and 6 individuals comprising 6 outgroup taxa) using BUSCO version 3.0.1 (93, 94) searches against the “tetrapoda_odb9” set of 3,950 genes from OrthoDB (95). BUSCO identified 3,633 of these genes as “complete and single copy” sequences in at least one individual. We produced alignments of each BUSCO locus using MAFFT version 7.313 (96, 97) with the GINSI algorithm to prevent overalignment: “mafft –adjustdirectionaccurately –allowshift –unalignlevel 0.8 –leavegappyregion –maxiterate 10.” Alignments were trimmed using trimAl version 1.4.15 (98) using the “automated1” setting, manually inspected alignments in Geneious version 10.2.4 (https://www.geneious.com), kept loci present in at least 50% of samples for phylogenetic analysis. This resulted in 1,525 loci totaling 1,997,137 bp.
We used RAxML version 8.2.10 (99) to infer a best tree estimate for each gene based on 100 independent searches and a GTRGAMMA model. Nodal support values for the best tree estimate were attained through 1,000 bootstrap replicates. The best tree for each gene was then used as input for species-tree inference in Astral-III version 5.6.1 (100). Gene tree nodes with less than 10% bootstrap support were collapsed to polytomies (101), and we incorporated multiple individuals per lineage in species-tree inference (102). To estimate divergence dates with credibility intervals, we dated our trees with the autocorrelated relaxed clock based on the penalized-likelihood approach as implemented in TreePL (103) (see SI Appendix, Methods for more details).
Measuring Toxin Transcriptomic Complexity.
We quantified expressed venom gene transcriptomic complexity by developing a -mer–based measure similar to those used in assessments of microbial community diversity (104), counting appearances of all unique k = 60 bp sequences in the subset of venom-gland RNA-seq reads mapping to venom gene transcripts. Venom -mer complexity is unaffected by known sources of bias in venom studies attempting to call paralogs versus alleles and filter chimeric assemblies in large, rapidly diverging multigene families (92). We first identified assembled contigs containing toxin sequences using Basic Local Alignment Search Tool (BLAST) (105) searches (blastn) (e = 10−8) against a curated set of 804 viperid toxin coding sequences (CDSs) spanning the known viperid venom gene families. We also used this curated set of coding sequences to calculate the mean sequence length of the SVMP, SVSP, , and CTL gene families. Portions of contigs with a BLAST hit to a toxin CDS were concatenated into a master database of all 191 adult snakes sampled, creating a comprehensive database of sampled toxin sequence diversity across Agkistrodon, Crotalus, and Sistrurus. We then used the mem algorithm within bwa (106) to align the cleaned reads from each sample to this master database, providing us with the set of reads that align to toxins for each snake. We randomly sampled 4 million toxin reads per snake using seqtk (107) to standardize sampled reads per individual and then used the count utility in Jellyfish version 2.2.1 (108) to count all unique 60-mers in the sampled toxin reads. Finally, we used the 60-mer counts to calculate expressed toxin sequence complexity as the Shannon Diversity Index [] of the -mer count table from Jellyfish, where i is the ith -mer and pi is the proportion of all -mers counted comprising -mer i. Shannon’s was translated to effective number of -mer as exp() for downstream analyses (109). These transcriptomic complexity measurements were robust to shorter and longer -mer sizes (SI Appendix, Fig. S8), and simulated transcriptomes showed they are sensitive to single mutations and small changes in expression and gene copy number (SI Appendix, Methods and Fig. S9). Variance in these transcriptome complexity scores was not clearly associated with sample size and showed significant variation among lineages (SI Appendix, Fig. S10). We therefore used the mean transcriptomic complexity value for each lineage in downstream comparative analyses, with sample size ranging from 1 to 17 individuals (mean = 2.8 individuals).
Measuring Venom Phenotypic Complexity.
