Significance
Genome reduction is commonly observed in bacteria of several phyla engaging in obligate nutritional symbioses with insects. In Actinobacteria, however, little is known about the process of genome evolution, despite their importance as prolific producers of antibiotics and their increasingly recognized role as defensive partners of insects and other organisms. Here, we show that “Streptomyces philanthi,” a defensive symbiont of digger wasps, has a G+C-enriched genome in the early stages of erosion, with inactivating mutations in a large proportion of genes, causing dependency on its hosts for certain nutrients, which was validated in axenic symbiont cultures. Additionally, overexpressed catabolic and biosynthetic pathways of the bacteria inside the host indicate host–symbiont metabolic integration for streamlining and control of antibiotic production.
Keywords: defensive symbiosis, protective mutualism, genome erosion, pseudogenization, Streptomyces
Abstract
Genome erosion is a frequently observed result of relaxed selection in insect nutritional symbionts, but it has rarely been studied in defensive mutualisms. Solitary beewolf wasps harbor an actinobacterial symbiont of the genus Streptomyces that provides protection to the developing offspring against pathogenic microorganisms. Here, we characterized the genomic architecture and functional gene content of this culturable symbiont using genomics, transcriptomics, and proteomics in combination with in vitro assays. Despite retaining a large linear chromosome (7.3 Mb), the wasp symbiont accumulated frameshift mutations in more than a third of its protein-coding genes, indicative of incipient genome erosion. Although many of the frameshifted genes were still expressed, the encoded proteins were not detected, indicating post-transcriptional regulation. Most pseudogenization events affected accessory genes, regulators, and transporters, but “Streptomyces philanthi” also experienced mutations in central metabolic pathways, resulting in auxotrophies for biotin, proline, and arginine that were confirmed experimentally in axenic culture. In contrast to the strong A+T bias in the genomes of most obligate symbionts, we observed a significant G+C enrichment in regions likely experiencing reduced selection. Differential expression analyses revealed that—compared to in vitro symbiont cultures—“S. philanthi” in beewolf antennae showed overexpression of genes for antibiotic biosynthesis, the uptake of host-provided nutrients and the metabolism of building blocks required for antibiotic production. Our results show unusual traits in the early stage of genome erosion in a defensive symbiont and suggest tight integration of host–symbiont metabolic pathways that effectively grants the host control over the antimicrobial activity of its bacterial partner.
Many insects engage in mutualistic relationships with bacteria that provide diverse adaptive benefits to their hosts, such as essential nutrients, digestive or detoxifying enzymes, or defense against predators, parasites, and pathogens (1, 2). These tightly insect-associated bacteria commonly experience substantial degenerative genome evolution (3, 4), reflected in massive gene loss, increased coding density, a strong nucleotide bias toward increased A+T content in DNA, and the accumulation of mutations that are assumed to be slightly deleterious (3). Because genes and pathways beneficial for symbiosis (e.g., genes for the biosynthesis of vitamins or amino acids) are retained in these reduced genomes, it is often possible to readily infer the functions of the bacterial partner (5–7). Genome erosion of symbiotic bacteria has been primarily described in intracellular nutritional symbionts in the phyla Proteobacteria, Tenericutes, and Bacteroidetes (3, 8) and more recently in extracellular Gammaproteobacterial mutualists providing nutritional supplements or digestive enzymes to the host (4, 9, 10). In contrast, defensive mutualists often retain larger genomes with more complete metabolic inventories (8). Exceptions to this rule are the drastically reduced genomes of the defensive symbiont of the Asian citrus psyllid Diaphorina citri, i.e. “Candidatus Profftella armatura” (11), as well as the protective Burkholderia symbiont of Lagria beetles (12).
Filamentous Actinobacteria are being discovered as defensive symbionts in an increasing number of insects and marine invertebrates (13–17). Unlike nutritional symbionts, most actinobacterial mutualists benefit their hosts by producing a range of bioactive secondary metabolites that have been shown to protect leaf-cutter ants (18–21), bark beetles (22, 23), and beewolf wasps (24–26) against pathogens. Due to their biotechnological potential, genomes of environmental Actinobacteria have been intensively sequenced over the last few decades with emphasis on antibiotic biosynthetic pathway discovery (27). They generally exhibit large, G+C-rich genomes encoding a versatile set of primary metabolic pathways as well as a plethora of secondary metabolite gene clusters (28–30). Despite these characteristic genomic traits, however, the genome evolution of symbiotic members in this clade has thus far received little attention (13).
“Streptomyces philanthi” are defensive bacterial symbionts that have been living in association with beewolf digger wasps of the genera Philanthus, Philanthinus, and Trachypus (Hymenoptera, Crabronidae) for ∼68 My (31). Beewolf females hunt and paralyze bees or wasps to provision them to their offspring in subterranean brood chambers. Throughout the life cycle of their hosts, the bacterial symbionts experience three different environments: 1) In the antennae of adult female wasps, the bacteria are cultivated in gland reservoirs that are likely nutritionally rich. 2) After deposition by the female wasp into the brood cell, 3) symbionts are transferred by the larvae to their cocoons, where the symbionts do not grow but produce a “mixture” of antibiotics (mainly piericidins and streptochlorin) (32, 33). These antibiotics protect beewolf larvae from fungal infection during the 9 to 10 mo period of diapause in the prepupal stage including hibernation (25, 33). An analysis of 22 symbiont biovars isolated in pure culture from diverse beewolf species collected in Eurasia, Africa, and North and South America showed broad variation in nutritional demands, with an African/Eurasian symbiont clade being particularly fastidious and only culturable on complex media imitating insect hemolymph (34). Because of their complex life cycle, defensive function, metabolic constraints, and their taxonomic affiliation within the Actinobacteria, the beewolf mutualists provide an interesting case to study the impact of a symbiotic lifestyle on genome evolution in a clade of bacteria with large, linear, and G+C-rich genomes.
Here, we used a combination of genomic, transcriptomic, and proteomic approaches to characterize “S. philanthi biovar triangulum” strain 23Af2, isolated from the European beewolf Philanthus triangulum, and shed light on metabolic interactions between host and defensive symbiont. In contrast to the symbionts of some other beewolf species, the European beewolf symbiont shows signs of an obligate association with its host based on its nutritional demands (34) and strictly vertical transmission route (31). The possibility of culturing it in vitro allowed for experimental approaches that are commonly not available for obligate symbionts in other insects. Comparative functional genome analysis complemented by in vitro physiological assays and proteomic and transcriptomic analyses reveal a genome in an early stage of erosion. Frameshift mutations inactivating about one-third of the protein-coding genes and thereby interrupting metabolic pathways resulted in the dependency on the host for fulfillment of the symbiont’s nutritional requirements. In contrast to most other cases of genome reduction in bacteria, we observed a bias toward increased G+C, rather than A+T, content in genomic DNA. Comparative analysis of gene expression within the antennal reservoirs and in vitro offers insights into host-provided nutrients and indicates streamlining of the symbionts’ metabolism for the defensive function.
Results
General Genome Characteristics: A Large, Eroding, G+C-Rich Genome.
The combination of 454 (shotgun and 8 kb paired-end libraries), Illumina, Sanger, and PacBio sequencing technologies followed by independent assemblies of the reads and a subsequent hybrid assembly of the resulting contigs with the verification of results by optical mapping resulted in one complete bacterial chromosome of 7,341,895 base pairs (bp) for “S. philanthi biovar triangulum” strain 23Af2. General characteristics of the “S. philanthi biovar triangulum” genome were similar to those of free-living Streptomyces species, such as a linear chromosome with high G+C content (71.8%) and a comparable number of transfer RNA genes and ribosomal RNA operons (Table 1 and Fig. 1A). However, compared to free-living Streptomyces, the S. philanthi genome showed some unusual characteristics such as low coding density (78%) and overall low average gene length (718 bp, Table 1). A feature that is typical for genomes of the genus Streptomyces is the presence of a “core” region around the center of the linear chromosome with high conservation and two “arms” at the chromosomal periphery that show little synteny among species and are prone to large chromosomal rearrangements (35). The genome of “S. philanthi,” however, appears to lack most of the left arm and to have the right arm shortened in comparison with sequenced genomes of free-living Streptomyces in the same clade (SI Appendix, Fig. S1), all of which have chromosomes over 10 million base pairs (SI Appendix, Fig. S2).
Table 1.
Comparison of the “S. philanthi biovar triangulum” 23Af2 (Sphi) genome with reference genomes of other Streptomyces species (Scoe: S. coelicolor A3(2), Sgri: S. griseus IFO13350, Save: S. avermitilis MA-4680, Ssca: S. scabiei 87.22, Sbin: S. bingchenggensis BCW-1, Shyg: S. hygroscopicus 5008)
| Category | Sphi | Scoe | Sgri | Save | Ssca | Sbin | Shyg |
| Genome size, bp | 7,341,895 | 8,667,507 | 8,545,929 | 9,025,608 | 10,148,695 | 11,936,683 | 10,145,833 |
| Chromosome type | linear | linear | linear | linear | linear | linear | linear |
| Genes, total | 8,011 | 7,911 | 7,138 | 7,669 | 8,708 | 10,107 | 8,936 |
| CDS, total | 7,939 | 7,825 | 7,138 | 7,582 | 8,530 | 10,023 | 8,849 |
| Secondary metabolite clusters, total | 23 | 20 | 34 | 25 | 34 | 47 | 29 |
| Transfer RNAs, total | 61 | 63 | 66 | 68 | 75 | 66 | 68 |
| Ribosomal RNA operons, total | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| Average gene length, bp | 723 | 991 | 1,055 | 1,027 | 1,005 | 1,031 | 952 |
| G+C content, % | 71.8 | 72.1 | 72.2 | 70.7 | 71.5 | 70.8 | 71.9 |
| Coding density, %* | 78 | 88.9 | 88.1 | 86.3 | 86.2 | 86.6 | 83.2 |
| Pseudogenes | 2916 | 55 | 95 | 291 | 155 | 810 | 186 |
| Genes G+C content, % | 72.26 | 72.34 | 72.43 | 71.09 | 71.75 | 71.19 | 72.17 |
| IR G+C content, % | 72.89 | 69.71 | 70.99 | 68.37 | 69.61 | 67.53 | 70.02 |
| Prophage regions | 4 | 2 | 1 | 1 | 3 | 1 | 3 |
| Genomic islands | 38 | 101 | 104 | 21 | 52 | 29 | 172 |
Proportion of nucleotides in coding sequences out of the total number of nucleotides in the whole genome.
