Nitrofurantoin (NIT) is a broad-spectrum bactericidal antibiotic used in the treatment of urinary tract infections. It is a prodrug that once activated by nitroreductases goes on to inhibit bacterial DNA, RNA, cell wall, and protein synthesis.
KEYWORDS: Escherichia coli, nitrofurantoin, urinary tract infection
ABSTRACT
Nitrofurantoin (NIT) is a broad-spectrum bactericidal antibiotic used in the treatment of urinary tract infections. It is a prodrug that once activated by nitroreductases goes on to inhibit bacterial DNA, RNA, cell wall, and protein synthesis. Previous work has suggested that NIT retains considerable activity against nongrowing bacteria. Here, we have found that Escherichia coli grown to stationary phase in minimal or artificial urine medium is not susceptible to NIT. Supplementation with glucose under conditions where cells remained nongrowing (other essential nutrients were absent) sensitized cultures to NIT. We conceptualized NIT sensitivity as a multi-input AND gate and lack of susceptibility as an insufficiency in one or more of those inputs. The inputs considered were an activating enzyme, cytoplasmic abundance of NIT, and reducing equivalents required for NIT activation. We systematically assessed the contribution of each of these inputs and found that NIT import and the level of activating enzyme were not contributing factors to the lack of susceptibility. Rather, evidence suggested that the low abundance of reducing equivalents is why stationary-phase E. coli are not killed by NIT and catabolites can resensitize those cells. We found that this phenomenon also occurred when using nitrofurazone, which established generality to the nitrofuran antibiotic class. In addition, we observed that NIT activity against stationary-phase uropathogenic E. coli (UPEC) could also be potentiated through metabolite supplementation. These findings suggest that the combination of nitrofurans with specific metabolites could improve the outcome of uncomplicated urinary tract infections.
INTRODUCTION
Bacteria that have transiently lost their susceptibility to antibiotics constitute a major cause of infection relapse (1–3) and a source for resistant mutations to evolve (4–6). Urinary tract infections (UTIs), which are most often caused by specific uropathogenic Escherichia coli (UPEC) sublineages (7), are common among women and characterized by their frequent recurrence (8, 9). Nitrofurantoin (NIT) is a nitrofuran antibiotic indicated for the treatment of UTIs that has been recently repositioned as first-line therapy for the treatment of uncomplicated UTIs (10–12). It accumulates in the urine and enters the bacterial cell as a prodrug (12). Once NIT is activated by cytosolic nitroreductase enzymes, it goes on to inhibit bacterial DNA, RNA, cell wall, and protein synthesis (12, 13). The major nitroreductase that activates NIT is NfsA and minor activators are NfsB and NfsC (14), although NfsA and NfsB are sufficient for maximal nitroreductase activity (15). Accordingly, in a recent study, NIT-resistant mutants of E. coli clinical isolates were associated with mutations in nfsA and nfsB (16). Both of these nitroreductases are oxygen insensitive and use a reduced pyridine nucleotide to catalyze the reduction of nitrocompounds; NfsA uses NADPH, whereas NfsB can use either NADH or NADPH for activity (14).
In general, antibiotics are more effective against growing bacteria than their nongrowing counterparts, with some drugs completely losing their bactericidal activity against growth-inhibited populations (17, 18). Previous work asserted that NIT is active against both growing and nongrowing bacteria (19–21). In a study of Mycobacterium bovis BCG, NIT reduced culturability of dormant bacilli by approximately 100-fold over 48 h (21). In a study of Listeria monocytogenes, NIT was as bactericidal in a nutrient-rich medium (brain heart infusion broth) as in nutrient-poor conditions (0.9% saline with 0.1% peptone) (19). In addition, Fleck and colleagues observed approximately a 10,000-fold difference in survival for E. coli grown to stationary phase in rich medium when treated for 24 h with 200 μg/ml NIT compared to that of untreated controls (20). This activity against growth-inhibited bacteria makes NIT a particularly interesting therapeutic option for hard-to-treat infections, which often contain nongrowing bacteria (22).
Based on the preceding work, we began this study with the anticipation of investigating how NIT kills nongrowing bacteria within the context of many other antibiotics failing to do so (17, 18). However, with E. coli populations grown to stationary phase in minimal or artificial urine media, we observed a lack of bacterial killing with NIT, which was unexpected. Drawing inspiration from a previous study where specific metabolites could restore the bactericidal activity of aminoglycosides without restoring growth (23), we tested whether metabolites could potentiate the activity of NIT in nongrowing cultures. Interestingly, metabolites did facilitate killing by NIT and were able to do so in environments unable to support growth (e.g., saline). To investigate this phenomenon, we conceptualized NIT sensitivity as an AND gate consisting of three inputs: NIT-activating enzymes, cytoplasmic levels of NIT, and availability of reducing equivalents, where loss of susceptibility was an insufficiency in one or more of those inputs. Notably, metabolites had the potential to facilitate all three inputs, since they could boost translation rates of starved cells, improve the activity of import mechanisms that rely on proton motive force or ATP, and increase reducing equivalent supply. We investigated the contribution of each of these inputs and found that insufficient levels of activating enzymes and NIT unavailability in the cytoplasm were not responsible for the lack of susceptibility to NIT. Rather, results suggested that low reducing equivalent levels protected stationary-phase E. coli from NIT and that metabolites stimulated killing by providing electrons to activate the prodrug, which is a mechanism distinct from the aminoglycoside potentiation studied previously, where metabolites facilitated import (23). Further, we found that our observations were generalizable to the nitrofuran class of antibiotics by carrying out experiments with nitrofurazone (NFZ) and that glucose and mannitol could potentiate the activity of NIT against stationary-phase UPEC. Notably, the ability to potentiate killing of UPEC with NIT and mannitol is promising for the treatment of recurrent UTIs because both normally accumulate in the urine (12, 24).
RESULTS
Lack of susceptibility to NIT conceptualized as failure of a multi-input AND gate.
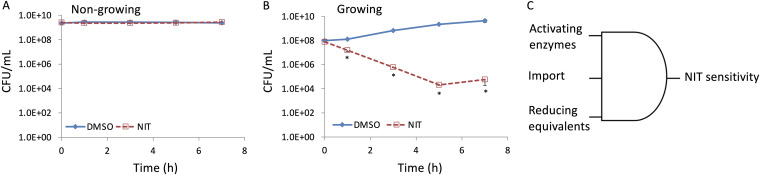
Nitrofurans are one class of antibiotics that are considered to be active against growing and nongrowing bacteria (19–21). Interested in the activity of this drug class against growth-inhibited bacteria, we performed assays with NIT and stationary-phase E. coli that had been grown in minimal glucose medium. We observed that such cultures were not susceptible to 250 μg/ml NIT (MIC = 4 μg/ml) (Fig. 1A), while cultures actively growing in the same medium were decimated by NIT (Fig. 1B). Given that NIT is a prodrug that is activated by nitroreductases, we conceptualized this phenotype as failure in a multi-input AND gate (Fig. 1C). Specifically, for NIT to be bactericidal, it needs (i) to penetrate bacterial membranes and be inside cells, (ii) nitroreductases to be present, and (iii) reducing equivalents in the form of NAD(P)H to be available. We reasoned that one or more of the inputs was insufficient and thus NIT was not being activated.
FIG 1.
Insensitivity of nongrowing E. coli to NIT. (A) Stationary-phase cultures of E. coli were treated with DMSO (solvent for NIT) or 250 μg/ml of NIT. CFU per milliliter were monitored at the indicated time points. (B) Overnight cultures were diluted into fresh MOPS plus glucose medium and incubated for 3 h before treatment with 250 μg/ml NIT or DMSO. CFU per milliliter were monitored at the indicated time points. (C) To be sensitive to NIT, E. coli must (i) contain enzymes that can activate NIT, (ii) contain NIT in the cytoplasm where activation occurs, and (iii) have sufficient reducing equivalents to activate NIT. *, P < 0.05 (t test), comparing DMSO with NIT. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
Metabolite-mediated resensitization to NIT.
