Significance
The myelin sheath surrounding human neurons acts as insulation and increases their rate of signal transmission. Peripheral myelin protein (PMP22) is a key transmembrane protein involved in neuron myelination, the dysfunction of which can lead to significant human neuropathies. This report provides direct evidence that disease-associated PMP22 mutants are unstable and can self-associate to form dimers to a greater extent than wild-type protein, providing mechanistic insight into diseases such as Charcot-Marie-Tooth and Dejerine–Sottas syndrome. To accomplish this, we link mass-selected gas-phase protein stability measurements to in vivo protein trafficking data, paving the way for the future use of such data in understanding such complex biochemical processes.
Keywords: membrane protein, mass spectrometry, protein misfolding
Abstract
Peripheral myelin protein (PMP22) is an integral membrane protein that traffics inefficiently even in wild-type (WT) form, with only 20% of the WT protein reaching its final plasma membrane destination in myelinating Schwann cells. Misfolding of PMP22 has been identified as a key factor in multiple peripheral neuropathies, including Charcot-Marie-Tooth disease and Dejerine–Sottas syndrome. While biophysical analyses of disease-associated PMP22 mutants show altered protein stabilities, leading to reduced surface trafficking and loss of PMP22 function, it remains unclear how destabilization of PMP22 mutations causes mistrafficking. Here, native ion mobility–mass spectrometry (IM-MS) is used to compare the gas phase stabilities and abundances for an array of mutant PM22 complexes. We find key differences in the PMP22 mutant stabilities and propensities to form homodimeric complexes. Of particular note, we observe that severely destabilized forms of PMP22 exhibit a higher propensity to dimerize than WT PMP22. Furthermore, we employ lipid raft–mimicking SCOR bicelles to study PMP22 mutants, and find that the differences in dimer abundances are amplified in this medium when compared to micelle-based data, with disease mutants exhibiting up to 4 times more dimer than WT when liberated from SCOR bicelles. We combine our findings with previous cellular data to propose that the formation of PMP22 dimers from destabilized monomers is a key element of PMP22 mistrafficking.
The misfolding of membrane proteins is implicated in the mechanisms of multiple debilitating diseases such as cystic fibrosis and retinitis pigmentosa (1–4). Specific membrane protein mutations are often associated with disease states, with variant forms exhibiting altered stability and cellular trafficking (5). Unfortunately, due to the challenges associated with preparing and handling pure, highly concentrated membrane protein samples, detailed structural information on such targets is often lacking, especially for disease mutant forms. Furthermore, as some membrane proteins associated with misfolding-based diseases have hundreds of mutations of interest (3), there is a clear need for high-throughput methods to assess disease mutation-induced changes in membrane protein stability and structure.
Peripheral myelin protein 22 (PMP22) is such a membrane protein, for which misfolding and trafficking of mutant variants have been implicated in disease (6). PMP22 is a tetra-span integral membrane glycoprotein predominately expressed in Schwann cells, which are the principal glial cells of the peripheral nervous system (PNS), where they produce myelin (7–9). In addition to accounting for ∼5% of the protein found in the myelin sheath surrounding PNS nerve axons, PMP22 is thought to regulate intracellular Ca2+ levels (10), apoptosis (11), linkage of the actin cytoskeleton with lipid rafts (12), formation of epithelial intercellular junctions (13), myelin formation (14), lipid metabolism, and cholesterol trafficking (15). Dysregulation and misfolding of PMP22 has been identified as a key factor in multiple neurodegenerative disorders, such as Charcot-Marie-Tooth disease types 1A and E, as well as Dejerine–Sottas syndrome (6, 16–18). Like a number of other disease-linked membrane proteins (19), the trafficking of PMP22 is known to be inefficient, with only 20% of the wild-type (WT) protein reaching its final plasma membrane destination in Schwann cells (16, 20). Previously, it has been shown through a range of biophysical analyses that disease-associated PMP22 mutations lower thermodynamic protein stability as the root cause of reduced trafficking and loss of protein function; however, the mechanism by which destabilization of PMP22 causes mistrafficking is still not well understood (6). Additionally, a high-resolution structure of PMP22 has not yet been published.
Native mass spectrometry (MS) has recently been demonstrated to overcome sample purity and concentration barriers to reveal critical details of membrane protein structure and function (21–23). Through the use of nano-electrospray (nESI), intact membrane proteins are ionized within detergent micelles or other membrane mimetics (24–27), which can then be removed from the membrane protein ions within the instrument. This method has been used to elucidate oligomeric state (28–30), complex organization (31, 32), and lipid interactions (33–35) of diverse membrane proteins. The addition of ion-mobility separation–mass spectrometry (IM-MS) provides data on the orientationally averaged size of analytes (36) and enables collision induced unfolding (CIU) experiments (37). In CIU, the energies experienced by gas-phase protein ions are increased in a stepwise fashion causing gas-phase protein unfolding to occur. These dynamic measurements have been shown to be sensitive to ligand binding (38, 39), glycosylation (40, 41), and disulfide bonding (40) in soluble proteins, as well as selective lipid and small molecule binding in membrane proteins (42–45). While CIU can clearly capture subtle structural changes in membrane proteins (43, 45, 46) and soluble mutant protein variants (47, 48) its ability to characterize membrane protein variants is only beginning to be explored.
Here, we demonstrate the ability of native MS and CIU to detect key differences in the gas-phase stability and homodimer complex formation of PMP22 variants, together leading to insights into the mechanism of PMP22 dysregulation in disease. We quantify the propensity of PMP22 to dimerize across WT and seven disease-associated point mutations. We find that mutations associated with severe disease states form significantly more dimer than WT. Through CIU, we quantify the stability of gas-phase monomeric and dimeric PMP22 and find that variants bearing mutations associated with severe neuropathy exhibit the lowest relative monomer conformational stability. Interestingly, we also observe that dimers formed by various disease mutant forms of PMP22 are all more stable than WT PMP22 dimeric complexes. We continue by comparing our results to previously published biophysical datasets and find that our monomeric PMP22 gas-phase stability values correlate well with cellular trafficking data (6). Finally, we probe the effects of solubilization agents on PMP22 by characterizing its dimerization within sphingomyelin and cholesterol rich (SCOR) bicelles (49). We find that dimeric PMP22 complexes persist within SCOR bicelles and that the mutants resulting in the most severe disease phenotypes form higher population of dimer than WT. We conclude by describing a possible mechanism of PMP22 dysregulation in severe neurodegenerative diseases by which PMP22 monomers are destabilized, leading to dimers that traffic much less efficiently to the plasma membrane than WT PMP22.