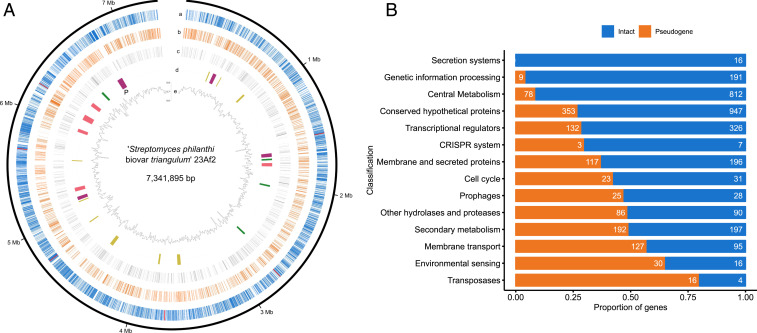
Fig. 1.
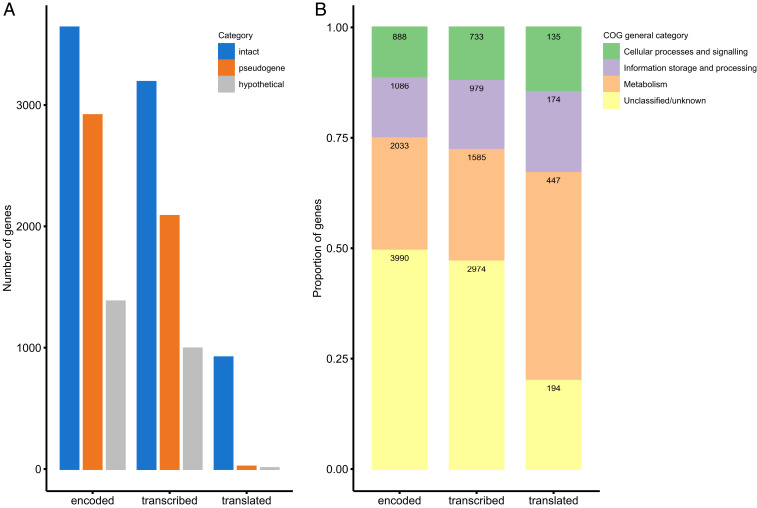
General genomic characteristics of “S. philanthi biovar triangulum.” (A) Genome of “S. philanthi.” a) Intact coding sequences (blue) and ribosomal RNA operons (red); b) pseudogenes; c) hypothetical genes. d) Secondary metabolite gene clusters: polyketide synthases (purple), terpenes (green), nonribosomal peptide synthetases (pink), and other (yellow). The biosynthetic gene cluster for piericidin is marked with “P”. e) G+C content variation for nonoverlapping 10 kb windows. (B) Proportion of intact genes (blue) versus pseudogenes (orange) grouped by functionality according to their KEGG categories.
Manual genome annotation and comparative genome analyses revealed massive gene decay within the “S. philanthi” genome. Overall, genes could be classified into three categories: 1) intact genes with high sequence identity and gene length similar to reference genes; this group includes both genes with known functions and conserved hypothetical genes; 2) pseudogenes and genes in the process of pseudogenization, most with frameshift mutation(s) affecting the integrity of coding sequences, typically causing gene shortening or gene splitting but occasionally also gene extension or even gene fusion; 3) hypothetical genes that share very low (<30%) sequence identity as a whole sequence with genes from the National Center for Biotechnology Information (NCBI) nr (nonredundant) database; this last category likely includes three groups: orphan genes that appeared during coevolution with the host, artifacts of the gene prediction procedure, and pseudogenes that have decayed beyond the point of recognition. Pseudogenes and hypothetical genes encompassed a large proportion of genes (36.7% and 17.5%, respectively) and were responsible for the low coding density and average gene length (average gene length is 922 bp for intact genes but 722 bp for pseudogenes and 407 bp for hypothetical genes).
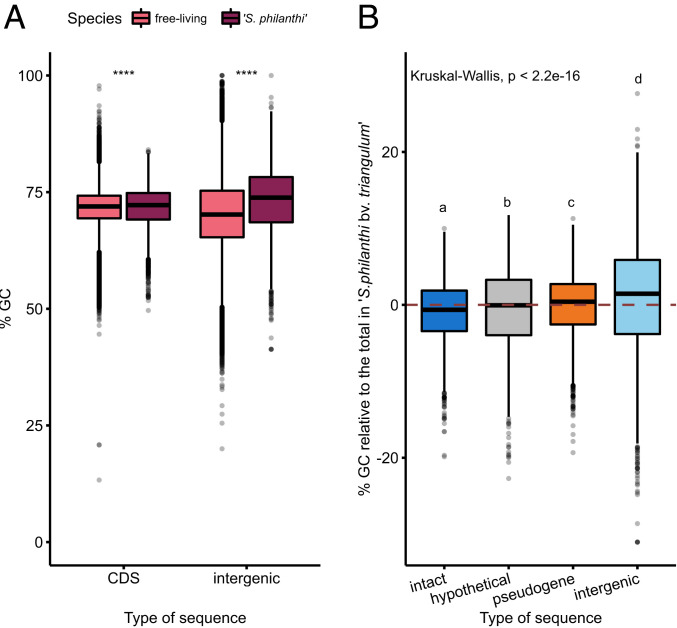
The overall G+C content in the symbiont strain 23Af2 was within the range of other streptomycetes (Table 1). However, when comparing the nucleotide composition of the individual coding sequences (CDS) and intergenic regions, the sequenced strain of “S. philanthi” showed a higher G+C content in both categories in comparison to free-living Streptomyces (Fig. 2) (two-sided Wilcoxon test, P < 1e-05). In addition, G+C is significantly higher in intergenic regions and pseudogenes of “S. philanthi” compared to intact protein-coding genes (Fig. 2B) (Kruskal–Wallis test: P < 2e-16, post hoc Dunn test: P < 0.01), whereas the G+C content of intergenic regions is reduced compared to CDS in free-living Streptomyces (two-sided Wilcoxon test, P < 1e-05). This suggests that there is a G+C mutational bias in “S. philanthi” in contrast to the A+T bias that has been reported in most other symbionts (3, 36) and pathogens (37–39) experiencing extensive degeneration, including the pathogenic Actinobacterium Mycobacterium leprae (40). Notable exceptions to the commonly observed A+T bias are the highly eroded but G+C-rich genomes of Hodgkinia and Tremblaya, the intracellular proteobacterial symbionts of cicadas and mealybugs, respectively (41, 42).
Fig. 2.
G+C bias in intergenic regions versus coding sequences of the “S. philanthi” genome. (A) G+C content of coding sequences and intergenic regions from the free-living Streptomyces species included in the phylogenetic tree from SI Appendix, Fig. S2 in comparison to “S. philanthi.” A two-sided Wilcoxon test results in P < 1e-05 for both comparisons. (B) G+C content of intact protein-coding genes, hypothetical protein-coding genes, pseudogenes, and intergenic regions in the “S. philanthi” genome. Lowercase letters indicate results from a post hoc Dunn test of statistical significance for difference of the means between groups where P < 0.01. The dashed line indicates the total genomic G+C content of “S. philanthi.”
Core Metabolic Functions Are Mostly Intact, but Regulatory and Accessory Genes Are Decaying.
Even though pseudogenes were randomly distributed across the genome (Fig. 1A), further genome analysis showed that cellular systems were differentially affected by frameshift mutations, with the following functions being heavily eroded (>25% and up to 88% of predicted genes): membrane transport, cell cycle (including proteins involved in sporulation), transcriptional regulation, environmental sensing (two-component systems), genome plasticity (transposases), secondary metabolism as well as secreted and conserved hypothetical proteins, lipoproteins, glycoside hydrolases, and protease-encoding genes (Fig. 1B). In contrast, protein secretion systems and central metabolic and genetic information processing pathways were much less eroded (Figs. 1B and 3). A notable exception of core functions with a high amount of pseudogenes was observed in DNA repair functions such as base excision repair (five out of 14 unique genes with frameshift mutations, lig, alkA, mug, nei, and tag), homologous recombination (two out of 13 genes with frameshift mutations, recA and recO), and nonhomologous end joining systems (one out of two genes with frameshift mutations, DNA-binding protein Ku); these genes remain intact in most free-living Streptomyces species but are often lost in obligate mutualists and pathogens (36), likely accelerating genome erosion by impairing DNA repair mechanisms. Other DNA repair functions remained apparently functional in “S. philanthi,” including the genes mutM and mutY, which are necessary to prevent GC to TA transversions (43). Within the central metabolism, incomplete pathways are found for biotin, arginine, and proline biosynthesis (Figs. 3 and 4A). Regarding transport systems, most genes encoding amino acid transporters were intact, but several involved in carbohydrate transport were affected by frameshift mutations.
