Significance
The global distribution of different Mtb lineages is very structured, with a few lineages that are globally widespread, whereas many others are restricted to particular geographic regions. Although local adaptation has been a favored model for explaining the sympatric relationship between the bacteria and host in these geographic regions, this has been difficult to study. Human migration has led to intermixing of ethnicities and has disturbed the bacterial population structure, thereby masking the putative genetic determinants that may have evolved from selection pressures in a given region. Here, by analyzing the genomes of hundreds of Mtb strains sampled from the relatively isolated population of Tibetan, we provide genetic evidence that Mtb can evolve to adapt to local populations and environments.
Keywords: Mycobacterium tuberculosis, evolution, local adaptation
Abstract
During its global dispersal, Mycobacterium tuberculosis (Mtb) has encountered varied geographic environments and host populations. Although local adaptation seems to be a plausible model for describing long-term host–pathogen interactions, genetic evidence for this model is lacking. Here, we analyzed 576 whole-genome sequences of Mtb strains sampled from different regions of high-altitude Tibet. Our results show that, after sequential introduction of a few ancestral strains, the Tibetan Mtb population diversified locally while maintaining strict separation from the Mtb populations on the lower altitude plain regions of China. The current population structure and estimated past population dynamics suggest that the modern Beijing sublineage strains, which expanded over most of China and other global regions, did not show an expansion advantage in Tibet. The mutations in the Tibetan strains showed a higher proportion of A > G/T > C transitions than strains from the plain regions, and genes encoding DNA repair enzymes showed evidence of positive selection. Moreover, the long-term Tibetan exclusive selection for truncating mutations in the thiol-oxidoreductase encoding sseA gene suggests that Mtb was subjected to local selective pressures associated with oxidative stress. Collectively, the population genomics of Mtb strains in the relatively isolated population of Tibet provides genetic evidence that Mtb has adapted to local environments.
The Mycobacterium tuberculosis (Mtb) strains circulating today are thought to have evolved from a common ancestral strain that originated in East Africa (1). During its out-of-Africa migration and global dispersal, Mtb encountered diverse natural environments, each with its own host population (1–4). Studies of the global distribution of Mtb show that there are a few lineages with widespread distribution but many others that are restricted to particular geographic regions (5, 6). This has led to the speculation that the prolonged interaction between the bacteria and host in specific geographic areas has allowed sublineages to evolve and become specialized at causing disease in their local human populations (2, 4, 7). One hypothesis proposed that different sublineages might have different T cell epitopes, but genome-wide purifying selection and the hyperconservation of T cell epitopes in Mtb populations argued against this possibility (8, 9). Though genomic studies have demonstrated that positive selection shaped the evolution of the Mtb population (2, 10, 11), and it has been postulated that geographically restricted Mtb populations have adapted to the local human hosts, there is a lack of genetic evidence to support the concept of local Mtb evolution (2, 12). While it seems plausible that Mtb sublineages could have evolved by genetic selection to optimally exploit the genetic composition of the local host population or adapt to the local environment, this model has yet to be proven.
Local selection is hard to demonstrate, in part because human migration has caused intermixing of ethnicities and disturbed the sympatric pattern between the bacterial population and its host populations (5, 13, 14), thereby masking the putative genetic determinants that evolved in response to the selection pressures in a particular host population. It therefore seemed that the best chance of finding evidence of local Mtb evolutionary selection would be in regions such as Tibet, where the population has remained relatively isolated. The Tibetan Plateau appears to have been inhabited for ∼25,000 y, with permanent settlements established at elevations of 3,500 to 4,500 m above sea level (15–17). Human habitation in the region was impeded by the physiological constraints of cold stress and hypoxia, which led to strong positive selective pressure on the hypoxia-inducible factor oxygen-signaling pathway of Tibetan highlanders (18). While other populations inhabiting high altitudes have adapted to a lower oxygen tension with high hemoglobin concentrations, the Tibetans have normal hemoglobin concentrations, but their hemoglobin has a higher affinity for oxygen (19). Tibetans are also able to inhale a greater volume of air with each breath and breathe more rapidly than either sea level dwellers or other high-altitude populations (e.g., Andeans) (20). These adaptive features have been associated with mutations in a transcription factor termed the endothelial Per-Arnt-Sim domain protein 1 (21), which is involved in the response to hypoxia. As a result, Tibetans are less susceptible to chronic mountain sickness than other populations living at high altitudes (22). In addition to low-oxygen adaptation, polymorphisms in the SP110 and PMP22 genes that are associated with increased tuberculosis (TB) risk have been described in a Tibetan population (23), as have polymorphisms in the HLA-DQB1 locus of the major histocompatibility complex (24).
The Tibetan population has a high TB burden, with an incidence up to 10 times the average in China (25, 26), and we wondered whether the Mtb strains circulating in Tibet might constitute a bacterial population that is distinct from the bacterial populations circulating in the plain regions of China. We reasoned that the extreme natural environment, particularly the reduced oxygen tension on the Tibetan Plateau, might shape the within-host environments encountered by Mtb. It is also possible that there are characteristics of the immune composition of the isolated Tibetan population, mentioned above, that could also have exerted selective pressures on Tibetan Mtb strains. To look for evidence of these selective pressures, we explored the population genetics of the Mtb strains circulating on the Tibetan Plateau. We found that several Mtb strains were introduced into Tibet and then diversified locally as these strains became endemic. The genomic evolution of these Tibetan Mtb strains included a preference for A > G/T > C mutations and a long-term selection for the loss of the thiol oxidoreductase encoded by sseA, which we believe are evidence that the Mtb strains in Tibet adapted to the local population and environment.
Results
Population Structure of Tibetan Mtb.