Materials and Methods
Membrane Protein Sample Preparation.
PMP22 WT and mutant variants (S22F, A67T, T118M, G93R, L16P, and H12Q) were expressed in Escherichia coli using protocols adapted from Schlebach et al. (50) Octaethylene glycol monododecyl ether (C12E8) and n-Dodecyl-β-D-melibioside (DDMB) were purchased from Anatrace. Ammonium acetate was purchased from Sigma Aldrich. Detailed information on the purification of PMP22, preparation of SCOR bicelles, and incorporation of PMP22 into SCOR bicelles can be found in the supporting information. For detergent-based native MS experiments, 50 μM PMP22 was buffer- and detergent-exchanged simultaneously from 50 mM Tris buffer pH 8.0, 0.15% β-n-decyl maltoside (DM), 15 mM imidazole, and 1 mM TCEP buffer and 0.1% β-n-dodecylmaltoside (DDM) to 0.02% C12E8 (∼4 × critical micelle concentration) to a final protein concentration of less than 25 μM in 200 mM ammonium acetate pH 8.0 using 10 kDa Amicon Ultra 0.5 centrifugal filter units (MilliporeSigma). Detergent screening was performed following established protocols (22). For SCOR bicelles native MS experiments (49), 40 μM PMP22 in 10 mM acetate buffer (pH 5.0) containing 100 mM NaCl, 0.2% SCOR bicelles (q = 0.33), 1 mM ethylenediaminetetraacetic acid, 5 mM TCEP, and 0.3 mM DDMB was exchanged into 200 mM ammonium acetate pH 8.0 using 10 kDa Amicon Ultra 0.5 centrifugal filter units to a final protein concentration of less than 20 μM.
Native IM-MS.
All IM-MS data were collected using a Synapt G2 HDMS IMQToF mass spectrometer (Waters) with a direct infusion nESI source set to positive ion mode. Instrument settings were tuned to dissociate solubilization agents with minimal perturbation to protein structure prior to the IM separator, including appropriately tuned settings for the source temperature (30 °C), source gas flow (50 mL/min), and the sampling cone (120 V). Trapping cell wave velocity and height were 116 m/s and 0.1 V. IMS wave velocity and height were 250 m/s and 15 V. Transfer cell wave velocity and height were 300 m/s and 10 V, with an accelerating potential of 70 V used to dissociate empty micelles and bicelles. Collision cross-section (CCS) analysis was performed by using IMSCal-19v4 (51), a semiautomated CCS calibration program developed in the Ruotolo laboratory in collaboration with Waters Corp using native bovine serum albumin, insulin, beta-lactoglobulin, and polyalanine ions as calibrants. Calibration was conducted using a database of known CCS values in He. Theoretical CCSs of monomeric PMP22 were calculated using a previously published homology model (52) and IMPACT in He gas (36, 53, 54). Percent dimer calculations were performed at trap collision voltages of 80 V to minimize the impact of surfactant-related noise. Arrival time distributions of charge states that were known to be uniquely related to monomer or dimer were extracted using to a text-based format using TWIMExtract (55). For data where excess detergent and lipid noise was present at low arrival times, the extraction window was set to begin at the drift times indicated in SI Appendix, Fig. S3. To calculate the percent dimer, the sum of the extracted dimers is divided by the monomer sum added to the dimer sum. All CIU analyses were performed in triplicate by increasing the trap collision voltage in a stepwise manner from 5 to 150 V in 5 V increments. There was no notable decrease in dimer-associated signals across this voltage range, supporting the specificity of the complexes observed. CIU data from the 9+ and 13+ charge states of monomeric and dimeric PMP22, respectively, were extracted using TWIMExtract (55) with extraction window widths of ∼90 m/z depending on the charge state interrogated, then processed and analyzed using CIUSuite 2 (56). Data processing included three rounds of two-dimensional Savitzky–Golay smoothing with a window of five bins and interpolation of the collision voltage axis by a factor of four. IM-MS plots shown were exported in gray scale and then shaded regions were added in Adobe Illustrator (Adobe Inc.) for ease of visualization.
High-Resolution MS.
Mutant S22F PMP22 was prepared as described above for IM-MS experiments. High-resolution MS (HRMS) experiments were performed on an Orbitrap Fusion Lumos Tribid MS (ThermoFisher). Instrument settings were optimized to dissociate the detergent micelle to facilitate detection of the PMP22 signal. The in-source fragmentation voltages were enabled and set to 100 V. The transfer tube temperature was also increased to 325°. The ions were subjected to higher-energy C-Trap Dissociation at an energy of 22.5% to further liberate the PMP22 from the detergent micelle. All HRMS spectra were collected at a resolution 120,000 FWHM at 200 Th and with the radio frequency set to 30%.
Results and Discussion
Quantifying the Abundance of PMP22 Dimers.
