Molecular genotyping holds tremendous potential to detect antimalarial drug resistance (ADR) related to single nucleotide polymorphisms (SNPs). However, it relies on the use of complicated procedures and expensive instruments.
KEYWORDS: single nucleotide polymorphisms (SNPs), point-of-care testing (POCT), allele-specific PCR, lateral flow biosensor, antimalarial drug resistance
ABSTRACT
Molecular genotyping holds tremendous potential to detect antimalarial drug resistance (ADR) related to single nucleotide polymorphisms (SNPs). However, it relies on the use of complicated procedures and expensive instruments. Thus, rapid point-of-care testing (POCT) molecular tools are urgently needed for field survey and clinical use. Herein, a POCT platform consisting of multiple-allele-specific PCR (AS-PCR) and a gold nanoparticle (AuNP)-based lateral flow biosensor was designed and developed for SNP detection of the Plasmodium falciparum dihydrofolate reductase (pfdhfr) gene related to pyrimethamine resistance. The multiple-AS-PCR utilized 3′ terminal artificial antepenultimate mismatch and double phosphorothioate-modified allele-specific primers. The duplex PCR amplicons with 5′ terminal labeled with biotin and digoxin are recognized by streptavidin (SA)-AuNPs on the conjugate pad and then captured by anti-digoxin antibody through immunoreactions on the test line to produce a golden red line for detection. The system was applied to analyze SNPs in Pfdhfr N51I, C59R, and S108N of 98 clinical isolates from uncomplicated P. falciparum malaria patients. Compared with the results from nested PCR followed by Sanger DNA sequencing, the sensitivity was 97.96% (96/98) for N51I, C59R, and S108N. For specificity, the values were 100% (98/98), 95.92% (94/98), and 100% (98/98) for N51I, C59R, and S108N, respectively. The limit of detection is approximately 200 fg/μl for plasmid DNA as the template and 100 parasites/μl for blood filter paper. The established platform not only offers a powerful tool for molecular surveillance of ADR but also is easily extended to interrelated SNP profiles for infectious diseases and genetic diseases.
INTRODUCTION
Human malaria induced by Plasmodium species parasites, particularly Plasmodium falciparum, is a serious public health problem. It is mainly prevalent in the tropics and subtropics, particularly sub-Saharan Africa, as well as in Southeast Asia (SEA) and South America. In 2019, there were an estimated 229 million new malaria cases, which were responsible for approximately 409,000 deaths globally (1). Furthermore, pregnant women and children less than 5 years old in sub-Saharan Africa were the primary victims. Currently, the prevalence of P. falciparum parasites with antimalarial drug resistance (ADR) decreases antimalarial drug efficacy, and the emergence and transmission of ADR are recognized as potentially major obstacles for malaria control globally (2). During the 1980s, chloroquine (CQ) was replaced by sulfadoxine-pyrimethamine (SP), because widespread CQ resistance developed in sub-Saharan Africa. Subsequently, SP was replaced by artemisinin-based combination therapy (ACTs) for to the same reason. However, SP is still used for intermittent preventive treatment in infants (IPTi) and pregnant women (IPTp) in regions of malaria endemicity, according to WHO guidance (3). Aimed at P. falciparum enzymes dihydropteroate synthase (Pfdhps) and dihydrofolate reductase (Pfdhfr), SP acts as a synergistic inhibitor of folate in Plasmodium species parasites (4, 5). Studies in vitro and in vivo found that SP resistance (SPR) is primarily conferred by amino acid mutations in Pfdhfr (N51I, C59R, S108N, and I164L) and Pfdhps (S436A, A437G, K540E, A581G, and A613S) (6, 7). These mutations, particularly S108N and A437G, are suggested to mediate a gradually increasing trend of SPR (8). Compared with mutations of Pfdhps, a Pfdhfr triple mutant (N51I, C59R, and S108N) has been documented more frequently in African regions where malaria is found (9).
Several PCR-based assays have been reported for single nucleotide polymorphism (SNP) detection in SPR genes (7, 10–16). Although DNA sequencing of PCR amplicons is considered the gold standard for SNP detection, it is expensive, time consuming, and requires trained professionals to report the results (17). Besides, the advantage of sequencing is to search for unknown SNPs rather than well-known SNPs. For other techniques, the multistep procedures, high cost, and need for precise devices limit their application (18, 19). The tremendous potential for SNPs in ADR genes of P. falciparum demands instantaneous and effective screening tools to meet the necessities of a point-of-care testing (POCT) strategy (20). Thus, it is necessary to develop a rapid, user-friendly, and POCT molecular tool for the monitoring of drug-resistant P. falciparum parasites. A lateral flow assay (LFA) can be a desirable choice because it saves time and is inexpensive (21). As one of the indicators in LFA, gold nanoparticles (AuNPs) show unique advantages for visualizing PCR products and enabling signal enhancement for increased sensitivity of detection. AuNPs present remarkable stableness and compatibility, making them biocompatible with biomacromolecules (22). Furthermore, they demonstrate potential serving as diagnostic or therapeutic agents in various medical applications. However, multiple-SNP detection using a combined allele-specific PCR (AS-PCR) and AuNP-LFA for mutations (N51I, C59R, and S108N) of the pfdhfr gene remains unclear.
In the present study, the combination of an AS-PCR and an AuNP-based lateral flow biosensor was developed, which can be a field-deployable tool to allow multiple-SNP screening (Fig. 1). The proposed protocol was performed for double-labeled products, thereby enabling analysis by a gel or visual biosensor. The method could be deployed in POCT, on-site screening, personalized medicine, and epidemiological surveillance. The developed AS-PCR-LFA system was applied to screening the SNPs (N51I, C59R, and S108N) in pfdhfr related to pyrimethamine resistance from clinical isolates. It will provide promising POCT in the detection of multiple-SNP-caused ADR and can easily be extended to other SNP profiles for infectious diseases and genetic diseases.