To measure phenotypic complexity, we subjected 155 whole venom samples to HPLC on a Shimadzu Prominence HPLC System (Shimadzu Scientific Instruments). We measured absorbance at 220 nm for 140 min (see SI Appendix, Methods for full analytical run parameters) and used the Chromatopac feature in the Lab Solutions version 5.92 software (Shimadzu) to call HPLC peaks between 10 and 125 min, where peaks generally represent separate venom protein classes or isoforms. We retained all called peaks contributing at least 0.2% of total peak area and then recalculated the proportional area of each peak on the final peak set. Venom protein complexity was then calculated by using the area of each HPLC peak as input into the equation for Shannon’s Index, where pi was the proportional area of peak i, and converting to effective number of protein peaks as exp(). We used the mean protein complexity value for each lineage in downstream analyses, with sample size ranging from 1 to 5 individuals (mean = 2.1).
Measuring Snake Diet Diversity.
We measured diet diversity by collating published gut content data from Agkistrodon, Crotalus, and Sistrurus (Dataset S1 A and B). We tabulated counts of prey items identified to the species level. If prey were identified only to the genus level or higher, the counts of these prey items were proportionally distributed among the prey identified to species. For example, if a diet study included 3 entries—3 Peromyscus sp., 10 Peromyscus leucopus, and 5 Peromyscus maniculatus—we would proportionally assign the 3 genus-level observations among the species-level observations, resulting in 12 P. leucopus and 6 P. maniculatus as input to later analysis. Some sampled diet species were not present in available vertebrate phylogenies, and counts of these prey species were assigned to the nearest congener present in the phylogeny. To control for different prey sample size in diet studies, we generated 1,000 random subsamples for 15 prey items. These species and count data were used to calculate Shannon’s Index of snake diet diversity, where pi was the proportion of all diet items composed of species i, and was converted to effective number of prey species as exp(). The mean effective number of species from the 1,000 subsamples was used as the prey species diversity measure for each snake lineage. We required at least 10 diet items to be reported in the literature to consider a snake lineage for comparative analyses, resulting in N = 29 lineages for hypotheses testing.
We incorporated evolutionary distance as a second measure of diet diversity by calculating abundance-weighted MPD of snake diet diversity, using the ses.mpd function from picante (110). To incorporate phylogeny we generated a combined phylogeny of all listed prey species using published, dated supertrees for mammals (50), squamate reptiles (53), and amphibians (51), as well as median node-depth consensus phylogenies for the archosaur and invertebrate taxa produced using www.timetree.org (52). We united these separate trees into one at the median node depth for their most recent common ancestors, again provided by www.timetree.org.
Statistical Analyses.
All analyses were conducted in R version 3.6.1 (111). To calculate phylogenetic signal for our venom complexity metrics, we used our Astral-III species tree (Fig. 1 and the pmc function in the pmc R package) (112) with 1,000 bootstraps for confidence intervals. To assess relationships between each diet complexity measure and each venom complexity measure, we used pgls analyses as implemented in the pgls function of the caper R package (113), with lambda set to “ML,” various phylogenetic trees named in Table 1 to accommodate alternative phylogenetic hypotheses, and venom transcriptomic or proteomic complexity as the dependent variable. The independent variables were prey species diversity or MPD. We conducted our pgls analyses both with and without the South American rattlesnake C. durissus terrificus in the dataset, as it presented a large standardized residual (−2.8) and high leverage in having both the simplest transcriptome (SD = −3.0), simplest proteome (SD = −2.4), and the third lowest MPD of any lineage in our pgls dataset. We thus sought to ensure that our pgls results were not driven by this datapoint. Regression results were robust to its removal (SI Appendix, Fig. S11). We present the relationships between venom complexity and diet diversity in the Results section with this lineage removed. These results are thus unbiased by potential altered relationships between dietary diversity and venom evolution associated with the recent arrival of the C. durissus complex as the lone South American lineage of rattlesnakes (114).