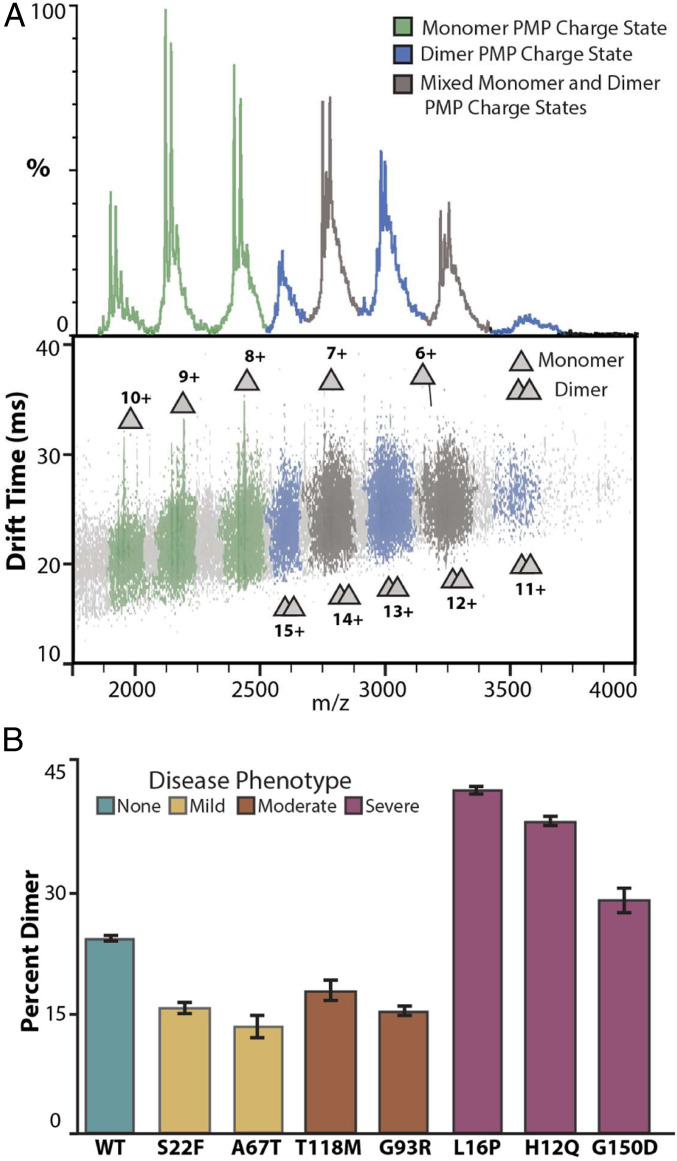
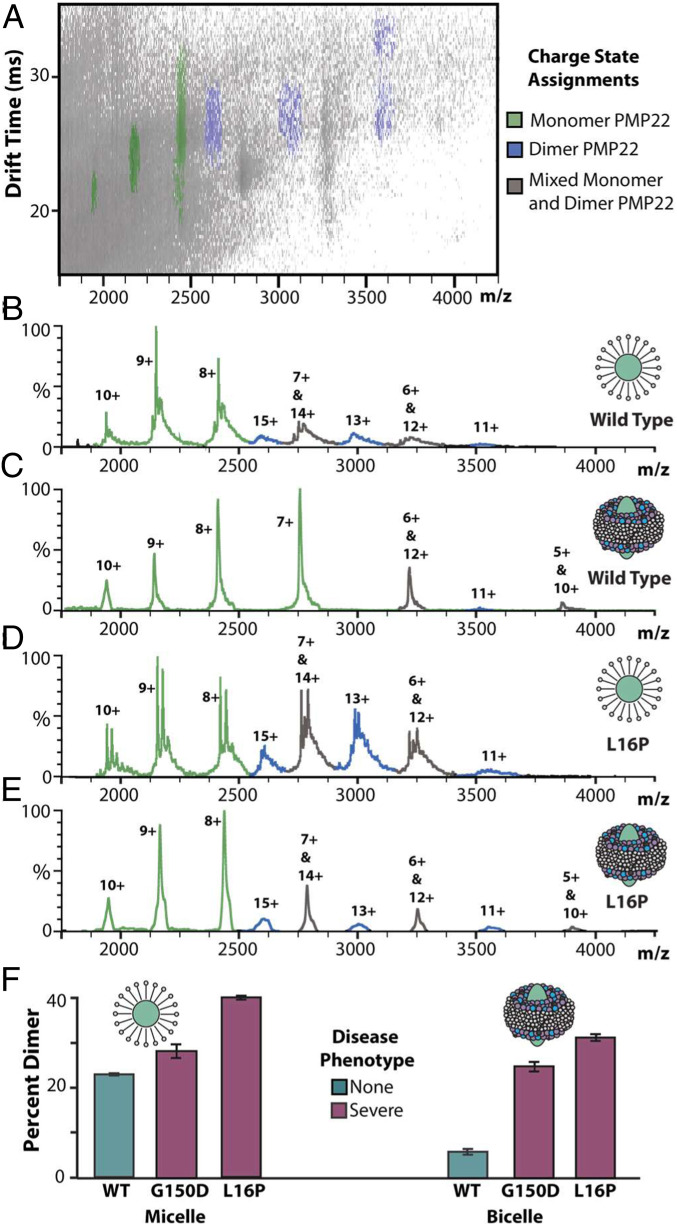
The existence of PMP22 homodimers formed by WT and mutant PMP22 variants (50, 57), as well as higher-order oligomeric species (58–60), has been inferred from prior assays. Therefore, we endeavored to use native MS, which enables the direct characterization of protein oligomeric state. While PMP22 has not previously been studied via native MS, we find that WT PMP22 liberated from C12E8 detergent micelles reveals two distributions of protein-related signals, as shown in Fig. 1A. The two distributions indicate that multiple protein oligomeric states are present within our PMP22 samples, with charge states ranging from 10+ to 6+ for the first distribution and 15+ to 11+ for the second, leading to intact masses of 19.4 and 39.1 kDa, respectively. These measured masses correspond well to those expected for monomeric and dimeric PMP22 (SI Appendix, Table S1). The data presented here confirm the existence of and directly charactize a PMP22 homodimer. Notably, the calculated CCS for 7+ monomeric PMP22 in these experiments was 1,595 ± 5 Å2, which agrees well with the theoretical CCS estimated for folded PMP22 based on a previously published homology model (1,525 ± 3 Å2) (52). The CCS agreement, as well as the range of charge states observed, indicate that PMP22 monomers retain compact, native-like conformations when subjected to IM-MS analysis.
Fig. 1.