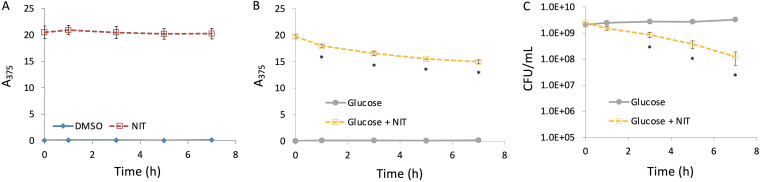
Consistent with our conceptualization of NIT inactivity, when we measured NIT reduction, we failed to see appreciable activation in stationary-phase cultures (Fig. 2A). To begin to unravel this phenomenon, we explored the possibility of metabolites facilitating NIT killing of those populations. We recognized that starvation for specific catabolites could limit antibiotic uptake (if it is energy dependent) (25), severely depress translation rates (26, 27), and drain pools of reducing equivalents (27). Therefore, metabolite supplementation had the capacity to impact each of the inputs in the NIT activity AND gate (Fig. 1C). Importantly, we also recognized that metabolites could stimulate growth, whose contribution we assess in the next section. We chose to supplement with glucose, and a significant reduction of NIT was observed (Fig. 2B), which was accompanied by an approximate 20-fold decrease in CFU after 7 h (Fig. 2C). In the presence of glucose and absence of NIT, CFU did not change appreciably over 7 h, which suggested that glucose supplementation was not stimulating growth (Fig. 2C). Further, lack of susceptibility in stationary-phase cultures and glucose restoration of sensitivity were also observed for a distinct nitrofuran, NFZ, which suggested that the phenomenon is applicable to the nitrofuran antibiotic class (see Fig. S1 in the supplemental material).
FIG 2.
Resensitization of nongrowing E. coli to NIT. Stationary-phase cultures of E. coli were treated with 250 μg/ml NIT or DMSO (A) or 10 mM glucose with or without 250 μg/ml NIT (B), and absorbance at 375 nm (A375) was determined at the indicated time points. (C) Stationary-phase cultures of E. coli were treated with 10 mM glucose with or without 250 μg/ml NIT, and CFU per milliliter were monitored at the indicated time points. *, P < 0.05 (t test), comparing glucose plus NIT at 1, 3, 5, and 7 h with t = 0 in panel B, and glucose with glucose plus NIT in panel C. Data represent at least three biological replicates. Each data point was denoted as mean ± SE. A similar phenomenon was observed when plating on MOPS plus glucose agar rather than LB agar, although the potentiation was slightly less pronounced (see Fig. S16 in the supplemental material).
Metabolite potentiation of NIT activity is independent of growth.
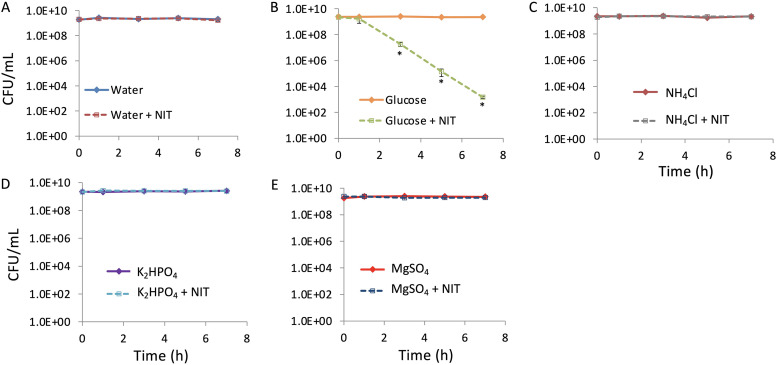
Although growth resumption was not observed to occur under the conditions of our potentiation assay (Fig. 2C), we recognized that the medium that we used was carbon limited and sought to obtain more definitive evidence of the involvement of growth or lack thereof in the resensitization to NIT that we observed. To do this, we adopted two approaches. In the first, we transferred stationary-phase populations to environments that could not support growth (saline solution), allowed them to acclimate for 1 h at 37°C, and then added carbon, nitrogen, phosphorus, or sulfur individually in the presence or absence of NIT (Fig. 3). Notably, only cultures supplemented with glucose showed potentiation of NIT activity (Fig. 3B), which confirmed that resensitization was independent of growth resumption. We confirmed that none of the environments assayed here could support growth due to the absence of essential nutrients by measuring growth from low-density cultures after 24 h of incubation (see Fig. S2 in the supplemental material). The second method that we employed to assess whether glucose was stimulating growth in the stationary-phase cultures used here was to assess sensitivity to ampicillin. β-lactam activity is closely linked to growth-rate, with nongrowing cultures losing susceptibility to this drug class (17, 23). When ampicillin challenge experiments were conducted, culturability of the stationary-phase E. coli cultures did not decline with ampicillin by itself or with cotreatment of ampicillin and glucose (see Fig. S3 in the supplemental material). These data support the results of the saline experiments (Fig. 3), and both together provide strong evidence that growth resumption did not contribute to the potentiation of NIT by glucose.
FIG 3.
Glucose resensitization mechanism is independent of growth resumption. Stationary-phase cultures were washed with, resuspended in, and further incubated in saline for 1 h before addition of water (A), 10 mM glucose (B), 9.5 mM NH4Cl (C), 1.32 mM K2HPO4 (D), or 0.286 mM MgSO4 (E). Concentrations were based on those found in minimal M9 medium (53). Following treatments, cultures were incubated for 7 h and CFUs were determined at the indicated time points. *, P < 0.05 (t test), comparing glucose with glucose plus NIT. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
NIT resensitization requires catabolism.
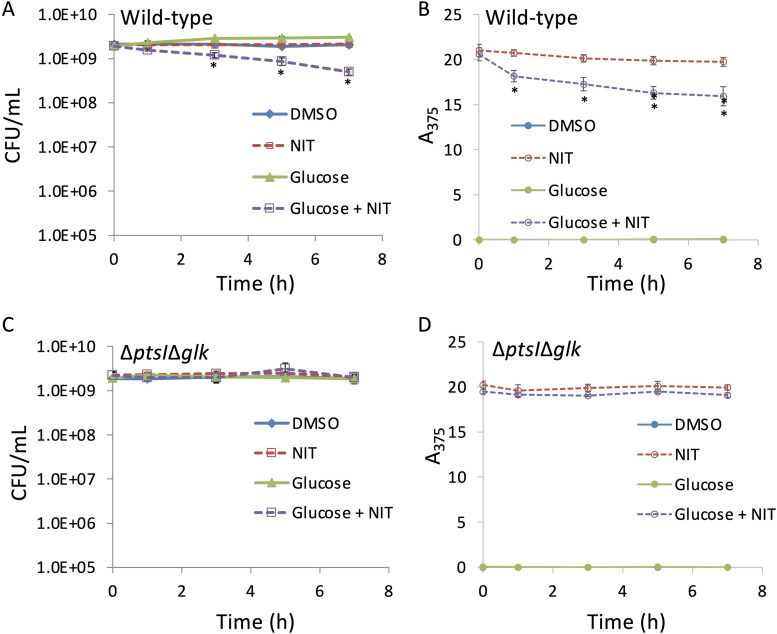
For glucose to restore the AND gate and potentiate NIT in nongrowing populations, we reasoned that it would have to be catabolized. We measured glucose consumption in stationary-phase cultures following the addition of glucose by monitoring extracellular glucose concentrations. As shown in Fig. S4 in the supplemental material, glucose was consumed by stationary-phase cultures under our experimental conditions. Next, we tested whether glucose consumption was required for potentiation. The main glucose import mechanism in E. coli is the phosphotransferase system (PTS) (28), which results in glucose phosphorylation to glucose-6-phosphate and constitutes the first step in glucose catabolism. We reasoned that inactivation of this system would eliminate glucose-mediated resensitization of NIT. Cells inactivated for the PTS component encoded by ptsI (EI enzyme) had poor growth in media with glucose as the sole carbon source (2.5-fold increase in optical density at 600 nm [OD600] after 24 h of incubation in morpholinepropanesulfonic acid [MOPS] plus glucose compared to a 170-fold increase for the wild type under the same conditions, which was in line with previous reports from other E. coli strains [29]), and therefore, cells were cultured to stationary phase in media with gluconate as the sole carbon source. Prior to assessing NIT resensitization by glucose in this mutant, we assessed whether the phenotypes that we had observed in cultures grown on glucose were conserved for the wild type in these altered culturing conditions. As depicted in Fig. 4A and B, the wild type showed a significant decrease in culturability and a significant increase in NIT reduction after treatment with NIT and glucose compared to controls when cells had been propagated to stationary phase in gluconate minimal medium, which confirmed that the phenotypes were conserved between assay conditions. Although it is worth noting that the reduced potentiation under these conditions compared to those with cells grown to stationary phase in minimal glucose medium (∼4-fold in Fig. 4A compared to ∼20-fold in Fig. 2C) likely reflects the different conditioning of cells grown in the different media. When the ΔptsI strain was assayed in a similar manner, little difference was observed between samples and controls until the 7-h time point, which was when a significant decrease in the ΔptsI strain culturability following treatment with NIT and glucose was present (see Fig. S5A in the supplemental material). These observations suggested that although EI inactivation in the ΔptsI strain impacted the phenotype, the mutation was not sufficient to abolish resensitization. In the absence of PTS, glucose can begin to be catabolized through phosphorylation by glucokinase, which is encoded by glk (30). Due to only partial attenuation of the phenotype in the ΔptsI strain, we tested glk involvement in NIT potentiation. We found that inactivation of both ptsI and glk (ΔptsI Δglk) resulted in a loss of susceptibility to NIT treatment supplemented with glucose and a lack of NIT reduction (Fig. 4C and D). Inactivation of only glk (Δglk) did not have much of an impact (Fig. S5B). These data demonstrated that the restoration of NIT sensitivity of stationary-phase populations requires glucose catabolism.
