The continued emergence of drug-resistant Plasmodium falciparum parasites hinders global attempts to eradicate malaria, emphasizing the need to identify new antimalarial drugs. Attractive targets for chemotherapeutic intervention are the cytochrome (cyt) bc1 complex, which is an essential component of the mitochondrial electron transport chain (mtETC) required for ubiquinone recycling and mitochondrially localized dihydroorotate dehydrogenase (DHODH) critical for de novo pyrimidine synthesis.
KEYWORDS: Plasmodium, Plasmodium falciparum, antimalarial agents, drug resistance, malaria, metabolism
ABSTRACT
The continued emergence of drug-resistant Plasmodium falciparum parasites hinders global attempts to eradicate malaria, emphasizing the need to identify new antimalarial drugs. Attractive targets for chemotherapeutic intervention are the cytochrome (cyt) bc1 complex, which is an essential component of the mitochondrial electron transport chain (mtETC) required for ubiquinone recycling and mitochondrially localized dihydroorotate dehydrogenase (DHODH) critical for de novo pyrimidine synthesis. Despite the essentiality of this complex, resistance to a novel acridone class of compounds targeting cyt bc1 was readily attained, resulting in a parasite strain (SB1-A6) that was panresistant to both mtETC and DHODH inhibitors. Here, we describe the molecular mechanism behind the resistance of the SB1-A6 parasite line, which lacks the common cyt bc1 point mutations characteristic of resistance to mtETC inhibitors. Using Illumina whole-genome sequencing, we have identified both a copy number variation (∼2×) and a single-nucleotide polymorphism (C276F) associated with pfdhodh in SB1-A6. We have characterized the role of both genetic lesions by mimicking the copy number variation via episomal expression of pfdhodh and introducing the identified single nucleotide polymorphism (SNP) using CRISPR-Cas9 and assessed their contributions to drug resistance. Although both of these genetic polymorphisms have been previously identified as contributing to both DSM-1 and atovaquone resistance, SB1-A6 represents a unique genotype in which both alterations are present in a single line, suggesting that the combination contributes to the panresistant phenotype. This novel mechanism of resistance to mtETC inhibition has critical implications for the development of future drugs targeting the bc1 complex or de novo pyrimidine synthesis that could help guide future antimalarial combination therapies and reduce the rapid development of drug resistance in the field.
INTRODUCTION
Infection by the human malaria parasite Plasmodium falciparum rapidly leads to the clinical symptoms associated with the 48-h asexual replicative cycle of the parasite within the blood of the host (1). P. falciparum malaria continues to present an enormous global public health burden due to the lack of an effective long-term vaccine (2) and the emergence of resistance to front-line antimalarial chemotherapies (3). This underscores the continued need to maintain a robust drug development pipeline that produces novel and effective antimalarial compounds for future deployment.
To accelerate novel antimalarial compound identification, independent laboratories and global consortia have rapidly identified numerous chemical entities that are efficacious against multiple Plasmodium species and developmental stages. Despite the diverse chemical scaffolds in the compounds tested, most effective candidates tend to have similar modes of action in the malaria parasite due to the paucity of molecular targets that are significantly divergent from the human host (4–6, 56). As such, it is not surprising that a large and diverse set of compounds selectively target the highly divergent Plasmodium mitochondrion (4), which is essential for de novo pyrimidine synthesis during the blood stages of parasite development (7, 8).
Although the Plasmodium mitochondrion maintains the canonical tricarboxylic acid cycle pathway of central carbon metabolism (9–12), these enzymes are largely dispensable for blood-stage development (12). We have previously shown that the main role of the parasite mitochondrion during the blood-stage is the provision of precursors for de novo pyrimidine synthesis (7), a process requiring a ubiquinone-dependent, mitochondrial dihydroorotate dehydrogenase (DHODH; PF3D7_0603300) and a functioning electron transport chain (ETC) to maintain turnover of ubiquinol, regenerating ubiquinone. This sole essential function of the mitochondrion was established by genetic supplementation with an alternative DHODH from Saccharomyces cerevisiae (yDHODH), which is localized to the cytoplasm and utilizes fumarate as an electron acceptor rather than ubiquinone (7). Therefore, parasites expressing yDHODH from an episome, bypassing the mitochondrially localized DHODH from P. falciparum (PfDHODH), are highly resistant to all chemical inhibitors of the mitochondrial electron transport chain (mtETC). Interestingly, the addition of the biguanide prodrug proguanil, combined with the mtETC cytochrome b inhibitor atovaquone in the antimalarial drug Malarone, reversed the panresistant phenotype of yDHODH expressing P. falciparum strain D10 (D10::yDHODH) (7). Although the mode of action of atovaquone, as well as other mtETC inhibitors, is well-understood, the molecular target of proguanil remains largely unknown. While the exact target of proguanil remains undefined, recent work has suggested it is a slow-acting compound that targets the mitochondrion independent of the previously characterized synergistic electron transport inhibition (13).
Due to the efficacy of disrupting mitochondrial function in the malaria parasite as a strategy for chemotherapeutic intervention, these pathways are the focus of comprehensive antimitochondrial drug development programs, and significant efforts have been made to identify novel compounds that target the mtETC both directly (cytochrome b) and indirectly (DHODH). To this end, there are several antimalarials, some of which are in the advanced stages of clinical trials, whose proposed modes of action include interaction with the ubiquinone binding pocket(s) of DHODH (DSM-1 and DSM265) (14–16) or cytochrome b (ELQ-300 and 6-NH2Ac) (17, 18). Despite the essentiality of both components of the mtETC, resistance to these compounds readily arose, and genotyping of resistant parasites revealed either amplification of or point mutations in DHODH as well as point mutations within cytochrome b (14, 19–23). However, parasites resistant to 6-NH2Ac, an acridone compound thought to target the mtETC bc1 complex (18), did not exhibit traditional bc1 resistance mutations (21). The acridone-resistant parasite line SB1-A6 was not only resistant to 6-NH2Ac but also exhibited panresistance to all mitochondrial inhibitors to which the parent strain, D6, was susceptible (21). Again, this resistance was abrogated by the addition of proguanil (21), similar to P. falciparum parasites that express yDHODH, confirming the target of acridones to be the P. falciparum mtETC, although the provenance of the phenotypic similarities in resistance to bc1 complex inhibitors between SB1-A6 and the yDHODH transgenic parasite line was unclear (7).
In this study, we present a clear genotype for the P. falciparum SB1-A6 acridone-resistant clonal parasite strain and, through a combination of targeted and whole-cell methods, establish that the mechanism of resistance to both cytochrome bc1 and DHODH inhibitors results from the contribution of multiple genetic polymorphisms. We find that P. falciparum SB1-A6 accumulates both a copy number variation and a specific mutation in PfDHODH, and both of these genetic polymorphisms contribute to the panresistant phenotype. This study also uncovers a mechanism of cross-resistance between PfDHODH and mtETC inhibitors and serves as a cautionary note to future antimalarial combination therapy formulations containing such drugs.
RESULTS
Strain SB1-A6 displays panresistance to mtETC inhibitors.