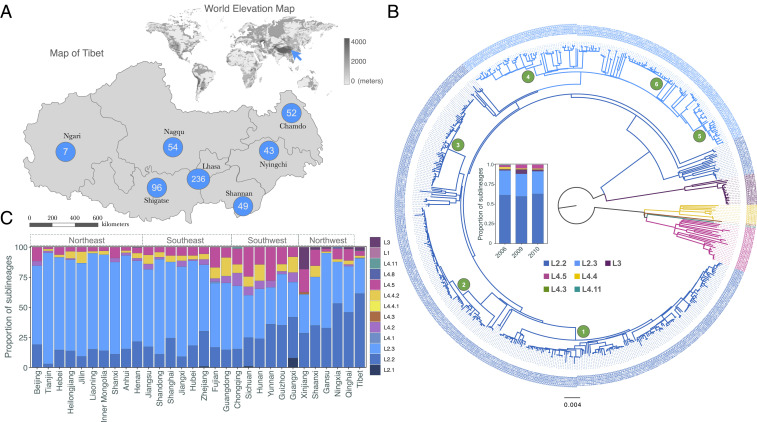
To characterize the population structure of the Mtb strains that are circulating in Tibet, a total of 576 Mtb isolates were sampled from seven municipal regions of Tibet in 2006, 2009, and 2010. Among these seven municipal regions, the provincial capital, Lhasa, contributed the most isolates (236 isolates), Ngari the fewest (7 isolates), and the other five regions contributed from 43 to 96 isolates (Fig. 1A). All 576 isolates were whole-genome sequenced, with an average sequencing depth of 53.1x (16.1x to 105.4x) and a mean genome coverage of 97.5% (91.2% to 98.6%). A maximum likelihood phylogenetic tree was constructed for the Tibetan Mtb strains (Fig. 1B), and barcode-SNP–based genotyping showed that 91% (514/576) belonged to Lineage 2 (L2), while only 6.3% (36/576) belonged to Lineage 4 and 2.8% (16/576) to Lineage 3. Among the L2 strains, 67.7% (355/524) belonged, unexpectedly, to the ancient Beijing sublineage (L2.2), while strains of the modern Beijing sublineage (L2.3) were the minority (32.3%; 169/524). This distribution is the opposite of what is found in most areas of China, where L2.3 strains expanded markedly over the past few hundred y (14). The predominance of L2.2 was not the result of sampling bias because it was consistent in the strains collected in 2006, 2009, and 2010 (Fig. 1B). Among all 30 Chinese provinces with Mtb population genotyped (14), Tibet has the highest prevalence of L2.2 strains (Fig. 1C).
Fig. 1.
Genetic structure of Tibetan Mtb population. (A) The numbers of Mtb isolates that were sampled from different municipal regions in Tibet; the blue arrow in the world elevation map refers to the location of the Tibetan Plateau. (B) A maximum likelihood phylogenetic tree of 576 Tibetan Mtb strains; the bar plot in the middle showed the constitutions of different sublineages for each of the 3 y. The branches of the tree were colored according to the sublineages, and the annotated numbers (1–6) refer to the Tibetan clades in Fig. 2. (C) Population structure of Mtb strains in 30 provinces of China; the data for other provinces were from a previous countrywide genotyping study (14).
Tibetan Indigenous Clades.
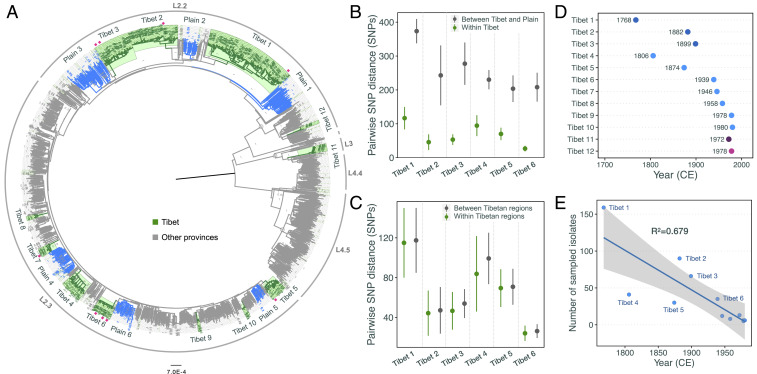
To trace the evolutionary history of Tibetan Mtb strains, we reconstructed a second maximum likelihood phylogenetic tree that included an additional 1,159 Mtb isolates collected from other provinces in China (Fig. 2A). This tree showed that the Tibetan strains nested within the Chinese Mtb population but were clustered in clades with restricted diversification (highlighted clades in Fig. 2A) that contained few strains from outside Tibet. This geographic restriction suggests that the clades diversified locally after their ancestral strains were introduced into the region. We defined 12 “Tibet clades,” each containing 6 to 159 isolates, that together accounted for 83.2% (479/576) of the total Tibetan Mtb strains. Six of these clades showed deeper evolutionary roots and contained more isolates, suggesting they were introduced into the region earlier than the other clades.
Fig. 2.
Local diversification and origin times of Tibetan Mtb strains. (A) A maximum likelihood phylogenetic tree of 576 Tibetan strains together with 1,159 plain Mtb strains; Tibetan strains are highlighted in green and plain strains in gray. The clades colored in blue represent the plain clades that were selected for comparison with Tibet clades. A total of 12 Tibetan-specific clades are marked, and the pink diamonds indicate the strains that were sampled from plain regions but nested within Tibetan clades. The outer circle marks the global lineages/sublineages. The numbers at each branching site indicate the bootstrap values. (B) Pairwise SNP distances between strains of Tibetan clades and the relative plain clades (gray), and pairwise SNP distances between strains within each Tibetan clade. (C) Pairwise SNP distance between strains from each Tibet municipal region and between different municipal regions. (D) Estimated origin time (median value) for the ancestor strains of each Tibetan clade. (E) A dot plot showing the correlation between origin time and current population size; the gray shading indicates the 95% CI of the linear regression.
To explore the differences between Mtb strains in the Tibet clades and strains from other provinces, we defined six paired “plains clades” from the strains that were isolated in other Chinese provinces (Fig. 2A). Although the plains clades we selected for comparison were from the phylogenetic branches closest to the Tibet clades, they were separated from the Tibet clades by relatively large pairwise SNP distances (Fig. 2B). In contrast, the Tibet strains were quite homogeneous. Within each Tibet clade, the SNP differences between strains isolated from different Tibetan regions were similar to the SNP differences between strains isolated from the same municipality (Fig. 2C). This suggests that while the local clades spread throughout Tibet, there was limited transmission of strains between Tibet and the plain regions, and therefore the indigenous Tibet clades could be appropriate for studying local Mtb evolution. We also calculated within-clade pairwise SNP distances for both Tibetan clades and plains clades and found that the Tibetan clades had larger SNP distances than their paired plains clades (SI Appendix, Fig. S1), suggesting that the Tibetan clades diversified more recently than the paired plains clades.
Absence of L2.3s Expansion Advantage.
