Emergence and selection of antibiotic resistance following exposure to high antibiotic concentrations have been repeatedly shown in clinical and agricultural settings, whereas the role of the weak selective pressures exerted by antibiotic levels below the MIC (sub-MIC) in aquatic environments due to anthropogenic contamination remains unclear. Here, we studied how exposure to sub-MIC levels of ciprofloxacin enriches for Escherichia coli with reduced susceptibility to ciprofloxacin using a mallard colonization model.
KEYWORDS: antibiotic contamination, antibiotic resistance, subinhibitory concentration, water environment, intestinal microbiota
ABSTRACT
Emergence and selection of antibiotic resistance following exposure to high antibiotic concentrations have been repeatedly shown in clinical and agricultural settings, whereas the role of the weak selective pressures exerted by antibiotic levels below the MIC (sub-MIC) in aquatic environments due to anthropogenic contamination remains unclear. Here, we studied how exposure to sub-MIC levels of ciprofloxacin enriches for Escherichia coli with reduced susceptibility to ciprofloxacin using a mallard colonization model. Mallards were inoculated with two isogenic extended-spectrum-β-lactamase (ESBL)-encoding E. coli strains, differing only by a gyrA mutation that results in increased MICs of ciprofloxacin, and exposed to different levels of ciprofloxacin in their swimming water. Changes in the ratios of mutant to parental strains excreted in feces over time and ESBL plasmid spread within the gut microbiota from individual birds were investigated. Results show that in vivo selection of gyrA mutants occurred in mallards during exposure to ciprofloxacin at concentrations previously found in aquatic environments. During colonization, resistance plasmids were readily transferred between strains in the intestines of the mallards, but conjugation frequencies were not affected by ciprofloxacin exposure. Our results highlight the potential for enrichment of resistant bacteria in wildlife and underline the importance of reducing antibiotic pollution in the environment.
INTRODUCTION
Multidrug-resistant Escherichia coli constitutes an increasing problem in clinical settings and a threat to modern health care worldwide (1). Emergence and spread of antibiotic resistance following antibiotic exposure have typically been assumed to occur at concentrations above or close to the MIC of susceptible bacteria (2). However, the importance of the selective pressure exerted by antibiotic levels below the MIC of susceptible bacteria (sub-MIC) has not been addressed in the same detail with regard to their role in enrichment of resistant bacteria in complex systems. Previous in vitro studies have shown that sub-MIC levels of antibiotics can enrich for preexisting resistant strains as well as selecting for highly resistant mutants de novo from a susceptible strain (3–6). The term “minimal selective concentration” (MSC) refers to the lowest antibiotic concentration that provides a selective benefit to a resistant bacterium over a susceptible counterpart, and it has been shown to be up to 200-fold below the MIC of the respective susceptible strain (4).
The outer environment can be contaminated with antibiotics originating from pharmaceutical industry effluents, human or animal wastewater (especially from hospitals), aquaculture, and soils treated with manure (7). Some antibiotics, such as fluoroquinolones, are chemically very stable and can accumulate and persist in the environment (8), and their occurrence has been detected in aqueous environmental matrices worldwide (9, 10). Antibiotic residues and antibiotic-resistant bacteria of human origin are considered a form of environmental pollution from anthropogenic activity, and wildlife, such as aquatic birds, is exposed to these antibiotic residues and bacteria of human origin (11–13). The spread and accumulation of antibiotic-resistant bacteria in the environment and wildlife have been repeatedly confirmed (14), but the magnitude of resistance development and selection that take place in nature remains poorly defined.
Mobile genetic elements with the potential to transfer between bacterial cells, e.g., plasmids and conjugative transposons, are central vectors for the dissemination of antibiotic resistance genes and multiresistant E. coli isolates often harbor one or several conjugative plasmids encoding multiple resistance genes (15). In some settings, sub-MIC levels of antibiotics have been shown to increase the rate of plasmid conjugation (16); however, other studies suggest that this effect has been overestimated (17).
The aim of this project was to investigate whether exposure to sub-MIC levels of ciprofloxacin (Cip) enriches for the most common mutations that result in reduced susceptibility to ciprofloxacin in E. coli in the intestines of water-residing birds, using mallards as a model. We also investigated the spread of resistance plasmids between different E. coli strains in the gut of the birds during colonization. Mallards were chosen as a representative of wild bird species because they reside in aquatic environments (18) and have been found to carry multidrug-resistant bacteria in the intestine in several studies (19–21). By oral administration of isogenic strains of ESBL-producing E. coli isolates of bird origin differing only by constructed single gyrA mutations, we showed that very low concentrations of ciprofloxacin in the birds' drinking and swimming water selected for the mutant strains. Plasmid transfer between inoculated bacterial strains and the endogenous intestinal E. coli of the birds was detected during colonization, but the conjugation frequency was not affected by ciprofloxacin exposure. We conclude that sub-MICs of antibiotics frequently present in water environments where birds reside can enrich for resistant bacteria in their intestinal microflora, something that merits evaluation of actions to reduce such exposure to minimize the risk of enriching resistant bacteria in wild animals.
RESULTS
To determine if sub-MIC levels of ciprofloxacin could select for resistant mutants during intestinal colonization of mallards, we constructed two different isogenic strain pairs in which one strain in each pair carried a gyrA mutation leading to reduced ciprofloxacin susceptibility but the strains were otherwise genetically identical, as verified by whole-genome sequencing. Streptomycin (Str) resistance was first selected in the E. coli SP132 genetic background as a resistance marker to allow us to select and distinguish the colonizing strains from endogenous E. coli of the mallard gut microbiota. The two isogenic strain pairs were DA26187/DA26192 (GyrAwt/GyrAD87Y) and DA28261/DA28400 (GyrAwt/GyrAS83L) (Table 1). The gyrA mutations chosen are among the most commonly found in clinical fluoroquinolone-resistant E. coli isolates (22).
TABLE 1.
Characteristics of constructed strainsa
| Strain pair | Strain | RpsL | GyrA | Fitness cost of gyrA mutation (%) | MIC (mg/liter) |
|||
|---|---|---|---|---|---|---|---|---|
| Str | Cip | Ctx | Tet | |||||
| GyrAwt | DA26187 | K43T | wt | 1,024 | 0.016 | 8 | 1 | |
| GyrAD87Y | DA26192 | K43T | D87Y | 1.4 | 1,024 | 0.125 | 8 | 0.5 |
| GyrAwt | DA28261 | K43R | wt | 1,024 | 0.016 | 16 | 1 | |
| GyrAS83L | DA28400 | K43R | S83L | 0.2 | 1,024 | 0.25 | 16 | 2 |
Str, streptomycin; Cip, ciprofloxacin; Ctx, cefotaxime; Tet, tetracycline; wt, wild type.