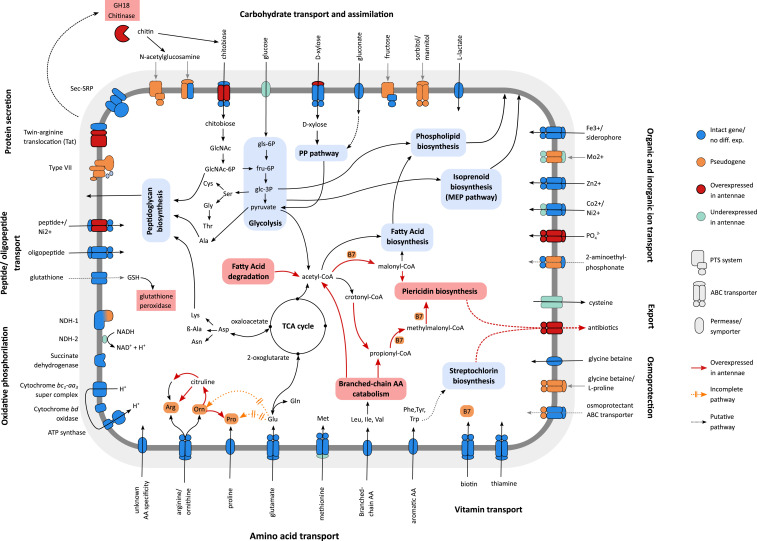
Fig. 3.
Reconstruction of metabolic pathways and transport systems of “S. philanthi.” Interrupted pathways and pseudogenes are shown in orange, genes and pathways significantly up-regulated in antenna versus in vitro culture (threshold: greater than twofold expression difference, q-value < 0.1) are highlighted in red, and down-regulated genes are shown in pale blue.
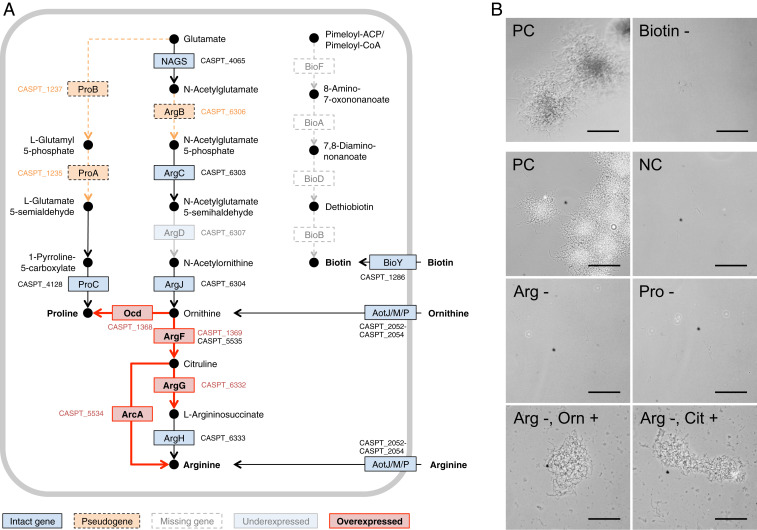
Fig. 4.
Effect of genome erosion on amino acid requirements of “S. philanthi biovar triangulum” strain 23Af2. (A) Proline, arginine, and biotin biosynthetic pathways. Pseudogenes are indicated in an orange color, and dashed lines denote putatively interrupted enzymatic steps. Missing genes are indicated in white boxes. Intact genes with no differential expression between antenna and in vitro cultures are in blue. Genes overexpressed (log2 > 1) in antennal samples are highlighted in red, and those underexpressed (log2 < −1) are in light blue. All differentially expressed genes shown had an adjusted P value (q-value) < 0.1, with the exception of CSP_1369. (B) Phase-contrast microscopy images of in vitro cultures of “S. philanthi biovar triangulum” without biotin, arginine, or proline show no growth. Arginine auxotrophy can be bypassed by the addition of precursors ornithine (Orn) or citrulline (Cit). NC, negative control (Grace’s based medium without added amino acids or vitamins); PC, positive control (complete Grace’s medium); Arg, arginine; Pro, proline. (Scale bar corresponds to 100 µm.)
Since “S. philanthi” is a defensive symbiont that produces antibiotics protecting beewolf larvae against fungal infection, the genomic basis of secondary metabolite biosynthesis was of special interest. In the “S. philanthi” genome, 26 secondary metabolism gene clusters were identified—a comparable number to that typically found in free-living Streptomyces (44, 45) (SI Appendix, Table S1)—but many of the genes contained in these clusters were frameshifted (Fig. 1B). However, clusters responsible for the biosynthesis of piericidins and actinopyrones (26), two other polyketides, a nonribosomal peptide, and a terpene remain intact (SI Appendix, Table S1). The biosynthesis of streptochlorin, one of the bioactive compounds present in the symbiont-produced antibiotic mixture (26, 33), remains elusive.
In Vitro Cultures of “S. philanthi” Confirm Amino Acid Auxotrophies.
In our previous work, we reported conditions for in vitro culturing of several isolates of beewolf symbionts, a rare feature in insect symbionts, and showed that “S. philanthi biovar triangulum” has specific nutritional demands, consistent with multiple auxotrophies (34). Our genome analysis indicates that central biosynthetic systems remain generally intact, with the exception of biotin, arginine, and proline biosynthesis. Therefore, detailed in vitro experiments on the physiology of strain 23Af2 were performed to complement the in silico predictions. This bacterium could grow on Grace’s-based medium containing the complete set of amino acids (34). Further tests using this medium lacking one out of 21 amino acids confirmed the auxotrophies for arginine and proline (Fig. 4B). The auxotrophy for arginine was consistent with the detected frameshift mutations in argB (CSP_6306, acetylglutamate kinase; EC 2.7.2.8). As predicted, supplementation of the medium with the intermediate metabolites citrulline or ornithine allowed for growth in the absence of arginine (Fig. 4B). Genes proA (CSP_1235) and proB (CSP_1237) also carried frameshift mutations, explaining the observed proline auxotrophy.
Remarkably, four genes (EC 2.3.1.47, 2.6.1.62, 6.3.3.3, and 2.8.1.6) of the biotin biosynthesis pathway were fully absent from the genome, even though these genes are present in all reference genomes of free-living Streptomyces, suggesting that they were lost during the genome reduction and that “S. philanthi” is dependent on its host for this vitamin. The strain 23Af2 was indeed unable to grow after iterative inoculation in biotin-free Grace’s medium (Fig. 4B). Accordingly, the biotin ATP-binding cassette (ABC) transporter remains intact in the genome and likely serves to import biotin provided by the host (Figs. 3 and 4A).
Pseudogenes Are Transcribed but not Translated.
Transcriptome analysis with RNA sequencing (RNA-seq) was performed on “S. philanthi” harvested from cultures in Grace’s medium and from female beewolf antennal reservoirs in order to explore the consequences of genome decay for gene expression in vivo and in vitro. The analysis of gene expression was complemented by a proteomic analysis on the in vitro cultures to assess the impact of both transcriptional and post-transcriptional regulation on protein abundances.
Generally, a large fraction of “S. philanthi” genes were transcribed in both beewolf antennae and in vitro cultures (Fig. 5 and SI Appendix, Fig. S3), yielding transcripts of almost all intact protein-coding genes (99.9%). Many hypothetical genes (88.5%) and pseudogenes (85.6%) were also transcribed, although the latter two categories were significantly underrepresented among the set of genes for which transcripts were detected (X2 = 20.18, degrees of freedom [df] = 2, P = 4.1e-05). Even when using a strict transcripts per million (TPM) cutoff of 10 to limit spurious detection of transcripts, a considerable number of pseudogenes were found to be transcriptionally active (71.6% of pseudogenes in contrast to the 87.7% of intact genes had TPM values > 10 in RNA samples from in vitro cultures).
Fig. 5.
Transcription and translation of pseudogenes and hypothetical genes in comparison to intact genes. (A) Number of intact genes, pseudogenes, and hypothetical genes found in the genome, their transcripts identified by RNA-seq from in vitro samples (with TPM > 10), and their corresponding peptides detected in the proteome of “S. philanthi.” (B) Classification of all protein-coding genes in the genome of “S. philanthi” according to their general Clusters of Orthologous Genes (COG) categories in comparison to the subset of genes which are transcribed or translated. The specific COG categories in each class are shown in SI Appendix, Fig. S5. The absolute number of genes in each category is indicated inside the bars.
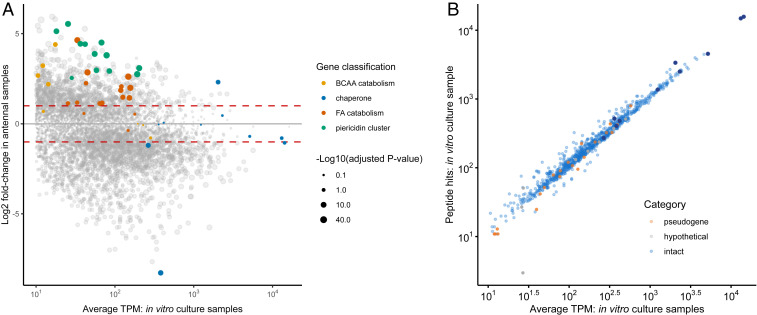
Interestingly, however, the proteome was strongly dominated by proteins encoded by intact genes (97% of the individual proteins detected) and almost completely depleted of pseudogenes or hypothetical proteins (only 21 and seven proteins out of 950, respectively) (Fig. 5A). Among the proteins detected, we observed a striking correlation between the messenger RNA (mRNA) expression levels and the peptide hits against protein databases from the proteome analysis (Fig. 6B). A χ2 homogeneity test confirms the overrepresentation of intact genes in the transcriptome (X2 = 20.18, df = 2, P = 4.1e-05) and proteome (X2 = 890.46, df = 2, P < 2.2e-16) in comparison to the full set of predicted coding sequences. Intact protein-coding genes also showed significantly higher mean expression levels in comparison to both pseudogenes and hypothetical genes (SI Appendix, Fig. S3, P < 1e-05). Both gene expression level and intactness of the CDS were significant predictors for the probability of a protein to be detected in the proteome (binomial logistic regression, P < 2e-16, SI Appendix, Fig. S4).