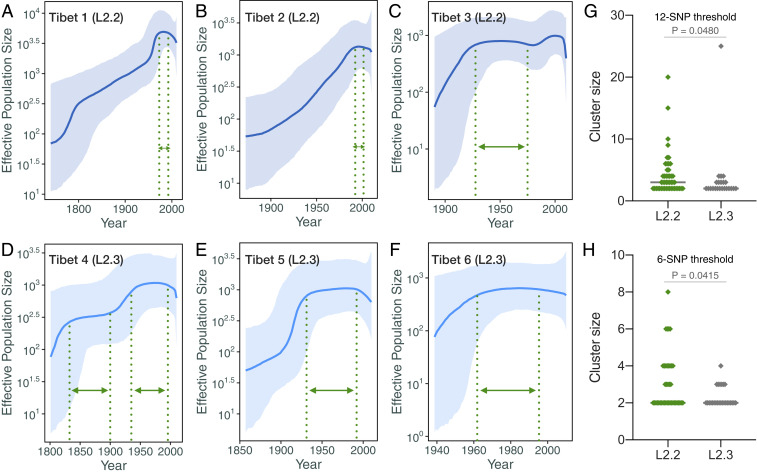
Bayesian-based coalescent analysis estimated that the first Mtb strain (clade Tibet 1) was introduced into Tibet in the second half of the eighteenth century, followed by four strains introduced in the nineteenth century (Fig. 2D) and more frequent introductions in the twentieth century (Fig. 2D). Regression analysis showed a linear correlation between the current size of the Tibetan clades and the time since they were introduced into the region (Fig. 2E). Surprisingly, clades Tibet 4 and Tibet 5, which belong to the L2.3 sublineage, were introduced earlier but had fewer isolates than L2.2 clades Tibet 2 and Tibet 3 (Fig. 2E). This is distinct from the population dynamics of L2.2 and L2.3 in most of China and the rest of the world, where L2.3 rapidly outcompeted L2.2 to become more prevalent (14). To compare the effective population size changes between the six Tibetan clades, we used Bayesian skyline plots, a method that uses a sample of molecular sequences to estimate the history of population dynamics over time (27). Bayesian skyline plots of the six major Tibet clades (Fig. 3 A–F) showed that clades Tibet 1 and Tibet 2 expanded continuously since they were introduced into the region and only entered a plateau phase in the 1980s. The expansions of the other clades were distinct. Tibet 4 and Tibet 5 expanded intermittently, with one or two long plateaus. Tibet 3 and Tibet 6 had similar patterns, but Tibet 3 had a sharper growth early after its introduction and a second expansion around the 1970s. A comparison of the size of transmission clusters showed that L2.2 strains had more large clusters than L2.3 strains using either 12-SNP or 6-SNP differences as the thresholds for defining clusters (Fig. 3 G and H; P = 0.0485 and P = 0.0415, respectively; Mann–Whitney U test), suggesting that in Tibet, L2.2 strains were more successful in causing ongoing transmission than L2.3 strains. The estimated expansion histories and cluster size comparisons are consistent with the population sizes of L2.2 and L2.3 in our sample and suggest that the relative expansion advantage of L2.3 over L2.2 that has been seen elsewhere was not operative in Tibet.
Fig. 3.
Bayesian skyline plots of six Tibetan clades. (A–F) Bayesian skyline plots for Tibet 1 to 6 clades; the color ribbons represent the 95% highest posterior density. The dashed green lines highlight the interval when Mtb population appeared to plateau. Effective population size (Ne) refers to the estimated number of breeding individuals in the given population. (G) Comparison of cluster size between clusters from L2.2 and L2.3 sublineages when using 12-SNP distance as the threshold. (H) Comparison of cluster size between clusters from L2.2 and L2.3 sublineages when using 6-SNP distance as the threshold. P values were given by Mann–Whitney U test.
A Mutagenesis Shift to A > G/T > C.
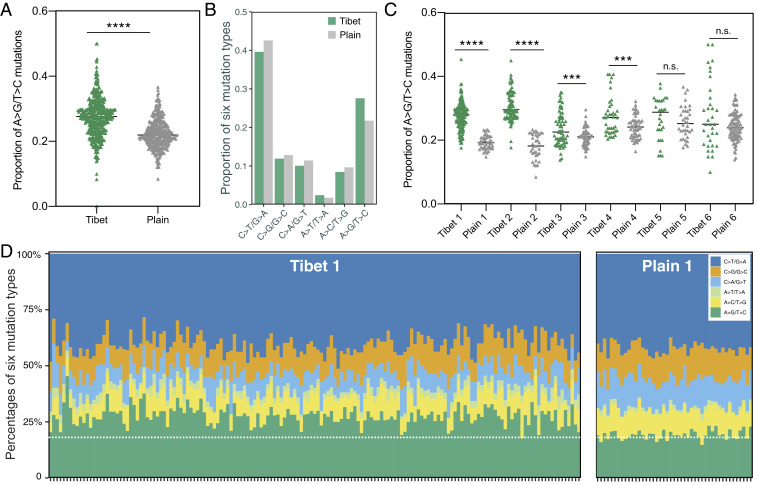
Host immune environments may modulate the mutagenesis patterns in Mtb (28), so we tested whether the mutations accumulated by the Tibetan strains might be different from those found in isolates from the Chinese plain regions. For each Tibetan clade, we considered only the mutations that occurred after the introduction of the common ancestral founder of the clade into Tibet. When these mutations from all of our Tibetan strains were grouped together and compared with the mutations from the plain strains, the Tibetan strains had a higher percentage of A > G/T > C mutations (27.6%, 95% CI: 17.0 to ∼28.1%) than the strains from the plain regions (21.8%, 95% CI: 21.3 to ∼22.3%) (P < 0.0001, Fig. 4A). All other mutation types were slightly decreased in the Tibetan strains, except for a slight increase of 1.8 to 2.4% for A > T/T > A (Fig. 4B and SI Appendix, Fig. S2). A comparison of individual Tibetan clades with their phylogenetically closest plain clades showed that four Tibetan clades (Tibet 1 to ∼4) consistently contained higher ratios of A > G/T > C mutations than the paired plains clades (Fig. 4C). This difference is also apparent when the A > G/T > C ratios of each single isolate in the Tibet 1 clade are compared with the isolates of the Plain 1 clade (Fig. 4D). For Tibet 5 and Tibet 6, however, although they contained more strains with higher proportions of A > G/T > C mutations, the total grouped mutations from these clades were not significantly different from those of their paired plain clades (Fig. 4C). Because nucleotide transitions can be biased by the evolution of drug resistance (29), we repeated the comparison either excluding the drug-resistant isolates or removing the mutations in drug resistance–associated genes and found that the Tibetan strains still had higher ratios of A > G/T > C mutations than the plain isolates (SI Appendix, Fig. S3). In addition, after removing the mutations from genes with evidence of positive selection (see the section Genetic Evidence of Local Selection below), the ratios of A > G/T > C mutations in the Tibetan strains were still higher than in the plain isolates (SI Appendix, Fig. S3).
Fig. 4.
Tibetan Mtb strains had higher ratio of A > G/T > C mutations. (A) Comparison of the ratio of A > G/T > C mutations between Tibetan strains and plain strains. (B) The proportion of six mutation types in all mutations from Tibet and plain strains, respectively. (C) Comparison of the ratio of A > G/T > C mutations between each Tibetan clade and the relative plain clade. (D) A bar plot showing the mutational composition for each individual strain from Tibet 1 and plain clades; the dashed white line indicates the mean level of A > G/T > C mutations in the plain group. ****P < 0.0001 and ***P < 0.001; n.s. refers to no significance (given by t test).
Genetic Evidence of Local Selection.