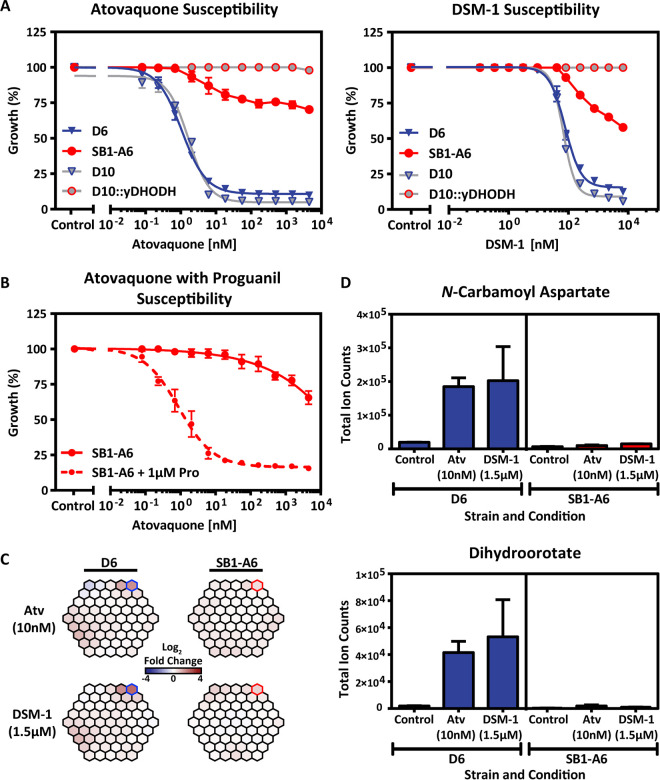
To confirm the resistance phenotype of P. falciparum SB1-A6 (21) and the line’s similarity to P. falciparum D10::yDHODH (7), we first tested the susceptibility of both strains to the classic malaria parasite cytochrome bc1 inhibitor, atovaquone, using a 3H-hypoxanthine incorporation assay to determine growth inhibition (Fig. 1A; see also Table S1 in the supplemental material). As expected, both SB1-A6 and D10::yDHODH displayed resistance to atovaquone, while the parental strains (D6 and D10, respectively) were susceptible (Fig. 1A; see also Table S1) as previously published (21). It has been well established that the expression of yDHODH in P. falciparum results in resistance to the PfDHODH inhibitor DSM-1 (24, 25). Therefore, we tested the susceptibility of SB1-A6 to direct inhibition of PfDHODH with DSM-1 (15) and found that this parasite line was resistant to both DHODH as well as cytochrome bc1 inhibitors (Fig. 1B; see also Table S1), making it the first P. falciparum clone generated to exhibit such cross-resistance (21). However, more recent studies aimed at generating resistance to atovaquone by increasing drug concentration exposure stepwise have also resulted in cross-resistance to DSM-1 (20). Resistance to mtETC inhibitors due to an alternative source of pyrimidines is abolished by the addition of the prodrug proguanil (7). Likewise, resistance to atovaquone inhibition of the mtETC in SB1-A6 was reversed upon the addition of 1.5 μM proguanil (Fig. 1B; see also Table S1), suggesting that the mechanism of panresistance to mitochondrial inhibitors is linked to the function of PfDHODH as was previously demonstrated for D10::yDHODH (7).
FIG 1.
Drug-resistant phenotyping of P. falciparum strain SB1-A6. (A) Early ring-stage P. falciparum strain D6 (blue) and SB1-A6 (red) were exposed to titrating concentrations of atovaquone or DSM-1. Their susceptibility was directly compared to a phenocopy strain, D10:yDHODH (gray, red outline), and its parent, D10 (gray, blue outline). Parasite survival was determined using a traditional 3H-hypoxanthine incorporation growth inhibition assay and plotted as an average of three technical replicates (±standard deviation [SD]). (B) Reversal of the SB1-A6 phenotype is demonstrated by exposure of ring-stage parasites to titrating concentrations of atovaquone with (red circles, dashed line) or without the addition of 1.0 μM proguanil (red circles, solid line). Parasite survival was determined as in panel A. (C) Whole-cell metabolomic analysis was performed on trophozoite stages of both P. falciparum strains D6 and SB1-A6 after 2 h of 10× 1C50 exposure to either atovaquone (Atv) or DSM-1. Metabolic changes are displayed as a MetaPrint profile (5) and highlighted are the increasing (blue outline) metabolites in D6 compared to no change in SB1-A6 (red outline). (D) The increased metabolites highlighted (C) in P. falciparum strain D6 (blue) are plotted as absolute signal determined by the average total ion counts (biological duplicates in technical triplicate) for N-carbamoyl-l-aspartate and dihydroorotate in the presence of atovaquone (10 nM), DSM-1 (1.5 μM), or absence of drug relative to P. falciparum strain SB1-A6 (red).
Drug resistance mechanisms in both prokaryotes and eukaryotes can involve “rewiring” to overcome the inhibition of essential metabolic pathways by genetic variation, metabolic bypass, or efflux mechanisms. Metabolomics has become a useful tool for unraveling antiparasite drug mechanisms of action and resistance (5, 26, 27, 56). To elucidate the mechanism(s) of resistance in SB1-A6, we determined if any metabolic perturbations occurred in the SB1-A6 drug-resistant line due to drug treatment that could account for cross-resistance to both PfDHODH and cytochrome bc1 inhibition, using whole-cell metabolomics analysis. Trophozoite-stage D6 and SB1-A6 were exposed to ∼10× 50% inhibitory concentration (IC50) of either atovaquone (10 nM) or DSM-1 (1.5 μM) for 2 h, followed by whole-cell metabolite extraction, and quantitated by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (5). Metabolomic analysis showed that treatment of SB1-A6 with DSM-1 or atovaquone did not result in any significant metabolic perturbation (Fig. 1C and D; see also Table S1), whereas treatment of the wild-type D6 revealed an accumulation of de novo pyrimidine synthesis precursors compared to that of an untreated control (Fig. 1C and D). The accumulation of pyrimidine precursors in the parental line upon treatment with either atovaquone or DSM-1 and lack of accumulation in the treated mutant line (Fig. 1D), in neither a stage- nor time-dependent manner (see Fig. S1 in the supplemental material), is consistent with the involvement of DHODH in the mechanism of resistance.
Genetic determinants of panresistance to mtETC and pyrimidine synthesis inhibition in SB1-A6.