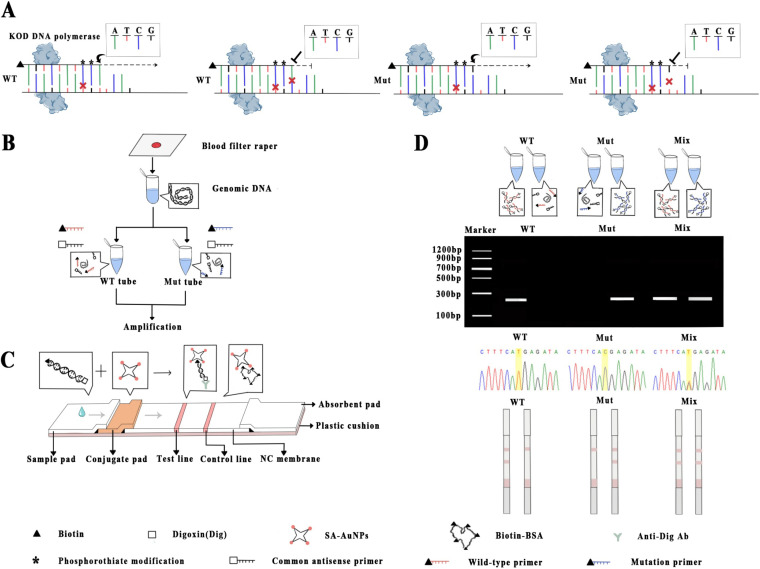
FIG 1.
Schematic illustration of the allele-specific PCR (AS-PCR) combined with lateral flow assay (LFA) system. (A) Principle of AS-PCR for every single nuclear polymorphism. (B) Sample preparation and target amplification. (C) Structure of labeled lateral flow device. (D) Results were analyzed based on the signal read-out by visual interpretation in LFA, agarose gel electrophoresis, and DNA sequencing.
RESULTS AND DISCUSSION
Principle of the AS-PCR-LFA system.
The schematic of allele-specific PCR combined with the AuNP-based lateral flow assay system is illustrated in Fig. 1. The PCR amplification is performed in a thermal cycling instrument for 4 h, and the products are then transferred to the LF strip for visual study. The principle of AS-PCR is shown in Fig. 1A. For each SNP, we set two pairs of allele-specific primers, which contain an antepenultimate artificial mismatch base and double phosphorothioate modification at antepenultimate and penultimate bases. Aided by KOD DNA polymerase, the amplification process of AS-PCR will proceed successfully when the wild-type (WT) tube includes the WT primer and WT template, and the mutant (Mut) tube includes Mut primer and Mut template. The amplification process will be blocked when the WT tube includes the WT primer and Mut template, and the Mut tube includes the Mut primer and WT template. As exhibited in Fig. 1B, genomic DNA (gDNA) is extracted from dried filter blood spots (DBS) followed by amplification through AS-PCR with specifically designed primers. The two sets of primers are used to amplify target fragments in independent tubes (WT tube and Mut tube) separately. Matched nucleotides in one tube will successfully amplify, but amplification will be blocked by KOD DNA polymerase in the other tube, which contains two mismatches; after amplification, both are added to the sample pad of the strip.
The SNP genotyping by LFA is performed over a strip, which is composed of five overlapping pads, i.e., sample pad, conjugate pad, nitrocellulose (NC) membrane, absorbent pad, and plastic cushion (Fig. 1C). As shown in Fig. 1C, the PCR products are added to the sample pads of LFA after amplification, and allele-specific primers labeled with biotin on the 5′-end can be recognized and captured by streptavidin-immobilized (SA)-AuNPs on the conjugate pad. Driven by capillary force, the PCR product-SA-AuNP composites migrate along the strip and aggregate on the T line through conjugation between T line-immobilized anti-digoxin monoclonal antibody (Ab) and the digoxin-label on the 5′ end of common primers, which make the T line a golden red band due to the bathochromic effect of AuNPs. The rest of the SA-AuNPs keep moving and are captured by biotin-bovine serum albumin (BSA) on the C line, which presents another red band that confirms the efficacy of the lateral flow system (Fig. 1C). In the absence of target PCR products, no red band is observed on the T line (Fig. 1D). The result can be visually read according to the coloration of the T line. As shown in Fig. 1D, the SNP genotyping results can be visually interpreted by a lateral flow dipstick (LFD) in 15 min without the need for any devices. They also can be interpreted via agarose gel electrophoresis and DNA sequencing (Fig. 1D).
Construction of recombinant plasmids.
The target sequences were amplified by nested PCR and inserted into the pEASY-T1 cloning vector to generate the recombinant plasmids, including pEASY-T1/Pfdhfr N51-C59-S108 (wild-type template) and pEASY-T1/Pfdhfr 51I-59R-108N (mutation template). These two constructed recombinant plasmids were confirmed by double digestion with restriction endonuclease BamHI and XhoI (see Fig. S1A in the supplemental material). Then, two fragments of approximately 3,900 bp and 680 bp were detected, in agreement with the bands for the vector and the target sequence, respectively (Fig. S1A). Subsequently, the recombinant plasmids were sequenced with M13F primer to be the same as the matching target sequences, indicating successful establishment of the recombinant plasmids (Fig. S1B).
Primer screening.