Fig. 6.
Gene expression in vitro and in the antennal reservoirs. (A) Comparison of gene expression in in vitro cultures versus antennal samples. Genes in the piericidin biosynthetic cluster are indicated in green, genes in fatty acid (FA) catabolism are indicated in orange, genes involved in branched-chain amino acid (BCAA) catabolism are in yellow, and molecular chaperones are in blue. Other genes are presented in gray. The area of the dots is proportional to -Log10(q-value). Only coding sequences with TPM values > 10 are displayed. Dashed red lines indicate fold change = +/− 2. (B) Correlation between proteome and transcriptome data. Peptide hits to identified proteins were used as an estimate of relative protein abundance to contrast with average TPM values of the corresponding coding sequences from in vitro culture samples. Intact genes are shown in blue, pseudogenes are shown in orange, and hypothetical proteins are shown in gray. Molecular chaperones are highlighted in dark blue. Coding sequences with TPM values < 10 or not detected experimentally in the proteome are not shown.
Core Metabolic Functions and Molecular Chaperones Dominate the Proteome.
An analysis of functions detected in the proteome shows a disproportionate enrichment in core metabolic processes in comparison to the full genome (Fig. 5B and SI Appendix, Fig. S5). This is in agreement with the high degree of conservation of these genes and their lower levels of inactivating mutations in comparison to other regulatory and accessory functions (Fig. 1B), which may be either translated at lower levels or expressed only under certain circumstances.
A category of proteins detected in the proteome with remarkable abundance were molecular chaperones. In fact, the three proteins with the most peptide hits were GroES and two copies of GroEL (taken together, these three proteins comprise 9.2% of all peptide matches against databases), and all molecular chaperones in the proteome had above median hits (Fig. 6 and SI Appendix, Fig. S6). High expression levels of chaperones are common in symbionts with extreme genome reduction (46, 47). It has been postulated that these proteins can help to stabilize the proteome after slightly deleterious mutations accumulate and are fixed due to the strong bottlenecks associated to vertical transmission, leading to proteins that may require increased assistance in folding (47–49).
Differential Expression in the Antennal Reservoir: Transport, Nutrient Catabolism, and Antibiotic Biosynthesis.
In order to gain insights into the molecular basis of host–symbiont interactions, we compared transcript levels between the antennal samples of “S. philanthi” and those grown in in vitro cultures. Despite the strong decay in transport systems, which is one of the cellular functions with the highest proportion of pseudogenes (Fig. 1B), several permeases and ABC transporters were significantly overexpressed in antennal samples (Fig. 3), highlighting the need to import nutrients from the host inside the antennal reservoirs. Among the overexpressed genes, we found transporters for inorganic phosphate and the sugar chitobiose. The former may indicate that cells are under phosphate limitation in the antennal reservoir milieu, while the latter is of interest, as we also found overexpression in the antennae of a GH18 domain protein, a possible chitinase, which could be excreted to degrade chitin into N-acetylglucosamine or chitobiose and/or may contribute to the antifungal properties of the symbionts.
Among the metabolic pathways, we found the transcripts of several of the terminal genes in arginine biosynthesis to be up-regulated in antennae, suggesting a metabolic bypass of the interrupted pathway through provisioning of ornithine by the host (Fig. 4). Additionally, many of the overexpressed genes in antennae are part of the catabolism of branched-chain amino acids and fatty acids, the products of which are acetyl-CoA, propionyl-CoA, malonyl-CoA, and methylmalonyl-CoA, all intermediate metabolites required for piericidin biosynthesis (26) (Figs. 3 and 6A and SI Appendix, Fig. S7). Malonyl-CoA and methylmalonyl-CoA in particular are the main chain extension units for the polyketide synthase involved in this pathway and derive mostly from the catabolism of isoleucine and valine. Furthermore, genes in the piericidin cluster itself are among the most highly overexpressed genes in antennal samples, all with more than fourfold increased transcript levels as compared to in vitro culture conditions (Fig. 6A). This expression pattern highlights the host–symbiont metabolic interaction, beginning with the transport of branched-chain amino acids from the antennal reservoirs into the symbiont cells, followed by the catabolism into short-chain fatty acids and their usage for piericidin and actinopyrone biosynthesis, resulting in the transport of antibiotics back into the antennal gland reservoirs. Concordantly, a putative antibiotic exporter is also overexpressed in antennal samples (Fig. 3). Thus, we observed a potential metabolic link between specific host-provided nutrients and the symbionts’ production of defensive metabolites that are beneficial for the host.
Discussion
Erosion of a Large, Linear, GC-Rich Genome.
Genome reduction is a common phenomenon observed in vertically transmitted symbionts (3, 8, 36). While “S. philanthi” retains a large genome in comparison to most obligate symbionts and even many free-living bacteria, a massive gene decay seems to be occurring in this strain. Concordantly, comparisons with the genome sizes of the closest free-living Streptomyces indicate that “S. philanthi” has already lost up to a third of its chromosome due to deletions. In addition, around one-third of its remaining CDS seem to be undergoing pseudogenization initiated by frameshift mutations. In contrast, other known actinobacterial symbionts show very minor (if any) signs of pseudogenization (16, 50), with only the obligate pathogen Mycobacterium leprae displaying a pattern of genomic decay comparable to “S. philanthi” (40).
A peculiar characteristic of Streptomyces bacteria is the linear genome topology, with a conserved central core region that contains most essential genes and a pair of arms toward the telomere regions that are prone to large-scale rearrangements due to their enrichment with transposable and horizontally acquired elements (44, 51). To the best of our knowledge, extensive genome erosion has not yet been described in bacteria with a linear genome. We observed different dynamics of genomic decay among these regions, with loss of genetic material heavily biased toward the arms as opposed to the core of the chromosome, to the point that one of the arms is almost completely absent in comparison to genomes from closely related free-living Streptomyces (SI Appendix, Fig. S1). However, we did not observe any atypical accumulation of prophage sequences or genomic islands (Table 1) in “S. philanthi,” so these mobile elements do not seem to be contributing to the genomic decay observed in this strain.
Another remarkable feature of the “S. philanthi” genome is the apparent bias toward G+C accumulation, especially in intergenic regions that are expected to experience relaxed selection. This is contrary to what is seen in most other cases of genome erosion (3), with the exception of the highly eroded but G+C-rich genomes of the cicada and mealybug symbionts Hodgkinia and Tremblaya, respectively (41, 42). While the G+C bias during genome erosion of S. philanthi could be interpreted as a feature of high GC bacteria due to the higher availability of G+C nucleotides in the cell (52), the only other reported case of genome erosion in high GC bacteria revealed a reduced G+C content in M. leprae [57.8% G+C (40)] as compared to the congeneric M. tuberculosis [65.6% G+C (53)]. In addition, mutations in bacteria seem to be universally biased toward A+T (54, 55), and this mutational bias has also been reported for the G+C-rich and eroded genome of Hodgkinia (56). In this nutritional symbiont of cicadas, hitherto unknown selective forces have been suggested to maintain a high genomic G+C content (56), given the absence of DNA repair and recombination pathways that could result in biased gene conversion as an alternative mechanism to counterbalance the mutational bias (42). However, as the beewolf symbiont exhibits an increased G+C content particularly in the intergenic regions, selection appears to be an unlikely driving force. Alternatively, an unusual mutational bias toward G+C may provide a possible explanation for the high genomic G+C content, putatively resulting from erosion of some of the DNA repair pathways while maintaining functional copies of mutM and mutY, which are known to cause an elevated rate of CG to AT transversions when mutated (43).
Interestingly, many of the pseudogenes in the genome of “S. philanthi biovar triangulum” were still transcribed, but the corresponding proteins were not detected in the proteome, indicating the apparent silencing of pseudogene-derived transcripts at the post-transcriptional, translational, or post-translational stage. This seems to imply that the costs of deleterious mutations are predominantly manifested at the level of the proteome, rather than the utilization of nucleotides in DNA or RNA or the costs of replication or transcriptional processes. Presumably, the accumulation of misfolded peptides because of interrupted and frameshifted coding sequences poses a large metabolic burden on the symbiont cell, which is in line with the observed abundance of molecular chaperones in the proteome. Concurrent observations have been recently reported in the facultative endosymbiont Sodalis glossinidius, which also shows a strong accumulation of pseudogenes (∼40% of the core genome) with active or residual transcription for around half of them (57). This suggests a transitional state during genome erosion in symbionts in which the transcription of pseudogenes is maintained, but the deleterious effects of accumulation of malfunctional proteins is kept at bay through post-transcriptional regulatory mechanisms.