Determination of minimal selective concentration of ciprofloxacin in vitro.
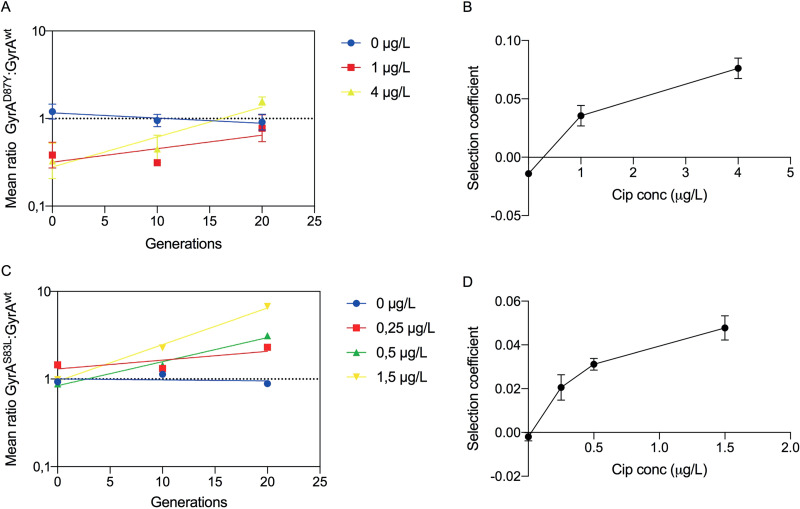
In vitro competition experiments in the absence of ciprofloxacin were used to determine the fitness cost of the gyrA mutations, and in vitro competition experiments in the presence of sub-MICs of ciprofloxacin were done to determine at which concentrations of drug the fitness cost of the respective mutation could be balanced by antibiotic selection (i.e., the minimal selective concentration [MSC]) (4). As illustrated in Fig. 1A, the mean logarithmic ratio of GyrAmut to GyrAwt strains changes as a function of the number of generations of growth. The slope of each exponential trend line is a measure of the selection coefficient (s) per generation. A negative slope indicates that the gyrA mutant has a disadvantage and a positive slope that the mutant is enriched. When the s values obtained from these experiments are plotted as a function of antibiotic concentration, the intercept with the y axis represents the fitness cost in the absence of antibiotics, while the intercept with the x axis (s = 0) represents the MSC where the fitness cost of the mutation is balanced by the antibiotic-conferred selection (4, 23). In the absence of ciprofloxacin, the GyrAD87Y strain was outcompeted by the GyrAwt strain and the cost of the D87Y substitution was estimated at 1.4% per generation, but at 1 µg/liter and above, it was enriched and the MSC was 0.25 µg/liter (Fig. 1B). This is 64-fold below the MIC of the wild-type strain. The GyrA S83L substitution had an even lower cost, only 0.2% per generation, in the absence of antibiotic and thereby an even lower MSC, below 0.1 µg/liter, in accordance with previously published data (4).
FIG 1.
Competition experiments to determine in vitro minimal selective concentrations. Linear regression of the ratios of GyrAD87Y/GyrAwt (A) and GyrAS83L/GyrAwt (C) strains at different concentrations of ciprofloxacin. Plots of the selection coefficients derived from A and C at each specific antibiotic concentration in GyrAD87Y/GyrAwt (B) and GyrAS83L/GyrAwt (D) with inserted trend lines and standard errors of the slope (calculated from the linear slopes in panels A and C). All concentrations are in micrograms per liter.
Determination of MSC during intestinal colonization.
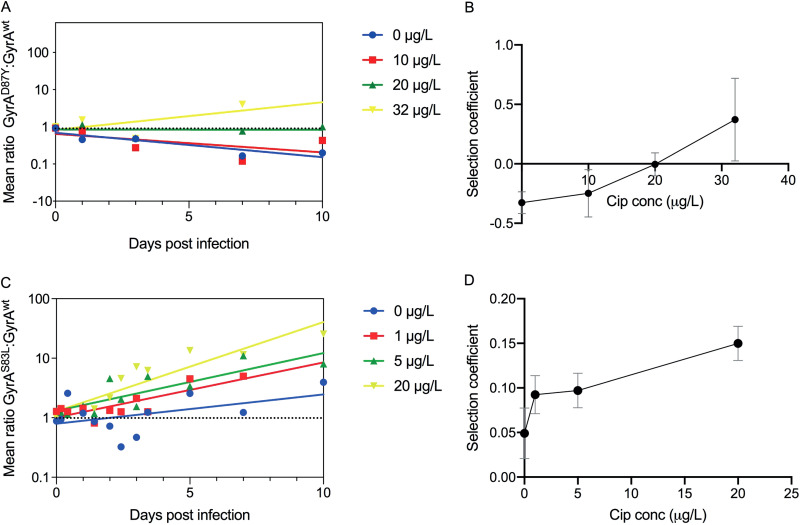
We inoculated two different sets of mallards intraesophageally with a 1:1 mix of the isogenic pairs and added ciprofloxacin at different concentrations to the water pool in the experimental room to determine at what concentrations gyrA mutants with reduced susceptibility to ciprofloxacin can be selected in the gut of animals residing in contaminated water environments. Measurements of the concentration of ciprofloxacin after 24 h showed that it did not deviate from the initial concentration (data not shown), indicating that it was stably maintained during the experiment. Individual fecal samples were retrieved daily from the mallards and plated on selective media to determine the ratio of wild-type and mutant bacteria. The GyrAwt/GyrAD87Y experiment comprised exposures with concentrations of ciprofloxacin in the water of 0, 10, 20, and 32 µg/liter. The number of recovered cells of the inoculated streptomycin-resistant strains ranged from 100 to 8,000 CFU per plating on day 1 postinoculation and thereafter declined until day 10 postinoculation, when a majority of the birds were negative for both strains (see Fig. S1 in the supplemental material). In the absence of ciprofloxacin, the GyrAD87Y strain was outcompeted by the GyrAwt strain, but at ciprofloxacin concentrations above 20 µg/liter of water, it was enriched and the MSC in the water was 21 µg/liter (Fig. 2A and B). Note that since the bacterial generation time in the bird gut is unknown, the selection coefficients were calculated per day instead of per generation. All raw data and ratio calculations are provided in Table S1A and B in the supplemental material.
FIG 2.