Native IM-MS of PMP22 reveals differences in dimeric abundances. (A) Mass spectra and IM-MS data set of PMP22 liberated from detergent micelles produces populations of monomer and dimer protein at charge state ranges of 6 to 10+ and 11 to 15+, respectively. Charge states were identified as containing only monomer (green), only dimer (blue), or a mix of monomer and dimer (gray). (Trap = 80 V, exponential intensity scale for IM-MS dataset). (B) Intensities for PMP22 signals identified as monomer or dimer were used to calculate the percent dimer in WT PMP22, as well as seven mutant PMP22 variants, n = 3, SD shown as error bars. Notably, L16P, H12Q, and G150D mutants correlated with severe disease phenotypes and exhibit significantly more dimer (41.0 ± 0.4%, 37.3 ± 0.6%, 28.1 ± 1.5%) than WT (23.2 ± 0.3%).
The observation of a PMP22 homodimer prompted us to quantify its relative abundance across a range of disease-associated PMP22 mutants. As there is some overlap between the IM-MS signals corresponding to PMP22 monomer and dimer, in order to calculate the abundance of PMP22 dimers, we filtered all signals detected using both IM and MS data, leaving only those signals that could be identified as uniquely resulting from monomer or dimer PMP22 (Fig. 1A). For example, our measurements indicate a 23.2 ± 0.3% dimer population for WT PMP22.
Of the 44 pathogenic missense mutations known to exist for PMP22, we chose 7 mutations occurring throughout its sequence based on their prevalence, disease phenotype relationship, and previous biophysical assay behavior (SI Appendix, Fig. S1). Two mutants, S22F and A67T, are associated with hereditary neuropathy with liability for pressure palsies, which is considered a mild neuropathy (17, 61). Previous assays have found that S22F and A67T have surface trafficking efficiencies and folding stabilities comparable to WT PMP22 [where trafficking efficiency is the fraction of total cellular PMP22 that reaches the cell surface—its functional location (6)]. Another set of mutants, T118M and G93R, are associated with Charcot-Marie-Tooth disease (CMT), a moderate form of neuropathy (16, 62). These mutations were observed to cause a reduction in cellular trafficking and micellar-phase thermodynamic stability relative to WT (6), and recent computational modeling of T118M PMP22 indicate that the mutation causes changes to structural stability (63). Relative quantitation of the dimer states for these four PMP22 mutations exhibit abundances within error of each other but significantly lower than that of WT PMP22 (Fig. 1B).
The final three mutants analyzed in this work, L16P, H12Q, and G150D, are associated with Dejerine–Sottas syndrome, a severe neuropathic condition. These mutations were previously found to cause a dramatic reduction in cellular surface trafficking for PMP22, as well as decreased solution-phase thermodynamic stability (6, 63, 64). Strikingly, we observed that all three of these mutant PMP22 forms exhibited a significantly larger amount of dimer than WT PMP22 or mutations corresponding to mild or moderate disease phenotypes (Fig. 1B). The severe disease mutations H12Q and L16P exhibited 41.0 ± 0.4 and 37.3 ± 0.6% dimer, respectively, while G150D shows a more moderate increase in dimer relative to WT, at 28.1 ± 1.5%. As H12Q and L16P occur in the first transmembrane-spanning helix and G150D in the fourth transmembrane-spanning helix (SI Appendix, Fig. S1), these differences in dimeric abundance may indicate that the residues Q12 and P16 are associated with the PMP22 homodimer interface.
CIU Reveals Differential Stabilities of Monomeric PMP22 Mutants.
The folding and stability of PMP22 mutants have been linked to their mistrafficking and role in neuropathies (4, 6). However, as a high-resolution structure of PMP22 has not yet been determined, measurements related to the folding of PMP22 have largely been made from micellar or bicellar solutions (49), biochemical, biophysical and cellular assays (6), and molecular dynamic simulations (51, 63). While chemical modification has been used to specifically study WT PMP22 dimers (50), many reports convolute the various oligomeric states of PMP22. The gas-phase unfolding of membrane proteins has been previously reported to inform on the conformational dynamics and stability of protein structures (43, 45), and it allows for independent analysis of discrete protein forms that are separated in m/z. Advances in CIU analysis software have also enabled the automated identification of features and the fitting of sigmoidal transitions between such identified features (56). We proceeded to use this approach to evaluate the influence of mutations on the gas-phase stabilities of monomeric and dimeric PMP22 ions.
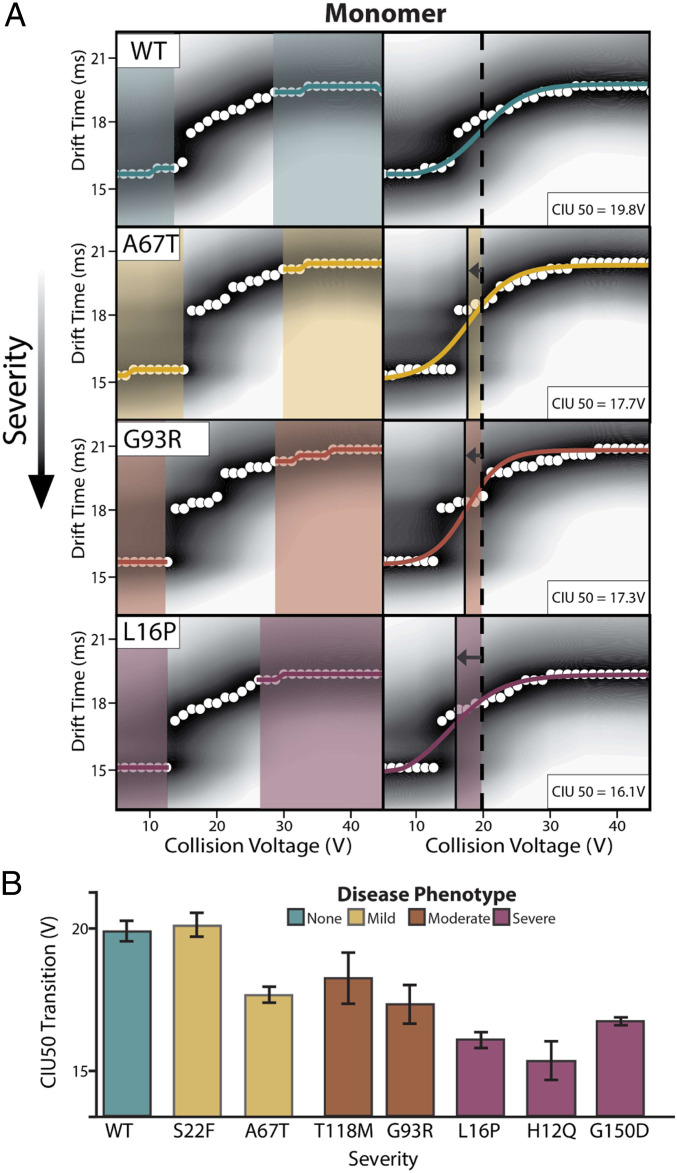
The feature observed at the lowest drift time and collision voltage in CIU plots represents the most folded and native-like conformational family observed for the PMP22 monomer in our experiments, while features appearing at longer IM arrival times correspond to a more unfolded PMP22 conformations. Sigmoidal curves fit to the transitions between CIU features allow us to compute inflection points, or CIU50 values, that we use to compare the relative stabilities of PMP22 variants (45, 56).