Considering that the beewolf–“S. philanthi” symbiosis dates back to the late Cretaceous (∼68 Mya) (31), it seems surprising that its genome presents signatures of an early stage of erosion, relative to many other insect symbionts (58). By comparison, Buchnera, the obligate nutritional endosymbiont of aphids, lost an estimated 85% of its genomic content over the course of 200 to 250 My (59). Although in the uni- or bivoltine beewolf hosts (60) the number of symbiont generations per year is probably substantially lower than for Buchnera in aphids, this difference seems insufficient to explain the retention of a much larger proportion of the genome over the course of 68 My, particularly since mutations in the base excision repair, homologous recombination, and nonhomologous end joining systems suggest that the genome may be prone to accumulate mutations at a fast pace. In addition, the beewolf symbionts experience particularly severe population bottlenecks during vertical transmission, likely exacerbating genomic decay (32). Hence, it seems possible that the process of genome erosion has started more recently than the inferred origin of the symbiotic relationship between Streptomyces and beewolves. Consistent with this idea are previous observations that the symbionts of North American beewolves appear to be metabolically more versatile (34) and can occasionally be transmitted horizontally between host species (31, 34). This suggests that “S. philanthi” symbionts have only recently transitioned to an obligate insect association and some strains may even still retain the ability to live outside of the host. A mixed lifestyle would be expected to impose a stronger selection for the retention of genes that are necessary for an autonomous life. Comparative genomic analyses of different beewolf symbionts are necessary to test this hypothesis and shed light on the onset and dynamics of genome erosion in these defensive actinobacterial symbionts.
Functional Implications of Genome Erosion in “S. philanthi.”
Defensive mutualists often retain more complete metabolic pathways than obligate nutritional symbionts, but their biosynthetic capabilities nevertheless show reductions and adaptations to their host environment (61, 62). In “S. philanthi,”, most of the essential metabolic functions appear to be still intact despite the ongoing genome erosion, except for auxotrophies for proline, arginine, and biotin, which were predicted in silico and corroborated in vitro (Fig. 4). These auxotrophies indicate dependence on the host for nutrition. Interestingly, both arginine and biotin are essential nutrients that insects cannot synthesize themselves (63), so beewolves likely have to meet their own and the symbionts’ demands for these metabolites from the diet.
One of the functional categories in the “S. philanthi” genome that is most severely affected by frameshift mutations is regulatory proteins, which may contribute to the surprisingly high correlation between gene expression and protein abundance for the intact genes in in vitro cultures (Fig. 6B), which stands in contrast with the weak correlation between transcript and protein levels observed in other bacteria (64, 65). Obligate intracellular symbionts often lose most or all of their regulatory genes (59, 66, 67), whereas free-living Streptomyces, similar to other soil-dwelling bacteria, encode dozens of sigma factors and hundreds of other regulatory proteins (44, 68). While not as drastic as in obligate symbionts, the reduced repertoire of regulators in “S. philanthi” fits with the notion that this bacterium occupies a niche that has little variation in nutritional and environmental conditions.
Symbionts with extreme genome reduction commonly show high expression levels of molecular chaperones (8, 46, 47), and it has been postulated that these proteins help to stabilize the proteome after slightly deleterious mutations accumulate and are fixed due to the strong bottlenecks associated with vertical transmission (47–49, 69). Due to the lack of axenic cultures for most insect symbionts, however, the expression of chaperones has been mostly assessed within the host, so stress associated with the host internal environment (e.g., due to the host’s immune response or the physicochemical environment) remained an alternative explanation for their high abundance in the proteome. However, in the case of “S. philanthi,” we observed a high constitutive expression of most chaperones and chaperonins, under both in vitro cultures and in vivo conditions (Fig. 6A), providing additional evidence that this is a compensatory mechanism to alleviate the mutational load in “S. philanthi” and maintain the structural stability of its proteome. Alternatively, the observed reduction in the number of transcriptional regulators may have led to the hardwiring of chaperone expression, preventing the symbionts from reacting to changes in their environment imposed by the artificial transfer into an in vitro culture.
Metabolic Interactions between Beewolves and Their Symbionts.
Consistent with the previous detection of antibiotics in beewolf antennae (26), our results revealed an overexpression of genes in the symbionts’ piericidin/actinopyrone biosynthetic cluster in the antennal reservoirs in comparison to in vitro cultures. In addition, transporters and catabolic pathways for branched-chain amino acids as well as enzymes involved in fatty acid catabolism were up-regulated in beewolf antennae, which provide the precursors (especially malonyl-CoA and methylmalonyl-CoA) for piericidin and actinopyrone biosynthesis (26). Hence, the host could exert control of antibiotic production by supplying both the substrates and the necessary cofactor (biotin) for polyketide biosynthesis. However, a nutritional role of these differentially expressed genes cannot be ruled out. Furthermore, it remains unclear why antibiotics are produced by the symbionts in the antennal glands in the first place, as a defensive benefit has thus far only been demonstrated for the beewolf cocoon (25). It seems possible, however, that the antibiotics are also used to defend the symbionts’ niche in the antennal reservoirs from microbial invaders or protect them from antagonists after secretion into the brood cell. Alternatively, the frequently observed grooming of antennae in female beewolves may serve to spread the antibiotics over the body surface, granting protection from fungal pathogens to the adult female, akin to what has been observed in attine ants that are defended by their Pseudonocardia symbionts from entomopathogens (70).
Overexpression in the antennal reservoirs of genes encoding a putative chitinase and a chitobiose transporter provides clues to other possible nutritional interactions between “S. philanthi” and its host as well as to the evolutionary origin of this mutualism. Conceivably, Streptomyces bacteria were initially growing directly on the wasp cuticle, using chitinase as a protection from antagonistic fungi and/or a virulence factor to acquire chitin (71) from the wasp cuticle as a carbon and nitrogen source before the antennal glands evolved as the relationship became more intimate (72). Even after 68 My of host–symbiont coevolution, chitin may have remained a major source of nutrition for the beewolf symbionts, as indicated by the high expression of the symbionts’ chitinase and the chitobiose transporter in female antennae. Interestingly, chitin and its derivatives have been found to be relevant for other symbiotic interactions as well, such as, for example, providing a potential nutritional source in a tortoise leaf beetle’s extracellular pectinolytic symbiont Candidatus Stammera capleta (73). Furthermore, in the squid–Aliivibrio fischeri symbiosis, chitobiose is known to play a key role in the colonization of the squid’s light organ (74), and the availability of chitin alters the metabolism of A. fischeri, presumably helping the host modulate the physiological state of its symbionts for luminescence (75). Given that chitin is an abundant macromolecule in the cuticle of many invertebrates, it may more generally play a key role for the regulation and nutrition of extracellular symbionts.
Conclusions
Since the late Cretaceous, beewolves engaged in a symbiosis with Streptomyces bacteria that provide protection from antagonistic fungi to the developing beewolf in the cocoon by producing antimicrobial compounds. Despite this evolutionarily old symbiotic alliance, we show that the genome of the European beewolf symbiont “S. philanthi biovar triangulum” is in an early stage of genome erosion relative to many other insect symbionts. A large proportion of this strain’s genes bear possibly inactivating frameshift mutations, but most of these pseudogenes were found to be transcribed, even if at lower levels than intact genes. Nevertheless, proteins encoded by pseudogenes were hardly detected by proteomic analysis, suggesting the fitness cost of producing nonfunctional or misfolded products from eroding genes. Finally, functional genomic analysis, transcriptomics, and proteomics uncover that the host’s supply of essential nutrients and cofactors not only compensates for the symbionts’ auxotrophies because of genomic decay but also might enable control of antibiotic production in the antennal reservoirs. Thus, the beewolf–Streptomyces symbiosis presents an experimentally tractable system that provides valuable insights into the metabolic integration and genomic evolution of extracellular defensive symbionts in insects.
Materials and Methods
Beewolf Specimens and Bacterial Cultures.
Adult European beewolves (Philanthus triangulum) were collected from field populations in Würzburg and Erlangen, Germany, and antennae of female specimens were pooled for subsequent nucleic acids extractions and genome and transcriptome sequencing. Additionally, axenic cultures of “S. philanthi biovar triangulum” strain 23Af2 were grown in Grace’s medium and used for sequencing. This strain was originally isolated from the antennae of a P. triangulum female collected in Berlin, Germany (34). Despite the successful cultivation, the strain has not yet been validly described, so the species name is given in quotation marks in accordance with the List of Prokaryotic names with Standing in Nomenclature (76).
Total DNA Extraction and Purification.
Total DNA was extracted from beewolf antennal samples using the MasterPure Complete DNA and RNA Purification Kit (Epicentre Biotechnologies) as described previously (31). Additionally, genomic DNA was extracted from a pure culture of “S. philanthi.” Briefly, bacteria were lysed in TE25S buffer (25 mM Tris [pH 8.0], 25 mM ethylenediaminetetraacetic acid [EDTA; pH 8.0], and 0.3 M sucrose) with lysozyme (2 mg/mL), proteinase K (20 mg/mL), and sodium dodecyl sulfate (SDS) (0.6%); proteins were precipitated from the lysate using Protein Precipitation Solution (Qiagen) followed by centrifugation at >16,000 × g for 10 min at 4 °C. Nucleic acids were precipitated with an equal volume of isopropanol followed by centrifugation at ≥16,000 × g for 10 min. The pellet was then washed twice with EtOH (70%), air dried, and resuspended in elution buffer (EB) buffer; later, the extracted nucleic acid solution was treated with RNase A (Epicentre), and the DNA was precipitated using isopropanol as described above.
Genome Sequencing, Assembly, and Assembly Validation.
Initially, the genome of symbiotic “S. philanthi” was sequenced from pooled European beewolf (Philanthus triangulum) antennal samples from field populations in Würzburg and Erlangen, Germany, using Illumina HiSeq 2000 (Fasteris SA) and Sanger technologies. However, very few (about 3,900) Sanger reads remained after trimming. Illumina reads were trimmed and assembled with CLC Genomic Workbench (CLC Bio); further analysis of the contigs conducted with the program MetaLingvo (http://seqword.bi.up.ac.za/metalingvo/index.html) demonstrated that only 10% of all contigs (in total 1,963 contigs, comprising 6.5 million nucleotides) potentially originated from Streptomyces bacteria, while the majority of the remaining 90% originated from the host according to blastn and blastx results that revealed similarities to insect sequences. Coverage of Sanger and Illumina reads across the genome were 1.4× and 731.8×, respectively.