FIG 4.
Inactivation of glucose metabolism abolishes resensitization to NIT. Cultures of the E. coli wild type (A and B) or the ΔptsI Δglk double mutant (C and D) strains were grown to stationary phase in gluconate minimal medium and treated with DMSO (solvent for NIT), 250 μg/ml NIT, 10 mM glucose, or NIT supplemented with glucose. At the indicated time points, samples were processed for both CFU determination (A and C) and absorbance at 375 nm (A375) (B and D). *, P < 0.05 (t test), comparing NIT with glucose plus NIT. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
Lack of NIT activity is not associated with insufficient abundances of nitroreductases.
Glucose can stimulate all inputs of the AND gate because it could improve the activity of import mechanisms, boost translation rates of starved cells, and increase reducing equivalent supply. To begin to assess the contribution of NfsA and NfsB to NIT inactivity and potentiation under nongrowing conditions, we assayed the ΔnfsA ΔnfsB strain for NIT sensitivity and reduction and found that the enzymes were required to observe those phenotypes (see Fig. S6A and B in the supplemental material). We note that the ΔnfsA ΔnfsB strain consumes glucose similarly to the wild type under our experimental conditions (see Fig. S6C and S4 in the supplemental material), even though it does not activate NIT (Fig. S6B). These data established that nfsA or nfsB is needed to observe metabolite potentiation of NIT activity in stationary-phase populations.
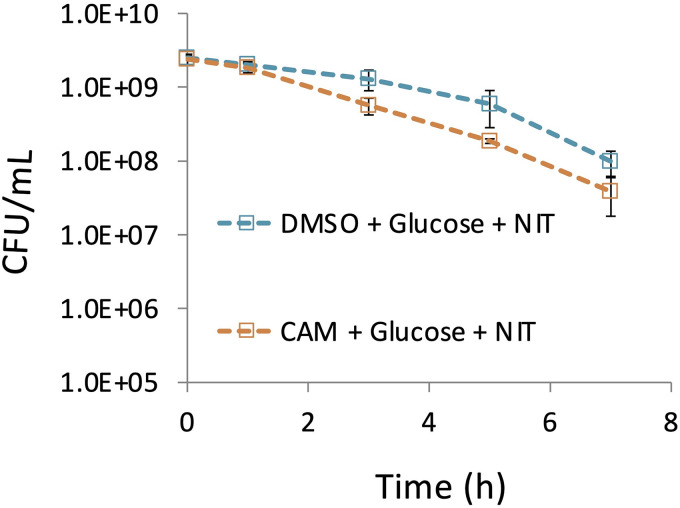
We next investigated whether cells under nongrowing conditions had sufficient levels of NfsA or NfsB to facilitate NIT killing or whether limited availability of these enzymes represents a reason for the failure of the AND gate. We carried out treatments in the absence or presence of a protein synthesis inhibitor (chloramphenicol [CAM]) that prevents any further protein synthesis that would be stimulated by glucose (see Fig. S7 in the supplemental material). We observed that cells did not need to synthesize any additional proteins during NIT treatment in order to observe metabolite potentiation because the CAM-treated samples, which were also treated with NIT and glucose, had comparable culturability losses to those that were treated with only NIT and glucose (Fig. 5; see also Fig. S8 in the supplemental material). These data suggested that the AND input associated with activating enzyme abundance was sufficient in the growth-inhibited conditions considered here.
FIG 5.
NIT kills in the absence of translation. Nongrowing cultures of E. coli were treated with 100 μg/ml CAM or DMSO (solvent for CAM) and incubated for 1 h. After 1 h of incubation, cultures were treated with 10 mM glucose and 250 μg/ml NIT. CFU were determined at the indicated time points. Additional controls are presented in Fig. S8 in the supplemental material. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
NIT inactivity is not affected by membrane permeabilization.
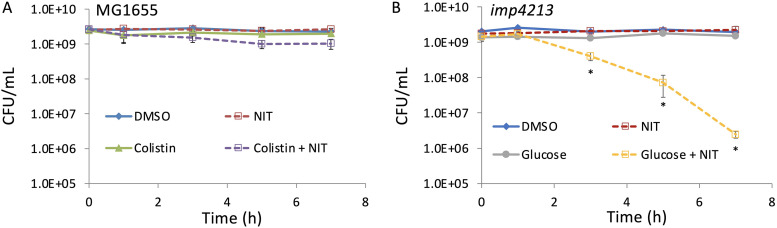
The second potential contributing factor that we evaluated was whether a lack of NIT import contributed to the loss of susceptibility observed. To assess the plausibility of import issues limiting NIT activity in our conditions, we employed two complementary methods. In the first, we treated cells with the membrane-permeabilizing agent colistin (31) at concentrations that we determined to permeate membranes by allowing penetration of bile salts (see Fig. S9 in the supplemental material), which are larger than NIT and excluded by the E. coli outer membrane (32). No impact on NIT susceptibility was observed after permeabilizing E. coli with colistin when compared to NIT only (Fig. 6A), and NIT potentiation by glucose was preserved in colistin-treated cultures (see Fig. S10 in the supplemental material). In the second approach, we used a genetic mutant, imp4213, that has a mutation to lptD, which increases penetration of many compounds (33) (see Fig. S11B in the supplemental material). Indeed, when the MIC of imp4213 for NIT was measured, it was found to be 2-fold lower than the wild type (Fig. S11A), which suggested that intracellular penetration can limit NIT activity. As depicted in Fig. 6B, stationary-phase cultures of imp4213 were not susceptible to NIT, just as stationary-phase cultures of wild-type were not susceptible (Fig. 1). Further, glucose potentiation of NIT in stationary-phase cultures of imp4213 demonstrated that the mutant can be killed by NIT under the conditions considered here (Fig. 6B). The lack of NIT potentiation when membranes were permeabilized by both chemical (colistin) and genetic (imp4213) approaches suggested that facilitating NIT import was not a critical aspect of the ability of metabolites to resensitize stationary-phase cultures to NIT.