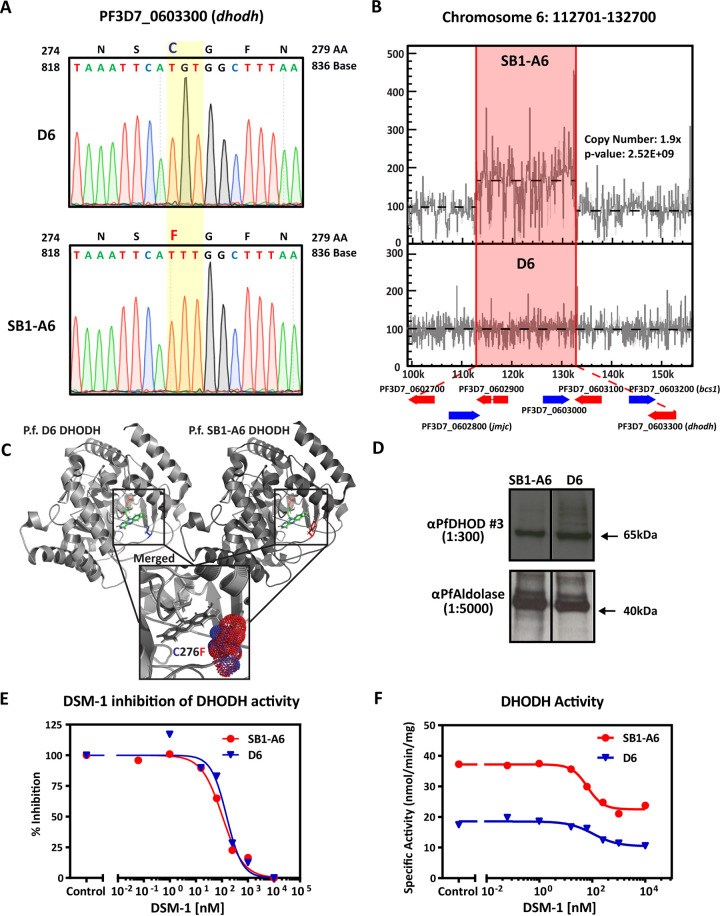
Previous studies have demonstrated that genetic mechanisms of resistance resulting in the ability of the parasite to overcome mtETC and de novo pyrimidine synthesis inhibition include mutations in either cytochrome b (28, 29) or pfdhodh (19, 30) or copy number variation of pfdhodh (20, 31). Analysis of the SB1-A6 cytochrome b sequence by Smilkstein et al. (21) did not reveal any mutations. Therefore, to identify any genetic polymorphisms that could lead to a molecular mechanism of resistance, we examined the genomes of both the D6 wild-type and SB1-A6 drug-resistant parasites using Illumina-based next-generation DNA sequencing. Whole-genome sequencing (WGS) revealed a nonsynonymous G→T point mutation in the coding sequence of PfDHODH, resulting in the amino acid change C276F (Fig. 2A; see also Table S2 in the supplemental material). Amino acid C276 is localized adjacent to the flavin mononucleotide binding pocket (Fig. 2C), and the substitution to a phenylalanine has been demonstrated to reduce the size of the binding pocket where DSM-1 binds (19), resulting in a binding pocket structure resembling that of the human DHODH (30). Although this mutation was not the only change found in SB1-A6 (Table S2), it was the only change in an annotated gene known to be directly linked to pyrimidine synthesis and/or mitochondrial function.
FIG 2.
Genetic and molecular differences in P. falciparum strain SB1-A6. (A) Whole-genome sequencing revealed a point mutation in PF3D7_0603300 (pfdhodh), and this mutation was verified by Sanger sequencing. A trace of the PfDHODH coding sequence is shown from nucleotide 818 to 836 and the single nucleotide polymorphism change from the parental D6 to SB1-A6 at nucleotide G827T (yellow highlight), which results in the amino acid change C276F. Additionally, WGS revealed a 1.9× copy number increase of an ∼20-kb region (red highlight) on chromosome 6, which includes PF3D7_0603300 (pfdhodh) (B). Copy number increase was calculated and graphically represented using Intansv (version 0.99.3). (C) Structural comparisons of wild-type (PDB accession number 4RX0, light gray, C276 highlighted in blue) and mutant (PDB accession number 6E0B, dark gray, C276F highlighted in red) PfDHODH bound with flavin mononucleotide (FMN) as well as the merged structure surrounding amino acid 276 (inset, cysteine in blue and phenylalanine in red, represent the isosurface of each sidechain) were made using PyMOL 2.3.4. (D) Western blot analysis of the PfDHODH expression in both D6 and SB1-A6 probed with mouse anti-PfDHODH antibody (1:300) compared to PfAldolase (mouse anti-PfAldolase-HRP, 1:5,000) as a loading control. Percent inhibition of PfDHODH activity (E) and capture of total PfDHODH activity (F) in isolated mitochondria from SB1-A6 (red) and D6 (blue) in the presence of titrating concentrations of DSM-1.
In addition, we identified an ∼20-kb, 2-fold copy number variation (CNV) in the SB1-A6 genome, including the coding sequence for PfDHODH, as well as six other protein-coding genes (Fig. 2B). We further confirmed the presence of this copy number increase by DNA microarray comparative genome hybridization (see Table S3 in the supplemental material). This region has previously been shown to increase in copy number in P. falciparum selected for resistance to DSM-1 (31), resulting in cross-resistance to atovaquone (20). Although each of these genetic polymorphisms has individually been previously identified as contributing significantly to both DSM-1 (19) and atovaquone resistance (20, 31), SB1-A6 possesses a unique genotype in which both alterations are present in a single parasite line, suggesting that the combination contributes to the panresistant phenotype of this line. Intriguingly, the increased CNV of PfDHODH did not directly translate to higher protein levels (Fig. 2D), nor did it produce any obvious mislocalization (see Fig. S2 in the supplemental material) of the protein in the parasite.
Due to previous observations that linked the PfDHODH C276F mutation and CNV to both DSM-1 and atovaquone resistance, respectively (20, 31), we examined the function of PfDHODH in mitochondria isolated from D6 and SB1-A6 to determine if these polymorphisms could account for the reduced sensitivity of SB1-A6 parasites to mitochondrial inhibition. Measurement of DHODH activity in isolated mitochondria from trophozoite stages of D6 and SB1-A6 revealed that the enzymes of both strains are equally susceptible to inhibition by DSM-1 (Fig. 2E), confirming the results from purified wild-type and C276F mutant PfDHODH activity assays (19). Interestingly, measurement of the specific activity of DHODH from SB1-A6-isolated mitochondria revealed a 2-fold increase compared to D6 (Fig. 2F), possibly due to the increase in copy number of PfDHODH. Although the C276F mutation does not lead to a decrease in the ability of DSM-1 to inhibit PfDHODH, these data suggest that the SB1-A6 parasite line has gained the ability to withstand either mtETC or pyrimidine biosynthesis inhibition by increasing the overall activity of PfDHODH, allowing sufficient levels of nucleotide synthesis for parasite survival.
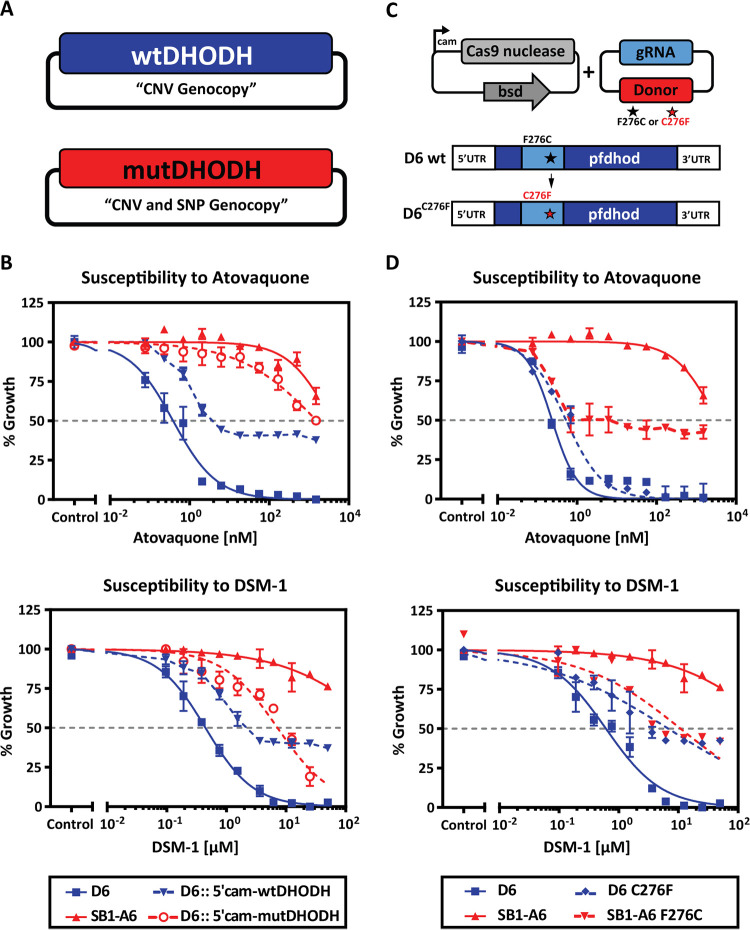
Genetic supplementation of DHODH increases copy number and CRISPR-Cas9 introduction of C276F point mutation.