Additionally, genomic DNA extracted from the subsequently achieved axenic bacterial culture of “Streptomyces philanthi biovar triangulum” strain 23Af2 was sequenced with GS FLX+ Titanium (454 Life Sciences) from shotgun (LGC Genomics) and 8 kb paired-end libraries (Eurofins MWG Operon, Inc.). De novo genome assembly of the reads was performed using Newbler software package v 2.7 (454 Life Sciences, Roche) that resulted in one scaffold of 261 contigs and two free contigs, with an average coverage of 35.8×. Trimmed Sanger reads and Illumina contigs were used to assemble two free contigs to the larger scaffold applying Geneious version 6.0.5 [http://www.geneious.com (77)].
Optical mapping (Eurofins MWG Operon, Inc.) with restriction endonuclease KpnI followed by the visualization of results with MapSolver (OpGen) verified the genome assembly but also helped to fix a mis-assembly of the ∼100 kb-long terminal regions of the bacterial chromosome. Next, the genomic DNA was sequenced using InView de novo Genome 2.0 with PacBio RS II technology followed by assembly service (GATC Biotech AG) that delivered eight contigs, with a mean coverage of 91.8×.
Finally, the 261 pyrosequencing contigs were assembled with the eight PacBio contigs using Geneious software followed by the manual curation of the assembled sequence, resulting in the complete bacterial chromosome in a single contig. This bacterial chromosome was compared and additionally verified against the scaffold constructed using pyrosequencing contigs, as well as Sanger reads and Illumina contigs obtained from the assembly of reads generated from antennal samples as described above. However, for the subsequent genome analysis we used the chromosome assembled from the pyrosequencing and PacBio contigs as they are originated from the same pure bacterial culture.
Genome Annotation and Analysis.
Gene prediction in the bacterial chromosome and automated genome annotation were conducted using GenDB system (v 2.4) (78) followed by manual curation of machine annotation. Gene categories were defined as follows: 1) intact genes with high sequence identity (>50%) and high length similarity (>80%) to sequences in the NCBI nr database, 2) genes in the process of pseudogenization with high sequence identity (>50%) but with one or more detected frameshift mutations, and 3) hypothetical genes with low sequence identity (≤35%) of the whole sequence. Sites of frameshift mutations were predicted using FrameD (79) and GeneTack (80, 81) software with S. bingchenggensis and S. avermitilis precalculated models, respectively. Furthermore, coverage at frameshift mutation sites was calculated with the trimmed 454 and Sanger reads using Geneious; mismatching nucleotides were ignored during the counts.
Analysis of “S. philanthi” central metabolic, genetic information processing and some other genes/pathways was done using individual gene categories via the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server (KAAS, v2.1) (82). Amino acid sequences of the predicted genes were run against the KAAS database using the bidirectional best hit method (complete or draft genome) for the subset of intact genes and the single-directional best hit method (partial genome) for the subset of frameshifted genes in order to receive reconstructed KEGG pathway maps and visualize intact pathways and those affected by frameshift mutations. The following species of streptomycetes available from the database were used as references: S. coelicolor, S. avermitilis, S. griseus, S. scabiei, and S. noursei. In addition to the automatic annotation, manual curation was performed by blastn and blastx against the NCBI nr database within the Basic Local Alignment Search Tool (BLAST) online tool (83). Secondary metabolite clusters were predicted using antiSMASH 5.0 (84) followed by a manual curation of the results. Prophages were identified using the PHAge Search Tool Enhanced Release (PHASTER, http://phaster.ca/, prophage database last updated December 2020) (85). Genomic islands were identified using SeqWord Sniffer (86) and the integrated predictions from IslandViewer 4 (https://www.pathogenomics.sfu.ca/islandviewer/, reference database last updated December 2020) (87).
The annotated genome of “S. philanthi biovar triangulum” strain 23Af2 is archived at NCBI database with BioProject accession number PRJNA37727.
Frameshift Mutation Validation.
On average, a frameshifted gene contained a single predicted frameshift mutation site with high (32.8×) average coverage with 454 reads fully consented at this position; 18% of the predicted frameshift sites were also covered with Sanger reads (average coverage 1.4×) matching at this position with 454 reads as well. Generally, coverage of 454 reads on frameshift sites overall complied with average read coverage across the genome (35.8×), indicating that the predicted frameshift mutations were not artifacts arising from genome sequencing errors or genome mis-assembly.
RNA-Seq Analysis.
RNA-seq analysis was conducted on the “S. philanthi” harvested after growing for 1 wk at 30 °C in solid Grace’s medium with added 10% fetal bovine serum (FBS; three replicates) and from in vivo samples (antennal reservoirs, two replicates of pooled antennae from six female Philanthus triangulum). Nucleic acids were extracted and harvested as described above; total DNA was then removed using deoxyribonuclease I. Total RNA was precipitated with an equal volume of isopropanol followed by centrifugation at ≥16,000 × g for 10 min at 4 °C. The pellet was then washed twice with EtOH (70%), air dried, and resuspended in EB buffer. Library preparation and sequencing of the antennal samples was done using the Illumina HiSeq 2000 platform (100 bp paired-end reads) by Fasteris SA. The in vitro samples were sequenced at the Max Planck Genome Center on the Illumina HiSeq 2500 platform. In both cases, library preparation included the depletion of ribosomal RNA and polyA depletion. Raw data were deposited in the NCBI Sequence Read Archive database under the BioProjects with accession numbers PRJNA542283 (antennal samples) and PRJNA701492 (in vitro cultures).
Sequenced reads were quality checked and trimmed using the Trimommatic implementation in KBase (v1.2.14, https://www.kbase.us) (88), the alignment of the reads to the reference genome was performed with Bowtie 2 (v2.3.2) (89), and aligned reads were assembled using StringTie (v1.3.3) (90). The differential expression analysis was done using DESeq2 (v1.20.0) (91). Statistical analysis and graphs were performed using R (v. 3.3.3) (92) implemented in RStudio (v. 1.1.383, https://www.rstudio.com/).
Protein Extraction and Separation.
For the characterization of the soluble proteome, bacterial biomass was harvested after 5 d of growth at 30 °C grown in liquid Grace’s medium with added 10% FBS. Proteins were precipitated by the addition of 10% (wt/vol) trichloroacetic acid. After centrifugation at 13,000 × g for 10 min at 4 °C, the supernatant was discarded, the pellet was washed twice in 100% acetone (−20 °C), and it was dried at room temperature. For SDS-polyacrylamide gel electrophoresis (PAGE), the pellet was resuspended in 5% (wt/vol) SDS solution, and an aliquot of 20 µg protein was solubilized with SDS-PAGE sample buffer (125 mM Tris HCl, pH 6.8, 20% vol/vol glycerol, 10% wt/vol SDS, 5% vol/vol b-mercaptoethanol, and bromophenol blue).
In order to characterize membrane-bound proteins, bacterial biomass was harvested after growth as indicated in the previous section by centrifugation (8,000 × g for 10 min at 4 °C). The cell pellet was washed twice with buffer (50 mM Tris and 150 mM NaCl, pH 7.5), and the cells were lysed using a beat beater (Precellys 24 Homogenizer, Peqlab). The lysate was centrifuged (13,000 × g for 20 min at 4 °C), and the collected supernatant was subjected to ultracentrifugation (100,000 × g for 1 h at 4 °C). The resulting pellet was resuspended in 4 mL carbonate buffer (100 mM Na2CO3, 100 mM NaCl, and 10 mM EDTA, pH 11), incubated in an overhead shaker for 1 h at 4 °C and ultracentrifuged (100,000 × g for 1 h at 4 °C). The pellet was homogenized in high salt buffer (1 M NaCl, 20 mM Tris HCl, and 10 mM EDTA, pH 7.5). After ultracentrifugation, the purified membrane pellet was resuspended in 5% SDS, and an aliquot of 20 µg protein was solubilized with SDS-PAGE sample buffer and separated by electrophoresis (93).
For two-dimensional (2D) gel electrophoresis, 200 μL rehydration buffer was added to the sample (7 M urea, 2 M thiourea, 2% Chaps, 0.75% IPG buffer 3–11, and bromophenol blue) and was immediately adsorbed onto 11 cm immobilized pH 3–11 gradient (immobilized pH gradient [IPG]) strips (GE Healthcare Bio-Sciences) and rehydrated overnight. Isoelectric focusing was performed on an Ettan IPGphor II (Amersham plc) by using the following program: 500 V for 1 h, 500 to 1,000 V for 1 h, 1,000 to 6,000 V for 2 h, and 6,000 V for 40 min. After focusing, the IPG strips were equilibrated for 15 min in 10 mL equilibration buffer containing 6 M urea, 30% [vol/vol] glycerol, 2% [wt/vol] SDS, 75 mM Tris HCl, 0.002% [wt/vol] bromophenol blue, and 1% (wt/vol) dithiothreitol (DTT). Then, the strips were incubated for 15 min in the same buffer containing 2.5% (wt/vol) iodoacetamide instead of DTT. For the separation of proteins in the second dimension, a 12.5% Criterion Tris HCl Gel (Bio-Rad) was used. The proteins were separated for 3 h at 100 V and stained with Roti-Blue (Carl Roth) and visualized by a densitometer (Bio-Rad GS-800).