FIG 6.
Membrane permeabilization does not potentiate NIT killing. (A) Stationary-phase cultures of E. coli were grown in MOPS plus glucose as described in Materials and Methods and treated with DMSO (control), 250 μg/ml NIT, 5 μg/ml colistin, or colistin plus NIT. Additional controls are shown in Fig. S10 in the supplemental material. (B) Stationary-phase cultures of the imp4213 strain were treated with DMSO, 250 μg/ml NIT, 10 mM glucose, or NIT supplemented with glucose. CFU were determined at the indicated time points. *, P < 0.05 (t test), comparing glucose plus NIT with NIT in panel B. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
Lack of reducing equivalents underlies lack of NIT susceptibility of stationary-phase E. coli.
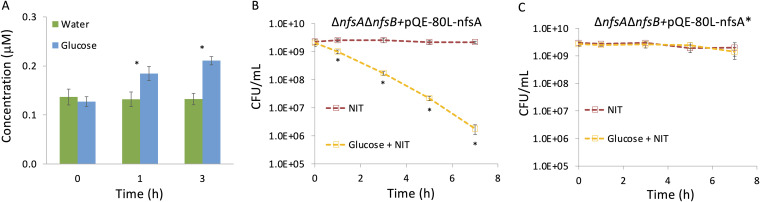
The last input to the AND gate to assess as a root cause for NIT inactivity was the availability of reducing equivalents for NfsA and NfsB to activate NIT. NADH and NADPH are known to increase in concentration upon the addition of glucose to glucose-limited E. coli cultures (34), and we confirmed that glucose supplementation significantly increased NADPH levels under the stationary-phase conditions used here (Fig. 7A). Because genetic modulation of NADPH levels is difficult due to the many reactions that consume and produce it, which includes the pyridine nucleotide transhydrogenases (30), we chose to assess the role of NADPH in NIT potentiation with a genetic approach that focused on NfsA variants with different affinity for NADPH in a ΔnfsA ΔnfsB background strain. Specifically, we used wild-type NfsA (apparent Km for NADPH, 8 μM) and an NfsA mutant (NfsA*), which has reduced affinity for NADPH (apparent Km for NADPH, 256 μM) but equivalent binding affinity for nitrofurans (35). Importantly, NfsA* retains intracellular activity, as reflected in an MIC of 8 μg/ml for a ΔnfsA ΔnfsB strain expressing NfsA* from a plasmid, which is only 2-fold higher than an NfsA-expressing control (MIC of 4 μg/ml) and 8-fold lower than an empty-vector control (MIC of 64 μg/ml) in the same genetic background (see Fig. S12 in the supplemental material). We expressed NfsA or NfsA* in the ΔnfsA ΔnfsB strain, which did not experience metabolite stimulation of NIT killing due to the absence of the relevant nitroreductases (Fig. S6A) and found that ΔnfsA ΔnfsB strain cells expressing NfsA* remained insensitive to NIT in the presence of glucose, whereas NfsA expression restored the ability of glucose to resensitize stationary-phase populations of the ΔnfsA ΔnfsB strain to NIT (Fig. 7B and C). Uninduced controls are shown in Fig. S13A and B in the supplemental material. Induction with isopropyl-β-d-thiogalactopyranoside (IPTG) did not impact stationary-phase culturability (Fig. S13C). These data support a mechanism where low levels of reducing equivalents underlie the loss of susceptibility of stationary-phase E. coli to NIT.
FIG 7.
NADPH availability underlies NIT insensitivity in nongrowing cells. (A) NADPH levels in stationary-phase E. coli cell lysates were measured just before and at 1 h and 3 h following treatment with 10 mM glucose. One milliliter of stationary-phase culture (∼109 cells) was used for lysate preparation, and the y axis reflects the concentration of NADPH in the lysate. (B) The ΔnfsA ΔnfsB strain carrying a plasmid with an IPTG-inducible NfsA was treated with 250 μg/ml NIT or NIT and 10 mM glucose in the presence of 1 mM IPTG (inducer). (C) The ΔnfsA ΔnfsB strain carrying a plasmid with an IPTG-inducible NfsA*, a mutant NfsA with reduced affinity for NADPH, was treated as in panel B. CFU were determined at the indicated time points. *, P < 0.05 (t test), comparing glucose and water treatments (A) or NIT with glucose + NIT (B and C). Additional controls are presented in Fig. S13 in the supplemental material. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
Metabolite-mediated potentiation of NIT with different carbon sources.
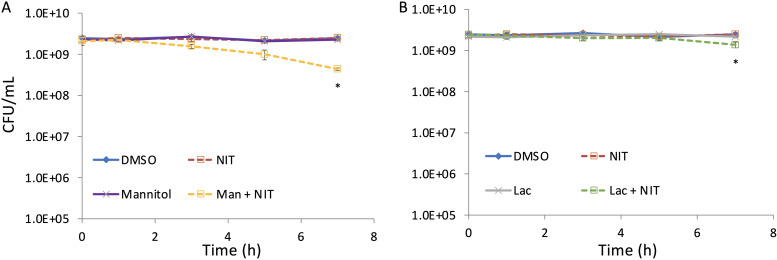
To assess the generality of this phenomenon, we tested whether carbon sources other than glucose could potentiate NIT activity. When mannitol was used, a significant ∼6-fold reduction in survival was observed for cultures treated with mannitol and NIT compared to that of NIT alone (Fig. 8A). When lactate was used, a significant ∼2-fold reduction in survival was observed for cultures treated with lactate and NIT compared to that of NIT alone (Fig. 8B). These data demonstrate that the level of NIT potentiation will depend on the metabolite provided.
FIG 8.
Metabolite-mediated potentiation of NIT activity with different carbon sources. Cultures of E. coli grown to stationary phase in MOPS plus glucose were treated with DMSO (control), 250 μg/ml NIT, and 10 mM mannitol with or without NIT (A) or 10 mM lactate with or without NIT (B). CFU per milliliter were determined at the indicated time points. *, P < 0.05 (t test), comparing NIT with mannitol plus NIT (A) or NIT with lactate plus NIT (B).
Metabolite-mediated potentiation of NIT with different stationary-phase cultures.
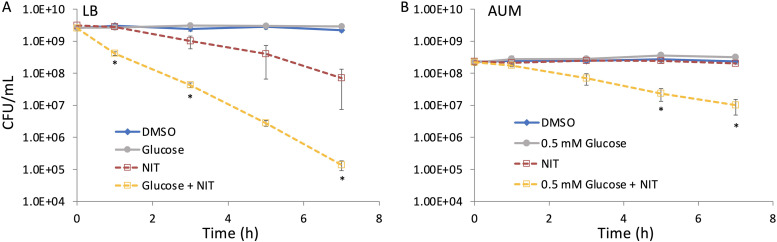
To assess whether this phenomenon would be observed with bacteria grown under different conditions, we performed NIT potentiation assays with E. coli grown to stationary phase in LB, a rich medium, and artificial urine medium (AUM), a more nutrient-limited medium that mimics the composition of urine (36). When grown in LB for 20 h, significant potentiation of NIT activity was observed with glucose supplementation, though NIT on its own did exhibit some killing (Fig. 9A). When the growth period was extended to 24 h, treatment with NIT alone did not deviate significantly from the dimethyl sulfoxide (DMSO) control, whereas glucose and NIT treatment exhibited a similar reduction in survival as that seen for the cultures grown for 20 h (see Fig. S14 in the supplemental material). When cultures were grown to stationary phase in AUM, cultures were not susceptible to NIT, whereas glucose plus NIT produced a significant 20-fold reduction in survival compared to that of NIT alone (Fig. 9B). We note that 0.5 mM glucose was used with AUM because 10 mM glucose produced significant growth in our assay conditions (see Fig. S15 in the supplemental material) due to the lower stationary-phase cell densities that could be obtained in AUM (2.34 × 108 ± 1.0 × 107 CFU/ml).