Previous studies have established that P. falciparum grown in the presence of stepwise increasing concentrations of DSM-1 leads to an increase in copy number of a chromosomal region that includes PfDHODH, leading to resistance to both DSM-1 (31) and atovaquone (20). To test the hypothesis that a copy number increase in PfDHODH also results in decreased sensitivity of SB1-A6 parasites to inhibition of electron transport or de novo pyrimidine synthesis, we introduced either wild-type or mutant (C276F) PfDHODH expressed from an episomal vector under the control of a constitutive promoter in the drug-susceptible strain D6 (Fig. 3A). After selection of a parasite population containing the mutant or wild-type PfDHODH, we assessed the susceptibility of these parasites to atovaquone or DSM-1 using a standard SYBR green growth inhibition assay. The introduction of additional copies of PfDHODH resulted in decreased susceptibility to DSM-1 and atovaquone (Fig. 3B). Notably, the episomal expression of the mutant PfDHODH also results in almost complete resistance to atovaquone (Fig. 3B), phenocopying the genotype of SB1-A6. Episomal increase of wild-type PfDHODH results in a 10- to 100-fold decrease in susceptibility to DSM-1 (Fig. 3B), confirming the phenotype established by Guler et al. of DSM-1 and atovaquone-resistant parasites with an increased copy number of PfDHODH (20, 31).
FIG 3.
Phenotyping individual genetic contributions to P. falciparum SB1-A6 resistance. (A) Strategy for phenotyping a mimic of the PfDHODH genotype in the wild-type P. falciparum D6 strain. Episomal expression of an additional copy of wild-type PfDHODH (blue) aims to genocopy a CNV, and episomal expression of the mutant PfDHODH (red) genocopies a CNV of the mutation to capture the contribution of these genotypes to the resistance phenotype. (B) These transgenic lines, D6::wtDHODH (blue triangle, dashed) and D6::mutDHODH (red open circle, dashed), were assessed for their susceptibility to titrating concentrations of both atovaquone and DSM-1 in comparison to the parental strains, A6 (red solid triangle) and D6 (blue solid square). Parasite survival was determined using a traditional 48-h SYBR green growth inhibition assay and plotted as an average of three technical replicates (±SD). (C) Graphic representation of the CRISPR-Cas9-mediated mutational strategy for modification of PfDHODH in both the wild-type D6 and mutant strain SB1-A6. A plasmid expressing Cas9 was cotransfected with a plasmid that contains the DHODH repair template that encodes for C276 (wild type) or F276 (mutant), which are provided to introduce a single point mutation converting strain D6 to C276F and SB1-A6 to F276C, respectively. The CRISPR-Cas9-mutated strains D6C276F (blue solid diamond, dashed) and SB1-A6F276C (red solid triangle, dashed) were then assayed for their susceptibility to titrating concentrations of both atovaquone and DSM-1 in comparison to the parental strains A6 (red solid triangle) and D6 (blue solid square). Parasite survival was determined using a traditional 48-h SYBR green growth inhibition assay and plotted as an average of three technical replicates (±SD).
Since the discovery that P. falciparum survival is dependent upon de novo pyrimidine synthesis (7), PfDHODH has become a popular target for chemical intervention strategies (15, 16, 32, 33). For example, discovery efforts have led to the development of triazolopyrimidine-based inhibitors for PfDHODH, which are currently undergoing clinical trials for safety and efficacy (14, 34). In vitro selection of P. falciparum for resistance to compound DSM265 led to the identification of various mutations within PfDHODH (19). Of these mutations, C276F was determined to reduce the binding capacity of chemical inhibitors using an in vitro expression system (19) as well as computational docking studies (30). To test the hypothesis that the same observed point mutation in SB1-A6 PfDHODH contributes to P. falciparum resistance to both PfDHODH and cytochrome bc1 inhibitors, we utilized CRISPR-Cas9 (35) to genetically introduce the C276F mutations into this gene in D6 (D6C276F) and also repair it in SB1-A6 (A6F276C) (Fig. 3C; see also Fig. S3A and B in the supplemental material). After confirming the introduction or repair of the mutation at amino acid 276 (Fig. S3A and B), we then performed SYBR green drug susceptibility assays to quantitate the contribution of this mutation to the panresistant phenotype (Table S1). While we were able to reverse this mutation to the wild-type background in A6F276C, decreasing the overall resistance to both atovaquone and DSM-1 (Fig. 3D; see also Table S1), there remains an increased CNV of PfDHODH, which is likely contributing to the higher IC50 than demonstrated for D6 (Fig. 3D; see also Table S1). Conversely, susceptibility to atovaquone is only decreased by 2-fold due to the introduction of C276F into strain D6 (Fig. 3D; see also Table S1). Interestingly, these assays show that this mutation results in a 10-fold increase in the IC50 of DSM-1 in D6C276F (Fig. 3D) and a 40-fold decrease in A6F276C (Fig. 3D). These results suggest that the point mutation directly contributes to the decreased efficacy of direct inhibition of PfDHODH and enhances the ability of P. falciparum to overcome mtETC inhibition (Table S1). Taken together, these data demonstrate the coordination between genetic polymorphisms of the PfDHODH SNP and CNV in the SB1-A6 parasite line that mediates the panresistance phenotype.
DISCUSSION
In this study, we have demonstrated the coordination of two genetic polymorphisms that result in resistance to compounds that target the mtETC. First, the copy number increase of DHODH results in reduced susceptibility to bc1 complex and de novo pyrimidine synthesis inhibition. However, a 2-fold increase in DHODH, by itself, was not sufficient for complete resistance to mtETC inhibitors, potentially due to a reliance on other genes within the CNV amplicon. Aside from DHODH, this amplicon encompasses seven genes, a majority of which are unannotated, except for PF3D7_0603200 (bcs1, mitochondrial chaperone, putative) and PF3D7_0602800 (jmjc, jumanji-C demethylase domain containing), the gene products of which could contribute to the drug-resistant phenotype. This is supported by previous studies that demonstrated that a similar amplicon results in resistance to both atovaquone and DSM-1 (20, 31). Further work is necessary to determine if the proteins encoded on the amplicon are involved in reducing the parasite’s susceptibility to mtETC and de novo pyrimidine synthesis inhibition.
The implications of the SB1-A6 phenotype should be emphasized in consideration of the mtETC-pyrimidine-biosynthesis axis as an antimalarial drug target. While the adaptation that led to SB1-A6 seems to have been a unique event, both the CNV and C276F point mutations have each been derived independently from exposure to atovaquone and DSM265, a next-generation PfDHODH inhibitor currently in clinical trials. Therefore, the possibility that adaptations similar to those found in SB1-A6 may arise during clinical trials or deployment of new bc1 complex or DHODH inhibitors deserves consideration. While concerns regarding atovaquone-resistant mutations in the cytb gene on mitochondrial DNA (mtDNA) have been largely allayed due to lack of inheritance during transmission within the mosquito vector (36), we have no reason to discount the transmissibility of nuclear-encoded pfdhodh polymorphisms at present. Since these adaptations can be overcome by the addition of proguanil, the continued inclusion of proguanil or an effective replacement should be considered in future combinations. In addition to the question of transmissibility and choice of partner drugs, the ultimate significance of the SB1-A6 phenotype to future antimalaria drug choices may also differ between treatment use and prophylaxis. During treatment, parasite number is high, increasing the risk for selection of both preexisting and de novo resistant parasites, whereas, in causal prophylactic use, in the absence of preexisting resistance, the risk would be expected to be negligible. In either case, assessing the risk and monitoring for the emergence of the SB1-A6 phenotype should be done.