Competition of isogenic strains in mallards. Linear regression of the ratios of GyrAD87Y/GyrAwt (A) and GyrAS83L/GyrAwt (C) strains at different concentrations of ciprofloxacin. Plots of the selection coefficient per day at each specific antibiotic concentration for GyrAD87Y/GyrAwt (B) and GyrAS83L/GyrAwt (D) with inserted trend lines and standard errors of the slopes (calculated from the linear slopes in panels A and C). All concentrations are in micrograms per liter.
For the less costly GyrA S83L substitution, the experiment comprised exposures to ciprofloxacin concentrations of 0, 1, 5, and 20 µg/liter. Interestingly, GyrAS83L was weakly selected for even in the absence of ciprofloxacin and increasingly selected for at all ciprofloxacin concentrations tested (Fig. 2C). As GyrAS83L conferred no cost but rather a small benefit in the absence of ciprofloxacin, no MSC value could be determined, but all tested antibiotic concentrations led to further enrichment of the mutant, indicating a positive selective effect of the antibiotic down to at least 1 µg/liter in the water.
To determine if the selection of the gyrA mutants was an effect of growth inside the gut of the mallards or if there was a selection effect by growth or death in the pool water, we performed in vitro selection experiments with the isogenic pairs at the same concentrations of ciprofloxacin in water instead of growth medium. As expected, this showed no net growth or net death of bacteria during 24 h (data not shown). Due to the strong dilution effect in the 170 liters of water (170,000-fold per g of feces) and daily change of the water, we therefore conclude that any enrichment mainly occurred during growth inside the mallards and not in the outer environment.
Extensive plasmid transfer between bacterial strains in the intestines of mallards.
In the GyrAwt/GyrAS83L experiment, we also included a third strain of a different genetic origin to test if conjugation of resistance plasmids occurred during gut colonization and if sub-MICs of ciprofloxacin would affect the conjugation frequency. The competing strains, derived from SP132, contained an IncI1 plasmid encoding the cefotaxime resistance gene blaCTX-M-1, while the third strain, DA26200 (cefotaxime susceptible, streptomycin susceptible, ciprofloxacin resistant, tetracycline resistant), derived from SP163, contained an IncF plasmid harboring the tetracycline resistance gene tetA. This meant that both plasmid-containing strains could potentially function as donors and recipients of the plasmids. Before the experiment started, some mallards were found to excrete tetracycline-resistant E. coli in the feces, referred to here as endogenous Tetr E. coli. In contrast to the experimental strains, the endogenous tetracycline-resistant E. coli strains were susceptible to ciprofloxacin, cefotaxime, and streptomycin. By selecting for transconjugants on cefotaxime and tetracycline, we could therefore extend the search and also include transfer of the ESBL plasmid to endogenous gut E. coli or transfer of the tetracycline resistance determinants if they were mobile. All strains could subsequently be differentiated from each other by selective plating, and the emergence of new combinations of resistance could be identified.
In total, 91 putative transconjugants were found in nine birds from all four experiments. These were further grouped into possible independent transfer events by experiment, resistance profile, and PCR genotype. One isolate from each of the resulting groups was subjected to whole-genome sequencing and analyzed regarding multilocus sequence type (MLST), resistance genes, plasmid replicons, and virulence genes (Table 2). In total, a minimum of eight different transfer events were identified; the SP132 derivatives served as recipients of an IncHI2 plasmid carrying tet(B) from endogenous ST409 Tetr E. coli in one event and as donors of the IncI1 CTX-M-1 plasmid in seven events (2 to SP163 and 5 to endogenous Tetr E. coli). The endogenous Tetr E. coli strains acting as recipients belonged to three different sequence types (SP48, SP165, and ST11511), and all have a tet(A) gene. These tet(A) genes were most likely carried on the chromosome in all but one transconjugant, as judged by lack of additional plasmid replicons. However, the sequenced ST48 transconjugant contained an additional IncFII replicon together with several additional resistance genes that coassembled on the same contig as the tet(A) gene. A clear dominance in number of isolated transconjugants was found in the experiment with 1 µg/liter ciprofloxacin, but this could be due to expansion of transconjugants from the same conjugation event. Most transconjugants were also detected in samples from the index birds obtained within the first 48 h after inoculation. No association between exposure to ciprofloxacin and the number of individual conjugation events was observed, likely due to the low total number of events.
TABLE 2.
Characteristics of transconjugants
| Strain | Cip concn (µg/liter) | na | Resistanceb |
MLSTc | Resistance genes (% identity) | Resistance mutations | Virulence gene(s) | Plasmid replicons | Transfer event | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cip | Str | |||||||||
| SP132 | S/R | R | ST409 | ctx-m-1, mdf(A) | wt or gyrA S83L | gad | IncI1 (100%) | |||
| SP163 | R | S | ST359 | tet(A), mdf(A) | gyrA S83L, D87N, parC S80I | cba, cma, iroN, iss, lpfA, mchF | IncFIB (AP001918), IncFIB (pLF82), IncFIC (FII) | |||
| Transconjugants | 0 | 4 | R | S | ST359 | tet(A), ctx-m-1, mdf(A) | gyrA S83L, D87N, parC S80I | cba, cma, gad, iroN, iss, lpfA, mchF | IncI1, IncFIB (AP001918), IncFIB (pLF82), IncFIC (FII) | SP163 recipient; SP132 donor |
| 5 | S | S | ST11511 | tet(A) (99%), ctx-m-1 | parC S57T, parE I355T | gad | IncI1, p0111 | Endogenous Tetr recipient; SP132 donor | ||
| 1 | 5 | S | S | ST11511 | tet(A) (99%), ctx-m-1 | parC S57T, parE I355T | gad | IncI1, p0111 | Endogenous Tetr recipient; SP132 donor | |
| 17 | S | S | ST48 | tet(A), ctx-m-1, mdf(A), aadA5, sul2, dfrA17 | No mutations | gad | IncI1, IncFIB (AP001918), IncFIB (pLF82), IncFII (pHN7A8), p0111 | Endogenous Tetr recipient; SP132 donor | ||
| 12 | R | S | ST359 | tet(A), ctx-m-1, mdf(A) | gyrA S83L, D87N, parC S80I | cba, cma, gad, iroN, iss, lpfA, mchF | IncI1, IncFIB (AP001918), IncFIB (pLF82), IncFIC (FII) | SP163 recipient; SP132 donor | ||
| 5 | 5 | S | S | ST165 | tet(A), ctx-m-1, mdf(A) | No mutations | gad | IncI1 | Endogenous Tetr recipient; SP132 donor | |
| 2 | R | R | ST409 | tet(B), ctx-m-1, mdf(A) | gyrA S83L, rpsL K43R | gad | IncI1, IncHI2, IncHI2A | SP132 recipient; endogenous Tetr donor | ||
| 20 | 2 | S | S | ST165 | tet(A), ctx-m-1, mdf(A) | No mutations | gad | IncI1 | Endogenous Tetr recipient; SP132 donor | |
n = number of transconjugants.