For primer screening, the primer candidates without 5′-terminal modification are listed in Table S1. Only one common primer was designed for Pfdhfr N51I. For C59R and S108N, a common primer is used. For amplification of C59R, four wild-type-specific primers and six mutation-specific primers with different mismatch positions and phosphorothioate modifications were designed, synthesized, and screened. According to the AS-PCR results (Fig. S2A), the primer named W1 only contains a penultimate phosphorothioate modification that has a false positive for the amplification using a mutation template. The primers (W2, W3, and W4) with the mismatch at a penultimate or antepenultimate base show higher differentiation than the primer (W1) without any mismatch. Furthermore, primers W2 and W4 contain the antepenultimate mismatch that has more specificity than primer W3 with a penultimate mismatch. Comparing W2 and W4, the primer with double phosphorothioate modification is more sensitive than the primer with a single modification (Fig. S2A). For screening the mutation-specific primers of C59R (Fig. S2A), the AS-PCR with wild-type and mutation templates was carried out concurrently. Compared to M1 without any mismatch, M2 with a penultimate mismatch can increase the primer specificity but reduces the sensitivity. Comparing M2 and M3, the primer with double phosphorothioate modification can further increase specificity but decrease the sensitivity. Comparing M3 and M4, the primer with a antepenultimate mismatch showed more amplification efficiency than the primer with a penultimate mismatch (Fig. S2A). Traditionally, the incorporation of mismatch nucleotides enhances allelic discrimination in AS-PCR (23), but it fails to completely suppress nonspecific amplification. A previous study found a single 3′-terminal mismatch primer is not optimal for certain types of SNPs (24). Thus, we developed several strategies to discriminate DNA mismatches. To improve the specificity of allele-specific primers, a modification of phosphorothioate was introduced at the 3′ terminus of an allele-specific primer; it can be against nonmatched alleles in the terms of exo+ polymerase (25, 26). Based on this strategy, the SNPs can be distinguished effectively.
Subsequently, a mutation-specific primer of Pfdhfr C59R with different base types at the antepenultimate mismatch was considered and identified by AS-PCR. The results showed there is no distinction among different mismatch base types, including T, C, and G (Fig. S2B). This demonstrated that the base types are not a major concern compared with the mismatch position. Similar to a previous study (27), an expected result was that mismatch nucleotides placed in a antepenultimate position of specific primers are more successful than a penultimate mismatch, because poor amplification was detected when the mismatched base was close to the 3′-terminal end. However, the alteration of mismatch type at the antepenultimate position does not result in significant distinction, in contrast to previously published AS-PCR protocol in which several kinds of mismatches influence the level of SNP primer specificity in the presence of Taq DNA polymerase (28). This effect might be attributed to exo+ polymerase that harbors the 3′ to 5′ exonuclease activity to recognize fewer than 8 mismatched bases from the 3′ end (29) and the position of additional mismatches near SNP sites (30, 31). The phosphorothioate modification primer possesses exonuclease resistance, which could inhibit mismatch amplification in the presence of exo+ polymerases (26). Taq DNA polymerase exhibits good amplification efficiencies, superior to most DNA polymerases, but its low proofreading activity results in occasional mismatch extension during PCR performance. In the present study, the advantage of using KOD DNA polymerase is that it has similar proofreading activity to that of Pfu DNA polymerase but much better than Taq DNA polymerase and an extension rate superior to that of Pfu DNA polymerase (32). Thus, it is more accurate and time saving. The primers W4 and M4 for C59R wild-type and mutation, respectively, were selected for developing the AS-PCR-LFA system.
Based on the aforementioned strategies, the mutation primers for Pfdhfr S108N and N51I were also designed and tested. According to the results (Fig. S2C), the primers W3 and M3 for S108N were successfully selected. Ascribed to the success of C59R and S108N, only one pair of allele-specific primers, named W and M, that bind to the putative SNP of N51I were designed and confirmed (see Table S1; Fig. S2D). According to the results of electrophoresis in Fig. S2D, a reduced concentration of the plasmid (pDNA) template was accompanied by a gradual decrease in the brightness of bands in a concentration-dependent manner. The selected primers can distinguish the WT and Mut for each SNP. This demonstrates that the predesigned strategy is accurate and meets the demand of AS-PCR. Subsequently, all selected allele-specific primers were labeled with biotin at the 5′-terminal and renamed as listed in Table 1. Meanwhile, the common antisense primers were modified with a 5′ digoxin (Table 1). All the labeled primers were used to develop the AS-PCR-LFA system for each SNP genotyping.
TABLE 1.
Selected and labeled primers for single nuclear polymorphisms detection in the pfdhfr genea
| Primer nameb | Sequence and modification (5′→3′)c | Descriptiond |
|---|---|---|
| Dig-Pfdhfr-N51I_Rev | Dig-GCTTTCCCAGCTTGTTCTTCCC | Common primer |
| Bio-Pfdhfr-N51_Fwd_WT (W') | Bio-AGGAGTATTACCATGGAAATGT*A*A | Allele-specific primer |
| Bio-Pfdhfr-51I_Fwd_Mut (M') | Bio-AGGAGTATTACCATGGAAATGT*A*T | |
| Dig-Pfdhfr-59_108_Fwd | Dig-GATGGAACAAGTCTGCGACGT | Common primer |
| Bio-Pfdhfr-C59_Rev4_WT (W4') | Bio-TCATTCACATATGTTGTAACTGCT*C*A | Allele-specific primer |
| Bio-Pfdhfr-59R_Rev4_Mut (M4') | Bio-TCATTCACATATGTTGTAACTGCT*C*G | |
| Bio-Pfdhfr-S108_Rev3_WT (W3') | Bio-TTTTGGAATGCTTTCCCT*G*C | Allele-specific primer |
| Bio-Pfdhfr-108N_Rev3_Mut (M3') | Bio-TTTTGGAATGCTTTCCCT*G*T |
Gene identifier PF3D7_0417200.