For SDS-PAGE, the protein sample was diluted in Roti-Load buffer (Carl Roth) and loaded on a 12.5% Criterion Tris HCl precast one-dimensional (1D) gel (Bio-Rad) according to manufacturer’s instructions. Proteins were allowed to migrate for 1 h at a constant voltage of 200 V. After migration, gels were stained for overnight with Roti-Blue (Carl Roth) staining solution. Gels were rinsed twice with deionized water, destained in 25% methanol, and scanned using a densitometer (Bio-Rad GS-800).
Protein Digestion and Liquid Chromatography–Mass Spectrometry Analysis.
Protein bands/spots were cut from the gel matrix and tryptic digestion was carried out as described previously (94). For liquid chromatography–mass spectrometry (LC–MS) analysis, the extracted tryptic peptides were reconstructed in 10 µL aqueous 1% formic acid. Depending on staining intensity, 1 to 5 µL of sample were injected into the LC–tandem mass spectrometry (MS/MS) system. The samples were acquired on a nanoACQUITY UPLC System online connected to a QTOF Synapt HDMS mass spectrometer (Waters). The peptides were concentrated on a Symmetry C18 Trap Column (20 × 0.18 mm, 5 µm particle size, Waters) using a mobile phase of 0.1% aqueous formic acid at a flow rate of 15 µL ⋅ min−1 and separated on a nanoACQUITY C18 column (200 mm × 75 µm ID, C18 BEH 130 material, 1.7 µm particle size, Waters) by inline gradient elution at a flow rate of 0.350 µl ⋅ min−1 using the following gradient: 1 to 40% B over 40 min, 40 to 80% B over 10 min, 80 to 95% B over 1 min, isocratic at 95% B for 2 min, and a return to 1% B (buffers: A, 0.1% formic acid in water; B, 100% acetonitrile in 0.1% formic acid). Data were acquired using data-dependent acquisition (DDA) and data-independent acquisition (referred to as enhanced MSE). The acquisition cycle for DDA analysis consisted of a survey scan covering the range of m/z 400 to 1,800 Da followed by MS/MS fragmentation of the five most intense precursor ions collected at 1.5 s intervals in the range of 50 to 2,000 m/z. Dynamic exclusion was applied to minimize multiple fragmentations for the same precursor ions.
For LC–MSE analyses, full-scan LC–MS data were collected using an alternating mode of acquisition: low energy (MS) and elevated energy (MSE) mode at 1.5 s in the range m/z of 50 to 1,900. The collision energy of low energy MS mode and high energy mode (MSE) were set to 4 eV and 20 to 40 eV energy ramping, respectively. A reference compound, human Glu-Fibrinopeptide B [650 fmol/mL in 0.1% formic acid/acetonitrile (vol/vol, 1:1)], was infused through a reference sprayer at 30 s intervals for external calibration. The data acquisition was controlled by MassLynx v4.1 software (Waters).
Data Processing and Protein Identification.
DDA raw data were processed and searched against a sub-database containing common contaminants (human keratins and trypsin) using ProteinLynx Global Server (PLGS) version 2.4 (Waters). The following search parameters were applied: fixed precursor ion mass tolerance of 15 ppm for survey peptide, fragment ion mass tolerance of 0.1 Da, estimated calibration error of 0.002 Da, one missed cleavage, fixed carbamidomethylation of cysteines, and possible oxidation of methionine. Spectra that remained unmatched by database searching were interpreted de novo to yield peptide sequences and subjected for homology-based searching using MS BLAST program (95) installed on a local server. MS BLAST searches were performed against a “Streptomyces philanthi” sub-database obtained from in silico translation of the “Streptomyces philanthi” transcriptome and against the bacterial genomes from the NCBI nr database downloaded from https://www.ncbi.nlm.nih.gov/ on May 12, 2012. In parallel, pkl files of MS/MS spectra were generated and searched against the “Streptomyces philanthi” sub-database combined with NCBI nr database) using Mascot software version 2.3 using the search parameters described above (Matrix Science Ltd).
The acquired continuum LC–MSE data were processed using PLGS version 2.5.2 (Waters). The thresholds for low/high energy scan ions and peptide intensity were set at 150, 30, and 750 counts, respectively. The processed data were searched against a Streptomyces protein sub-database combined with a Swiss-Prot database downloaded from https://www.uniprot.org/ (database downloaded in February 2012). The database search was performed at a false discovery rate of 4%, with the following search parameters for the minimum numbers of product ion matches per peptide (3), product ion matches per protein (7), peptide matches (1), and maximum number of missed tryptic cleavage sites (1). Searches were restricted to tryptic peptides with a fixed carbamidomethyl modification for Cys residues. In total, 951 individual proteins representing the main metabolic pathways were identified from 1D and 2D gels resolving soluble and membrane protein fractions. The amount of peptide matches to identified proteins was used as a semiquantitative proxy to gauge their relative abundance.
Statistical Methods.
To evaluate the effects of coding sequence intactness on gene expression, several statistical tests were performed on the transcriptome and proteome data sets. Each CDS was assigned to a gene category (intact, pseudogene, hypothetical) as described in Genome Annotation and Analysis. Values of TPM from the RNA-seq dataset and binary data indicating whether the encoded protein was identified in the proteomic experiments were included in a single table. All analysis was performed using R (v. 3.3.3) (92) implemented in RStudio (v. 1.1.383). Pearson’s χ2 test for homogeneity (chisq.test function from package stats) was used to evaluate whether the proportions of intact, frameshifted, and hypothetical genes differed between the genome, transcriptome, and proteome in pairwise comparisons. To discern whether the probability of protein detection was influenced by the mRNA abundance and/or the intactness of the coding sequence, a binomial logistic regression was used as follows. A generalized linear model was fitted using the binomial family and logit link function (glm function from package stats). The explanatory variables evaluated were TPM values and intactness category, and the response variable was detection in the proteome. Models were considered with and without interaction between the explanatory variables, but the interaction did not significantly reduce the deviance (analysis of deviance using the anova.glm function, χ2 test, P = 0.595) and was therefore discarded in favor of the simpler model without interaction. It was then evaluated whether the individual explanatory variables contributed to the model fit, using the anova function. Both TPM values and intactness category significantly reduced the deviance of the model (Wald’s χ2 test, P < 2e-16), but intactness category caused the largest drop in deviance in comparison to the null model. The package “effects” (96) was used to plot the effects of both predictors (SI Appendix, Fig. S4). All other graphs were created using ggplot2 and ggpubr (97).
Supplementary Material
Acknowledgments
We thank Alex Aoyagi and Andrew von Niederhausen for help with the sequencing, Johanna Rüllich for assistance with the in vitro assays, and Yvonne Hupfer for protein extraction, protein separation, and sample preparation for LC–MS analysis. We acknowledge assistance for genome analysis by the Bioinformatics Core Facility/Professorship of Systems Biology at Justus Liebig University Giessen and access to resources financially supported by the Federal Ministry of Education and Research Grant FKZ 031A533 to the Bielefeld-Gießen center within the German Network for Bioinformatics Infrastructure network. We gratefully acknowledge funding from the German Research Foundation (DFG KA2846/2-1 and KA2846/2-2, to M.K.), the Volkswagen Foundation (to M.K.), and the Max Planck Society (to T.Y.N., T.E., and M.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023047118/-/DCSupplemental.
Data Availability
Genome and transcriptome sequencing data have been deposited in the NCBI database, BioProject, with the accession numbers PRJNA37727 (98), PRJNA542283 (99), and PRJNA701492 (100). Proteomic data are available from the Open Research Data Repository of the Max Planck Society (https://dx.doi.org/10.17617/3.5r) (101).