Cip, ciprofloxacin; Str, streptomycin; S, sensitive; R, resistant according to EUCAST breakpoints.
MLST is reported according to the Achtman scheme. Identity to the reference is given when <100%.
DISCUSSION
By subjecting isogenic pairs of bacteria to competition in a mallard model for oral colonization, we showed that selection for ESBL-producing E. coli mutants with reduced susceptibility to ciprofloxacin occurs in the gut of the birds when they are exposed to sub-MICs of ciprofloxacin in their water environment. A ciprofloxacin concentration of 1 µg/liter in the drinking/swimming water conferred increased selection of the least costly mutation, gyrA S83L, in the gut of the birds. The MIC of the corresponding parental strain was 16 µg/liter, and thus, selection was observed when the drinking water contained ciprofloxacin concentrations >10-fold below the MIC. However, this was the lowest concentration tested and resulted in a significant positive selection of the gyrA mutant, so the MCS is likely to be even lower. Previous in vitro studies have shown selection for GyrA S83L at substantially lower ciprofloxacin concentrations, i.e., 1/230 of the MIC of susceptible cells, corresponding to 0.1 µg/liter, in accordance with what we found here for in vitro competitions (4). Since the aim of this study was to investigate at what concentrations of contaminating ciprofloxacin in the water environments less susceptible mutants were selected, we measured only the concentration of ciprofloxacin in the water and not in the birds. Therefore, the MSC is based on the water concentration, and the concentration of ciprofloxacin in the bird intestine is likely much lower than that in the drinking water if pharmacokinetics similar to those in humans apply (24). In addition, it is well established that fluoroquinolones bind strongly to particulate matter (e.g., fecal matter), further complicating determination of the absolute selective concentrations of the drug (25). Nonetheless, since fluoroquinolone concentrations exceeding the MSC found here have been repeatedly shown in hospital wastewater and wastewater treatment plant (WWTP) influent (1 to 80 µg/liter) (10), there is a clear risk that selection in water-dwelling birds occurs in environments that receive these effluents.
Interestingly, the GyrA S83L substitution was weakly selected for in vivo even in the absence of antibiotics, although it conferred a small fitness reduction in vitro. This particular mutation is the most commonly detected mutation entailing clinical resistance to ciprofloxacin and has also been frequently found in the environment regardless of the presence of fluoroquinolones (22, 26). Similar to our findings, a GyrA T86I substitution increased the fitness of Campylobacter jejuni in chickens without antibiotic exposure, although the effect was dependent on the genetic background (27), and GyrA S91F in Neisseria gonorrhoeae was found to increase bacterial fitness in a mouse genital tract infection experiment, even though there was a fitness cost in vitro (28). Thus, it has now been demonstrated in three different animal models and bacterial species that specific gyrA mutations may increase in vivo fitness, which may well maintain fluoroquinolone resistance even in the absence of antibiotics and serve as a basis for selection of further increase in resistance upon continued antibiotic exposure.
Several other studies of the fate of inoculated resistant bacteria, or changes in preexisting gut flora, in response to antibiotic treatment in different animal models have been performed (mice, rats, zebrafish, calves, and piglets) (29–35). Most studies used therapeutic antibiotic concentrations, but some included enrichment of resistant bacteria in response to low antibiotic concentrations. For example, zebrafish were experimentally infected by isogenic pairs of resistant and susceptible Staphylococcus aureus or Pseudomonas aeruginosa strains and exposed to subcurative concentrations of tetracycline, oxacillin, or erythromycin, which led to enrichment of the resistant strain (32). Enrichment for resistant E. coli was found to occur when calves were fed milk containing sub-MICs of tetracycline (0.3 mg/liter), ceftiofur (0.1 mg/liter), and ampicillin (0.01 mg/liter), corresponding to concentrations 53-, 80-, and 3,200-fold below the respective clinical breakpoint for resistant E. coli (33). Exposing piglets to ceftiofur or enrofloxacin at concentrations 1/10 of the therapeutic dose led to a significant enrichment of ceftiofur- and enrofloxacin-resistant E. coli in the feces, in comparison to both the control group and the group exposed to therapeutic doses (34). A study on rats showed that orally administered subtherapeutic doses of enrofloxacin (1/10, 1/100, and 1/1,000 the therapeutic dose) select for less susceptible E. coli strains without target mutations and E. coli strains with de novo target mutations in gyrA and parC (35). In contrast to our findings, chickens exposed to therapeutic or subtherapeutic concentrations (75% and 2.5% of the therapeutic dose) of enrofloxacin did not result in an increase of enrofloxacin-resistant E. coli in feces (36). However, most of these studies used antibiotic concentrations relative to the clinical dosing and not relative to the MIC of susceptible bacteria, and none of these studies estimated the MSC, making it hard to set breakpoints regarding safe concentrations below which selection of resistance does not occur. Thus, there is a need for further studies focusing on the effects of sub-MIC levels of antibiotic to determine the MSCs for various antibiotics in natural settings.
There are some important limitations in our study. Despite the initial high inoculum, CFU counts were very low at the end of the experiment. The E. coli strains used in our study were originally isolated from yellow-legged gulls and are probably less well adapted to growth in the mallard gut, even though we showed previously that the original strains could colonize mallards for at least 1 month (37). Thus, low numbers of recovered bacteria and short recovery times negatively affect the accuracy of the determination of the strength of selection at different drug concentrations. Furthermore, the birds in each ciprofloxacin concentration group roamed freely in the same room and shared the same drinking/swimming pool and, thus, shared bacteria among themselves as they do in the wild. As we have to assume that selection for the mutant strains is the result of the enrichment in the group of birds as a whole, each group was considered only one biological replicate. This led to decreased power and more uncertainty in the measured selection coefficients. Future experiments should consider using several separate groups of birds. It would also be interesting to determine the concentration of ciprofloxacin in the guts of the birds to get a better understanding of the pharmacokinetics of the drug when administered through the water environment.