This study presents the first identification of the acquisition of multiple genetic polymorphisms in response to a bc1 complex inhibitor that results in panresistance to both direct and indirect inhibitors of DHODH. Fully elucidating the molecular mechanisms of this panresistance is essential to inform future chemotherapeutic targeting strategies for malaria elimination. The emergence of resistance to many of the drugs in the antimalarial arsenal has highlighted the urgency of developing future medicines with modes of action distinct from those of the current therapies. More recently, considerations have been made for combination therapies that target a single essential metabolic pathway via different modes of action. In light of the potential development of cross-resistance to a single inhibitor due to its combinatorial partner, the molecular mechanisms of resistance for future therapeutic strategies should be thoroughly investigated prior to approval for use in the clinical setting.
Recently, the essentiality of DHODH has been demonstrated in cell cycle-arrested cancer cells without mitochondria, wherein reactivation of growth occurs upon the acquisition of mtETC-linked DHODH (37), suggesting that the lack of de novo pyrimidine synthesis is a major obstacle for in vivo growth of respiration-compromised tumor cells. The combined detrimental effects of both dysfunctional mitochondrial respiration and mtETC-linked de novo pyrimidine synthesis on cell growth strengthen their relevance as drug targets against both parasite development and metastatic cell growth. This further emphasizes the continued benefit of identifying novel inhibitors of DHODH that could ultimately be appropriated for both parasite elimination and perhaps extended to treatment targeting metastatic cells within the human host.
MATERIALS AND METHODS
Parasite strains and culture.
Plasmodium falciparum strains D6 and SB1-A6 were provided by Michael Riscoe and Martin Smilkstein (21). All parasites obtained from other sources or generated in this study were maintained under standard conditions (38) at 2% hematocrit with O+ human erythrocytes in RPMI 1640 (Thermo Fisher Scientific) containing hypoxanthine, NaH2CO3 (Sigma-Aldrich), HEPES (Sigma-Aldrich), glutamine (Sigma-Aldrich), and 2.5 g/liter AlbuMax II (Thermo Fisher Scientific). Intraerythrocytic developing parasites were synchronized with 5% sorbitol (Sigma-Aldrich) over three subsequent developmental cycles to synchronize parasites within 4 to 6 h as previously described (39).
Parasite cultures were maintained mycoplasma-free and were confirmed prior to metabolomic analysis. Separate mycoplasma-free culture apparatuses and solutions were used for all metabolomics culturing to prevent contamination. Cultures were checked for mycoplasma contamination weekly using IntronBio e-Myco, mycoplasma PCR detection kit (Boca Scientific; catalog no. 25235). If parasite cultures tested positive for mycoplasma, cultures were treated as per manufacturer’s instructions with Mycoplasma Removal Agent (MP Biomedicals, LLC; catalog no. 30-500-44).
Drug inhibition assays.
All parasite growth inhibition assays were performed in 96-well plates as described by Desjardin et al. (40) and adapted for SYBR green I (41). Drugs were prepared in dimethyl sulfoxide (DMSO) as a 500× dilution series (typically a 2- or 3-fold dilution series was used, yielding a final concentration in media ranging from 0.001 to 30 μM depending on the cell line) and were then diluted 1:50 into medium to yield a final DMSO concentration of 0.2%. Parasites were diluted to 1% parasitemia and 1% hematocrit in each well of a 96-well plate and then propagated for 48 h, and growth was assessed using the SYBR green method. SYBR green fluorescence was measured (excitation/emission, 485/535 nm), and data analysis was performed using GraphPad Prism 7. For transgenic P. falciparum strains generated in this study, parasites were propagated for three intraerythrocytic cycles in medium lacking drug before plating. All data were collected in triplicate and biological duplicate.
Mitochondrial activity assays.
P. falciparum cultures were synchronized at least twice by treatment with alanine (42), expanded, and harvested at 8 to 15% parasitemia in the mid- to late-trophozoite stages. A hemozoin-depleted mitochondrial fraction was isolated from the parasites using nitrogen cavitation, magnetic separation, and differential centrifugation as previously described (43).
DHODH activity assays were conducted essentially as previously described (16, 43) with minor modifications, utilizing the ubiquinone analog decylubiquinone as the primary electron acceptor, coupled to the redox dye 2,6-dichloroindophenol to take advantage of the latter’s higher extinction coefficient and absorbance in the visible range. Assays contained 1 mM l-dihydroorotate, 100 μM decylubiquinone, 60 μM 2,6-dichloroindophenol, 50 mM sodium malonate, 2 mM potassium cyanide, 100 mM potassium chloride, 0.05% Triton X-100, 10% glycerol, and 0.1 M HEPES (pH 7.6) in a final volume of 1 ml. Five-microliter aliquots of diluted inhibitor or vehicle (DMSO) were added, and each assay was initiated by addition of a 7-μl aliquot of mitochondrial preparation (50 to 110 μg protein) and recorded with a modified SLM-AMINCO DW2C dual-wavelength spectrophotometer (On-Line Instrument Systems, Inc., Bogart, GA, USA) in dual mode (600 nm to 528 nm) at 30°C.
Inhibitors.
The antimalaria compound atovaquone was a gift from Glaxo Wellcome, Research Triangle Park, NC. Proguanil was kindly provided by the Jacobus Pharmaceutical Company, Princeton, NJ, USA. Both ELQ-300 and 6-NH2Ac were contributed to this study by Michael Riscoe, Oregon Health Sciences University, Portland, OR, USA. DSM-1 (15) and DSM265 (14) were provided by Margaret Phillips, University of Texas Southwestern, Dallas, TX, USA. Chloroquine (catalog no. C6628) was purchased from Sigma-Aldrich, St. Louis, MO, USA.
Whole-cell metabolomics.
Metabolite extractions, data collection, and analysis were performed as described previously (5, 6, 44). Briefly, an ∼20-μl pellet of cells was resuspended in 1 ml of prechilled 90:10 methanol/water and placed at 4°C. The internal standard 13C4-15N1-aspartate was spiked into the extraction methanol solution to control for sample preparation and handling. Samples were vortexed, resuspended, and centrifuged for 10 min at 15,000 rpm and 4°C. Supernatants were collected and stored at −80°C or dried down immediately under nitrogen flow. The dried metabolites were resuspended in high-pressure liquid chromatography (HPLC)-grade water (Sigma-Aldrich; Chromasolv) to between 1 × 105 and 1 × 106 cells/μl based on hemocytometer counts of purified parasites. All samples were processed in technical triplicate with method blanks to reduce unwanted variation and account for background signal, respectively. Samples were randomized, and 10 μl of resuspended metabolite extract or method blank was injected for ultrahigh-performance liquid chromatography-mass spectrometry (HPLC-MS) analysis.
Extracts were analyzed using HPLC-MS on a Thermo Exactive Plus Orbitrap. Metabolite separation was performed with a C18 column (Phenomenex Hydro-RP; catalog no. 00D-4387-B0) using a 25 min gradient of (i) 3% aqueous methanol/15 mM acetic acid/10 mM tributylamine ion-pairing agent and (ii) 100% MeOH (45). Metabolite detection was performed using a scan range of 85 to 1,000 m/z and a resolution of 140,000 at m/z 200 in negative ion mode. The detection of cellular metabolites was aided by the generation of a database from 292 pure metabolite standards using the same instrument and method to determine detection capability, mass/charge ratio (m/z), and retention time for each metabolite. Analysis of data was carried out as previously described (5), and all spectral data and analytical metadata from this study have been deposited into the NIH Metabolomics Workbench (study ID no. ST001652).