Bio, biotin; Dig, digoxin; Fwd, forward; Rev, reverse; WT, wild type; Mut, mutant.
Artificial mismatches are in boldface font. *, the location of phosphorothioate modification.
The same common primer utilized in allele-specific amplification of C59R and S108N of Pfdhfr.
Development and optimization of the AS-PCR-LFA system.
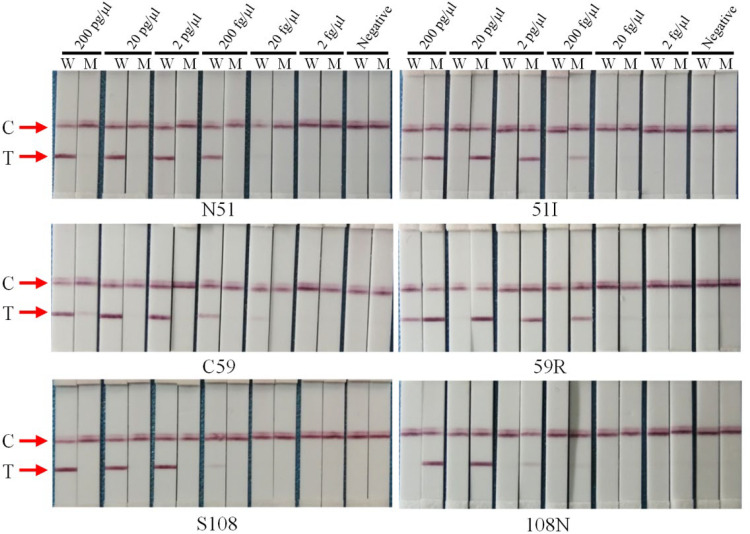
To assess the sensitivity and specificity of the selected allele-specific primers with 5′-terminal modification, AS-PCR-LFA was developed and assessed. The constructed recombinant plasmids were diluted with different gradients and used as the template. In the 20-μl PCR system, the final concentrations of plasmid were from 200 pg/μl to 2 fg/μl. For evaluation of sensitivity and specificity, all the allele-specific primers for the wild type and mutation were detectable at 200 fg/μl in the gel and strip without cross-reaction (Fig. 2 and Fig. S3). Furthermore, the wild-type primer for N51I and C59R and the mutation primer for C59R were detectable at 20 fg/μl. However, the false positive was also detected with the wild-type primer of C59R and mutation primers of N51I and C59R at a high concentration (200 pg/μl). This phenomenon illustrates that the template concentration should be considered in a reasonable range for its optimal performance. Compared to the results of agarose gel electrophoresis, the strip signals of the T line were weak. This might be attributed to the loading amount. For the LFA, only 2 μl PCR product is added compared to 5 μl for electrophoresis. If the loading amount is increased to the same amount used for electrophoresis, the strip displays clearer bands.
FIG 2.
Sensitivity and specificity evaluation of selected primers for pfdhfr gene with AS-PCR-LFA system through visualized interpretation. The final concentrations of serial dilutions of plasmid pEASY-T1/Pfdhfr in the PCR system (20 μl) were from 200 pg/μl to 2 fg/μl. W and M on the sample lanes represent the wild-type and mutation primer, respectively; C and T represent the control line and test line, respectively.
Under high-purity conditions, the pDNA did not noticeably influence the amplification process. However, the composition of the extracted gDNA from the blood filter paper was complicated and contained various PCR inhibitors. All these may have affected the amplification efficiency. To reduce nonspecific amplification, AuNPs were added to the AS-PCR system and evaluated. In the study, Pfdhfr S108N was taken as an example for illustration. The results show that reactions with 1 ng/ml AuNPs decrease the nonspecific amplification compared to that for the reaction without AuNPs (see Fig. S4). Furthermore, the cycle number of AS-PCR at different parasite densities (parasitemia) was also demonstrated by Pfdhfr S108N. For easy comparison, samples were divided into three groups based on the parasitemia (parasites/μl) (Table S2). Thus, the optimal cycle number for AS-PCR-LFA was determined. As shown in Fig. S5, the optimal numbers of cycles for the groups with middle and high parasitemia were 20 and 15, respectively. However, nonspecific amplification was also found in these two groups at 25 cycles. This indicates that the suitable number of cycles for amplification is less than 25. For the low-parasitemia group, 25 cycles were selected (Fig. S5).
Assessment of the AS-PCR-LFA system with clinical isolates.