Fig. 2A displays averaged CIU fingerprints, n = 3, with detected features (left) and sigmoidal fits (right) for 9+ monomeric PMP22 WT, as well as example mutants from each disease severity phenotype. While a CIU50 of 19.8 ± 0.4 V is observed for WT PMP22, variants of PMP22 possessing the A67T, G93R, and L16P mutations all exhibit a range of CIU50 values significantly lower than that observed for WT. These lower CIU50 values indicate that PMP22 mutants are destabilized in a manner correlated with the severity of the neuropathy they induce. A clear example of this trend is apparent upon comparing CIU data collected for WT PMP22 (teal) and L16P PMP22 (pink), where the feature appearing at longer drift times in the L16P PMP22 data begins at significantly decreased collision voltage values when compared to the analogous feature appearing in the WT fingerprint, resulting in an average CIU50 of 16.1 ± 0.3 V for the mutant protein.
Fig. 2.
CIU data for monomeric PMP22. (A) Analysis of 9+ monomer WT PMP22 and all mutant PMP22 forms tested identify two features between 5 and 45 V. Fitting a sigmoid to these features allows for CIU50 determinations. (B) CIU50 values recorded for monomeric WT and mutant PMP22, n = 3, ordered by the disease phenotype severity engendered by the indicated mutant with SD shown as error bars.
Destabilization is observed for all PMP22 mutants studied here, except for S22F, where each mutant protein monomer exhibits an average CIU50 value significantly below the lower bound of error defined for our WT CIU50 measurement (Fig. 2B and SI Appendix, Fig. S4). The observation that monomer S22F PMP22 exhibits a gas-phase stability comparable to WT PMP22 is interesting in light of previously published data concluding that this variant may be trafficked more efficiently than WT and exhibits unique metal-binding properties (6). We note that the most destabilized class of PMP22 variants are those associated with severe neuropathy (pink). Specifically, H12Q and L16P PMP22 exhibit CIU50 values of 15.4 ± 0.7, and 16.1 ± 0.3 V, respectively, which are both significantly less than the stability we record for the WT PMP22. This observation agrees well with previous data recorded for these mutants (6, 52, 63); however, this study represents direct stability measurements of mutant PMP22 monomers excluded from other oligomeric states.
Mutant PMP22 Dimeric Complexes Are More Stable Than WT Dimers.
While the mistrafficking of PMP22 leading to loss of its functions is a common theme in its proposed roles in neuropathic disease (4, 57, 65), the underlying mechanism involved, as well as the fate of proteins that do not get properly trafficked, are still active areas of research. Proposed as either a cause or a consequence of mistrafficking, PMP22 oligomerization has been linked to the etiology and possible treatment modalities for its associated peripheral neuropathies (58–60, 66, 67). Recognizing the potential importance of PMP22 oligomers to its dysregulation, we assessed the gas-phase stabilities of disease mutant forms of PMP22 homodimers.
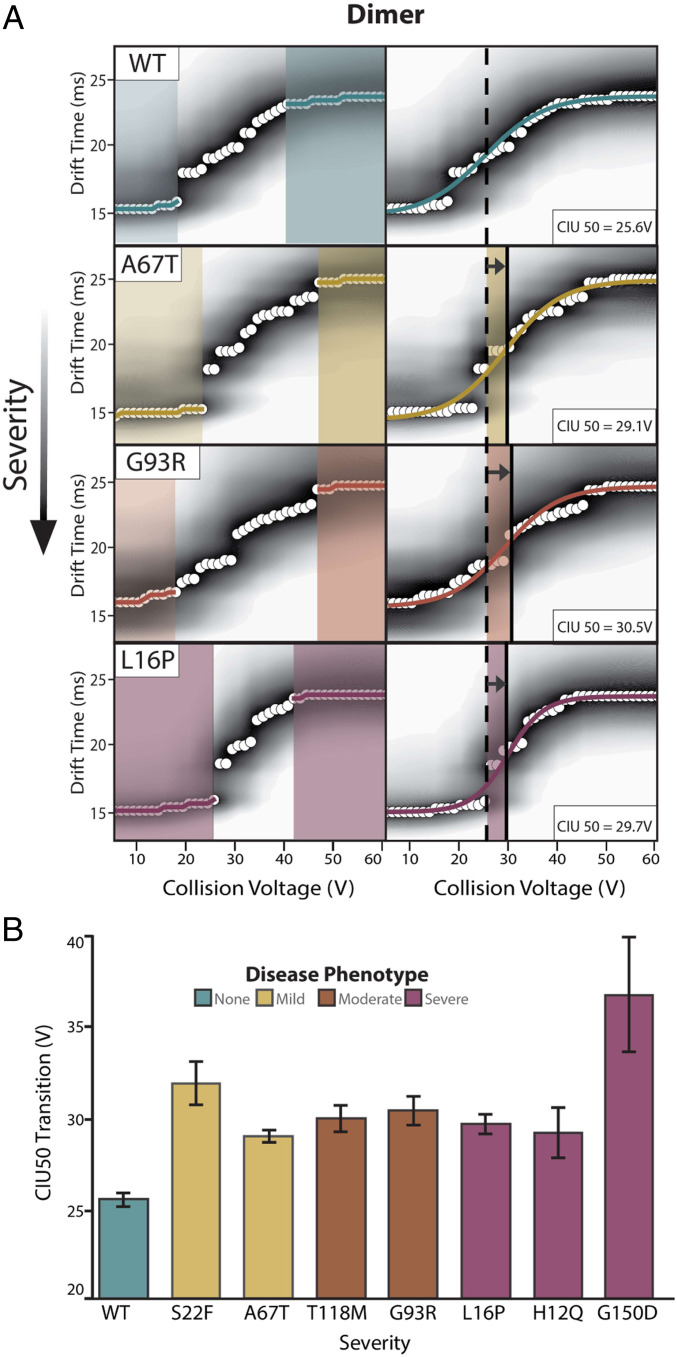
To study the CIU of PMP22 dimers, we selected the 13+ charge state, as it appears across all datasets and cannot overlap with monomeric PMP22 ions. Automated feature analysis detected two conformational populations between 5 and 60 V for all WT and mutant PMP22 dimer fingerprints. Sigmoidal fits between these two states yielded reproducible CIU50 values (Fig. 3A). We interpreted these data similarly to the monomeric fingerprints described in Fig. 2, where CIU50 values were compared across dimeric fingerprints for all PMP22 variants (Fig. 3B and SI Appendix, Fig. S4).