Our results show that conjugation of plasmids readily takes place between E. coli strains in the guts of mallards. At least 8 individual transfer events occurred during the experiments, most of which were identified at the beginning of the experiment, when bacterial counts in the intestines were the highest. However, the frequency of plasmid transfer was not high enough to reliably draw any conclusions regarding the potential impact of ciprofloxacin exposure on conjugation. Other studies have found that antibiotic exposure enhances the opportunities for conjugative transfer in gnotobiotic rats (38, 39) and that the dissemination of blaCTX-M-bearing plasmids increased in the intestines of piglets exposed to subtherapeutic concentrations of ceftiofur or enrofloxacin (34). Furthermore, the conjugation frequency of a multidrug resistance plasmid from E. coli to enteric bacteria present in sludge from wastewater treatment plants was increased during exposure to sub-MIC tetracycline concentrations (40). These differences could very well reflect variation in the response to antibiotic exposure of different plasmids.
Fluoroquinolones are highly stable molecules and end up in the environment (e.g., soil and water) as a result of human and animal treatment as well as from wastewater from the manufacturing industry (41–45). Since fluoroquinolones are frequently used antibiotics worldwide, this steady-state pollution with stable antibiotics leads to substantial contamination of surface waters. In relation to this, our finding that concentrations of ciprofloxacin as low as 1 µg/liter can selectively enrich the most commonly found clinical mutations with reduced susceptibility in water-dwelling birds highlights the need for efforts to reduce the contaminating concentrations in water environments at least below this concentration, but preferably substantially further. Mallards are found in both fresh- and salt-water wetlands, including parks, ponds, rivers, lakes, shallow inlets, and open sea near the coastline (18), and they are attracted to bodies of water with aquatic vegetation (46). Ciprofloxacin in the environment binds to soil, water sediment, and fecal matter and is absorbed and accumulated in plants (10, 47). The binding limits its bioavailability (10) but might also lead to high ciprofloxacin exposure in mallards that graze plants and seek food in the ground sediments of water. It is therefore reasonable to believe that wild mallards are frequently exposed to concentrations shown to be selective in this study. Very high frequencies of resistant bacteria have also been found in mallard flocks (19). Although the majority of multidrug-resistant bacteria detected in wild birds are likely a result of direct environmental pollution by human feces (48), our results imply that selection of antibiotic-resistant bacteria could occur in wildlife. Spread of resistance under such selective conditions may be a route for novel resistance combinations to occur and perhaps find their way back to humans. Our results underline the importance of reducing overall selective pressure of antibiotics and implementing methods to reduce environmental pollution with antibiotics.
MATERIALS AND METHODS
Bacterial strains.
The extended-spectrum-β-lactamase (ESBL)-producing E. coli strains used in the experiment (SP132 and SP163) were originally isolated from yellow-legged gulls (Larus michaelis) in Spain in 2009 (49). The strains had been subjected to whole-genome sequencing and phenotypically characterized (37) and were selected based on their wild-bird origin and different phenotypic traits that make them easily distinguishable from each other and the normal gut flora of the mallards (Table 1). Isogenic pairs of streptomycin-resistant mutants with different mutations yielding reduced susceptibility to ciprofloxacin were selected from SP132 by first selecting spontaneous streptomycin-resistant mutants on Luria-Bertani (LB) agar plates (LA) supplemented with 500 mg/liter of streptomycin. Streptomycin-resistant mutants were subsequently used to select spontaneous mutants with reduced susceptibility to ciprofloxacin on LA plates supplemented with 0.023 mg/liter of ciprofloxacin. Mutants were verified by whole-genome sequencing. The selection was performed twice, resulting in two isogenic pairs with different rpsL (Str) and gyrA (Cip) mutations. The pairs were named DA26187/DA26192 (GyrAwt/GyrAD87Y) and DA28261/DA28400 (GyrAwt/GyrAS83L).
For plasmid conjugation studies, strain SP163 containing a tetA gene on a conjugative IncF plasmid and blaCTX-M-1 on a conjugative IncK plasmid was cultured in LB broth for 3 days with daily passage to lose the plasmid conveying cefotaxime resistance. Four lineages were passaged at 1/100 dilutions daily in LB medium without selection. Dilutions were plated daily on nonselective medium, and around 200 colonies/lineage were replica plated on plates containing 10 mg/liter of cefotaxime. At day 3, several cefotaxime-sensitive colonies had appeared, and whole-genome sequencing identified the loss of the entire IncK plasmid compared to the parental strain. The strain retained the plasmid carrying the tetracycline resistance gene tet(A) and was named DA26200. All constructed strains were subjected to whole-genome sequencing to verify that no additional genetic changes were present.
In vitro competitions.
The isogenic pairs were subjected to competition by transferring 1 µl (approximately 106 CFU) of each of the respective strain from overnight cultures into 1 ml LB broth. The cultures were incubated at 37°C with shaking at 200 rpm. Each day, 1 µl of overnight culture was passaged to 1 ml of fresh medium, resulting in a 1,000-fold dilution of the bacteria, corresponding to approximately 10 generations of growth per cycle. To determine the ratio of the gyrA mutant to the wild-type strain, 10 µl of the culture was diluted 1,000-fold, and 100 µl was plated on LA plates supplemented with 500 mg/liter streptomycin and incubated overnight at 37°C. Individual colonies were screened by either replica plating or streaking 100 individual colonies to LA plates supplemented with 0.04 mg/liter ciprofloxacin and the ratio of growing versus nongrowing colonies was calculated. To determine the fitness cost of the gyrA mutations, the strains were subjected to competition without antibiotics, and for measurement of the minimal selective concentration, the strains were subjected to competition in the presence of different sub-MICs of ciprofloxacin. To control for potential selection in water, dilutions of overnight cultures of the respective isogenic pairs were inoculated in 20 ml sterile water to a final concentration of 104 CFU/ml for each strain. The concentrations of ciprofloxacin used in the mallard experiments were added to the water. The mixes were incubated at room temperature, and samples were obtained 24 h postinoculation and plated as described above.
Mallard model.
We previously used the mallard model in bacterial colonization experiments (37). The mallards were kept at the laboratory animal facility of the Swedish National Veterinary Institute in accordance with ethical guidelines and approvals from Uppsala Ethical Committee on Animal Experiments (approvals C201/11, C125/12, and C63/13). One-day-old mallards were purchased from a commercial breeder and housed indoors until they were included in the experiments at the age of 2 to 6 months and a mass of approximately 1 kg. They were fed nonmedicated, clean (but not sterilized) feed. Before inclusion in the experiments, all birds were screened for the presence of resistant E. coli by plating of feces on selective eosin–methylene blue–1% lactose (EMBL) plates to detect any E. coli resistant to cefotaxime (10 mg/liter), nalidixic acid (10 mg/liter), or streptomycin (50 mg/liter) that could interfere with detection of the experimental strains. The room used for the birds was approximately 12 m2, kept at room temperature (approximately 20°C), and contained a 1-m2 pool filled with 170 liters of water, which the mallards used for swimming and as their only drinking water source. For experiments with antibiotic selection, ciprofloxacin (Sigma Aldrich) was added to the pool by first dissolving the required amount (depending on experimental final concentration) in 50 ml of water and subsequently adding this stock to 5 liters of water, which was mixed with the pool water. The water was changed daily to avoid accumulation of bacteria in the water, and fresh ciprofloxacin was added to maintain the correct antibiotic concentration. The birds had continuous access to feed and a bedding of dust-free wood shavings and hay. At the end of each experiment, the mallards were euthanized with 100 mg/kg sodium pentobarbital intravenously (pentobarbital veterinary, 100 g/liter; Apotek Produktion & Laboratorier AB, Sweden).