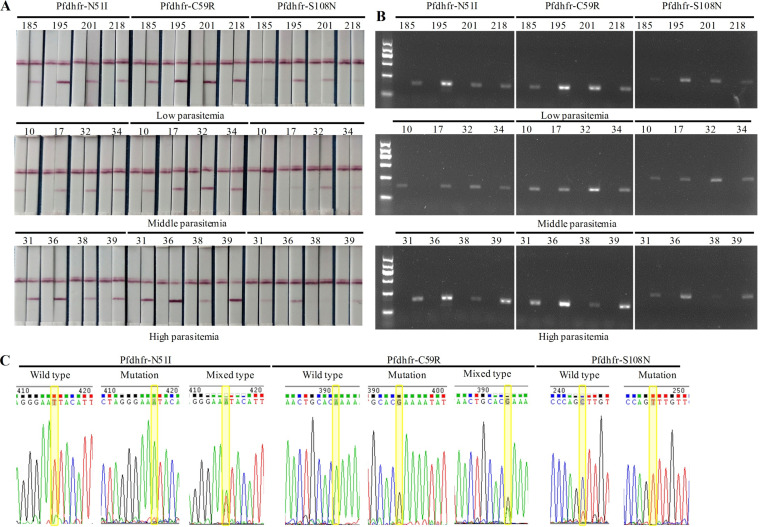
To evaluate the reliability of the optimized AS-PCR-LFA system, the accuracy was further verified with clinical samples. A final genotyping result of an allele was visually interpreted by golden-red color development on the T lines of both strips (Fig. 3A). For the mutant allele, a distinct red band was observable on the T line of the strip used only for the Mut tube but not for the WT tube. In contrast, for the wild-type allele, the red band was detected solely on the strip receiving the WT tube and not the Mut tube. However, the presence of red bands on the T lines of both strips indicated a mixed type. Furthermore, the SNP genotyping results detected by agarose gel electrophoresis (Fig. 3B) and DNA sequencing (Fig. 3C) are displayed as a comparison, which indicated the same accuracy of the AS-PCR-LFA system as a golden standard of genotyping methods.
FIG 3.
Genotyping result (take partial samples as an example) provided by the AS-PCR-LFA system through visualized interpretation (A), agarose gel electrophoresis (B), and DNA sequencing (reverse sequencing) (C).
The three SNPs (N51I, C59R, and S108N) in 98 individuals were fully genotyped by nested-PCR and sequencing and then tested by the AS-PCR-LFA (Table S2). In this study, mixed types were detected for N51I and C59R. However, two samples with false positives were also detected for C59R but not for N51I and S108N. The frequencies of different mutant alleles are displayed in Table S3, and the frequencies of mutant alleles in Pfdhfr provided with the AS-PCR-LFA system were in accordance with the results from gel electrophoresis and nested PCR followed by sequencing. As shown in Table S3, the sensitivity was 97.96% (96/98) for N51I, C59R, and S108N. For specificity, the values were 100% (98/98), 95.92% (94/98), and 100% (98/98) for N51I, C59R, and S108N, respectively. The results demonstrate that the developed AS-PCR-LFA system is comparable to the nested PCR followed by DNA sequencing for Pfdhfr genotyping. For evaluation of false negatives, N51I and S108N were present at 2.04%. Regarding C59R, 4.08% of samples appeared to result in false positives. The limit of detection (LOD) of the system validated with clinical samples is 100 parasites/μl of blood filter paper. Two samples with low parasitemia (200 parasites/μl and 400 parasites/μl) were detected as negative for all three SNPs according to the results of agarose gel electrophoresis and LFA. Of the 96 samples successfully genotyped for the pfdhfr gene by the AS-PCR-LFA system, 90.63% (87/96) harbored the mutant allele N51I, while 1 sample (1.04%) had a mixed type. For Pfdhfr C59R, 87.5% (84/96) of the samples had the mutant allele 59R, and 4 (4.17%) had mixed genotypes. At codon 108, 95.83% (92/96) harbored the mutation, and no mixed infection was found.
Compared to conventional PCR-based techniques, the proposed assay is a well-suited platform to monitor multiple SNPs, opening a way to be deployable in clinical analysis for treatment guidance. The limited resources in regions of malaria endemicity increase the demand for a convenient tool for diagnosis. With AuNPs, genotyping results can be provided both quantificationally and qualitatively by using visual inspection of colors on the T and C lines. For clinical sample examination, it should be noted that nested PCR was employed, as one round of PCR was unable to amplify target sequences within the A+T rich genome of P. falciparum (33). Furthermore, nested-PCR is the primary technique for molecular detection of Plasmodium parasite ADR genes (7, 9). Although pDNA was detectable at 200 fg/μl, clinical samples containing low parasite density at 100 parasites/μl were identified effectively. This shows that the AS-PCR-LFA system is efficient and accurate for clinical samples with different parasitemia, thus offering an ideal molecular diagnostic tool in clinical practice. The established system for drug-resistant SNP detection in Pfdhfr exhibits high specificity and sensitivity and can be reconfigured for the genotypes of other genetic-related diseases.
In the present study, several shortcomings should not be neglected and need to be improved in further study. First, the gDNA extraction method is still complex. Thus, a simplified gDNA extraction method should be considered. Second, the amplification with the PCR-based method takes at least 2 h. Several isothermal amplification methods, including loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) could reduce the reaction time (34, 35). Last but not least, the LOD in the present study can still be improved. The recently developed gene-editing technique CRISPR/Cas should be considered. Thus, if the platform combined the above-mentioned techniques, a powerful tool would be established to support the molecular surveillance of ADR genes.
Conclusions.
In summary, we presented the AS-PCR-LFA system to detect multiple SNPs related to antimalarial pyrimethamine resistance. The study reports a multiple-SNP detection system combining AS-PCR with AuNP-LFA for Pfdhfr N51I, C59R, and S108N genotyping. The current data demonstrate that the established AS-PCR-LFA system could potentially lead to multiple-SNP detection, opening a way to a near-to-patient molecular test for treatment guidance. The system can be further employed for the detection of other genes with multiple SNPs that are associated with ADR. It also offers a potential platform for the development of SNP profiles for applications in infectious diseases and genetic diseases.
MATERIALS AND METHODS
Materials and reagents.