Fig. 3.
CIU data for dimeric PMP22. (A) Analysis of the 13+ WT PMP22 dimer and all mutant PMP22 forms exhibit two features between 5 and 60 V. (B) CIU50 values for WT and mutant PMP22 dimers, n = 3, ordered by disease phenotype severity with SD shown as error bars.
In contrast to monomeric data, we find that all disease variant dimers exhibit enhanced gas-phase stabilities relative to the WT dimer. Specifically, we record a WT PMP22 dimer CIU50 of 25.6 ± 0.4 V, while all mutant variants studied here exhibit significantly larger values, ranging from 29.3 ± 1.4 to 36.8 ± 3.0 V. Note that equal levels of homodimer stabilization are observed for all mutant variants, despite the abundance differences detected between mutants (Fig. 1B). Additionally, the stability measurements reported by the CIU50s in Fig. 3 reflect the unfolding of the intact dimeric complexes, not dissociation events. However, no clear differences between CIU50s of dimeric PMP22 mutants associated with different disease phenotypes is observed. Given the literature precedence for PMP22 oligomers (58–60), our findings clearly support the notion that destabilized mutant PMP22 monomers are also more apt to form stabilized dimeric states, and this equilibrium may play a role in disease state severity. This insight is further supported by the general correlations we observe between gas-phase monomer and dimer stability, disease severity, and the quantitative amounts of PMP22 dimers detected in our experiments.
Gas Phase Stability Data Correlate with Cellular and In Vivo Assays.
To probe how our results fit with prior findings associated with PMP22 mutations, we performed a series of correlative analyses where we plotted our native MS and CIU data presented in Figs. 1 and 2 against previously published cellular and in vivo data (6). For these analyses, we included prior data tracking cellular trafficking efficiencies (which give the fraction of PMP22 present in the plasma membrane compared with the total PMP22 in the cell, the intracellular population of which is deemed misfolded), nerve conduction velocities (NCV), and Gibbs free energy of Zn(II) binding (ΔΔGapp,total) (which informs directly on the solution phase folding of variant PMP22 relative to WT). Previous data have shown that there is a linear correlation between PMP22 ΔΔGapp, total and cell surface trafficking efficiencies, as well as between PMP22 cell surface trafficking efficiencies and nerve conduction velocities (6).
We first assessed how our monomer gas-phase stability data and dimer abundance values correlated to patient NCVs, as such data presents the most direct link to disease phenotype. Previously, NCVs have been shown to correlate well with both trafficking efficiency (R2 = 0.88) and ΔΔGapp,total (R2 = 0.64) in an analysis of a larger set of 10 PMP22 mutants (6). While we find no significant correlation between NCVs and the abundance of dimeric complexes measured here (SI Appendix, Fig. S5B), we do observe a correlation between the CIU50 values recorded for monomeric PMP22 and patient NCVs (R2 = 0.57, SI Appendix, Fig. S5A). The nature of this relationship is proportional, where a lower monomeric CIU50 value is associated with slower nerve conduction velocities. We next explored relationships between the ΔΔGapp,total and our measurements but found no significant correlations for gas-phase stability values recorded for PMP22 monomers or our abundance measurements for PMP22 dimers (SI Appendix, Fig. S5 D and E). We interpret this result as stemming from the foundational differences underpinning the two measurements analyzed in these separate studies. First, the prior data relies specifically on Zn(II) binding to PMP22, and Zn(II) was not present in our native IM-MS experiments. Secondly, in the CIU experiments, we are extracting the gas-phase stability information from specific oligomeric species, where the solution-phase experiments are likely sampling complex mixtures of PMP22 oligomers.
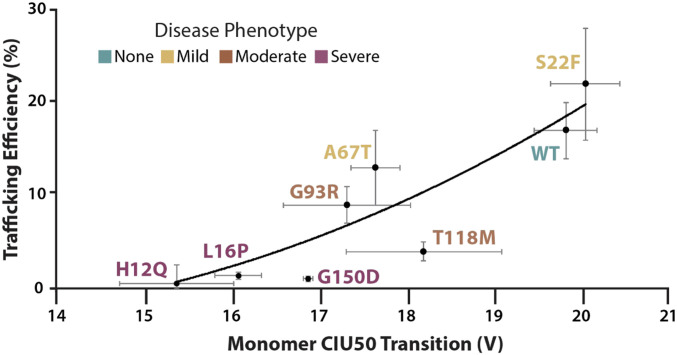
In contrast to the results described above, we find that cellular trafficking efficiency data correlates strongly with our monomer CIU50 values (R2 = 0.80, Fig. 4). The quadratic regression shown in Fig. 4 exhibits a relationship wherein less stable PMP22 monomers are associated with reduced cellular trafficking. Additionally, we find that PMP22 trafficking efficiency is anticorrelated with the dimer abundance values recorded from our native IM-MS measurements (R2 = 0.66, SI Appendix, Fig. S5F). Notably, the mutants associated with severe forms of neuropathy, H12Q, L16P, and G150D, cluster at the extremes of these plots, all exhibiting low trafficking efficiencies, low monomeric stabilities, slow NCV values, and abundant dimers. The above correlations clearly indicate the importance of PMP22 dimers, an oligomeric state our data indicates is ultimately fed by the instability of the preceding monomers, in predicting PMP22-associated disease phenotypes.