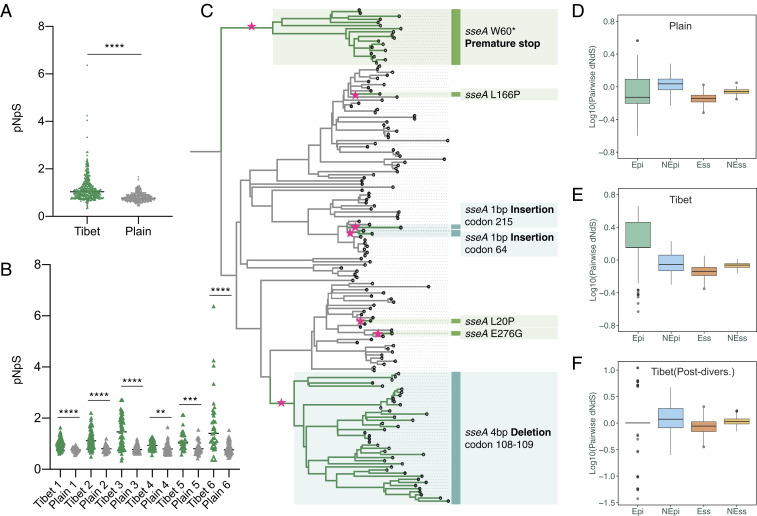
We looked for evidence that the Tibetan Mtb population had been subjected to a selection process by calculating the ratio of nonsynonymous to synonymous mutations (pNpS, similar to dN/dS) that were accumulated by each individual Mtb strain since the introduction of its clade ancestor into Tibet. We found the pNpS in the Tibetan Mtb population (1.16, 95% CI: 1.1 to ∼1.22) was overall higher than in the plain isolates (0.80, 95% CI: 0.78 to ∼0.82) (Fig. 5A). We then compared each Tibetan clade with its paired plains clade and again found that the Tibetan clades consistently had higher pNpS values (Fig. 5B). The large proportion of isolates with pNpS values greater than one suggested that the Tibetan Mtb genomes had been subject to a positive selection process.
Fig. 5.
Local adaptation of Tibetan Mtb strains. (A) Comparison of pNpS ratio for mutations accumulated by Tibetan and plain strains; each dot represents one Mtb isolate. (B) Comparison of pNpS ratio between each Tibetan clade and the relative plain clade. (C) A maximum likelihood phylogenetic tree for Tibet 1 clade with the mutational events of sseA highlighted. Each pink star represents where the relative mutation was accumulated. (D–F) Comparison of pairwise dNdS ratio of Epi, NEpi, Ess, and NEss; Tibet (Post-divers.) refers to the comparison that only considered the mutations accumulated after local diversification. ****P < 0.0001, ***P < 0.001, and ***P < 0.01; n.s. refers to no significance (given by t test).
We next sought to identify genes with evidence of positive selection in Tibet but not in the plain regions. We calculated both the pNpS and the dN/dS values for each individual gene based on the SNPs in the Tibetan strains and, separately, in the plain isolates. As expected, drug resistance–associated genes, such as katG, rpoB, rpoC, pncA, and embB, showed the strongest evidence of positive selection in strains from both Tibetan and the plains regions. We also found other genes with evidence of positive selection in both plains and Tibet strains (Table 1), including Rv1129c(prpR) and glpK, which have been associated with drug tolerance (30–32), and phoR, whiB6, and dnaA, which have been previously identified as targets of positive selection in other Mtb populations (30, 33).
Table 1.
Genes that were under selection in Tibet
| Gene | Plains | Tibet | pNpS (plains) | dN/dS (plains) | pNpS (Tibet) | dN/dS (Tibet) | Gene product | Categories | ||
| No. of NS* | No. of SY* | No. of NS | No. of SY | |||||||
| katG | 43 | 3 | 43 | 1 | 6.08 | 3.68 | 16.36 | 12.48 | Catalase-peroxidase-peroxynitritase T | Drug resistance |
| rpoB | 44 | 4 | 27 | 1 | 4.07 | 3.01 | 8.39 | 6.47 | RNA polymerase beta subunit | |
| rpoC | 42 | 10 | 24 | 1 | 2.12 | 1.65 | 11.91 | 13.82 | RNA polymerase beta' subunit | |
| embB | 29 | 7 | 23 | 0 | 2.08 | 1.75 | 10.69 | 6.86 | Arabinosylindolylacetylinositol synthase | |
| pncA | 47 | 0 | 22 | 0 | 25.92 | 78.82 | 11.99 | 28.84 | Pyrazinamidase/nicotinamidase | |
| Rv1129c | 13 | 1 | 15 | 0 | 6.64 | 3.71 | 9.60 | 8.59 | Transcriptional regulator prpR | |
| Rv0678 | 8 | 1 | 8 | 0 | 4.20 | 2.21 | 5.52 | 11.34 | Transcriptional regulator | |
| glpK | 15 | 0 | 8 | 0 | 7.55 | 7.30 | 4.49 | 3.77 | Glycerol kinase | |
| thyA | 15 | 3 | 7 | 1 | 1.62 | 1.92 | 2.30 | 3.35 | Probable thymidylate synthase | |
| gyrA | 13 | 7 | 7 | 3 | 0.94 | 0.72 | 1.38 | 1.18 | DNA gyrase (subunit A) | |
| ethA | 27 | 2 | 7 | 0 | 5.52 | 4.64 | 3.73 | 2.89 | Monooxygenase | |
| embA | 14 | 9 | 7 | 2 | 0.77 | 0.72 | 1.12 | 1.90 | Arabinosylindolylacetylinositol synthase | |
| phoR | 12 | 1 | 21 | 0 | 5.83 | 6.84 | 8.20 | 6.98 | Possible two-component system response sensor kinase membrane-associated PhoR | Common selection between Tibet and plains |
| whiB6 | 9 | 1 | 15 | 0 | 5.83 | 2.82 | 6.43 | 34.68 | Possible transcriptional regulatory protein WhiB-like WhiB6 | |
| Rv3645 | 16 | 1 | 13 | 0 | 7.63 | 8.54 | 6.77 | 5.80 | Probable conserved transmembrane protein | |
| pks15 | 19 | 2 | 13 | 3 | 2.46 | 3.23 | 1.77 | 2.01 | Probable polyketide synthase Pks15 | |
| Rv1194c | 6 | 1 | 9 | 1 | 2.33 | 1.67 | 3.61 | 4.95 | Conserved protein | |
| espK | 17 | 4 | 9 | 1 | 2.18 | 1.70 | 3.32 | 2.77 | ESX-1 secretion-associated protein EspK | |
| lppD | 7 | 1 | 8 | 2 | 3.08 | 4.08 | 2.81 | 1.65 | Possible lipoprotein LppD | |
| dnaA | 11 | 2 | 8 | 0 | 1.49 | 2.73 | 4.65 | 3.71 | Chromosomal replication initiator protein DnaA | |
| whiA | 8 | 2 | 7 | 2 | 1.60 | 1.26 | 1.54 | 2.02 | Probable transcriptional regulatory protein WhiA | |
| Rv2752c | 19 | 4 | 7 | 1 | 2.52 | 1.93 | 3.25 | 4.71 | Conserved hypothetical protein, similar to RNase J | |
| ribG | 5 | 1 | 7 | 0 | 1.73 | 2.59 | 3.86 | 4.64 | Probable bifunctional riboflavin biosynthesis protein RibG | |
| fadD34 | 15 | 3 | 7 | 2 | 1.90 | 1.97 | 1.14 | 1.41 | Probable fatty-acid-CoA ligase FadD34 | |
| sseA | 7 | 2 | 17 | 1 | 0.86 | 0.85 | 5.27 | 5.25 | Probable thiol-oxidoreductase | Tibet-specific selection |
| glcB | 5 | 5 | 9 | 1 | 0.50 | 0.43 | 3.79 | 2.42 | Malate synthase G | |
| Rv2084 | 5 | 2 | 8 | 0 | 0.97 | 0.96 | 2.23 | 1.68 | Hypothetical protein | |
| dnaE2 | 5 | 3 | 8 | 2 | 0.85 | 0.75 | 1.89 | 2.23 | DNA nucleotidyltransferase | |
| recB | 8 | 6 | 7 | 2 | 0.62 | 0.53 | 1.29 | 1.90 | Probable exonuclease V (beta chain) | |
| mfd | 5 | 7 | 7 | 1 | 0.35 | 0.28 | 2.91 | 3.67 | Probable transcription-repair coupling factor | |
| kdpD | 16 | 10 | 7 | 1 | 0.62 | 0.81 | 3.46 | 2.19 | Probable sensor protein | |
NS refers to nonsynonymous SNPs, and SY refers to synonymous SNPs.