All chemicals were of analytical grade and purchased from local reputable vendors. Buffers were prepared according to standard laboratory procedures. TIANamp blood spots DNA kit and TIAN prep mini plasmid kit were ordered from Tiangen Biotech Co., Ltd. (Beijing, China). Green PCR master mix was purchased from Hubei Jinmao Tech. Co., Ltd. (Wuhan, China). The pEASY-T1 Cloning kit, isopropyl-β-d-thiogalactopyranoside (IPTG), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were ordered from Beijing TransGen Co., Ltd. (Beijing, China). FastDigest BamHI and XhoI were purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). KOD-Plus-Neo and related PCR reagents were ordered from TOYOBO Co., Ltd. (Shanghai, China). The 1-mg/ml 10-nm AuNPs were purchased from Hualan Chemistry Science and Technology Co., Ltd. (Shanghai, China). The 0.4-mg/ml 35-nm streptavidin-immobilized gold nanoparticles (SA-AuNPs) were obtained from Beijing Biosynthesis Biotechnology Co., Ltd. (Beijing, China). Biotinylated bovine serum albumin (biotin-BSA; 2.5 mg/ml) was acquired from Solarbio (Beijing, China). A 1.3-mg/ml monoclonal antibody (Ab) against digoxin (Dig) was purchased from Jackson ImmunoResearch Laboratories, Inc. (Baltimore, MD, USA). Bovine serum albumin (BSA) was procured from Biosharp Life Sciences (Hefei, China). Glass fiber (RB65), nitrocellulose (NC) membrane (CN140), absorbent paper (CH37), and plastic backing (SM31-35) were ordered from Shanghai Kinbio Tech. Co., Ltd. (Shanghai, China).
P. falciparum isolate collection and genotyping.
The initial diagnoses of P. falciparum infection in patients were made by thick and thin smears stained with diluted Giemsa solution and later confirmed by rapid diagnostic test (RDT) and quantitative PCR (qPCR) (7). Subsequently, blood samples were spotted on filter paper for further use. Genomic DNA (gDNA) from P. falciparum isolates was extracted from dried filter blood spots (DBS) using TIANamp blood spots DNA kit, according to the manufacturer’s instructions. Briefly, approximately 130 μl whole blood was used for gDNA extraction and eluted with 40 μl elution buffer. The genotyping of the pfdhfr gene by nested-PCR with direct sequencing of PCR products was performed according to previously published procedures (7, 13). This study was approved by the ethics committees of the Hubei University of Medicine and Wuhan City Center for Disease Prevention and Control. Informed consent was obtained from all participating individuals.
Plasmids construction and identification.
To prepare reference DNA, amplified fragments containing wild-type or mutations of Pfdhfr N51I, C59R, and S108N were cloned into a pEASY-T1 Cloning vector according to the kit instructions. The recombinant plasmids were extracted from transformed Escherichia coli Trans1-T1 phage-resistant chemically competent cells using a TIAN prep mini plasmid kit. All plasmids were identified via restriction enzyme analysis and Sanger sequencing by Genewiz Biotechnology Ltd. (Soochow, China). The concentration and quality of recombinant plasmids were determined using Gene 5 (Thermo Fisher Scientific, Wilmington, DE, USA).
Design and screening of oligonucleotides.
Wild-type, mutation, and mixed-type SNPs are named here as WT, Mut, and Mix, respectively. To discriminate each allele, allele-specific primers corresponding to putative SNPs and a common antisense primer were designed manually and evaluated by using Oligo 7.0. The artificial mismatch was redesigned to increase the specificity of allele-specific mutation primers. The DNA sequencing of pfdhfr gene (gene identifier [ID] PF3D7_0417200) was downloaded from PlasmoDB (http://plasmodb.org/plasmo/) (7). Experiments were performed by using a Bio-Rad T100 thermal cycler (Bio-Rad, Hercules, CA, USA).
For screening the targeted primers, concentrations of wild-type or mutation plasmid DNA (pDNA) were used as the templates. The PCR system was performed at a 20 μl final volume which contained 2.0 μl of 10× buffer, 2 μl of 2 mΜ deoxynucleoside triphosphates (dNTPs), 1.5 μl of 25 mM MgSO4, 0.35 U of KOD-Plus-Neo, 1 μl of 2 μM common antisense primer, 1 μl of 2 μM allele-specific primer (Mut primer in Mut tube and WT primer in WT tube), and 1 μl pDNA. The reaction condition was as follows: 95°C for 3 min, 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s, and then 72°C for 5 min. The 5-μl PCR products were analyzed using 1.0% agarose gel electrophoresis at 120 V for 25 min.
Establishment and optimization of the AS-PCR-LFA platform.
A schematic diagram of the AS-PCR and lateral flow strip is shown in Fig. 1. To assess the sensitivity of optimized primers used for the AS-PCR-LFA platform, the selected primers were modified, including a 5′ digoxin-labeled common antisense primer and a 5′ biotin-labeled specific primer for wild type and mutation, respectively (Table 1; Fig. 1B). All labeled primers were synthesized by Genewiz. The AS-PCR system and conditions were optimized with small-scale clinical samples before clinical application. The methods for the first round of PCR are described in a previous study (7). The PCR products were stored at 4°C until use. The AS-PCR was performed with a 20-μl final volume which contained 2.0 μl of 10× buffer, 2 μl of 2 mΜ of dNTPs, 1.5 μl of 25 mM MgSO4, 0.35 U of KOD-Plus-Neo, 1 μl of 2 μM commonly targeted primer, 1 μl of 2 μM allele-specific primer (Mut primer in Mut tube and WT primer in WT tube), 1 μl amplified PCR product, 0 to 2.5 μl of 1 ng/ml AuNPs (size ,10 nm), and ultrapure H2O up to 20 μl. The reaction condition was as follows: 95°C for 3 min, 15 to 25 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s, and then 72°C for 10 min. Subsequently, the AS-PCR product was mixed with buffer and detected by LFA.