Fig. 4.
Correlation between monomer CIU50 stability values and PMP22 trafficking efficiencies, n = 3 (6). CIU50 values are from the 9+ monomeric charge state of PMP22 and describe the relative gas-phase stability of the protein forms. “Trafficking efficiency” is defined and the measured levels of PMP22 as the plasma membrane of model mammalian cell lines relative to the total cellular population, which include cell-internal PMP22 that is thought to be misfolded. Mutations associated with different disease severities are color coded. A quadratic regression (y = 0.3538x2 − 8.4629x + 47.218) fit is shown for the data, along with errors for each dataset calculated by SD, producing a correlation coefficient of 0.80, which shows a positive correlation between monomeric protein gas-phase stability and trafficking efficiency.
Encapsulation within SCOR Bicelles Engenders Mutant PMP22 Homodimer Formation.
Lipids are recognized as key regulators of membrane proteins through selective interactions or association with the membrane itself (29, 68–70). Many biochemical processes are thought to be driven by the partitioning of membrane proteins into regions containing saturated lipids, sphingolipids, and cholesterol (71, 72). The myelin membrane in which PMP22 natively resides is rich in cholesterol and sphingomyelin, and the protein is thought to play an important role in cholesterol homeostasis (15, 73, 74). Recently, a novel bicelle construct aiming to imitate the ordered membrane environment encountered natively by PMP22 has been described (49). These SCOR bicelles have been found to be compatible with a wide range of biophysical measurement techniques, including native IM-MS (49). To evaluate the effect of a more native-like lipid environment on the formation of PMP22 homodimers, we employed SCOR bicelles to prepare samples for native IM-MS analysis.
In Fig. 5A, an example plot of IM drift time versus m/z for L16P PMP22 liberated from SCOR bicelles is shown, and we observe signals associated with monomer (green), dimer (blue), and overlapping PMP22 oligomeric states (gray). Additionally, noise signals associated with lipid and detergent clusters can be observed in the lower IM drift times and m/z values (not shown). Due to this noise, the SCOR bicelle-related data shown in Fig. 5 C and E were background subtracted for visualization purposes, and prior to our quantitative analysis of the observed PMP22 dimers, data were de-noised (SI Appendix, Fig. S7). MS data collected for these samples exhibited evidence of enhanced levels of adduction (Fig. 5 B–E), which likely stems from the sample preparation procedures associated with SCOR bicelles used in these experiments (also observed in SI Appendix, Fig. S6).
Fig. 5.
WT and L16P PMP22 liberated from SCOR bicelles. (A) IM-MS of L16P liberated from SCOR bicelles reveals populations of monomer and dimer PMP22 as well as lipid- and detergent-related noise (low m/z noise not shown, trap = 80 V, exponential intensity scale). Such noise was systematically excluded from all further analysis (SI Appendix, Fig. S7). (B–E) Mass spectra of WT PMP22 liberated from either detergent micelles or SCOR bicelles (as indicated), followed by similar data collected for L16P PMP22 (trap = 80 V). Monomeric PMP22 (green) and dimeric PMP22 (blue) are present in each spectrum but in varying amounts. (F) Measured percent dimer values from native IM-MS, n = 3, SD shown as error bars.
Fig. 5F displays our quantitative analysis of PMP22 dimers for WT, L16P, and G150D variants liberated from micelles and SCOR bicelles. While minor differences exist in the amount of dimer observed for the mutants between our micelle and bicelle samples, we detect significant differences in the relative abundance of WT dimers ejected from SCOR bicelles when compared with our micelle data: 22.7 ± 0.3% PMP22 dimer in micelles and only 5.6 ± 0.6% dimer in SCOR bicelles. This data further demonstrates that the increased levels of dimer we detect are not an artifact associated with the location of the mutation studied but rather a function of the overall structure and stability of the PMP22 variant, which is higher in SCOR bicelles than in micelles. In general, these observations demonstrate the importance of the native membrane and lipid environment on PMP22 dimer formation and are further evidence that PMP22 homodimers complexes are strongly correlated with mutations that give rise to severe neuropathies.
Conclusions
The role of PMP22 in neuropathies is complex, with its folding, stability, trafficking, and aggregation all previously indicated as important predictive links to its role in disease (6, 58, 63—64, 66). Native IM-MS and CIU have allowed us to identify PMP22 homodimers as a candidate for inclusion among these factors. Our measurements enabled the gas-phase stabilities of PMP22 monomers to be deconvoluted from their associated oligomers, and they suggest that those mutations that engender the least stable monomers also produce the most stable dimers. These results indicate that those monomers that are least stable in the gas phase are the most likely to form dimers in solution and thus accumulate a significant dimeric population. The likelihood that these results bear relevance under native cellular environments is high, as our SCOR bicelle measurements reveal a more dramatic difference in dimer amounts in WT and disease-associated mutants. Furthermore, if these observations are combined with our analyses that highlight correlations between our results and previous measurements of PMP22 trafficking efficiency, a possible mechanistic link can be formed between protein misfolding, dimer production, and a loss of functional PMP22 within the cellular plasma membrane, leading to direct consequences in NCV and the overall severity of the neuropathy experienced by patients.