Whole-genome sequencing and analysis.
Genomic DNA (gDNA) was isolated from P. falciparum strains D6 and SB1-A6, as well as the two derived lines containing CRISPR edits. These gDNAs were prepared and sequenced as previously described (46). Briefly, a total of 10 μg of genomic DNA (gDNA) from each line was sheared to obtain a fragment size of ∼200 to 400 bp. The resulting sheared gDNA was size selected on a 2% (wt/vol) low-melting agarose gel and then purified and concentrated using MinElute purification columns followed by the QIAquick PCR purification kit (Qiagen). Barcoded libraries for Illumina TruSeq single-end sequencing were then constructed from the size-selected, sheared material and size selected using Agencourt AMPure XP magnetic beads (Agencourt Biosciences, Beckman Coulter). The quality of the final sequencing libraries was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies), and the concentration of each library was quantified using a Quant-iT double-stranded DNA (dsDNA) broad-range assay kit (Invitrogen). The final libraries were multiplexed, and 20% (vol/vol) PhiX control DNA (Illumina; catalog no. FC-110-3001) per lane was added per lane prior to sequencing by Illumina HiSeq 2500 Rapid Run (150 bp).
Sequencing outputs were uploaded into Galaxy (47), which is hosted locally at the Millennium Science Complex at Pennsylvania State University. Sequence reads were mapped to the P. falciparum 3D7 reference genome version 10.0 (http://plasmodb.org/common/downloads/release-10.0/Pfalciparum/) using the Burrows-Wheeler alignment tool while selecting and filtering unique reads for map quality > 30 (48). The aligned .bam files are available on Sequence Read Archive under accession no. PRJNA692491. Sequence variations were detected by Freebayes (version 0.9.0.a) using stringent filtering parameters based on quality and read depth (49, 50). Then SnpEff (version 3.3) was applied to annotate and determine the statistical significance of each variant (51). Genome copy number variations were detected based upon local chromosomal read depth using CNVnator (version 0.3) and annotated with Intansv (version 0.99.3) (52).
DHODH Western blot analysis.
Total protein extracts were run on a 4 to 12% Bis-Tris gel (Thermo Fisher) in 2-(N-morpholino)ethanesulfonic acid (MES) buffer and then transferred to a nitrocellulose membrane. After blocking with 5% milk in phosphate-buffered saline with Tween 20 (PBS-T) for 30 min, membranes were incubated overnight with the primary antibody in 5% milk. Primary antibodies used were the following: 1:300 dilution of mouse anti-PfDHODH no. 3 (a kind gift from Margeret Phillips, University of Texas Southwestern) or 1/5,000 mouse anti-PfAldolase-horseradish peroxidase (HRP) (Abcam; catalog no. ab38905). The next day, membranes were washed three times with PBS-T and incubated for 2 h with the appropriate secondary antibody and then washed again. The secondary antibody that was used was 1/5,000 goat anti-mouse HRP conjugate (Pierce). Enhanced chemiluminescence (ECL) reagent (Pierce) was used to detect bound antibody.
DNA constructs.
To mutate DHODH using CRISPR-Cas9, complementary oligonucleotides encoding the guide RNA were inserted into pDC2-U6A-hDHFR (kind gift of Marcus Lee) using BbsI sites. A 515-bp homology region containing the mutation in PfDHODH (C276F) or the wild-type sequence from either SB1-A6 or D6 was cloned into StrataClone (Agilent) and then inserted using the NotI site of pDC2-U6A-hDHFR. The resulting plasmids are pDC2-C276F-hDHFR and pDC2-F276C-hDHFR, which were cotransfected with the Cas9-BSD plasmid from Jose-Juan Lopez-Rubio (35) into D6 and SB1-A6, respectively.
To introduce multiple copies of either the mutant or wild-type PfDHODH into D6 parasites, PfDHODH was amplified from SB1-A6 (mutant) or D6 (wild-type) genomic DNA including the stop codon, cloned into StrataClone (Agilent), sequence-verified, and then inserted into pLN-ENR-GFP (53) using the AvrII and BsiWI restriction sites. The resulting plasmid, pLN-A6DHODH, was transfected into D6 parasites.
Transfection of Plasmodium falciparum.
Transfection of P. falciparum was performed as previously described (54). Briefly, 5 to 7% ring-stage parasite cultures were washed three times with cytomix (55). The parasitized red blood cell (RBC) pellet was resuspended to 50% hematocrit in cytomix. The plasmid, isolated using a Qiagen maxikit and stored in 50-μg aliquots in ethanol, was centrifuged down and resuspended in 100 μl cytomix. The plasmid and 250 μl of the 50% parasitized RBC suspension were combined and transferred to a 0.2-cm cuvette on ice. Electroporations were carried out using a Bio-Rad Gene Pulser set at 0.31 kV, 960 μF. The electroporated cells were immediately transferred to a T-25 flask containing 0.2 ml uninfected 50% RBCs and 7 ml medium. To select for parasites containing plasmid, medium containing 5 nM WR99210 or 1.5 μg/ml Blasticidin was added at 48 h posttransfection. Cultures were maintained under constant 5 nM WR92210 pressure, splitting weekly, until viable parasites were observed. To confirm the successful mutation of PfDHODH, genomic DNA was purified using the Qiagen DNeasy blood and tissue kit, and the region of interest was PCR amplified and Sanger sequenced. Positive transfectants were cloned by limiting dilution to obtain a pure population of transgenic parasites.
Data availability.
All spectral data and analytical metadata from this study have been deposited into the NIH Metabolomics Workbench (study ID no. ST001652). The aligned .bam files are available on Sequence Read Archive under accession no. PRJNA692491.
Supplementary Material
ACKNOWLEDGMENTS
We thank the many people who have contributed to this project including Andrew D. Patterson and Philip B. Smith of the Penn State Huck Institutes of the Life Sciences Metabolomics Core Facility and Simon Cobbold, previously of the Llinás lab at Princeton University, for analytical expertise and technical oversight, Craig Praul of the Genomics Core Facility of the Penn State Huck Institutes of the Life Sciences, and the Genomics Core Facility at Princeton University and its staff, especially Donna Storton and Jessica Buckles Wiggins. We also thank Meg Phillips and Pradip Rathod for sharing reagents and critical comments. Lastly, we thank the many past and present members of the Llinás lab who supported this project by enhancing the evaluation of data and providing critical comments.
H.J.P., M.W.M., J.M.M., L.M.O., and C.L. designed and performed experiments and analyzed data. H.J.P. wrote the manuscript and generated figures. M.W.M., M.J.S., and M.K.R. provided reagents, critical comments, and edits. M.L. and A.B.V. designed the experiments, provided critical comments, edits, and oversight.
This work was supported by generous funding awarded to M.L. from the Bill and Melinda Gates Foundation Grand Challenges Grant (phase II - OPP1119049), an NIH Director’s New Innovators Award (1DP2OD001315-01), the Center for Quantitative Biology (P50 GM071508), and startup funding from the Pennsylvania State University. J.M.M., M.W.M., and A.B.V. were supported by NIH grant R01 AI028398. M.K.R. is a recipient of a Veterans Administration Research Career Scientist Award (14S-RCS001), and his group receives financial support from the Veterans Administration Merit Review Program (i01 BX003312). Research reported in this publication was also supported by the U.S. National Institutes of Health under award no. AI100569 (M.K.R., A.B.V., J.M.M., and M.W.M.), the U.S. Department of Defense Peer Reviewed Medical Research Program (log no. PR130649; contract no. W81XWH-14-1-0447) (M.K.R.), and the Intramural Research Program of the Center for Biologics Evaluation and Research, U.S. Food and Drug Administration (H.J.P.). The team also acknowledges valuable support provided by the Medicines for Malaria Venture.