Supplementary Material
Acknowledgments
We thank the following people for help in the field: H. Franz-Chávez, I. T. Ahumada-Carrillo, M. A. de la Torre Loranca, R. Ramirez- Chaparro, R. Solis, L. Badillo, B. La Forest, C. Rodriguez, Jeffrey Williams, J. Adams, H. Dahn, R. Govreau, T. Burkhardt, T. Dimler, B. Eaton, R. Engeldorf, M. Feldner, T. Fisher, R. Mayerhofer, E. McCormick, C. McMartin, J. McNally, D. Ortiz, T. Petty, M. Price, G. Salmon, J. Slone, D. Speckin, R. Swanson, E. Swanson, G. Territo, B. Townsend, C. Trumbower, K. VanSooy, I. Villalobos, D. Weber, Javier Ortiz, Marcos Millan, Bianca Sabido, W. J. Stark, C. J. Schmidt, J. J. Mead, C. Cochran, J. Aceves, M. Bernard, D. Deem, M. Linsalata, J. Lock, C. Mallery, C. May, S. May, A. Quillen, Flavio García, Esau Flores, Cristobal Moreno, Alan Salas, Juan Castañeda-Gaytán, K. Wray, E. Neri-Castro, K. Lawrence, L. Koffinas, R. Saul, A. Oliver, and S. Gable. We thank the following museums for cataloging specimens used in this study: Angelo State Natural History Collections, Arizona State University Natural History Collections, California Academy of Sciences, Coleccion Herpetologica Facultad de Ciencias Biologicas. Financial support was provided by NSF Grants DUE 1161228 (to C.L.P. and which funded J.L.S. and A.J.M.), DEB 1638879 and DEB 1822417 (to C.L.P.), DEB 1145978 and DEB 1638902 (to D.R.R.), DEB 1638872 (to H.L.G.), PRFB 1711141 (to M.L.H.); and Fundação de Amparo à Pesquisa do Estado de São Paulo Grant 2015/50127-5 (to F.G.G., A.M.M.-d.-S., I.L.M.J.-d.-A.). Additional support was provided by Fundação de Amparo à Pesquisa do Estado do Amazonas (PRO-ESTADO) (to A.M.M.-d.-S. as a Manaus Visiting Professor), Clemson University through faculty startup funds (C.L.P.), Consejo Nacional de Ciencia y Tecnologia Grant 247437 (to M.B.), Prairie Biotic Research Inc. (J.L.S.), Sigma Xi Grants-in-Aid-of-Research (J.L.S.), a SnakeDays Research Grant (J.L.S.), the Southwestern Association of Naturalists McCarley Research Grant (to J.L.S., A.J.M., and M.B.), and the Theodore Roosevelt Memorial Fund through the American Museum of Natural History (A.J.M. and J.L.S.). Parallel computing resources were provided by the Clemson Palmetto High-Performance Computing Cluster and supplemented with computing resources from the Clemson University Bioinformatics and Genomics Facility, which is supported by an Institutional Development Award from the National Institute of General Medical Sciences of the NIH (Grant P20GM109094). Helpful suggestions by S. Price, L. Alencar, L. Richards, and three anonymous reviewers improved the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015579118/-/DCSupplemental.
Data Availability
Short read data have been deposited in GenBank (BioProject PRJNA88989 and Biosample and Sequence Read Archive accession numbers in Dataset S2A). Alignments of nontoxin loci for phylogenetics have been deposited in the Dryad repository (https://doi.org/10.5061/dryad.mpg4f4qxt) (115). Other raw and processed data are provided in Datasets S1–S8.