FIG 9.
Metabolite-mediated potentiation of NIT with different stationary-phase cultures. Stationary-phase cultures of E. coli were grown in either LB and treated with DMSO, 250 μg/ml NIT, and 10 mM glucose with or without NIT (A) or AUM and treated with DMSO, 250 μg/ml NIT, and 0.5 mM glucose with or without NIT (B). CFU were monitored at indicated time points. *, P < 0.05 (t test), comparing glucose plus NIT with NIT. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
Metabolite-mediated potentiation of NIT against UPEC.
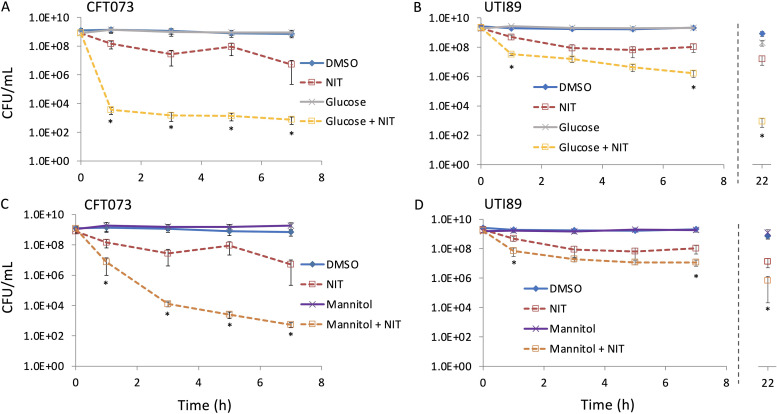
Since NIT is indicated for the treatment of UTIs, we treated stationary-phase UPEC cultures with NIT in the absence and presence of glucose. We found that treatment with NIT alone resulted in a significant drop in culturability for two different UPEC strains (Fig. 10), which was not observed for E. coli K-12 (Fig. 1B). Supplementation of NIT with glucose significantly increased the killing compared to that of NIT alone for both strains, although to varying degrees, which suggested that the phenomenon that we observed for E. coli K-12 was generalizable to UPEC. We then assessed the ability of a more clinically adaptable sugar, mannitol (24), and observed significant decreases in the culturability for UPEC that was cotreated with NIT and mannitol compared to that of samples that were treated only with NIT (Fig. 10C and D). Controls that were treated with DMSO and mannitol or glucose only did not lose culturability (Fig. 10). In addition, we inspected longer treatment times with UTI89 and observed that by 22 h, the activity of NIT continued to be enhanced by glucose and mannitol (Fig. 10B and D). Collectively, these data suggest that potentiation of NIT activity with metabolites, such as mannitol that accumulates in the urine by itself, could result in positive outcomes in the treatment of UTIs caused by UPEC.
FIG 10.
Metabolite-mediated potentiation of NIT activity against UPEC. Stationary-phase UPEC strains CFT073 and UTI89 were treated with DMSO, 250 μg/ml NIT, 10 mM glucose, or NIT supplemented with glucose (A and B). (C and D) Treatments were carried out as in panel A, except that 10 mM mannitol was used instead of glucose. CFU were determined at the indicated time points. *, P < 0.05 (t test), comparing NIT with either glucose plus NIT or mannitol plus NIT. Data represent at least three biological replicates. Each data point was denoted as mean ± SE.
DISCUSSION
UTIs are caused primarily by strains of UPEC (37), and recurrence is associated with the ability of UPEC to survive in extracellular biofilms as well as inside bladder epithelial cells, where they form biofilm-like inclusions in the cytosol or enter a quiescent state inside late endosomal compartments (38, 39). Interestingly, in 68% of chronic UTIs, bacteria that are isolated in recurrent episodes are identical to the initial isolate, which suggests that temporary loss of susceptibility rather than resistance underlies the propensity of those infections to relapse. In addition, susceptibility loss may take the form of persistence, where a small subpopulation exhibits lower susceptibility than the rest of the population (40), as we observed with the two UPEC isolates examined here (Fig. 10).
NIT is preferentially excreted into the lower urinary tract, which makes it useful to treat lower UTIs (10–12). Once activated, NIT has a unique mechanism of action that interferes with DNA, protein, and cell wall synthesis (12, 13), and clinically-resistant mutants, which often exhibit disturbed growth, are virtually nonexistent (13, 16). Further, NIT has a broad spectrum of activity and is effective against E. coli, Staphylococcus saprophyticus, and Enterococcus faecium (41). These qualities contribute to the repositioning of NIT as a first-line therapy for uncomplicated UTIs (42).
Transient loss of susceptibility to NIT in bacteria can be a source of treatment failure and relapse. In this study, we observed that starved/growth-inhibited E. coli K-12 was not killed by NIT (Fig. 1) and conceptualized that insensitivity as failure of a multi-input AND gate (Fig. 1C). We hypothesized that metabolite supplementation could restore NIT sensitivity by affecting one or more of those inputs and confirmed that hypothesis to be true (Fig. 2). Resensitization of E. coli to NIT by metabolite supplementation did not require growth but did require catabolism (Fig. 3 and 4), and further analyses revealed that nitroreductase availability and NIT penetration did not underlie the phenotype (Fig. 5 and 6), but rather, data suggested that stationary-phase E. coli was not killed by NIT due to insufficient levels of reducing equivalents (Fig. 7). In addition, we observed that the nutrient environment following NIT treatment had a modest impact on survival, with an increase of 2-fold when cells were plated on MOPS plus glucose agar compared to that on LB agar (see Fig. S16 in the supplemental material). We speculate that slower growth resumption during the recovery period provided cells with a longer time period to repair NIT-induced damage prior to replication and cell division, which is a phenomenon that has been observed for nongrowing E. coli treated with fluoroquinolones (43).
Metabolites have previously been found to resensitize bacterial populations to different antibiotics through a number of distinct mechanisms (23, 44, 45). For instance, in one study, metabolites potentiated aminoglycoside activity independent of growth resumption by generation of proton motive force, which in turn facilitated aminoglycoside uptake (23). In another study, metabolites sensitized bacterial populations (E. coli, Staphylococcus aureus, Mycobacterium smegmatis) to quinolones through stimulation of carbon and respiratory metabolism (44). In addition, killing of stationary-phase S. aureus by daptomycin was increased by supplementation with glucose, although the mechanism of potentiation remains to be determined (45). Taken together, a mounting body of evidence suggests that metabolite supplementation could be efficacious for the treatment of bacterial infections.
To explore the practical relevance of metabolite potentiation of NIT activity, we carried out NIT treatments on two UPEC strains. We found that although UPEC was more sensitive to NIT than E. coli K-12, supplementation with glucose led to significant potentiation (Fig. 10A and B). Because normal urine does not contain glucose and the main carbon sources are amino acids, lactate, and citrate (36), we chose to use mannitol to potentiate NIT activity. We reasoned that mannitol, as it is excreted in urine (24), would be a more clinically adaptable sugar. Similar to glucose, mannitol was able to potentiate the activity of NIT against stationary-phase populations of UPEC (Fig. 10C and D). Interestingly, mannitol has previously been shown to potentiate aminoglycoside activity in urinary catheter infections in mice (23), and it has been found to reach levels in the urine that are above or near those used in this study (46). Taken together, our findings along with other studies, suggest that metabolites could prove to be valuable adjuvants for the treatment of chronic infections, such as UTIs caused by UPEC.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains used are listed in Table S1 in the supplemental material. The wild-type strain used was E. coli K-12 MG1655, and all mutants were constructed in this background. Where indicated, mutant strains were generated by P1 transduction using the Keio collection (29) as the donor strains. In the case of the MG1655 ΔptsI::FRTΔglk::kan strain, ΔptsI::kan was moved by P1 transduction followed by curing of the kanamycin resistance cassette by FLP recombinase, and integration of glk::kan using the method of Datsenko and Wanner (47). For all instances where removal of the kanamycin resistance cassette was necessary, FLP recombinase was expressed from pCP20. Specifically, cells were transformed with pCP20 and grown at the permissive temperature (30°C) and then plated at 42°C. Strains were then tested for ampicillin and kanamycin sensitivity at 30°C and 37°C, respectively (to assess loss of pCP20, which harbors an ampicillin resistance selection marker, and loss of the kanamycin resistance cassette from the chromosome). Further confirmation of the antibiotic-resistance marker excision was carried out by PCR. UPEC strain CFT073 (WAM2267) (ATCC 700928) was purchased from the American Type Culture Collection (Manassas, Virginia), and sequencing of its rpoS gene revealed the presence of a frameshift mutation (Table S1). UPEC strain UTI89 was a generous gift from Matthew Mulvey.