REFERENCES
- 1.Miller LH, Ackerman HC, Su XZ, Wellems TE. 2013. Malaria biology and disease pathogenesis: insights for new treatments. Nat Med 19:156–167. doi: 10.1038/nm.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.RTS,S Clinical Trials Partnership. 2015. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet 386:31–45. doi: 10.1016/S0140-6736(15)60721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. 2015. Antimalarial drug resistance: literature review and activities and findings of the ICEMR network. Am J Trop Med Hyg 93:57–68. doi: 10.4269/ajtmh.15-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowell AN, Istvan ES, Lukens AK, Gomez-Lorenzo MG, Vanaerschot M, Sakata-Kato T, Flannery EL, Magistrado P, Owen E, Abraham M, LaMonte G, Painter HJ, Williams RM, Franco V, Linares M, Arriaga I, Bopp S, Corey VC, Gnadig NF, Coburn-Flynn O, Reimer C, Gupta P, Murithi JM, Moura PA, Fuchs O, Sasaki E, Kim SW, Teng CH, Wang LT, Akidil A, Adjalley S, Willis PA, Siegel D, Tanaseichuk O, Zhong Y, Zhou Y, Llinas M, Ottilie S, Gamo FJ, Lee MCS, Goldberg DE, Fidock DA, Wirth DF, Winzeler EA. 2018. Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359:191–199. doi: 10.1126/science.aan4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allman EL, Painter HJ, Samra J, Carrasquilla M, Llinas M. 2016. Metabolomic profiling of the malaria box reveals antimalarial target pathways. Antimicrob Agents Chemother 60:6635–6649. doi: 10.1128/AAC.01224-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobbold SA, Chua HH, Nijagal B, Creek DJ, Ralph SA, McConville MJ. 2016. Metabolic dysregulation induced in Plasmodium falciparum by dihydroartemisinin and other front-line antimalarial drugs. J Infect Dis 213:276–286. doi: 10.1093/infdis/jiv372. [DOI] [PubMed] [Google Scholar]
- 7.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. 2007. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature 446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 8.Painter HJ, Morrisey JM, Vaidya AB. 2010. Mitochondrial electron transport inhibition and viability of intraerythrocytic Plasmodium falciparum. Antimicrob Agents Chemother 54:5281–5287. doi: 10.1128/AAC.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaidya AB, Painter HJ, Morrisey JM, Mather MW. 2008. The validity of mitochondrial dehydrogenases as antimalarial drug targets. Trends Parasitol 24:8–9. doi: 10.1016/j.pt.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 10.MacRae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, Kenny S, Bottova I, Tilley L, McConville MJ. 2013. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol 11:67. doi: 10.1186/1741-7007-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, Fishbaugher M, Healer J, Cowman AF, Kappe SH, Llinas M. 2013. Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J Biol Chem 288:36338–36350. doi: 10.1074/jbc.M113.503557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke H, Lewis IA, Morrisey JM, McLean KJ, Ganesan SM, Painter HJ, Mather MW, Jacobs-Lorena M, Llinas M, Vaidya AB. 2015. Genetic investigation of tricarboxylic acid metabolism during the Plasmodium falciparum life cycle. Cell Rep 11:164–174. doi: 10.1016/j.celrep.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skinner-Adams TS, Fisher GM, Riches AG, Hutt OE, Jarvis KE, Wilson T, von Itzstein M, Chopra P, Antonova-Koch Y, Meister S, Winzeler EA, Clarke M, Fidock DA, Burrows JN, Ryan JH, Andrews KT. 2019. Cyclization-blocked proguanil as a strategy to improve the antimalarial activity of atovaquone. Commun Biol 2:166. doi: 10.1038/s42003-019-0397-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips MA, Lotharius J, Marsh K, White J, Dayan A, White KL, Njoroge JW, El Mazouni F, Lao Y, Kokkonda S, Tomchick DR, Deng X, Laird T, Bhatia SN, March S, Ng CL, Fidock DA, Wittlin S, Lafuente-Monasterio M, Benito FJ, Alonso LM, Martinez MS, Jimenez-Diaz MB, Bazaga SF, Angulo-Barturen I, Haselden JN, Louttit J, Cui Y, Sridhar A, Zeeman AM, Kocken C, Sauerwein R, Dechering K, Avery VM, Duffy S, Delves M, Sinden R, Ruecker A, Wickham KS, Rochford R, Gahagen J, Iyer L, Riccio E, Mirsalis J, Bathhurst I, Rueckle T, Ding X, Campo B, Leroy D, Rogers MJ, et al. 2015. A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci Transl Med 7:296ra111. doi: 10.1126/scitranslmed.aaa6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips MA, Gujjar R, Malmquist NA, White J, El Mazouni F, Baldwin J, Rathod PK. 2008. Triazolopyrimidine-based dihydroorotate dehydrogenase inhibitors with potent and selective activity against the malaria parasite Plasmodium falciparum. J Med Chem 51:3649–3653. doi: 10.1021/jm8001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldwin J, Michnoff CH, Malmquist NA, White J, Roth MG, Rathod PK, Phillips MA. 2005. High-throughput screening for potent and selective inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J Biol Chem 280:21847–21853. doi: 10.1074/jbc.M501100200. [DOI] [PubMed] [Google Scholar]
- 17.Nilsen A, LaCrue AN, White KL, Forquer IP, Cross RM, Marfurt J, Mather MW, Delves MJ, Shackleford DM, Saenz FE, Morrisey JM, Steuten J, Mutka T, Li Y, Wirjanata G, Ryan E, Duffy S, Kelly JX, Sebayang BF, Zeeman AM, Noviyanti R, Sinden RE, Kocken CH, Price RN, Avery VM, Angulo-Barturen I, Jimenez-Diaz MB, Ferrer S, Herreros E, Sanz LM, Gamo FJ, Bathurst I, Burrows JN, Siegl P, Guy RK, Winter RW, Vaidya AB, Charman SA, Kyle DE, Manetsch R, Riscoe MK. 2013. Quinolone-3-diarylethers: a new class of antimalarial drug. Sci Transl Med 5:177ra37. doi: 10.1126/scitranslmed.3005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter RW, Kelly JX, Smilkstein MJ, Dodean R, Bagby GC, Rathbun RK, Levin JI, Hinrichs D, Riscoe MK. 2006. Evaluation and lead optimization of anti-malarial acridones. Exp Parasitol 114:47–56. doi: 10.1016/j.exppara.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 19.White J, Dhingra SK, Deng X, El Mazouni F, Lee MCS, Afanador GA, Lawong A, Tomchick DR, Ng CL, Bath J, Rathod PK, Fidock DA, Phillips MA. 2019. Identification and mechanistic understanding of dihydroorotate dehydrogenase point mutations in Plasmodium falciparum that confer in vitro resistance to the clinical candidate DSM265. ACS Infect Dis 5:90–101. doi: 10.1021/acsinfecdis.8b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guler JL, White J, Phillips MA, Rathod PK. 2014. Atovaquone tolerance in Plasmodium falciparum parasites selected for high-level resistance to a dihydroorotate dehydrogenase inhibitor. Antimicrob Agents Chemother 59:686–689. doi: 10.1128/AAC.02347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smilkstein MJ, Forquer I, Kanazawa A, Kelly JX, Winter RW, Hinrichs DJ, Kramer DM, Riscoe MK. 2008. A drug-selected Plasmodium falciparum lacking the need for conventional electron transport. Mol Biochem Parasitol 159:64–68. doi: 10.1016/j.molbiopara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stickles AM, De Almeida MJ, Morrisey JM, Sheridan KA, Forquer IP, Nilsen A, Winter RW, Burrows JN, Fidock DA, Vaidya AB, Riscoe MK. 2015. Subtle changes in endochin-like quinolone structure alter the site of inhibition within the cytochrome bc1 complex of Plasmodium