Chemical analysis of water samples.
Water samples were taken from the pool in the experimental room to measure ciprofloxacin levels. An on-line solid-phase liquid extraction/liquid chromatography-tandem mass-spectrometry (SPE–LC-MS/MS) system, previously described in detail (50), was used to analyze the levels of ciprofloxacin in the water samples. Briefly, 1 ml of 10 ml prefiltered and acidified (0.1% of formic acid on a volume basis) samples was analyzed by the SPE–LC-MS/MS system. Samples were quantified using an internal standard method (deuterated ciprofloxacin was used as an internal standard) with four calibration points. The analyses confirmed the levels of ciprofloxacin; further results and discussion are given in the supplemental material.
In vivo competition in mallards.
(i) Bacterial inoculation. The suspensions of bacteria given to the mallards were prepared from separate concentrated overnight cultures in LB medium supplemented with 10 mg/liter of cefotaxime. First, the cells were pelleted and washed to get rid of medium and antibiotics and resuspended in phosphate-buffered saline (PBS). In order to determine the amounts of bacteria in the suspensions as well as the ratio of the bacterial strains in the mixed suspensions, a viable count with subsequent screening on appropriate selective plates was performed for every experiment as described below.
(ii) Experimental design. The GyrAwt/GyrAD87Y strain pair was used in four individual competitions in mallards, each with five inoculated birds and a concentration of ciprofloxacin in the water of either 0 µg/liter, 10 µg/liter, 20 µg/liter, or 32 µg/liter. Each mallard was inoculated intraesophageally with 1 ml of a 1:1 mix of DA26187 (GyrAwt) and DA26192 (GyrAD87Y) containing approximately 1010 CFU of each strain. Inoculation was performed using a blunt syringe that was inserted into the esophagus. The birds were then left to roam freely in the room. Fecal samples were obtained at 1, 3, 7, and 10 days postinoculation (dpi). The experiments were interrupted on day 10.
The GyrAwt/GyrAS83L strain pair was used in four individual competitions in mallards, each with six birds and a ciprofloxacin concentration of 0 µg/liter, 1 µg/liter, 5 µg/liter, or 20 µg/liter. Here, we inoculated three of the birds (index birds) intraesophageally with 1 ml of a 1:1 mix of DA28261 (GyrAwt) and DA28400 (GyrAS83L) containing approximately 1010 CFU of each strain and allowed the other birds in the room (transmission birds) to be inoculated by spread of the bacteria from the index birds through the natural fecal-oral route of transmission in a flock. We have previously shown that spread of bacteria between birds is very rapid under these conditions (37). After 4 h, the index birds were inoculated with a third, genetically different strain, DA26200 (cefotaxime susceptible, ciprofloxacin resistant, tetracycline resistant) containing an IncF tetracycline resistance plasmid to test for plasmid transfer between strains in the guts of the mallards. The index birds were euthanized 24 h after the inoculation, and only the three transmission birds remained and were sampled during the rest of the experiment. Fecal samples were obtained twice daily the first 4 days and once daily at 5, 7, and 10 dpi. The experiments were interrupted on day 10.
(iii) Bacterial sampling. Each bird was put in an individual, clean cardboard box and within a few minutes, feces were collected from the box using a sterile swab and stored in phosphate-buffered saline (PBS) with 20% dimethyl sulfoxide (DMSO) as freeze protectant. On the few occasions when the bird did not leave droppings in the box, a cloacal swab sample was obtained and treated in the same manner as the fecal samples. For analysis, 500 μl of each fecal sample was plated within 5 h after sampling, without enrichment, on EMBL plates supplemented with 500 mg/liter streptomycin, and the remaining sample was frozen at −80°C. The plates were incubated overnight at 37°C. Phenotypically distinct bacteria growing with big red-brown or green colonies on streptomycin containing EMBL plates were considered to be E. coli. 50 colonies from each streptomycin plate were plated on an EMBL plate containing 0.04 mg/liter ciprofloxacin, and the ratio of the GyrAmut and GyrAwt strains was calculated. The ratio was calculated for all colonies combined from each group and day. On one occasion, there was no GyrAmut detected in one group, and to allow ratio calculation, 0 CFU was changed to 1 CFU. The mean ratio of GyrAmut to GyrAwt strains in each group was plotted as a function of the number of days of growth at each concentration of antibiotic. In the GyrAwt/GyrAS83L experiment, three of the birds excreted streptomycin-resistant E. coli in their feces prior to the experiment. In contrast to the experimental strains, the endogenous streptomycin-resistant E. coli strains were not cefotaxime resistant and could therefore be excluded from the data by supplementing all EMBL plates with 10 mg/liter cefotaxime.
Transconjugant screening.
In the GyrAwt/GyrAS83L experiment, samples were also plated on EMBL plates containing 10 mg/liter tetracycline plus 10 mg/liter cefotaxime to detect transconjugants. Putative transconjugants were verified by streaking individual colonies on EMBL plates supplemented with 10 mg/liter tetracycline plus 10 mg/liter cefotaxime, on EMBL plates supplemented with 0,04 mg/liter ciprofloxacin, and on EMBL plates supplemented with 50 mg/liter streptomycin to distinguish SP132 derivatives. Selected transconjugants were screened by PCR targeting tet(A) (primers forward [5′-CAGTGCTCAGAATTACGATC-3′] and reverse [CGGTCCTTCAACGTTCCTG]), the ESBL plasmid from SP132 (primers forward1 [CAACAGTAGGCATCGACATG], reverse1 [CCTTCTTGATGACCTTCTAC], forward2 [ATC-GCCTTAGACGGCAAAAG], and reverse2 [GCAACAGCTACTTGTCCAAC]), and the tetracycline resistance plasmid from SP163 (primers forward [TACAGTCGTCCGAAAGTCAC] and reverse [CAGTTAACT-AGCGCTCTGAGM]). A set of transconjugants displaying different resistance profiles/genetic profiles and coming from different experimental runs were further analyzed by whole-genome sequencing to precisely define the donor/recipient combination.