Besides these, however, we found nine genes that were potentially under positive selection in the Tibetan strains but negative selection in the plains isolates (Table 1), and, among these, sseA had the strongest evidence for differential selection. In the Tibetan strains, there were a total of 18 unique mutations in the sseA gene, of which 17 were nonsynonymous (including two premature stop mutations) and only 1 was synonymous (pNpS: 5.25; Fig. 5C and SI Appendix, Fig. S4), while the pNpS in the plain strains was only 0.85 (7 nonsynonymous and 2 synonymous, Table 1). Two different nonsynonymous SNPs were found in codon 276 of the sseA encoded protein (E276K and E276G), suggesting diversifying selection (SI Appendix, Fig. S4). Consistently, sseA codon 276 was identified as a site of positive selection with a mean ω ratio of 9.02 by the site model implemented in CODEML (34). In addition to the SNPs, four different insertions/deletions (INDELs) in sseA were found in Tibetan strains (Fig. 5C and SI Appendix, Fig. S4). Taken together, nonsynonymous SNPs or INDELs affected the sseA gene in 45.1% (260/576) of the Tibetan strains and were distributed over all Tibetan clades, suggesting selective pressure in Tibet to eliminate the function of the sseA encoded protein. Because both ancestral and recent mutations were found in sseA (Fig. 5C), it appears that the selective pressure started early after the introduction of Mtb into Tibet and remains ongoing. In addition to sseA, the Tibetan strains also showed evidence of selective pressure in three genes whose encoded proteins are involved in DNA repair: dnaE2, recB, and mfd. One of the eight nonsynonymous mutations in dnaE2 and one of the seven nonsynonymous mutations in recB create premature stop codons (dnaE2 Q373* and recB Q101*), preventing translation of the full-length encoded proteins. We used SIFT to predict the effect of the other mutations found in these three genes (35); four of the other seven nonsynonymous mutations in dnaE2 and three of the eight nonsynonymous mutations in mfd were predicted to affect protein function, while the remaining mutations were predicted to be “tolerated,” with a small probability of affecting protein function. Collectively, these results indicate that the Mtb genomes were subject to an adaptive process, presumably to optimize their ability to replicate or cause disease in the Tibetan population.
As sequence diversity in T cell epitopes has been shown to be associated with the global distribution and prevalence of different Mtb sublineages (2), we asked whether the diversity in the T cell epitopes of the Tibetan strains could be different from that of the plain isolates. We calculated pairwise dN/dS for T cell epitopes (Epi) and nonepitope (NEpi) regions of T cell antigens and also for essential (Ess) and nonessential (NEss) genes. In the plain isolates, the Epi regions were more conserved than NEpi regions (Fig. 5D), consistent with previous reports that human T cell epitopes are hyperconserved (8, 9). Surprisingly, this pattern was reversed in the Tibetan strains, where the Epi regions showed higher sequence diversity than the NEpi regions (W = 152,395; P < 0.0001; Wilcoxon rank-sum test) (Fig. 5E). However, when we excluded the phylogenetic SNPs that were shared by all strains of each Tibetan clade and then recalculated the pairwise dN/dS, the Epi regions showed the expected lower diversity than the NEpi regions (W = 81,267; P = 0.03244; Wilcoxon rank-sum test) (Fig. 5F). Thus, the higher epitope diversity in Tibetan strains was not generated locally but rather was present in the ancestral strains when they were introduced into the region.
Discussion
Our study found that the Mtb strains currently circulating in the Tibetan Plateau were introduced repeatedly into the region between eighteenth and twentieth century and then diversified locally while remaining geographically isolated from the Mtb strains in the plain regions of China. This isolation made it possible to document evidence that local selection has shaped the evolution of the Mtb strains on the Tibetan Plateau. First, although most Tibetan Mtb strains belonged to the Beijing lineage, a striking two-thirds belonged to L2.2 in contrast to most other regions of China and the rest of the world where L2.3 strains are dominant. Second, SNP changes in the Tibetan strains showed a markedly higher proportion of A > G/T > C mutations than strains from other regions of China. Third, nine genes showed signatures of positive selection only in Tibetan Mtb strains. The gene with the strongest selective pressure, sseA, was affected by premature stop mutations or INDELS that would eliminate its encoded thiol oxidoreductase activity. Finally, the T cell epitopes in the Tibetan Mtb strains showed higher diversity than in the plain isolates, but the diversity was present in the original ancestral strains introduced into the region. These findings demonstrate that the relatively high TB incidence in Tibet was caused by an Mtb population that is distinct from the population in the plain regions. The high frequency of mutations in specific genes suggests that local pressures on the Tibetan Plateau selected for Mtb strains that had adapted to this environment.