References
- 1.Darwin C., On the Origin of Species by Means of Natural Selection, or Preservation of Favoured Races in the Struggle for Life (John Murray, 1859, London, UK, 1859). [Google Scholar]
- 2.Salgado-Luarte C., Gianoli E., Herbivores modify selection on plant functional traits in a temperate rainforest understory. Am. Nat. 180 (2), E42–E53 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Cohen J. E., Jonsson T., Carpenter S. R., Ecological community description using the food web, species abundance, and body size. Proc. Natl. Acad. Sci. U.S.A. 100 (4), 1781–1786 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koskella B., Lin D. M., Buckling A., Thompson J. N., The costs of evolving resistance in heterogeneous parasite environments. Proc. Natl. Acad. Sci. U.S.A. 279 (1735), 1896–1903 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lievens E. J. P., Perreau J., Agnew P., Michalakis Y., Lenormand T., Decomposing parasite fitness reveals the basis of specialization in a two-host, two-parasite system. Evol. Lett. 2 (4), 390–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friman V. P., Buckling A., Effects of predation on real-time host-parasite coevolutionary dynamics. Ecol. Lett. 16 (1), 39–46 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Broniewski J. M., Meaden S., Paterson S., Buckling A., Westra E. R., The effect of phage genetic diversity on bacterial resistance evolution. ISME J. 14 (3), 828–836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwao K., Rausher M. D., Evolution of plant resistance to multiple herbivores: Quantifying diffuse coevolution. Am. Nat. 149 (2), 316–335 (1997). [Google Scholar]
- 9.Betts A., Gifford D. R., MacLean R. C., King K. C., Parasite diversity drives rapid host dynamics and evolution of resistance in a bacteria-phage system. Evolution 70 (5), 969–978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright R. C. T., Friman V. P., Smith M. C. M., Brockhurst M. A., Cross-resistance is modular in bacteria–phage interactions. PLoS Biol. 16 (10), e2006057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonovics J., et al. , The origin of specificity by means of natural selection: Evolved and nonhost resistance in host-pathogen interactions. Evolution 67 (1), 1–9 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Betts A., Rafaluk C., King K. C., Host and parasite evolution in a tangled bank, Trends Parasitol. 32, 863–873 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lamb T. D., Collin S. P., Pugh E. N., Evolution of the vertebrate eye: Opsins, photoreceptors, retina and eye cup. Nat. Rev. Neurosci. 8 (12), 960–976 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf Y. I., Katsnelson M. I., Koonin E. V., Physical foundations of biological complexity. Proc. Natl. Acad. Sci. U.S.A. 115, E8678–E8687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adami C., Ofria C., Collier T. C., Evolution of biological complexity. Proc. Natl. Acad. Sci. U.S.A. 97 (9), 4463–4468 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards L. A., et al. , Phytochemical diversity drives plant–insect community diversity. Proc. Natl. Acad. Sci. U.S.A. 112, 10973–10978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salazar D., et al. , Origin and maintenance of chemical diversity in a species-rich tropical tree lineage. Nat. Ecol. Evol. 2 (6), 983–990 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Arbuckle K., From molecules to macroevolution: Venom as a model system for evolutionary biology across levels of life. Toxicon X 6, 100034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zancolli G., Casewell N. R., Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol. Biol. Evol. 37, 2777–2790 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Berenbaum M. R., Zangerl A. R., Chemical phenotype matching between a plant and its insect herbivore. Proc. Natl. Acad. Sci. U.S.A. 95 (23), 13743–13748 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson S., Wilson K., Pemberton J. M., Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.). Proc. Natl. Acad. Sci. U.S.A. 95 (7), 3714–3719 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manczinger M., et al. , Pathogen diversity drives the evolution of generalist MHC-II alleles in human populations. PLoS Biol. 17 (1), e3000131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casewell N. R., Wüster W., Vonk F. J., Harrison R. A., Fry B. G. (2012) Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 28:219–229. [DOI] [PubMed] [Google Scholar]
- 24.Phuong M. A., Mahardika G. N., Alfaro M. E., Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 17 (1), 401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pekár S., et al. , Venom gland size and venom complexity - Essential trophic adaptations of venomous predators: A case study using spiders. Mol. Ecol. 27, 4257–4269 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Davies E. L., Arbuckle K., Coevolution of snake venom toxic activities and diet: Evidence that ecological generalism favours toxicological diversity. Toxins 11 (12), 711 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Fry B. G., Kini R. M., Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J. Mol. Evol. 60 (1), 81–89 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Li M., Fry B. G., Manjunatha Kini R., Putting the brakes on snake venom evolution: The unique molecular evolutionary patterns of Aipysurus eydouxii (marbled sea snake) phospholipase A2 toxins. Mol. Biol. Evol. 22 (4), 934–941 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Smiley-Walters S. A., Farrell T. M., Gibbs H. L., The importance of species: Pygmy rattlesnake venom toxicity differs between native prey and related non-native species. Toxicon 144, 42–47 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Holding M., Biardi J., Gibbs H., Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc. Biol. Sci. 283 (1829), (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margres M. J., et al. , Quantity, not quality: Rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol. 34, 3099–3110 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Smiley-Walters S. A., Farrell T. M., Gibbs H. L., Evaluating local adaptation of a complex phenotype: Reciprocal tests of pigmy rattlesnake venoms on treefrog prey. Oecologia 184, 1–10 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Modahl C. M., Mrinalini, Frietze S., Mackessy S. P., Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proc. Biol. Sci. 285 (1884), 20181003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernardoni J. L., et al. , Functional variability of snake venom metalloproteinases: Adaptive advantages in targeting different prey and implications for human envenomation. PloS One 9 (10), e109651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doolittle R. F., Step-by-step evolution of vertebrate blood coagulation. Cold Spring Harbor Symp. Quant. Biol. 74, 35–40 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Davidson C. J., et al. , Molecular evolution of the vertebrate blood coagulation network. Thromb. Haemostasis 89 (3), 420–428 (2003). [PubMed] [Google Scholar]
- 37.Alencar L. R., Martins M., Greene H. W., Evolutionary History of Vipers, (John Wiley & Sons, Ltd, 2018). [Google Scholar]
- 38.Uetz P., Freed P., Hošek Je., “The reptile database” (2020). http://www.reptile-database.org. Accessed 22 November 2020.