Whole-genome sequencing.
Total bacterial DNA was prepared with the Epicentre MasterPure DNA purification kit (Illumina) according to the manufacturer’s instructions. Whole-genome sequencing was performed on an Illumina MiSeq using the Nextera XT library prep kit (Illumina) and sequenced with a paired-end read length of 100 bp. Sequences were analyzed with CLC Genomics Workbench v12 (Qiagen) by de novo assembly, and contigs were analyzed for resistance genes, plasmid replicons, and virulence genes by the ResFinder (51), PlasmidFinder (52), and VirulenceFinder (53) applications. Multilocus sequence typing was performed from de novo contigs using the CLC Microbial Genomics module.
Data availability.
The raw sequencing reads have been deposited at NCBI under BioProject number PRJNA645797, under which all sequence files can be found.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants to L.S. from the Swedish Research Council Formas (2016-00642), to D.I.A. from the Swedish Research Council (2017-01527) and the European Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) project Selection and Transmission of Antimicrobial Resistance in Complex Systems (STARCS), and to J.D.J. from the Swedish Research Council (2016-02606). The funders had no part in the design of the study or interpretation of the data.
We declare no competing interests.
REFERENCES
- 1.Pitout JD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 2.Canton R, Morosini MI. 2011. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev 35:977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 3.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5:e01918-14. doi: 10.1128/mBio.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gullberg E, Cao S, Berg OG, Ilback C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu A, Fong A, Becket E, Yuan J, Tamae C, Medrano L, Maiz M, Wahba C, Lee C, Lee K, Tran KP, Yang H, Hoffman RM, Salih A, Miller JH. 2011. Selective advantage of resistant strains at trace levels of antibiotics: a simple and ultrasensitive color test for detection of antibiotics and genotoxic agents. Antimicrob Agents Chemother 55:1204–1210. doi: 10.1128/AAC.01182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wistrand-Yuen E, Knopp M, Hjort K, Koskiniemi S, Berg OG, Andersson DI. 2018. Evolution of high-level resistance during low-level antibiotic exposure. Nat Commun 9:1599. doi: 10.1038/s41467-018-04059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grenni P, Ancona V, Caracciolo A. 2018. Ecological effects of antibiotics on natural ecosystems: a review. Microchemical J 136:25–39. doi: 10.1016/j.microc.2017.02.006. [DOI] [Google Scholar]
- 8.Lin JS, Pan HY, Liu SM, Lai HT. 2010. Effects of light and microbial activity on the degradation of two fluoroquinolone antibiotics in pond water and sediment. J Environ Sci Health B 45:456–465. doi: 10.1080/03601231003800222. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho IT, Santos L. 2016. Antibiotics in the aquatic environments: a review of the European scenario. Environ Int 94:736–757. doi: 10.1016/j.envint.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Janecko N, Pokludova L, Blahova J, Svobodova Z, Literak I. 2016. Implications of fluoroquinolone contamination for the aquatic environment—a review. Environ Toxicol Chem 35:2647–2656. doi: 10.1002/etc.3552. [DOI] [PubMed] [Google Scholar]
- 11.Guenther S, Ewers C, Wieler LH. 2011. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol 2:246. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atterby C, Borjesson S, Ny S, Jarhult JD, Byfors S, Bonnedahl J. 2017. ESBL-producing Escherichia coli in Swedish gulls—a case of environmental pollution from humans? PLoS One 12:e0190380. doi: 10.1371/journal.pone.0190380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atterby C, Ramey AM, Hall GG, Jarhult J, Borjesson S, Bonnedahl J. 2016. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect Ecol Epidemiol 6:32334. doi: 10.3402/iee.v6.32334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolejska M, Papagiannitsis CC. 2018. Plasmid-mediated resistance is going wild. Plasmid 99:99–111. doi: 10.1016/j.plasmid.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 17.Lopatkin AJ, Huang S, Smith RP, Srimani JK, Sysoeva TA, Bewick S, Karig DK, You L. 2016. Antibiotics as a selective driver for conjugation dynamics. Nat Microbiol 1:16044. doi: 10.1038/nmicrobiol.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton M, Burton R. 2002. International wildlife encyclopedia: leopard - marten. Marshall Cavendish, Singapore. [Google Scholar]
- 19.Hessman J, Atterby C, Olsen B, Jarhult JD. 2018. High prevalence and temporal variation of extended spectrum beta-lactamase-producing bacteria in urban Swedish mallards. Microb Drug Resist 24:822–829. doi: 10.1089/mdr.2017.0263. [DOI] [PubMed] [Google Scholar]
- 20.Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, Bzoma S, Cizek A. 2010. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microbiol 76:8126–8134. doi: 10.1128/AEM.01446-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tausova D, Dolejska M, Cizek A, Hanusova L, Hrusakova J, Svoboda O, Camlik G, Literak I. 2012. Escherichia coli with extended-spectrum beta-lactamase and plasmid-mediated quinolone resistance genes in great cormorants and mallards in Central Europe. J Antimicrob Chemother 67:1103–1107. doi: 10.1093/jac/dks017. [DOI] [PubMed] [Google Scholar]
- 22.Everett MJ, Jin YF, Ricci V, Piddock LJ. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother 40:2380–2386. doi: 10.1128/AAC.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes D, Andersson DI. 2012. Selection of resistance at lethal and non-lethal antibiotic concentrations. Curr Opin Microbiol 15:555–560. doi: 10.1016/j.mib.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Drusano GL, Standiford HC, Plaisance K, Forrest A, Leslie J, Caldwell J. 1986. Absolute oral bioavailability of ciprofloxacin. Antimicrob Agents Chemother 30:444–446. doi: 10.1128/aac.30.3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riaz L, Mahmood T, Khalid A, Rashid A, Ahmed Siddique MB, Kamal A, Coyne MS. 2018. Fluoroquinolones (FQs) in the environment: a review on their abundance, sorption and toxicity in soil. Chemosphere 191:704–720. doi: 10.1016/j.chemosphere.2017.10.092. [DOI] [PubMed] [Google Scholar]