Plasmid DNA construction.
Plasmids used are listed in Table S1. Primer sequences used for plasmid construction, mutations, and sequencing are listed in Table S2 in the supplemental material. For monitoring expression of green fluorescent protein (GFP), plasmid pQE-80L-gfp was used. This plasmid was previously constructed using a pQE-80L (Qiagen) variant containing a synthetic T5 promoter, lacI expression from the lacIq promoter, and a kanamycin resistance marker (48). Plasmid pQE-80L (Qiagen) carrying IPTG-inducible nfsA (pQE-80L-nfsA) was generated by double digestion of plasmid and nfsA insert with EcoRI and HindIII, followed by ligation with Quick Ligation kit (NEB). Plasmid pQE-80L-nfsA*, in which Arg-203 was mutated to an Ala in NfsA, was generated by site-directed mutagenesis using a Q5 site-directed mutagenesis kit (NEB). All plasmids constructed were confirmed by sequencing.
Chemicals and media.
In all experiments, we used deionized water purified to attain a resistivity of 18.2 MΩ · cm at 25°C using a Millipore Milli-Q lab water system (Burlington, MA). Unless noted, all chemicals were purchased from Fisher Scientific or Sigma-Aldrich. All chemical solutions were sterile filtered using a 0.22-μm syringe filter purchased from Merck Millipore, Ltd (Burlington, MA), except for those containing dimethyl sulfoxide (DMSO). DMSO was used to dissolve NIT and NFZ, and those solutions were prepared fresh before use and protected from light before and during treatment. Glucose, gluconate, and mannitol were dissolved in water and used at working concentrations of 10 mM, except where indicated. MOPS minimal medium included 10 mM glucose as the sole carbon source (herein referred to as MOPS plus glucose) (purchased from Teknova, Hollister, CA), and it was sterile filtered using a 0.22-μm filter. Chloramphenicol was dissolved in DMSO and used at a working concentration of 100 μg/ml for inhibition of protein synthesis. Ampicillin and kanamycin were dissolved in water and used at working concentrations of 100 μg/ml and 50 μg/ml, respectively, except where indicated. Bile salts were dissolved in water and used at a working concentration of 1.5 mg/ml. Colistin was dissolved in water. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology (St. Louis, MO) and dissolved in water. Artificial urine medium (AUM) was prepared based on the recipe provided in Table S3 in the supplemental material (36).
Determination of MICs.
Determination of MICs using the microdilution method was carried out as described previously (49). Optical density at 600 nm (OD600) was measured using a plate reader after 16 to 18 h of incubation in Mueller-Hinton Broth at 37°C with the antibiotics. The MIC was defined as the lowest concentration at which no significant growth was measured. For the determination of MIC of the ΔnfsA::FRTΔnfsB::FRT strain containing pQE-80L-nfsA, pQE-80L-nfsA*, or pQE-80L, 1 mM IPTG was added during the time of drug treatment for plasmid induction, and ampicillin was kept at a concentration of 100 μg/ml for plasmid maintenance. MIC data are provided in Fig. S11, S12, and S17 in the supplemental material.
NIT treatment assay.
A loop full of cells from a glycerol stock maintained at −80°C was used to inoculate 2 ml LB medium in a test tube. Cultures were incubated for approximately 4 h at 37°C with shaking (250 rpm). Following incubation, OD600 was measured, and an appropriate volume of culture was spun down to harvest the cells. Supernatant was removed, and cells were resuspended in 300 μl of fresh MOPS plus glucose and used to inoculate 50 ml of fresh MOPS plus glucose to an OD600 of ∼0.01 and incubated for 20 h at 37°C with shaking. After 20 h of incubation, treatments with 250 μg/ml NIT (MIC = 4 mg/liter), 10 mM glucose, 10 mM lactate, 10 mM mannitol, NIT plus glucose, NIT plus lactate, or NIT plus mannitol were carried out. We chose to use NIT at a clinically relevant concentration (approximately 250 μg/ml), which is the concentration found in the urine during NIT excretion into the lower urinary tract (50). DMSO (solvent for NIT) was added to the control culture. Baffled flasks (500 ml) containing the 50-ml cultures were incubated at 37°C with shaking in the dark following addition of NIT. Immediately before addition of the chemicals and after 1, 3, 5, and 7 h, samples (500 μl) were taken and washed twice by centrifugation to remove the chemicals. Each wash step was carried out by centrifugation at 15,000 rpm, removal of 450 μl of the supernatant, and resuspension of the cell pellet in 450 μl of phosphate-buffered saline (PBS). Two wash steps ensured that the final NIT concentration was below their MICs (both were 4 mg/liter). Ten microliters of washed cells was serially diluted in PBS. Ten microliters of each dilution was spotted on LB agar and incubated for 16 h at 37°C. Where indicated, samples were also plated on MOPS plus glucose agar and incubated for 39 h at 37°C (see Fig. S16 in the supplemental material). The NFZ treatment assay followed the same protocol as the NIT treatment assay except NFZ was used instead of NIT. For the imp4213 strain, to ensure the absence of spontaneous suppressor mutations, samples from before drug treatments were plated on LB agar plus 1.5 mg/ml bile salts, along with a wild-type control. In all instances, the imp4213 strain could not form colonies on bile salt plates, whereas the wild type did form colonies (suppressor mutations would have been those that allowed the imp4213 strain to grow on bile salt plates). In addition to cultures grown in MOPS plus glucose, NIT treatment assays were also conducted on cultures grown in LB medium and artificial urine medium (AUM). When grown to stationary phase in AUM, potentiation assays were performed with glucose at 0.5 mM rather than 10 mM, because significant bacterial growth was observed with 10 mM glucose supplementation in those cultures (see Fig. S15 in the supplemental material), whereas 0.5 mM glucose did not stimulate growth (Fig. 9B). The protocol for growth of UPEC strains CFT073 and UTI89 differed from that of E. coli K-12 in that UPEC was inoculated to a higher density than E. coli K-12 (OD600 equal to 0.1 for UPEC compared to 0.01 for E. coli K-12), and NIT treatment of UPEC was carried out after 24 h of incubation (OD600 equal to 1.30 ± 0.08 for CFT073 and 1.47 ± 0.05 for UTI89), while, as stated above, E. coli K-12 treatments were performed after 20 h of incubation (OD600 equal to 1.70 ± 0.03). These changes in density of the inoculum and extension of UPEC culture time prior to NIT treatment were to account for its slower growth kinetics in MOPS plus glucose medium. An additional time point at t = 22 h following drug treatment was taken for UPEC strain UTI89. For treatment of growing cultures, overnight cultures in MOPS plus glucose were generated as described above. Next, these overnight cultures were used to inoculate (100-fold dilution) fresh MOPS plus glucose medium and allowed to resume growth for 3 h before initiating NIT treatments as described above. Samples were collected, and CFU were determined as described above.
Ampicillin challenge assay.
The protocol for the ampicillin challenge assays follows the NIT treatment assay except that at the drug treatment step, 200 μg/ml ampicillin (MIC = 4 mg/liter) (49), 10 mM glucose, ampicillin plus glucose, or water (solvent for ampicillin) was added to the culture.