- 39.Clark R. W., Diet of the timber rattlesnake, Crotalus horridus. J. Herpetol. 36 (3), 494–499 (2002). [Google Scholar]
- 40.Gloyd H. K., Conant R. P., The Snakes of the Agkistrodon Complex: A Monographic Review (Society for the Study of Amphibians and Reptiles, Oxford, OH, 1990), p. 614. [Google Scholar]
- 41.Mackessy S. P., “Venom composition in rattlesnakes: Trends and biological significance” in Biology of the Rattlesnakes, Hayes W. K., Beaman K. R., Cardwell M. D., Bush S. P., Eds. (Loma Linda University Press, Loma Linda, CA, 2008), pp. 495–510. [Google Scholar]
- 42.Rokyta D. R., Lemmon A. R., Margres M. J., Aronow K., The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 13 (1), 312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dowell N. L., et al. , The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 26 (18), 2434–2445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittington A. C., Mason A. J., Rokyta D. R., A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Mol. Biol. Evol. 35 (4), 887–898 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Betts A., Gray C., Zelek M., MacLean R. C., King K. C., High parasite diversity accelerates host adaptation and diversification. Science 360 (6391), 907 LP–911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berenbaum M. R., Zangerl A. R., Parsnip webworms and host plants at home and abroad: Trophic complexity in a geographic mosaic. Ecology 87 (12), 3070–3081 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Goüy de Bellocq J., Charmonnel N., Morand S., Coevolutionary relationship between helminth diversity and MHC class II polymorphism in rodents. J. Evol. Biol. 21 (4), 1144–1150 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Barrow L. N., et al. , Deeply conserved susceptibility in a multi-host, multi-parasite system. Ecol. Lett. 22 (6), 987–998 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Pilosof S., et al. , Host-parasite network structure is associated with community-level immunogenetic diversity. Nat. Commun. 5 (1), 5172 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Bininda-Emonds O. R., et al. , The delayed rise of present-day mammals. Nature 446 (7135), 507–512 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Isaac N. J. B., Redding D. W., Meredith H. M., Safi K., Phylogenetically-informed priorities for Amphibian conservation. PloS One 7 (8), e43912 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34 (7), 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Pyron R., Burbrink F. T., Wiens J. J., A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13 (1), 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20 (2), 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Alencar L. R., et al. , Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 105, 50–62 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Blair C., Sánchez-Ramírez S., Diversity-dependent cladogenesis throughout western Mexico: Evolutionary biogeography of rattlesnakes (viperidae: Crotalinae: Crotalus and Sistrurus). Mol. Phylogenet. Evol. 97, 145–154 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Serrano S. M. T., The long road of research on snake venom serine proteinases. Toxicon 62, 19–26 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Holding M., Drabeck D., Jansa S., Gibbs H., Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr. Comp. Biol. 56 (5), (2016). [DOI] [PubMed] [Google Scholar]
- 59.Papareddy P., et al. , An ecoimmunological approach to study evolutionary and ancient links between coagulation, complement and Innate immunity. Virulence 9 (1), 724–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowe A. H., Xiao Y., Rowe M. P., Cummins T. R., Zakon H. H., Voltage-gated sodium channel in grasshopper mice defends against bark scorpion toxin. Science 342 (6157), 441–446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zdenek C. N., et al. , A taxon-specific and high-throughput method for measuring ligand binding to nicotinic acetylcholine receptors. Toxins 11 (10), 600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Healy K., Carbone C., Jackson A. L., Snake venom potency and yield are associated with prey-evolution, predator metabolism and habitat structure. Ecol. Lett. 22 (3), 527–537 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Pomento A., Perry B., Denton R., Gibbs H., Holding M., No safety in the trees: Local and species-level adaptation of an arboreal squirrel to the venom of sympatric rattlesnakes. Toxicon 118, 149–155 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Lyons K., Dugon M. M., Healy K., Diet