Three sublineages have been identified within Mtb L2: L2.1 (proto-Beijing), L2.2 (ancient Beijing), and L2.3 (modern Beijing) (3, 14). Strains of the L2.1 sublineage are rare and only occasionally found in southeast Asia and the surrounding regions (3, 36). L2.2 had a long population history and was widely distributed throughout China and much of Asia, while L2.3 emerged in China from within L2.2 only relatively recently and rapidly expanded to become the dominant L2 sublineage in China and many other countries (14). We found that the expansion advantage of the L2.3 apparently did not operate in the Tibetan Mtb population, where two-thirds of the Mtb strains belong to the L2.2 sublineage. Coincidently, L2.2 strains are also more prevalent than L2.3 in the two neighboring high plateau provinces of Ningxia and Qinghai (Fig. 1C), suggesting that L2.2 strains could have an intrinsic advantage in the environments of the high plateaus or may have adapted during their evolution there. Alternatively, the success of L2.3 correlates with increased population density and migration (14), so the lower mobility and relative isolation of the Tibetan host population could have diminished its transmission. This interesting but unexplained finding warrants further epidemiological or experimental studies.
The increase in the proportion of A > G/T > C transition mutations was significant for isolates in Tibetan clades 1 to 4 but was not significant for Tibet clades 5 and 6, which belong to L2.3. The opposite was noted in the plain isolates, where the L2.3 plains clades had overall higher ratios of A > G/T > C mutations than the three plains clades belonging to L2.2 (24.5% versus 19.9%, P < 0.0001). This suggests that the selective pressures in Tibet elicited a greater response in the L2.2 strains than in L2.3 strains. The increased percentage of A > G/T > C changes suggests that the Tibetan strains may have been exposed to conditions that induce misincorporation of deoxynucleoside triphosphates (dNTPs). Concordantly, we observed positive selection for mutations in DNA repair–associated genes (dnaE2, recB, and mfd), and the mutations in dnaE2 and recB tended to be loss-of-function mutations. dnaE2 encodes an error-prone DNA polymerase implicated in the error-prone bypass of DNA lesions (37), recB encodes a helicase/nuclease that prepares double-stranded DNA breaks for recombinational DNA repair (38), and mfd encodes a protein that couples transcription and DNA repair by recognizing RNA polymerase stalled at DNA lesions (39). Mismatch was caused by 4,6-diamino-5-formamidopyrimidine and 8-oxo-A can induce A > G/T > C mutations (40). Alternatively, a recent study found that A > G/T > C transition is the dominant mutation observed in the mutT4-deletion or mutM2-deletion mutant of Mycobacterium smegmatis (41). Apparently, A > G/T > C transition mutations can be induced by either DNA oxidative damage or misincorporation of oxidized nucleotides by a DNA polymerase (such as DnaE2). Determining which of these is more likely to have caused the increase of A > G/T > C mutations would warrant a further study.
In different Tibetan Mtb strains, SNPs in the sseA gene occurred 17 independent times, and 16 of these caused nonsynonymous changes, highlighting the strong positive selection on sseA in a variety of genetic backgrounds. In total, nearly half (45.1%) of all Tibetan strains contained mutations that tended to eliminate SseA activity, either by creating premature stop codons or with frameshift INDELS. In contrast, in the plain strains, the sseA mutations demonstrated purifying selection, indicating that the selection to eliminate SseA activity was specific for the strains in Tibet. The sseA gene is predicted to encode a thiol oxidoreductase (SseA), also termed a rhodanase. Recent studies suggest that SseA works in coordination with the superoxide-detoxifying enzyme SodA and the integral membrane protein DoxX to form a membrane-associated oxidoreductase complex (MRC) that coordinates reactive oxygen species (ROS) detoxification and thiol homeostasis during Mtb infections (42). Loss of any MRC component was associated with defective recycling of mycothiol and the accumulation of oxidative damage in the bacteria (42). If there was pressure to block MRC-mediated ROS detoxification, this could also occur through mutations in the other MRC components. Mutations would not be expected in the essential sodA gene, but doxX is not essential, and yet this gene was not mutated in the Tibetan strains. Therefore, the reason why the selective pressure only affected sseA requires further exploration. If the sseA mutations diminished or blocked ROS detoxification, one would expect the strains with sseA mutations to show an increase in the type of mutations associated with oxidative damage (C > T/G > A and C > A/G > T). Consistent with this speculation, the subclades with a premature stop or 4 bp deletion in sseA showed an increase in C > A/G > T mutations (SI Appendix, Fig. S5). Although the nature of the selective pressure remains elusive, it seems likely to be related to the inhospitable high-altitude environment, and the presence of both ancestral and recent sseA mutations in the Tibetan strains indicates that it has been influencing the evolution of Tibetan strains for hundreds of years.
Ideally, the paired Tibetan and plains clades would have similar ages and share a common genetic background before they separated and diversified. However, the plains clades appeared to be older than their paired Tibetan clades, as reflected by the lower within-clade pairwise SNP distances. We were unable to predict how the selective pressure on Mtb varies with different evolutionary time scales, but the possibility of such a variation introduces a measure of uncertainty into the paired analysis. Nevertheless, we believe that the findings from the paired analysis are valid for several reasons. First, the shift of mutational signature to A > G/T > C is robustly present if the comparison is either between all Tibetan isolates (Fig. 4A) and all plain isolates or between the Tibetan and paired plains clades (Fig. 4C). Second, this potential uncertainty should not affect the analysis of Tibetan-specific selection because we were looking for genes selected only in the Tibetan strains, a process that should be independent of any putative selection specific for the plain strains. Third, for the epitope diversity analysis, the Tibetan clades were grouped together and compared with the grouped plain isolates, without excluding the phylogenetic mutations, and therefore not dependent on the comparability of the paired clades.
In conclusion, our analysis of Mtb strains from the isolated Tibetan population revealed patterns of diversification and evolution that are evidence of past and ongoing adaptation to selective pressures in the local population.
Materials and Methods
Tibetan Mtb Isolates and Whole-Genome Sequencing.
A total of 643 Mtb isolates were collected in different municipal regions of Tibet by the Chinese Center for Disease Control and Prevention in 2006, 2009, and 2010. The initial purpose of this sample collection was to investigate the prevalence of drug resistance and the molecular epidemiology of Tibetan TB. From samples of the isolates that had been stored at −80 °C, aliquots were obtained, and adequate genomic DNA was isolated from 576 of the 643 isolates using the Cetyltrimethylammonium bromide-lysozyme method. The genomic DNA of the samples was then used for standard Illumina construction of 300 bp fragment length genomic libraries that were paired-end sequenced on an Illumina HiSeq. 2500 instrument.
SNPs and INDELs Calling.