falciparum. Antimicrob Agents Chemother 59:1977–1982. doi: 10.1128/AAC.04149-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandt REK, Lafuente-Monasterio MJ, Sakata-Kato T, Luth MR, Segura D, Pablos-Tanarro A, Viera S, Magan N, Ottilie S, Winzeler EA, Lukens AK, Gamo FJ, Wirth DF. 2019. In vitro selection predicts malaria parasite resistance to dihydroorotate dehydrogenase inhibitors in a mouse infection model. Sci Transl Med 11:eaav1636. doi: 10.1126/scitranslmed.aav1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke H, Morrisey JM, Ganesan SM, Painter HJ, Mather MW, Vaidya AB. 2011. Variation among Plasmodium falciparum strains in their reliance on mitochondrial electron transport chain function. Eukaryot Cell 10:1053–1061. doi: 10.1128/EC.05049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganesan SM, Morrisey JM, Ke H, Painter HJ, Laroiya K, Phillips MA, Rathod PK, Mather MW, Vaidya AB. 2011. Yeast dihydroorotate dehydrogenase as a new selectable marker for Plasmodium falciparum transfection. Mol Biochem Parasitol 177:29–34. doi: 10.1016/j.molbiopara.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent IM, Barrett MP. 2015. Metabolomic-based strategies for anti-parasite drug discovery. J Biomol Screen 20:44–55. doi: 10.1177/1087057114551519. [DOI] [PubMed] [Google Scholar]
- 27.Creek DJ, Barrett MP. 2014. Determination of antiprotozoal drug mechanisms by metabolomics approaches. Parasitology 141:83–92. doi: 10.1017/S0031182013000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross LS, Gamo FJ, Lafuente-Monasterio MJ, Singh OM, Rowland P, Wiegand RC, Wirth DF. 2014. In vitro resistance selections for Plasmodium falciparum dihydroorotate dehydrogenase inhibitors give mutants with multiple point mutations in the drug-binding site and altered growth. J Biol Chem 289:17980–17995. doi: 10.1074/jbc.M114.558353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. 1999. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol 33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 30.Bedingfield PTP, Cowen D, Acklam P, Cunningham F, Parsons MR, McConkey GA, Fishwick CWG, Johnson AP. 2012. Factors influencing the specificity of inhibitor binding to the human and malaria parasite dihydroorotate dehydrogenases. J Med Chem 55:5841–5850. doi: 10.1021/jm300157n. [DOI] [PubMed] [Google Scholar]
- 31.Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, DeRisi J, Rathod PK. 2013. Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog 9:e1003375. doi: 10.1371/journal.ppat.1003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munier-Lehmann H, Vidalain PO, Tangy F, Janin YL. 2013. On dihydroorotate dehydrogenases and their inhibitors and uses. J Med Chem 56:3148–3167. doi: 10.1021/jm301848w. [DOI] [PubMed] [Google Scholar]
- 33.Patel V, Booker M, Kramer M, Ross L, Celatka CA, Kennedy LM, Dvorin JD, Duraisingh MT, Sliz P, Wirth DF, Clardy J. 2008. Identification and characterization of small molecule inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J Biol Chem 283:35078–35085. doi: 10.1074/jbc.M804990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells TNC, Van Huijsduijnen RH, Van Voorhis WC. 2015. Malaria medicines: a glass half full? Nat Rev Drug Discov 14:424–442. doi: 10.1038/nrd4573. [DOI] [PubMed] [Google Scholar]
- 35.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio J-J. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 36.Goodman CD, Siregar JE, Mollard V, Vega-Rodriguez J, Syafruddin D, Matsuoka H, Matsuzaki M, Toyama T, Sturm A, Cozijnsen A, Jacobs-Lorena M, Kita K, Marzuki S, McFadden GI. 2016. Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science 352:349–353. doi: 10.1126/science.aad9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bajzikova M, Kovarova J, Coelho AR, Boukalova S, Oh S, Rohlenova K, Svec D, Hubackova S, Endaya B, Judasova K, Bezawork-Geleta A, Kluckova K, Chatre L, Zobalova R, Novakova A, Vanova K, Ezrova Z, Maghzal GJ, Magalhaes Novais S, Olsinova M, Krobova L, An YJ, Davidova E, Nahacka Z, Sobol M, Cunha-Oliveira T, Sandoval-Acuña C, Strnad H, Zhang T, Huynh T, Serafim TL, Hozak P, Sardao VA, Koopman WJH, Ricchetti M, Oliveira PJ, Kolar F, Kubista M, Truksa J, Dvorakova-Hortova K, Pacak K, Gurlich R, Stocker R, Zhou Y, Berridge MV, Park S, Dong L, Rohlena J, Neuzil J. 2019. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab 29:399–416. doi: 10.1016/j.cmet.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 39.Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65:418–420. doi: 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- 40.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother 16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806. doi: 10.1128/aac.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes JD, Moch JK. 2002. Automated synchronization of Plasmodium falciparum parasites by culture in a temperature-cycling incubator. Methods Mol Med 72:489–497. doi: 10.1385/1-59259-271-6:489. [DOI] [PubMed] [Google Scholar]
- 43.Mather MW, Morrisey JM, Vaidya AB. 2010. Hemozoin-free Plasmodium falciparum mitochondria for physiological and drug susceptibility studies. Mol Biochem Parasitol 174:150–153. doi: 10.1016/j.molbiopara.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olszewski KL, Llinas M. 2013. Extraction of hydrophilic metabolites from Plasmodium falciparum-infected erythrocytes for metabolomic analysis. Methods Mol Biol 923:259–266. doi: 10.1007/978-1-62703-026-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. 2010. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem 82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulcini S, Staines HM, Lee AH, Shafik SH, Bouyer G, Moore CM, Daley DA, Hoke MJ, Altenhofen LM, Painter HJ, Mu J, Ferguson DJ, Llinas M, Martin RE, Fidock DA, Cooper RA, Krishna S. 2015. Mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, enlarge the parasite's food vacuole and alter drug sensitivities. Sci Rep 5:14552. doi: 10.1038/srep14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goecks J, Nekrutenko A, Taylor J, Galaxy T, Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907v2 [q-bio.GN].
- 51.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. 2006. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods 3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fidock DA, Wellems TE. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A 94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Hoff MJ, Moorman AF, Lamers WH. 1992. Electroporation in 'intracellular' buffer increases cell survival. Nucleic Acids Res 20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Creek DJ, Chua HH, Cobbold SA, Nijagal B, MacRae JI, Dickerman BK, Gilson PR, Ralph SA, McConville MJ. 2016. Metabolomic-based screening of the Malaria Box reveals both novel and established mechanisms of action. Antimicrob Agents Chemother 60:6650–6663. doi: 10.1128/AAC.01226-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All spectral data and analytical metadata from this study have been deposited into the NIH Metabolomics Workbench (study ID no. ST001652). The aligned .bam files are available on Sequence Read Archive under accession no. PRJNA692491.