breadth mediates the prey specificity of venom potency in snakes. Toxins 12 (2), 74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devictor V., et al. , Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: The need for integrative conservation strategies in a changing world. Ecol. Lett. 13 (8), 1030–1040 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Tucker C. M., et al. , A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 92 (2), 698–715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson J. N., Schwind C., Guimarães P. R., Friberg M., Diversification through multitrait evolution in a coevolving interaction. Proc. Natl. Acad. Sci. U.S.A. 110 (28), 11487–11492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daltry J. C., Wüster W., Thorpe R. S., Diet and snake venom evolution. Nature 379, 537–540 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Holding M. L., Margres M. J., Rokyta D. R., Gibbs H. L., Local prey community composition and genetic distance predict venom divergence among populations of the northern Pacific rattlesnake ( Crotalus oreganus ). J. Evol. Biol. 31, 1513–1528 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Webber M. M., Jezkova T., Rodríguez-Robles J. A., Feeding ecology of sidewinder rattlesnakes, Crotalus cerastes (viperidae). Herpetologica 72 (4), 324–330 (2016). [Google Scholar]
- 71.Jansa S. A., Voss R. S., Adaptive evolution of the venom-targeted vWF protein in opossums that eat pitvipers. PloS One 6 (6), e20997 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams V., White J., Schwaner T. D., Sparrow A., Variation in venom proteins from isolated populations of tiger snakes (Notechis ater Niger, N. scutatus) in South Australia. Toxicon 26 (11), 1067–1075 (1988). [DOI] [PubMed] [Google Scholar]
- 73.Aird S. D., et al. , Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genom. Biol. Evol. 9 (10), 2640–2649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rokyta D. R., Wray K. P., Margres M. J., The genesis of an exceptionally lethal venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom-gland transcriptomics. BMC Genom. 14, 394–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zancolli G., et al. , Is hybridization a source of adaptive venom variation in rattlesnakes? A test, using a Crotalus scutulatus × viridis hybrid zone in southwestern New Mexico. Toxins 8 (6), 188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fry B. G., et al. , Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (caenophidia). Mol. Cell. Proteomics 7 (2), 215–246 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Giorgianni M. W., et al. , The origin and diversification of a novel protein family in venomous snakes. Proc. Natl. Acad. Sci. U.S.A. 117 (20), 201920011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.J. A. Eble, Structurally robust and functionally highly versatile—C-Type Lectin (-related) proteins in snake venoms. Toxins 11, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clemetson K. J., Navdaev A., Dörmann D., Du X. Y., Clemetson J. M., Multifunctional snake C-type lectins affecting platelets. Pathophysiol. Haemostasis Thrombosis 31 (3-6), 148–154 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Odokonyero D., et al. , Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family. Proc. Natl. Acad. Sci. U.S.A. 111 (23), 8535–8540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sikosek T., Chan H. S., Biophysics of protein evolution and evolutionary protein biophysics. J. R. Soc. Interf. 12, 20150915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schield D. R., et al. , The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29, 590–601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gibbs H. L., Chiucchi J. E., Deconstructing a complex molecular phenotype: Population-level variation in individual venom proteins in eastern massasauga rattlesnakes (Sistrurus c. catenatus). J. Mol. Evol. 72 (4), 383–397 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Mason A. J., et al. , Trait differentiation and modular toxin expression in palm-pitvipers. BMC Genom. 21 (1), 147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olivera B. M., “E.E. Just lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology” in Molecular Biology of the Cell (American Society for Cell Biology, 1997), Vol. 8, pp. 2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Casewell N. R., Huttley G. A., Wüster W., Dynamic evolution of venom proteins in squamate reptiles. Nat. Commun. 3, 1066 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Casewell N. R., et al. , Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 9205–9210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forister M. L., et al. , The global distribution of diet breadth in insect herbivores. Proc. Natl. Acad. Sci. U.S.A. 112 (2), 442–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schemske D. W., Mittelbach G. G., Cornell H. V., Sobel J. M., Roy K., Is there a latitudinal gradient in the importance of biotic interactions? Annu. Rev. Ecol. Evol. Syst. 40 (1), 245–269 (2009). [Google Scholar]