- 26.Johnning A, Kristiansson E, Fick J, Weijdegård B, Larsson DGJ. 2015. Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Front Microbiol 6:1355. doi: 10.3389/fmicb.2015.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci U S A 102:541–546. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunz AN, Begum AA, Wu H, D'Ambrozio JA, Robinson JM, Shafer WM, Bash MC, Jerse AE. 2012. Impact of fluoroquinolone resistance mutations on gonococcal fitness and in vivo selection for compensatory mutations. J Infect Dis 205:1821–1829. doi: 10.1093/infdis/jis277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertz FB, Lobner-Olesen A, Frimodt-Moller N. 2014. Antibiotic selection of Escherichia coli sequence type 131 in a mouse intestinal colonization model. Antimicrob Agents Chemother 58:6139–6144. doi: 10.1128/AAC.03021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakobsen L, Cattoir V, Jensen KS, Hammerum AM, Nordmann P, Frimodt-Moller N. 2012. Impact of low-level fluoroquinolone resistance genes qnrA1, qnrB19 and qnrS1 on ciprofloxacin treatment of isogenic Escherichia coli strains in a murine urinary tract infection model. J Antimicrob Chemother 67:2438–2444. doi: 10.1093/jac/dks224. [DOI] [PubMed] [Google Scholar]
- 31.Allou N, Cambau E, Massias L, Chau F, Fantin B. 2009. Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob Agents Chemother 53:4292–4297. doi: 10.1128/AAC.01664-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McVicker G, Prajsnar TK, Williams A, Wagner NL, Boots M, Renshaw SA, Foster SJ. 2014. Clonal expansion during Staphylococcus aureus infection dynamics reveals the effect of antibiotic intervention. PLoS Pathog 10:e1003959. doi: 10.1371/journal.ppat.1003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira RV, Siler JD, Bicalho RC, Warnick LD. 2014. In vivo selection of resistant E. coli after ingestion of milk with added drug residues. PLoS One 9:e115223. doi: 10.1371/journal.pone.0115223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin D, Chen K, Xie M, Ye L, Chan EW, Chen S. 2017. Effect of ceftiofur and enrofloxacin on E. coli sub-population in pig gastrointestinal tract. J Glob Antimicrob Resist 10:126–130. doi: 10.1016/j.jgar.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Lin D, Chen K, Li R, Liu L, Guo J, Yao W, Chen S. 2014. Selection of target mutation in rat gastrointestinal tract E. coli by minute dosage of enrofloxacin. Front Microbiol 5:468. doi: 10.3389/fmicb.2014.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Horst MA, Fabri TH, Schuurmans JM, Koenders BB, Brul S, ter Kuile BH. 2013. Effects of therapeutical and reduced levels of antibiotics on the fraction of antibiotic-resistant strains of Escherichia coli in the chicken gut. Foodborne Pathog Dis 10:55–61. doi: 10.1089/fpd.2012.1217. [DOI] [PubMed] [Google Scholar]
- 37.Sandegren L, Stedt J, Lustig U, Bonnedahl J, Andersson DI, Jarhult JD. 2018. Long-term carriage and rapid transmission of extended spectrum beta-lactamase-producing E. coli within a flock of mallards in the absence of antibiotic selection. Environ Microbiol Rep 10:576–582. doi: 10.1111/1758-2229.12681. [DOI] [PubMed] [Google Scholar]
- 38.Bahl MI, Sorensen SJ, Hansen LH, Licht TR. 2004. Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl Environ Microbiol 70:758–764. doi: 10.1128/aem.70.2.758-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feld L, Schjorring S, Hammer K, Licht TR, Danielsen M, Krogfelt K, Wilcks A. 2008. Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J Antimicrob Chemother 61:845–852. doi: 10.1093/jac/dkn033. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Yun Z, Ha UH, Lee S, Park H, Kwon EE, Cho Y, Choung S, Oh J, Medriano CA, Chandran K. 2014. Transfer of antibiotic resistance plasmids in pure and activated sludge cultures in the presence of environmentally representative micro-contaminant concentrations. Sci Total Environ 468–469:813–820. doi: 10.1016/j.scitotenv.2013.08.100. [DOI] [PubMed] [Google Scholar]
- 41.Al-Ahmad A, Daschner FD, Kummerer K. 1999. Biodegradability of cefotiam, ciprofloxacin, meropenem, penicillin G, and sulfamethoxazole and inhibition of waste water bacteria. Arch Environ Contam Toxicol 37:158–163. doi: 10.1007/s002449900501. [DOI] [PubMed] [Google Scholar]
- 42.Fick J, Soderstrom H, Lindberg RH, Phan C, Tysklind M, Larsson DG. 2009. Contamination of surface, ground, and drinking water from pharmaceutical production. Environ Toxicol Chem 28:2522–2527. doi: 10.1897/09-073.1. [DOI] [PubMed] [Google Scholar]
- 43.Halling-Sorensen B, Lutzhoft H, Andersen HR, Ingerslev F. 2000. Environmental risk assessment of antibiotics: comparison of mecillinam, trimethoprim and ciprofloxacin. J Antimicrob Chemother 46(Suppl A):53–58. doi: 10.1093/jac/46.suppl_1.53. [DOI] [PubMed] [Google Scholar]
- 44.Kummerer K, Al-Ahmad A, Mersch-Sundermann V. 2000. Biodegradability of some antibiotics, elimination of the genotoxicity and affection of wastewater bacteria in a simple test. Chemosphere 40:701–710. doi: 10.1016/S0045-6535(99)00439-7. [DOI] [PubMed] [Google Scholar]
- 45.Rutgersson C, Fick J, Marathe N, Kristiansson E, Janzon A, Angelin M, Johansson A, Shouche Y, Flach CF, Larsson DG. 2014. Fluoroquinolones and qnr genes in sediment, water, soil, and human fecal flora in an environment polluted by manufacturing discharges. Environ Sci Technol 48:7825–7832. doi: 10.1021/es501452a. [DOI] [PubMed] [Google Scholar]
- 46.Cramp S. 1994. Handbook of the birds of Europe, the Middle East and North Africa: the birds of the Western Palearctic, vol 9. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 47.Lillenberg M, Litvin SV, Nei L, Roasto M, Sepp K. 2010. Enrofloxacin and ciprofloxacin uptake by plants from soil. Agronomy Res 8:807–814. [Google Scholar]
- 48.Karkman A, Parnanen K, Larsson DGJ. 2019. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat Commun 10:80. doi: 10.1038/s41467-018-07992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stedt J, Bonnedahl J, Hernandez J, Waldenstrom J, McMahon BJ, Tolf C, Olsen B, Drobni M. 2015. Carriage of CTX-M type extended spectrum beta-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand 57:74. doi: 10.1186/s13028-015-0166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan GA, Lindberg R, Grabic R, Fick J. 2012. The development and application of a system for simultaneously determining anti-infectives and nasal decongestants using on-line solid-phase extraction and liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal 66:24–32. doi: 10.1016/j.jpba.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing reads have been deposited at NCBI under BioProject number PRJNA645797, under which all sequence files can be found.