We used a previously validated pipeline to map short sequencing reads to the reference genome (43). In brief, the Sickle tool was used for trimming whole-genome sequencing data (44). Sequencing reads with Phred base quality scores above 20 and read lengths longer than 30 were kept for analysis. The inferred ancestral genome of the most recent common ancestor of the Mycobacterium tuberculosis complex (MTBC) was used as the reference template for read mapping (1). Sequencing reads were mapped to the reference genome using Bowtie 2 (version 2.2.9) (45). SAMtools (v1.3.1) (46) was used for SNP calling with mapping quality greater than 30. Fixed mutations (frequency ≥ 75%) were identified using VarScan (v2.3.9) (47) with at least 10 supporting reads and the strand bias filter option on. SNPs in repetitive regions of the genome (e.g., PPE/PE-PGRS family genes, phage sequences, insertion, or mobile genetic elements) were excluded. Small INDELs were identified by VarScan (v2.3.9) (47) using the mpileup2indel function.
Phylogenetic Reconstruction.
The whole-genome sequencing data of 1,159 Mtb isolates from other provinces of China were downloaded from National Center for Biotechnology Information (NCBI) under Bioprojects SRA065095, PRJNA268900, PRJNA559678, and PRJNA522942 (30, 48–50). For phylogenetic reconstructions, all SNP locations for each isolate were combined into a nonredundant consensus list and recalled with the mpileup2cns function of VarScan (version 2.3.9) (47). Nucleotide positions with missing calls in more than 5% of the isolates were removed. An alignment of the remaining polymorphic positions from all strains was used for phylogeny reconstruction with MEGA 6.0 (51). For the estimation of phylogenies, the maximum likelihood method was applied under the general time reverse model with at least 100 replicates for bootstrapping confidence levels. Phylogeny trees were exported from MEGA 6.0 and visualized in FigTree (version 1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/). We adapted a recently described hierarchical nomenclature to define sublineage nodes and subclades within the tree (14, 52). For the subsequent analyses of mutational signatures, genes subject to selection and epitope diversity, six plains clades were selected and paired with Tibetan clades 1 to 6 based on their genetic proximity to the Tibetan clades.
Dating Analysis.
We selected 38 L2 strains from published studies to represent the major phylogenetic structure of L2. These 38 strains together with 72 Tibetan strains (4 to ∼12 strains for each of the Tibet 1 to 12 clades) were used for phylogenetic reconstruction. We estimated the dates of the most common recent ancestors of each Tibetan clade using BEAST (v1.8.0) (53). The XML input file was modified to specify the number of invariant sites in the MTBC genomes. For the MTBC genome substitution rate, we used a normal distribution with a mean of 4.6 × 10−8 substitutions per genome per site per year (3.0 × 10−8 to 6.2 × 10−8, 95% highest polar density interval) (54), which was calibrated by ancient DNA samples (54, 55). An uncorrelated lognormal distribution was used for the substitution rate and a constant population size for the tree priors. We ran three chains of 5 × 107 generations and sampled every 10,000 generations to assure independent convergence of the chains; we discarded the first 10% as a burn-in. Convergence was assessed using Tracer (v1.6.0), ensuring all relevant parameters reached an effective sample size >100.
Bayesian Skyline Plot.
Bayesian skyline plot was applied to estimate the past effective population size dynamics of Tibet clades 1 to 6 based on the substitution rate model, as described above, using all isolates from each Tibetan clade. Clade-based skyline analysis was performed, and the ages of the most recent common ancestors obtained from the dating analysis were used as the tree heights. In each case, three chains of 5 × 107 generations each were sampled every 10,000 generations to assure independent convergence of the chains.
Transmission Clusters.
The genomic locations of the fixed SNPs that differed between any two Mtb isolates were used to generate an SNP matrix for all the Tibetan Mtb strains. The allele type at each location for each Tibetan Mtb strain was validated through a consensus file generated by VarScan (v2.3.9) (47) using the mpileup2cns function. We used two SNP thresholds to identify transmission clusters, 6-SNPs and 12-SNPs. The 6-SNP threshold was defined as the range of SNP distances between paired isolates of the same strain obtained at different times from relapsed TB patients (56). The 12-SNP threshold was defined in a study examining the SNP distances between Mtb strains from TB patients with epidemiological links, suggesting that they belonged to the same transmission chain (49). The cluster size was defined as the number of Mtb isolates that are included within the threshold definition.
pNpS.
To test if the mutations accumulated in the Tibetan strains could be a result of positive selection, we used pNpS to evaluate the selective pressure in Tibetan strains and compared the values to those from the plain strains (28, 57). First, pNpS was calculated for each individual isolate at a whole-genome level, using the SNPs that accumulated after clade diversification. Second, pNpS was calculated for each individual gene by grouping all the mutations in that gene that were found in all the Tibetan or all the plain strains. To identify those genes with evidence of selective pressure in the Tibetan strains, the pNpS values from the genes in the Tibetan strains were then compared with the pNpS values of the same genes from the plain strains. The basic principle of the pNpS method is similar to that of dN/dS, but pNpS can be applied to concatenated sequences of all the mutations in the genomes. A codon substitution matrix was generated using a base substitution model that takes into account the proportion of guanine and cytosine in the genome (percentage GC content, 0.656). Briefly, for each variant codon, we used a custom Python script to simulate 50,000 individual introductions of a single mutation into the codon and scored the outcomes as either synonymous or nonsynonymous. We considered the average number of nonsynonymous outcomes of the simulations as an estimate of the probability that a mutation in the given codon would be nonsynonymous. The formula used to calculate the pNpS was described previously (57).
Site Selection.
In order to identify the positive selection sites in sseA, we applied the site model in the CODEML program of the Phylogenetic Analysis by Maximum Likelihood (PAML) package to estimate the ω ratio at individual sites (34). An alignment of sseA consensus sequences of 576 Tibetan strains was constructed, and 18 nonredundant sequences were kept after removing duplicated sequences. The likelihood ratio tests (LRTs) comparing the null models (M0, M1a, and M7: do not allow ω > 1) to the alternative models (M2a and M8: allow sites with ω > 1) were conducted using CODEML. The statistical significance of the difference between the two LRTs was tested using χ2 distribution. Positive selection sites were identified using the BEB approach to calculate the Bayesian posterior probabilities of specific codon sites.
T Cell Epitopes dN/dS Analysis.
Experimentally confirmed human T cell epitope sequences were retrieved from the Immune Epitope Database (http://www.iedb.org/) and processed under the criteria previously described (2). A total of 1,335 epitope sequences across 318 antigens were obtained in this study. Pairwise dN and dS values within each sublineage were calculated using the R package “seqinr” with the kaks function. A mean dN/dS was then calculated for the epitopes in each isolate by dividing its mean pairwise dN by its mean pairwise dS with respect to all other sequenced isolates within each sublineage. Wilcoxon rank-sum tests were performed in R Studio (3.4.0).
Ethics Statement.
The study obtained approval from the Ethics Committee of National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention.