The dry-reagent test strip (width, 4 mm) was fabricated from five components, including a sample pad (16 mm), a gold conjugate pad (8 mm), an NC membrane (25 mm), and an absorbent pad (17 mm), and these parts were fixed on plastic backing (60 mm) with a 2-mm overlap (Fig. 1A). Briefly, biotin-BSA and anti-Dig Ab were preimmobilized via an XYZ3010 dispensing platform in a control line (C line) and a defined test line (T line), respectively, on a porous NC membrane. The sample pad and conjugate pad were both made from glass fiber which was pretreated with suspending buffer containing 10 mM phosphate buffer (pH 7.4), 1% BSA, 0.5% Tween 20, and 0.5% sucrose. Subsequently, the solution containing SA-AuNPs was dispensed on the conjugate pad (Fig. 1A). Then, the strips were dried at 56°C for 4 h and stored in a sealed aluminum foil bag at room temperature until use. On the lateral flow dipstick (LFD), 2 μl of the PCR product was added into 70 μl running buffer (pH 7.4), and the whole solution was then loaded to the sample pad (Fig. 1C). The test results could be visualized within 15 min and determined by the presence of red lines.
Clinical application of the AS-PCR-LFA system.
For the genotyping of clinical samples, a two-step PCR was conducted. Each sample underwent two independent PCRs for SNP detection. PCR product detection was dependent on color changes of the LFA. The reference DBS (confirmed by sequencing) were used to validate the method (Fig. 1).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Department of Schistosomiasis and Endemic Diseases, Wuhan Center for Disease Prevention and Control, and all participants who contributed their blood samples.
The work was supported by the Foundation for Innovative Research Team of Hubei University of Medicine (FDFR201603), Foundation for Graduate Science and Technology Innovation Project of Hubei University of Medicine (YC2019015 and YC2020003), Basic Research Business Fund of Central Universities (FRF-TP-18-020A1), China Postdoctoral Science Foundation (2018 M631332), and National Natural Science Foundation of China (81802046).
REFERENCES
- 1.WHO. World malaria report. 2019. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Artimovich E, Schneider K, Taylor TE, Kublin JG, Dzinjalamala FK, Escalante AA, Plowe CV, Laufer MK, Takala-Harrison S. 2015. Persistence of sulfadoxine-pyrimethamine resistance despite reduction of drug pressure in Malawi. J Infect Dis 212:694–701. doi: 10.1093/infdis/jiv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Global Malaria Programme, WHO Department of Reproductive Health and Research, WHO Department of Maternal, Newborn, Child and Adolescent Health. 2013. WHO policy brief for the implementation of intermittent preventive treatment of malaria in pregnancy using sulfadoxine-pyrimethamine (IPTp-SP). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Zolg JW, Plitt JR, Chen GX, Palmer S. 1989. Point mutations in the dihydrofolate reductase-thymidylate synthase gene as the molecular basis for pyrimethamine resistance in Plasmodium falciparum. Mol Biochem Parasitol 36:253–262. doi: 10.1016/0166-6851(89)90173-4. [DOI] [PubMed] [Google Scholar]
- 5.Triglia T, Wang P, Sims PF, Hyde JE, Cowman AF. 1998. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J 17:3807–3815. doi: 10.1093/emboj/17.14.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koukouikila-Koussounda F, Bakoua D, Fesser A, Nkombo M, Vouvoungui C, Ntoumi F. 2015. High prevalence of sulphadoxine-pyrimethamine resistance-associated mutations in Plasmodium falciparum field isolates from pregnant women in Brazzaville, Republic of Congo. Infect Genet Evol 33:32–36. doi: 10.1016/j.meegid.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Jiang T, Chen J, Fu H, Wu K, Yao Y, Eyi JUM, Matesa RA, Obono MMO, Du W, Tan H, Lin M, Li J. 2019. High prevalence of Pfdhfr-Pfdhps quadruple mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Malar J 18:101. doi: 10.1186/s12936-019-2734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mita T, Ohashi J, Venkatesan M, Marma ASP, Nakamura M, Plowe CV, Tanabe K. 2014. Ordered accumulation of mutations conferring resistance to sulfadoxine-pyrimethamine in the Plasmodium falciparum parasite. J Infect Dis 209:130–139. doi: 10.1093/infdis/jit415. [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Pi L, Qin Y, Lu Y, Zeng W, Xiang Z, Qin P, Chen X, Li C, Zhang Y, Wang S, Si Y, Yang G, Rosenthal BM, Huang Y, Yang Z. 2020. Widespread resistance mutations to sulfadoxine-pyrimethamine in malaria parasites imported to China from Central and Western Africa. Int J Parasitol Drugs Drug Resist 12:1–6. doi: 10.1016/j.ijpddr.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolg JW, Chen GX, Plitt JR. 1990. Detection of pyrimethamine resistance in Plasmodium falciparum by mutation-specific polymerase chain reaction. Mol Biochem Parasitol 39:257–265. doi: 10.1016/0166-6851(90)90064-s. [DOI] [PubMed] [Google Scholar]
- 11.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- 12.Duraisingh MT, Curtis J, Warhurst DC. 1998. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol 89:1–8. doi: 10.1006/expr.1998.4274. [DOI] [PubMed] [Google Scholar]