NIT treatment in environments that do not support growth.
Two milliliters of a 20-h overnight culture of MG1655, grown in MOPS plus glucose as described for NIT treatment assays, were washed twice with saline (0.9% NaCl). Each wash step was carried out by removing approximately 2 ml of the supernatant after centrifugation and adding 2 ml of saline. Following washes, cell pellets were resuspended in 2 ml of saline before transfer to a test tube and incubated for 1 h for acclimation prior to treatments with chemicals (10 mM glucose, 9.5 mM NH4Cl, 1.32 mM K2HPO4, or 0.286 mM MgSO4) in the absence or presence of 250 μg/ml NIT. Samples were taken immediately before addition of the chemicals and after 1, 3, 5, and 7 h of incubation at 37°C with shaking (250 rpm). At each time point, samples were washed with saline and plated on LB agar plates as described for NIT treatment assays. Each wash step was carried out by removing 450 μl of the supernatant following centrifugation and adding 450 μl of saline. To make sure that these culture conditions do not support growth, we carried out experiments under the same conditions described above, except that we made a dilution (OD600 = 0.01) of the overnight cultures in saline following the washes in saline. A lower culture density allows more accurate assessment of growth. An extra treatment in which we included all of the nutrients and calcium (0.1 mM CaCl2) was used as a growth control. We measured OD600 at the time of inoculation (OD600, ∼0.01) and after 24 h of culture (t = 24 h) (see Fig. S2 in the supplemental material).
Determination of NIT reduction in cell cultures.
Absorbance at 375 nm of nitrofuran solutions has been shown to decline as nitrofurans are reduced, and this has been used previously to measure nitrofuran reduction by cell cultures (51, 52). Here, cells were cultured and treated as described for NIT treatment assays. Samples (500 μl) were taken immediately after the addition of NIT (t = 0) and at 1, 3, 5, and 7 h, centrifuged to pellet the cells, and supernatant was removed and filtered. Filtered supernatant was used to measure absorbance at 375 nm in a 96-well plate using a plate reader. In the original assay (52), cells were pelleted by centrifugation and the supernatant was used for absorbance measurements, whereas here we added a sterile filtration step. The rationale for the removal of cells is to stabilize the signal because the cells are the catalyst that reduces nitrofurans and thereby reduces the absorbance of the medium at 375 nm. The plots of nitrofuran reduction portrayed here reflect the loss of nitrofuran from the media by cellular activities.
Determination of NADPH.
NADPH levels in cell lysates were measured using the EnzyChrom NADP+/NADPH assay kit (ECNP-100). Cultures were grown to stationary phase as described for NIT treatment assays, and then 10 mM glucose or water was added. NADPH was measured in cultures just prior to glucose addition as well as after 1 and 3 h. For each time point, 1 ml of cultures was removed and processed according to the manufacturer’s instructions with the addition of a sonication step (20 s, 10% amplitude using a BF50 Fisherbrand Model 50 sonicator) before the heat extraction step to assist cell lysis.
Determination of colistin concentration necessary to permeate stationary-phase cells.
One milliliter of a 20-h overnight culture of MG1655 grown in MOPS plus glucose as described for NIT treatment assays was transferred to a test tube for treatment with increasing concentrations of colistin in the absence or presence of 1.5 mg/ml bile salts. Water in the absence or presence of 1.5 mg/ml bile salts was used as a control. Cultures were incubated 7 h before a sample (500 μl) was taken and washed twice by centrifugation to remove the chemicals. Each wash step was carried out by removing 450 μl of supernatant following centrifugation and adding 450 μl of PBS. Washed cells were 10-fold serially diluted in PBS. Ten microliters of each dilution was spotted on LB agar and incubated for 16 h at 37°C.
Green fluorescent protein expression assay in stationary-phase cultures.
MG1655 carrying plasmid pQE-80L-gfp was used. Kanamycin (50 μg/ml) was added to cultures for plasmid maintenance. Cell cultures were grown to stationary phase as described above for the NIT treatment assays. Protein synthesis was inhibited by the addition of chloramphenicol (100 μg/ml) at t = 19 h. A control culture was treated with DMSO, the solvent for CAM. After the addition of chloramphenicol or DMSO, incubation continued for another hour. At t = 20 h, IPTG (1 mM) was added to induce synthesis of GFP. Samples were taken immediately before and after 1, 3, 5, and 7 h addition of IPTG. Cells were washed with PBS twice by centrifugation and diluted to an OD600 of 0.1 in PBS for fluorescence reading (excitation = 485 nm; emission = 528 nm) using a plate reader. Each wash step was carried out by removing 450 μl of supernatant following centrifugation and adding 450 μl of PBS. Relative fluorescence was calculated by dividing the fluorescence signal at a specific time point by the fluorescence signal at the time of IPTG addition (t = 0).
Glucose concentration determination.
Glucose levels were determined using the commercial Amplex red glucose/glucose oxidase assay kit (Thermo Fisher Scientific). Cell cultures were grown to stationary phase (cultured for 20 h) as described above for NIT treatment assays. At t = 20 h, glucose was added at a concentration of 10 mM. As a control, an equal volume of water was added to a culture. Supernatant was collected by centrifugation and filtered. Supernatants were stored at 4°C for up to a few days until samples from all replicates were collected. Supernatants were diluted 100-fold for measurements following the manufacturer’s protocol.
Statistical analysis.
At least three biological replicates were performed for each experimental condition. Each data point depicts the mean value ± standard error (SE). A two-tailed t test with unequal variance was used for pairwise comparisons. For the culturability data, the log-transformed values of CFU per milliliter were utilized for statistical analysis. P values of ≤0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National BioResource Project (NIG, Japan) for distribution of the Keio collection, Matthew Mulvey and Amanda Richards for distribution of UTI89, and Joel Freundlich for his suggestions on this project.
This work was supported by the NIAID of the NIH (R01AI130293 to M.P.B.). The content is solely the responsibility of the authors, does not necessarily represent the official views of the funding agency, and the funder had no role in the design or implementation of the experiments or the decision to publish.