- 90.Kerkhoff A. J., Moriarty P. E., Weiser M. D., The latitudinal species richness gradient in New World woody angiosperms is consistent with the tropical conservatism hypothesis. Proc. Natl. Acad. Sci. U.S.A. 111 (22), 8125–8130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Safi K., et al. , Understanding global patterns of mammalian functional and phylogenetic diversity. Phil. Trans. Biol. Sci. 366 (1577), 2536–2544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holding M., Margres M., Mason A., Parkinson C., Rokyta D., Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins 10 (6), 249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31 (19), 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Waterhouse R. M., et al. , BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35 (3), 543–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kriventseva E. V., et al. , OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47 (D1), D807–D811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katoh K., Standley D. M., A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32 (13), 1933–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 (14), 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15), 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang C., Rabiee M., Sayyari E., Mirarab S., ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf. 19 (6), 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang C., Sayyari E., Mirarab S., “ASTRAL-III: Increased scalability and impacts of contracting low support branches” in Comparative Genomics, Meidanis J., Nakhleh L., Eds. (Springer International Publishing, Cham, Switzerland, 2017), pp. 53–75. [Google Scholar]
- 102.Rabiee M., Sayyari E., Mirarab S., Multi-allele species reconstruction using ASTRAL. Mol. Phylogenet. Evol. 130, 286–296 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Smith S. A., O’Meara B. C., treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28 (20), 2689–2690 (2012). [DOI] [PubMed] [Google Scholar]
- 104.Dubinkina V. B., Ischenko D. S., Ulyantsev V. I., Tyakht A. V., Alexeev D. G., Assessment of k-mer spectrum applicability for metagenomic dissimilarity analysis. BMC Bioinf. 17 (17), (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinf. 10 (1), 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25 (14), 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li H., seqtk Toolkit for processing sequences in FASTA/Q formats (2012). https://github.com/lh3/seqtk. Accessed 16 March 2020.
- 108.Marçais G., Kingsford C., A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27 (6), 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jost L., Entropy and diversity. Oikos 113 (2), 363–375 (2006). [Google Scholar]
- 110.Kembel S. W., et al. , Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26 (11), 1463–1464 (2010). [DOI] [PubMed] [Google Scholar]
- 111.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2019). [Google Scholar]
- 112.Boettiger C., Coop G., Ralph P., Is your phylogeny informative? Measuring the power of comparative methods. Evolution 66 (7), 2240–2251 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Orme D., et al. , Caper: Comparative Analyses of Phylogenetics and Evolution in R (2018). R package version 5.2: 1–36. [Google Scholar]
- 114.Wüster W., et al. , Tracing an invasion: Landbridges, refugia, and the phylogeography of the neotropical rattlesnake (serpentes: Viperidae: Crotalus durissus). Mol. Ecol. 14 (4), 1095–1108 (2005). [DOI] [PubMed] [Google Scholar]
- 115.Holding M. L., Parkinson C. L.. Data for “Phylogenetically diverse diets favor more complex venoms in North American pitvipers”. Dryad. 10.5061/dryad.mpg4f4qxt. Deposited 19 December 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Short read data have been deposited in GenBank (BioProject PRJNA88989 and Biosample and Sequence Read Archive accession numbers in Dataset S2A). Alignments of nontoxin loci for phylogenetics have been deposited in the Dryad repository (https://doi.org/10.5061/dryad.mpg4f4qxt) (115). Other raw and processed data are provided in Datasets S1–S8.