References
- 1.Feldhaar H., Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36, 533–543 (2011). [Google Scholar]
- 2.Douglas A. E., Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCutcheon J. P., Moran N. A., Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Salem H., et al., Drastic genome reduction in an herbivore’s pectinolytic symbiont. Cell 171, 1520–1531.e13 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bennett G. M., Moran N. A., Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol. Evol. 5, 1675–1688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-Madrigal S., Latorre A., Porcar M., Moya A., Gil R., Complete genome sequence of “Candidatus Tremblaya princeps” strain PCVAL, an intriguing translational machine below the living-cell status. J. Bacteriol. 193, 5587–5588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Łukasik P., et al., Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc. Natl. Acad. Sci. U.S.A. 115, E226–E235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran N. A., McCutcheon J. P., Nakabachi A., Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi Y., et al., Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7, 2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikoh N., Hosokawa T., Oshima K., Hattori M., Fukatsu T., Reductive evolution of bacterial genome in insect gut environment. Genome Biol. Evol. 3, 702–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakabachi A., et al., Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 23, 1478–1484 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Flórez L. V., et al., An antifungal polyketide associated with horizontally acquired genes supports symbiont-mediated defense in Lagria villosa beetles. Nat. Commun. 9, 2478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaltenpoth M., Actinobacteria as mutualists: General healthcare for insects? Trends Microbiol. 17, 529–535 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Seipke R. F., Kaltenpoth M., Hutchings M. I., Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 36, 862–876 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Selvin J., et al., Antibacterial potential of antagonistic Streptomyces sp. isolated from marine sponge Dendrilla nigra. FEMS Microbiol. Ecol. 50, 117–122 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Quezada M., et al., Diverse cone-snail species harbor closely related Streptomyces species with conserved chemical and genetic profiles, including polycyclic tetramic acid macrolactams. Front. Microbiol. 8, 2305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Z., et al., Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem. Sci. (Camb.) 8, 3218–3227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Currie C. R., Scott J. A., Summerbell R. C., Malloch D., Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398, 701 (1999). [Google Scholar]
- 19.Haeder S., Wirth R., Herz H., Spiteller D., Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4742–4746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen R., et al., Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. U.S.A. 106, 17805–17810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barke J., et al., A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 8, 109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulcr J., et al., Presence and diversity of Streptomyces in Dendroctonus and sympatric bark beetle galleries across North America. Microb. Ecol. 61, 759–768 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Scott J. J., et al., Bacterial protection of beetle-fungus mutualism. Science 322, 63 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaltenpoth M., et al., ‘Candidatus Streptomyces philanthi’, an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int. J. Syst. Evol. Microbiol. 56, 1403–1411 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Kaltenpoth M., Göttler W., Herzner G., Strohm E., Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15, 475–479 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Engl T., et al., Evolutionary stability of antibiotic protection in a defensive symbiosis. Proc. Natl. Acad. Sci. U.S.A. 115, E2020–E2029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Challis G. L., Hopwood D. A., Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U.S.A. 100 (suppl. 2), 14555–14561 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ventura M., et al., Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doroghazi J. R., Metcalf W. W., Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14, 611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barka E. A., et al., Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaltenpoth M., et al., Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc. Natl. Acad. Sci. U.S.A. 111, 6359–6364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaltenpoth M., Goettler W., Koehler S., Strohm E., Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol. Ecol. 24, 463–477 (2010). [Google Scholar]
- 33.Kroiss J., et al., Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6, 261–263 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Nechitaylo T. Y., Westermann M., Kaltenpoth M., Cultivation reveals physiological diversity among defensive ‘Streptomyces philanthi’ symbionts of beewolf digger wasps (Hymenoptera, Crabronidae). BMC Microbiol. 14, 202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirby R., Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol. Lett. 319, 1–10 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Wernegreen J. J., Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3, 850–861 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Casadevall A., Evolution of intracellular pathogens. Annu. Rev. Microbiol. 62, 19–33 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Darby A. C., Cho N.-H., Fuxelius H.-H., Westberg J., Andersson S. G., Intracellular pathogens go extreme: Genome evolution in the Rickettsiales. Trends Genet. 23, 511–520 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Moran N. A., Microbial minimalism: Genome reduction in bacterial pathogens. Cell 108, 583–586 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Cole S. T., et al., Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011 (2001). [DOI] [PubMed] [Google Scholar]
- 41.McCutcheon J. P., von Dohlen C. D., An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21, 1366–1372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCutcheon J. P., McDonald B. R., Moran N. A., Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lind P. A., Andersson D. I., Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17878–17883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bentley S. D., et al., Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Nett M., Ikeda H., Moore B. S., Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26, 1362–1384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan Y., Thompson J. W., Dubois L. G., Moseley M. A., Wernegreen J. J., Proteomic analysis of an unculturable bacterial endosymbiont (Blochmannia) reveals high abundance of chaperonins and biosynthetic enzymes. J. Proteome Res. 12, 704–718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kupper M., Gupta S. K., Feldhaar H., Gross R., Versatile roles of the chaperonin GroEL in microorganism-insect interactions. FEMS Microbiol. Lett. 353, 1–10 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Fares M. A., Moya A., Barrio E., GroEL and the maintenance of bacterial endosymbiosis. Trends Genet. 20, 413–416 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Moran N. A., Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 93, 2873–2878 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmes N. A., et al., Complete genome sequence of Streptomyces formicae KY5, the formicamycin producer. J. Biotechnol. 265, 116–118 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Chen C. W., Huang C.-H., Lee H.-H., Tsai H.-H., Kirby R., Once the circle has been broken: Dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18, 522–529 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Dietel A.-K., Merker H., Kaltenpoth M., Kost C., Selective advantages favour high genomic AT-contents in intracellular elements. PLoS Genet. 15, e1007778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole S. T., et al., Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Hershberg R., Petrov D. A., Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6, e1001115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hildebrand F., Meyer A., Eyre-Walker A., Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6, e1001107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Leuven J. T., McCutcheon J. P., An AT mutational bias in the tiny GC-rich endosymbiont genome of Hodgkinia. Genome Biol. Evol. 4, 24–27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodhead I., et al., Large-scale and significant expression from pseudogenes in Sodalis glossinidius–A facultative bacterial endosymbiont. Microb. Genom. 6, e000285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCutcheon J. P., Boyd B. M., Dale C., The life of an insect endosymbiont from the cradle to the grave. Curr. Biol. 29, R485–R495 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H., Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Evans H. E., O’Neill K. M., The Natural History and Behavior of North American Beewolves (Cornell University Press, 1988). [Google Scholar]
- 61.Toh H., et al., Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Degnan P. H., Yu Y., Sisneros N., Wing R. A., Moran N. A., Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl. Acad. Sci. U.S.A. 106, 9063–9068 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douglas A. E., The microbial dimension in insect nutritional ecology. Funct. Ecol. 23, 38–47 (2009). [Google Scholar]
- 64.Taniguchi Y., et al., Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkataramanan K. P., et al., Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol. Biofuels 8, 81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCutcheon J. P., Moran N. A., Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 19392–19397 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakabachi A., et al., The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Castro-Melchor M., Charaniya S., Karypis G., Takano E., Hu W.-S., Genome-wide inference of regulatory networks in Streptomyces coelicolor. BMC Genomics 11, 578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fares M. A., Ruiz-González M. X., Moya A., Elena S. F., Barrio E., Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417, 398 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Mattoso T. C., Moreira D. D., Samuels R. I., Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biol. Lett. 8, 461–464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Enomoto S., Chari A., Clayton A. L., Dale C., Quorum sensing attenuates virulence in Sodalis praecaptivus. Cell Host Microbe 21, 629–636.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaltenpoth M., Yildirim E., Gürbüz M. F., Herzner G., Strohm E., Refining the roots of the beewolf-Streptomyces symbiosis: Antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl. Environ. Microbiol. 78, 822–827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bauer E., Kaltenpoth M., Salem H., Minimal fermentative metabolism fuels extracellular symbiont in a leaf beetle. ISME J. 14, 866–870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandel M. J., et al., Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl. Environ. Microbiol. 78, 4620–4626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan M., Schwartzman J. A., Dunn A. K., Lu Z., Ruby E. G., A single host-derived glycan impacts key regulatory nodes of symbiont metabolism in a coevolved mutualism. MBio 6, e00811–e00815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parte A. C., Sardà Carbasse J., Meier-Kolthoff J. P., Reimer L. C., Göker M., List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kearse M., et al., Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyer F., et al., GenDB–An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31, 2187–2195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiex T., Gouzy J., Moisan A., de Oliveira Y., FrameD: A flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res. 31, 3738–3741 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Antonov I., Baranov P., Borodovsky M., GeneTack database: Genes with frameshifts in prokaryotic genomes and eukaryotic mRNA sequences. Nucleic Acids Res. 41, D152–D156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Antonov I., Borodovsky M., Genetack: Frameshift identification in protein-coding sequences by the Viterbi algorithm. J. Bioinform. Comput. Biol. 8, 535–551 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M., KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 84.Blin K., et al., antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arndt D., et al., PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ganesan H., Rakitianskaia A. S., Davenport C. F., Tümmler B., Reva O. N., The SeqWord genome browser: An online tool for the identification and visualization of atypical regions of bacterial genomes through oligonucleotide usage. BMC Bioinformatics 9, 333 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertelli C.et al.; Simon Fraser University Research Computing Group , IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arkin A. P., et al., KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pertea M., et al., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.R Core Team , R: A Language and Environment for Statistical Computing, v. 3.3.3 (R Foundation for Statistical Computing, Vienna, Austria, 2014). https://www.r-project.org/.
- 93.Hahne H., Wolff S., Hecker M., Becher D., From complementarity to comprehensiveness–Targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8, 4123–4136 (2008). [DOI] [PubMed] [Google Scholar]
- 94.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Shevchenko A., et al., Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926 (2001). [DOI] [PubMed] [Google Scholar]
- 96.Fox J., Effect displays in R for generalised linear models. J. Stat. Softw. 8, 27 (2003). [Google Scholar]
- 97.Kassambara A., ggpubr: ‘‘ggplot2’’ based publication ready plots (Version 0.2.1, R package, 2019). https://cran.r-project.org/web/packages/ggpubr/index.html. [Google Scholar]
- 98.Nechitaylo T. Y., et al., Streptomyces philanthi bv. triangulum 23Af2, complete genome sequence. NBCI GenBank. https://www.ncbi.nlm.nih.gov/bioproject/prjna37727. Deposited 9 April 2021.
- 99.Nechitaylo T. Y., Sandoval-Calderón M., Engl T., Strohm E., Kaltenpoth M., RNAseq reads from Philanthus triangulum and its symbiont, 'Candidatus Streptomyces philanthi.' NBCI Sequence Read Archive (SRA). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA542283. Deposited 4 June 2019.
- 100.Nechitaylo T. Y., Sandoval-Calderón M., Engl T., Strohm E., Kaltenpoth M., Streptomyces philanthi bv. triangulum transcriptome. NBCI Sequence Read Archive (SRA). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA701492. Deposited 16 February 2021.
- 101.Nechitaylo T. Y., et al., Proteomic data from Candidatus Streptomyces philanthi biovar triangulum. Open Research Data Repository of the Max Planck Society. 10.17617/3.5r. Deposited 9 March 2021. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome and transcriptome sequencing data have been deposited in the NCBI database, BioProject, with the accession numbers PRJNA37727 (98), PRJNA542283 (99), and PRJNA701492 (100). Proteomic data are available from the Open Research Data Repository of the Max Planck Society (https://dx.doi.org/10.17617/3.5r) (101).