Patent protection is pending.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 2.Tuomanen E, Durack DT, Tomasz A. 1986. Antibiotic tolerance among clinical isolates of bacteria. Antimicrob Agents Chemother 30:521–527. doi: 10.1128/aac.30.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 4.Windels EM, Michiels JE, Van den Bergh B, Fauvart M, Michiels J. 2019. Antibiotics: combatting tolerance to stop resistance. mBio 10:e02095-19. doi: 10.1128/mBio.02095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 6.Barrett TC, Mok WWK, Murawski AM, Brynildsen MP. 2019. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat Commun 10:1177. doi: 10.1038/s41467-019-09058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mobley HL, Donnenberg MS, Hagan EC. 21 December 2009. Uropathogenic Escherichia coli. EcoSal Plus 2013 doi: 10.1128/ecosalplus.8.6.1.3. [DOI] [PubMed] [Google Scholar]
- 8.Forsyth VS, Armbruster CE, Smith SN, Pirani A, Springman AC, Walters MS, Nielubowicz GR, Himpsl SD, Snitkin ES, Mobley HLT. 2018. Rapid growth of uropathogenic Escherichia coli during human urinary tract infection. mBio 9:e00186-18. doi: 10.1128/mBio.00186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terlizzi ME, Gribaudo G, Maffei ME. 2017. Uropathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol 8:1566. doi: 10.3389/fmicb.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guay DR. 2001. An update on the role of nitrofurans in the management of urinary tract infections. Drugs 61:353–364. doi: 10.2165/00003495-200161030-00004. [DOI] [PubMed] [Google Scholar]
- 11.Keys TF. 1977. Antimicrobials commonly used for urinary tract infection: sulfonamides, trimethoprim-sulfamethoxazole, nitrofurantoin, nalidixic acid. Mayo Clin Proc 52:680–682. [PubMed] [Google Scholar]
- 12.Huttner A, Verhaegh EM, Harbarth S, Muller AE, Theuretzbacher U, Mouton JW. 2015. Nitrofurantoin revisited: a systematic review and meta-analysis of controlled trials. J Antimicrob Chemother 70:2456–2464. doi: 10.1093/jac/dkv147. [DOI] [PubMed] [Google Scholar]
- 13.McOsker CC, Fitzpatrick PM. 1994. Nitrofurantoin: mechanism of action and implications for resistance development in common uropathogens. J Antimicrob Chemother 33(Suppl):23–30. doi: 10.1093/jac/33.suppl_a.23. [DOI] [PubMed] [Google Scholar]
- 14.Bryant DW, McCalla DR, Leeksma M, Laneuville P. 1981. Type I nitroreductases of Escherichia coli. Can J Microbiol 27:81–86. doi: 10.1139/m81-013. [DOI] [PubMed] [Google Scholar]
- 15.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol 180:5529–5539. doi: 10.1128/JB.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandegren L, Lindqvist A, Kahlmeter G, Andersson DI. 2008. Nitrofurantoin resistance mechanism and fitness cost in Escherichia coli. J Antimicrob Chemother 62:495–503. doi: 10.1093/jac/dkn222. [DOI] [PubMed] [Google Scholar]
- 17.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother 35:1824–1828. doi: 10.1128/aac.35.9.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 19.Knudsen GM, Ng Y, Gram L. 2013. Survival of bactericidal antibiotic treatment by a persister subpopulation of Listeria monocytogenes. Appl Environ Microbiol 79:7390–7397. doi: 10.1128/AEM.02184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleck LE, North EJ, Lee RE, Mulcahy LR, Casadei G, Lewis K. 2014. A screen for and validation of prodrug antimicrobials. Antimicrob Agents Chemother 58:1410–1419. doi: 10.1128/AAC.02136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murugasu-Oei B, Dick T. 2000. Bactericidal activity of nitrofurans against growing and dormant Mycobacterium bovis BCG. J Antimicrob Chemother 46:917–919. doi: 10.1093/jac/46.6.917. [DOI] [PubMed] [Google Scholar]
- 22.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mc Causland FR, Prior LM, Heher E, Waikar SS. 2012. Preservation of blood pressure stability with hypertonic mannitol during hemodialysis initiation. Am J Nephrol 36:168–174. doi: 10.1159/000341273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. 2017. Carbon Sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houser JR, Barnhart C, Boutz DR, Carroll SM, Dasgupta A, Michener JK, Needham BD, Papoulas O, Sridhara V, Sydykova DK, Marx CJ, Trent MS, Barrick JE, Marcotte EM, Wilke CO. 2015. Controlled measurement and comparative analysis of cellular components in E. coli reveals broad regulatory changes in response to glucose starvation. PLoS Comput Biol 11:e1004400. doi: 10.1371/journal.pcbi.1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adolfsen KJ, Brynildsen MP. 2015. A kinetic platform to determine the fate of hydrogen peroxide in Escherichia coli. PLoS Comput Biol 11:e1004562. doi: 10.1371/journal.pcbi.1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escalante A, Salinas Cervantes A, Gosset G, Bolivar F. 2012. Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: peculiarities of regulation and impact on growth and product formation. Appl Microbiol Biotechnol 94:1483–1494. doi: 10.1007/s00253-012-4101-5. [DOI] [PubMed] [Google Scholar]
- 29.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Peralta-Gil M, Subhraveti P, Velázquez-Ramírez DA, Weaver D, Collado-Vides J, Paulsen I, Karp PD. 2017. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui P, Niu H, Shi W, Zhang S, Zhang H, Margolick J, Zhang W, Zhang Y. 2016. Disruption of membrane by colistin kills uropathogenic Escherichia coli persisters and enhances killing of other antibiotics. Antimicrob Agents Chemother 60:6867–6871. doi: 10.1128/AAC.01481-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson BA, Misra R, Benson SA. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoque MA, Ushiyama H, Tomita M, Shimizu K. 2005. Dynamic responses of the intracellular metabolite concentrations of the wild type and pykA mutant Escherichia coli against pulse addition of glucose or NH3 under those limiting continuous cultures. Biochem Eng J 26:38–49. doi: 10.1016/j.bej.2005.05.012. [DOI] [Google Scholar]
- 35.Kobori T, Sasaki H, Lee WC, Zenno S, Saigo K, Murphy ME, Tanokura M. 2001. Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution. J Biol Chem 276:2816–2823. doi: 10.1074/jbc.M002617200. [DOI] [PubMed] [Google Scholar]
- 36.Brooks T, Keevil CW. 1997. A simple artificial urine for the growth of urinary pathogens. Lett Appl Microbiol 24:203–206. doi: 10.1046/j.1472-765x.1997.00378.x. [DOI] [PubMed] [Google Scholar]
- 37.Ipe DS, Sundac L, Benjamin WH, Jr, Moore KH, Ulett GC. 2013. Asymptomatic bacteriuria: prevalence rates of causal microorganisms, etiology of infection in different patient populations, and recent advances in molecular detection. FEMS Microbiol Lett 346:1–10. doi: 10.1111/1574-6968.12204. [DOI] [PubMed] [Google Scholar]
- 38.Blango MG, Mulvey MA. 2010. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother 54:1855–1863. doi: 10.1128/AAC.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 40.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komp Lindgren P, Klockars O, Malmberg C, Cars O. 2015. Pharmacodynamic studies of nitrofurantoin against common uropathogens. J Antimicrob Chemother 70:1076–1082. doi: 10.1093/jac/dku494. [DOI] [PubMed] [Google Scholar]
- 42.McKinnell JA, Stollenwerk NS, Jung CW, Miller LG. 2011. Nitrofurantoin compares favorably to recommended agents as empirical treatment of uncomplicated urinary tract infections in a decision and cost analysis. Mayo Clin Proc 86:480–488. doi: 10.4065/mcp.2010.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mok WWK, Brynildsen MP. 2018. Timing of DNA damage responses impacts persistence to fluoroquinolones. Proc Natl Acad Sci U S A 115:E6301–E6309. doi: 10.1073/pnas.1804218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutierrez A, Jain S, Bhargava P, Hamblin M, Lobritz MA, Collins JJ. 2017. Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol Cell 68:1147–1154. doi: 10.1016/j.molcel.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Prax M, Mechler L, Weidenmaier C, Bertram R. 2016. Glucose augments killing efficiency of daptomycin challenged Staphylococcus aureus persisters. PLoS One 11:e0150907. doi: 10.1371/journal.pone.0150907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobolevsky T, Ahrens B. 2019. Urinary concentrations of AICAR and mannitol in athlete population. Drug Test Anal 11:530–535. doi: 10.1002/dta.2557. [DOI] [PubMed] [Google Scholar]
- 47.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orman MA, Brynildsen MP. 2015. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat Commun 6:7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aedo SJ, Orman MA, Brynildsen MP. 2019. Stationary phase persister formation in Escherichia coli can be suppressed by piperacillin and PBP3 inhibition. BMC Microbiol 19:140. doi: 10.1186/s12866-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunha BA, Schoch PE, Hage JR. 2011. Nitrofurantoin: preferred empiric therapy for community-acquired lower urinary tract infections. Mayo Clin Proc 86:1243–1244. doi: 10.4065/mcp.2011.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breeze AS, Obaseiki-Ebor EE. 1983. Mutations to nitrofurantoin and nitrofurazone resistance in Escherichia coli K12. J Gen Microbiol 129:99–103. doi: 10.1099/00221287-129-1-99. [DOI] [PubMed] [Google Scholar]
- 52.McCalla DR, Reuvers A, Kaiser C. 1970. Mode of action of nitrofurazone. J Bacteriol 104:1126–1134. doi: 10.1128/JB.104.3.1126-1134.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chubukov V, Sauer U. 2014. Environmental dependence of stationary-phase metabolism in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol 80:2901–2909. doi: 10.1128/AEM.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.