- 13.Pearce RJ, Drakely C, Chandramohan D, Mosha F, Roper C. 2003. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob Agents Chemother 47:1347–1354. doi: 10.1128/aac.47.4.1347-1354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Monbrison F, Raynaud D, Latour-Fondanaiche C, Staal A, Favre S, Kaiser K, Peyron F, Picot S. 2003. Real-time PCR for chloroquine sensitivity assay and for pfmdr1-pfcrt single nucleotide polymorphisms in Plasmodium falciparum. J Microbiol Methods 54:391–401. doi: 10.1016/s0167-7012(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 15.Wilson PE, Alker AP, Meshnick SR. 2005. Real-time PCR methods for monitoring antimalarial drug resistance. Trends Parasitol 21:278–283. doi: 10.1016/j.pt.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Andriantsoanirina V, Lascombes V, Ratsimbasoa A, Bouchier C, Hoffman J, Tichit M, Rabarijaona LP, Durand R, Menard D. 2009. Rapid detection of point mutations in Plasmodium falciparum genes associated with antimalarial drugs resistance by using high-resolution melting analysis. J Microbiol Methods 78:165–170. doi: 10.1016/j.mimet.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Yao Y, Wu K, Xu M, Yang Y, Zhang Y, Yang W, Shang R, Du W, Tan H, Chen J, Lin M, Li J. 2018. Surveillance of genetic variations associated with antimalarial resistance of Plasmodium falciparum isolates from returned migrant workers in Wuhan, Central China. Antimicrob Agents Chemother 62:e02387-17. doi: 10.1128/AAC.02387-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaudet AL, Belmont JW. 2008. Array-based DNA diagnostics: let the revolution begin. Annu Rev Med 59:113–129. doi: 10.1146/annurev.med.59.012907.101800. [DOI] [PubMed] [Google Scholar]
- 19.Roberts DG, Morrison TB, Liu-Cordero SN, Cho J, Garcia J, Kanigan TS, Munnelly K, Brenan CJ. 2009. A nanoliter fluidic platform for large-scale single nucleotide polymorphism genotyping. Biotechniques 46:ix–xiii. doi: 10.2144/000112887. [DOI] [PubMed] [Google Scholar]
- 20.Tost J, Gut IG. 2005. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin Biochem 38:335–350. doi: 10.1016/j.clinbiochem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Quesada-Gonzalez D, Merkoci A. 2015. Nanoparticle-based lateral flow biosensors. Biosens Bioelectron 73:47–63. doi: 10.1016/j.bios.2015.05.050. [DOI] [PubMed] [Google Scholar]
- 22.Howes PD, Rana S, Stevens MM. 2014. Plasmonic nanomaterials for biodiagnostics. Chem Soc Rev 43:3835–3853. doi: 10.1039/c3cs60346f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma D, Lather M, Dykes CL, Dang AS, Adak T, Singh OP. 2016. Disagreement in genotyping results of drug resistance alleles of the Plasmodium falciparum dihydrofolate reductase (Pfdhfr) gene by allele-specific PCR (ASPCR) assays and Sanger sequencing. Parasitol Res 115:323–328. doi: 10.1007/s00436-015-4750-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Li K. 2003. On-off regulation of 3' exonuclease excision to DNA polymerization by Exo+ polymerase. J Biochem Mol Biol 36:525–528. doi: 10.5483/BMBRep.2003.36.6.525. [DOI] [PubMed] [Google Scholar]
- 26.Li K, Zhang J, Chen L, Sommer SS. 2005. Superb nucleotide discrimination by a novel on/off switch for DNA polymerization and its applications. Mol Biotechnol 29:93–100. doi: 10.1385/MB:29:2:093. [DOI] [PubMed] [Google Scholar]
- 27.Chen F, Zhao Y, Fan C, Zhao Y. 2015. Mismatch extension of DNA polymerases and high-accuracy single nucleotide polymorphism diagnostics by gold nanoparticle-improved isothermal amplification. Anal Chem 87:8718–8723. doi: 10.1021/acs.analchem.5b01545. [DOI] [PubMed] [Google Scholar]
- 28.Huang MM, Arnheim N, Goodman MF. 1992. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res 20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H-L, Jiang H-J, Fang W-Y, Xu Y-Y, Li K, Zhang J, Liao D-F, He F-C. 2005. High fidelity PCR with an off/on switch mediated by proofreading polymerases combining with phosphorothioate-modified primer. Biochem Biophys Res Commun 328:265–272. doi: 10.1016/j.bbrc.2004.12.159. [DOI] [PubMed] [Google Scholar]
- 30.Wu JH, Hong PY, Liu WT. 2009. Quantitative effects of position and type of single mismatch on single base primer extension. J Microbiol Methods 77:267–275. doi: 10.1016/j.mimet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Lefever S, Pattyn F, Hellemans J, Vandesompele J. 2013. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin Chem 59:1470–1480. doi: 10.1373/clinchem.2013.203653. [DOI] [PubMed] [Google Scholar]
- 32.Takagi M, Nishioka M, Kakihara H, Kitabayashi M, Inoue H, Kawakami B, Oka M, Imanaka T. 1997. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl Environ Microbiol 63:4504–4510. doi: 10.1128/AEM.63.11.4504-4510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malpartida-Cardenas K, Rodriguez-Manzano J, Yu LS, Delves MJ, Nguon C, Chotivanich K, Baum J, Georgiou P. 2018. Allele-specific isothermal amplification method using unmodified self-stabilizing competitive primers. Anal Chem 90:11972–11980. doi: 10.1021/acs.analchem.8b02416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G, Chen R, Ding S, Li M, Wang J, Zou J, Du F, Dong J, Cui X, Huang X, Deng Y, Tang Z. 2020. Recombinase assisted loop-mediated isothermal DNA amplification. Analyst 145:440–444. doi: 10.1039/c9an01701a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.