Supplementary Material
Acknowledgments
We thank Iñaki Comas for the insightful comments and helpful discussions. We also want to thank Xin Wang for helpful discussions during the writing of the paper. This work was supported by Natural Science Foundation of China (81661128043 and 81871625 to Q.G., 81701975 to Q.L., and 31970032 to L.D.L.), National Science and Technology Major Project of China (2017ZX10201302 and 2018ZX10715012-005 to Q.G., 2018ZX10101002 to K.W., and 2018ZX10302301 to L.D.L.), and Sanming project of Medicine in Shenzhen (SZSM201611030 to Q.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.D.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017831118/-/DCSupplemental.
Data Availability
Sequencing reads have been submitted to the NCBI or The European Bioinformatics Institute under study accession number PRJNA656167 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA656167). The analyzing scripts have been deposited at GitHub previously (14, 28, 58).
References
- 1.Comas I., et al., Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stucki D., et al., Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 48, 1535–1543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo T., et al., Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with han Chinese. Proc. Natl. Acad. Sci. U.S.A. 112, 8136–8141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brynildsrud O. B., et al., Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Sci. Adv. 4, eaat5869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill M. B., et al., Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia. Mol. Ecol. 28, 3241–3256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagneux S., Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Gagneux S., et al., Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2869–2873 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comas I., et al., Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42, 498–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coscolla M., et al., M. tuberculosis T cell epitope analysis reveals paucity of antigenic variation and identifies rare variable TB antigens. Cell Host Microbe 18, 538–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osório N. S., et al., Evidence for diversifying selection in a set of Mycobacterium tuberculosis genes in response to antibiotic- and nonantibiotic-related pressure. Mol. Biol. Evol. 30, 1326–1336 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Pepperell C. S., et al., The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 9, e1003543 (2013). Corrected in: PLoS Pathog.9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee R. S., et al., Population genomics of Mycobacterium tuberculosis in the inuit. Proc. Natl. Acad. Sci. U.S.A. 112, 13609–13614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulholland C. V., et al., Dispersal of Mycobacterium tuberculosis driven by historical European trade in the south pacific. Front. Microbiol. 10, 2778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q., et al., China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nat. Ecol. Evol. 2, 1982–1992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu D., et al., Ancestral origins and genetic history of Tibetan highlanders. Am. J. Hum. Genet. 99, 580–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi X., et al., Genetic evidence of paleolithic colonization and neolithic expansion of modern humans on the Tibetan plateau. Mol. Biol. Evol. 30, 1761–1778 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Meyer M. C., et al., Permanent human occupation of the central Tibetan Plateau in the early Holocene. Science 355, 64–67 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Bigham A. W., Lee F. S., Human high-altitude adaptation: Forward genetics meets the HIF pathway. Genes Dev. 28, 2189–2204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beall C. M., Two routes to functional adaptation: Tibetan and andean high-altitude natives. Proc. Natl. Acad. Sci. U.S.A. 104 (suppl. 1), 8655–8660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beall C. M., et al., Ventilation and hypoxic ventilatory response of Tibetan and Aymara high altitude natives. Am. J. Phys. Anthropol. 104, 427–447 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Beall C. M., et al., Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107, 11459–11464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore L. G., Measuring high-altitude adaptation. J. Appl. Physiol. 123, 1371–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren G., et al., SP110 and PMP22 polymorphisms are associated with tuberculosis risk in a Chinese-Tibetan population. Oncotarget 7, 66100–66108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., et al., Genetic signatures of high-altitude adaptation in Tibetans. Proc. Natl. Acad. Sci. U.S.A. 114, 4189–4194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dierberg K. L., et al., Improved detection of tuberculosis and multidrug-resistant tuberculosis among Tibetan refugees, India. Emerg. Infect. Dis. 22, 463–468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorjee K., et al., High prevalence of active and latent tuberculosis in children and adolescents in Tibetan schools in India: The Zero TB kids initiative in Tibetan refugee children. Clin. Infect. Dis. 69, 760–768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond A. J., Rambaut A., Shapiro B., Pybus O. G., Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Liu Q., et al., Mycobacterium tuberculosis clinical isolates carry mutational signatures of host immune environments. Sci. Adv. 6, eaba4901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne J. L., et al., Transition bias influences the evolution of antibiotic resistance in Mycobacterium tuberculosis. PLoS Biol. 17, e3000265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicks N. D., et al., Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3, 1032–1042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellerose M. M., et al., Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. MBio 10, e00663-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safi H., et al., Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc. Natl. Acad. Sci. U.S.A. 116, 19665–19674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiner-Oms Á., et al., Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex. Sci. Adv. 5, eaaw3307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Sim N. L., et al., SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holt K. E., et al., Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat. Genet. 50, 849–856 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boshoff H. I., Reed M. B., Barry C. E. 3rd, Mizrahi V., DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113, 183–193 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Malyarchuk S., et al., Expression of Mycobacterium tuberculosis ku and ligase D in Escherichia coli results in RecA and RecB-independent DNA end-joining at regions of microhomology. DNA Repair (Amst.) 6, 1413–1424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prabha S., Rao D. N., Nagaraja V., Distinct properties of hexameric but functionally conserved Mycobacterium tuberculosis transcription-repair coupling factor. PLoS One 6, e19131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boiteux S., Gajewski E., Laval J., Dizdaroglu M., Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): Excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry 31, 106–110 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Dupuy P., Howlader M., Glickman M. S., A multilayered repair system protects the mycobacterial chromosome from endogenous and antibiotic-induced oxidative damage. Proc. Natl. Acad. Sci. U.S.A. 117, 19517–19527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nambi S., et al., The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe 17, 829–837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., et al., Genetic features of Mycobacterium tuberculosis modern Beijing sublineage. Emerg. Microbes Infect. 5, e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joshi N., Fass J., Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files, Version 1.33 (2011).
- 45.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koboldt D. C., et al., VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Q., et al., Citywide transmission of multidrug-resistant tuberculosis under China’s rapid urbanization: A retrospective population-based genomic spatial epidemiological study. Clin. Infect. Dis. 71, 142–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C., et al., Transmission of multidrug-resistant Mycobacterium tuberculosis in shanghai, China: A retrospective observational study using whole-genome sequencing and epidemiological investigation. Lancet Infect. Dis. 17, 275–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., et al., Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45, 1255–1260 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coll F., et al., A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 5, 4812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bos K. I., et al., Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514, 494–497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabin S., et al., A seventeenth-century Mycobacterium tuberculosis genome supports a Neolithic emergence of the Mycobacterium tuberculosis complex. Genome Biol. 21, 201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryant J. M., et al., Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: A retrospective observational study. Lancet Respir. Med. 1, 786–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trauner A., et al., The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 18, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q., et al., Have compensatory mutations facilitated the current epidemic of multidrug-resistant tuberculosis? Emerg. Microbes Infect. 7, 98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads have been submitted to the NCBI or The European Bioinformatics Institute under study accession number PRJNA656167 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA656167). The analyzing scripts have been deposited at GitHub previously (14, 28, 58).