While we here present no mechanistic evidence that explains how the formation of PMP22 dimers may negatively impact their cellular trafficking, we can postulate multiple potential mechanisms based on what is known about membrane protein trafficking. The trafficking of WT PMP22 is known to be tightly regulated (20), and it is likely that the molecular machinery which recognizes properly folded PMP22 may not similarly interact with dimeric PMP22. Additionally, it has been shown that increased expression of PMP22 results in reduced cellular trafficking and increased formation of intracellular protein (75), most probably arising from overwhelming the endoplasmic reticulum quality control system, leading to formation of cytotoxic aggregates. PMP22 dimers may be the first step in a more extensive aggregation pathway leading to disruption of neuronal function, as PMP22 aggregates have been observed in mouse models with multiple copies of PMP22 and in dermal fibroblasts of patients with CMT1A (57, 76–78). Beyond the implications for PMP22 biology, we suggest that the methods described here could be more broadly applied to membrane proteins misfolding disorders, paving the way for future insights into a host of debilitating human ailments.
Supplementary Material
Acknowledgments
Membrane protein research in the B.T.R. laboratory is supported by the NIH under R01 GM105942. HRMS research on the Orbitrap Fusion Lumos was supported by the NIH under S10OD021619. This work was also supported by NIH R01 NS095989 granted to the C.R.S. and M.D.O. laboratories.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015331118/-/DCSupplemental.
Data Availability
Raw IM-MS data files have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.c.5271845.v1).
References
- 1.Ng D. P., Poulsen B. E., Deber C. M., Membrane protein misassembly in disease. Biochim. Biophys. Acta 1818, 1115–1122 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Farinha C. M., Canato S., From the endoplasmic reticulum to the plasma membrane: Mechanisms of CFTR folding and trafficking. Cell. Mol. Life Sci. 74, 39–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moeller H. B., Rittig S., Fenton R. A., Nephrogenic diabetes insipidus: Essential insights into the molecular background and potential therapies for treatment. Endocr. Rev. 34, 278–301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marinko J. T., et al., Folding and misfolding of human membrane proteins in health and disease: From single molecules to cellular proteostasis. Chem. Rev. 119, 5537–5606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H., et al., Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci. Adv. 4, eaar2631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlebach J. P., et al., Conformational stability and pathogenic misfolding of the integral membrane protein PMP22. J. Am. Chem. Soc. 137, 8758–8768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Parker B., Martyn C., Natarajan C., Guo J., The PMP22 gene and its related diseases. Mol. Neurobiol. 47, 673–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantera H., Shy M. E., Svaren J., Regulating PMP22 expression as a dosage sensitive neuropathy gene. Brain Res. 1726, 146491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jetten A. M., Suter U., The peripheral myelin protein 22 and epithelial membrane protein family. Prog. Nucleic Acid Res. Mol. Biol. 64, 97–129 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Vanoye C. G., et al., Peripheral myelin protein 22 modulates store-operated calcium channel activity, providing insights into Charcot-Marie-Tooth disease etiology. J. Biol. Chem. 294, 12054–12065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabbretti E., Edomi P., Brancolini C., Schneider C., Apoptotic phenotype induced by overexpression of wild-type gas3/PMP22: Its relation to the demyelinating peripheral neuropathy CMT1A. Genes Dev. 9, 1846–1856 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Lee S., et al., PMP22 is critical for actin-mediated cellular functions and for establishing lipid rafts. J. Neurosci. 34, 16140–16152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux K. J., Amici S. A., Fletcher B. S., Notterpek L., Modulation of epithelial morphology, monolayer permeability, and cell migration by growth arrest specific 3/peripheral myelin protein 22. Mol. Biol. Cell 16, 1142–1151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Urso D., Ehrhardt P., Müller H. W., Peripheral myelin protein 22 and protein zero: A novel association in peripheral nervous system myelin. J. Neurosci. 19, 3396–3403 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., et al., PMP22 regulates cholesterol trafficking and ABCA1-mediated cholesterol efflux. J. Neurosci. 39, 5404–5418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Paassen B. W., et al., PMP22 related neuropathies: Charcot-Marie-tooth disease type 1A and hereditary neuropathy with liability to pressure palsies. Orphanet J. Rare Dis. 9, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fusco C., et al., Hereditary neuropathy with liability to pressure palsy (HNPP): Report of a family with a new point mutation in PMP22 gene. Ital. J. Pediatr. 43, 97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roa B. B., Dyck P. J., Marks H. G., Chance P. F., Lupski J. R., Dejerine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat. Genet. 5, 269–273 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Sanders C. R., Myers J. K., Disease-related misassembly of membrane proteins. Annu. Rev. Biophys. Biomol. Struct. 33, 25–51 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Pareek S., et al., Neurons promote the translocation of peripheral myelin protein 22 into myelin. J. Neurosci. 17, 7754–7762 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrera N. P., Robinson C. V., Advances in the mass spectrometry of membrane proteins: From individual proteins to intact complexes. Annu. Rev. Biochem. 80, 247–271 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Laganowsky A., Reading E., Hopper J. T. S., Robinson C. V., Mass spectrometry of intact membrane protein complexes. Nat. Protoc. 8, 639–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konijnenberg A., van Dyck J. F., Kailing L. L., Sobott F., Extending native mass spectrometry approaches to integral membrane proteins. Biol. Chem. 396, 991–1002 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Borysik A. J., Robinson C. V., The ‘sticky business’ of cleaning gas-phase membrane proteins: A detergent oriented perspective. Phys. Chem. Chem. Phys. 14, 14439–14449 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Calabrese A. N., Watkinson T. G., Henderson P. J. F., Radford S. E., Ashcroft A. E., Amphipols outperform dodecylmaltoside micelles in stabilizing membrane protein structure in the gas phase. Anal. Chem. 87, 1118–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marty M. T., Hoi K. K., Robinson C. V., Interfacing membrane mimetics with mass spectrometry. Acc. Chem. Res. 49, 2459–2467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chorev D. S., et al., The use of sonicated lipid vesicles for mass spectrometry of membrane protein complexes. Nat. Protoc. 15, 1690–1706 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keener J. E., et al., Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J. Am. Chem. Soc. 141, 1054–1061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta K., et al., The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvey S. R., Liu Y., Liu W., Wysocki V. H., Laganowsky A., Surface induced dissociation as a tool to study membrane protein complexes. Chem. Commun. (Camb.) 53, 3106–3109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrich E., et al., Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. eLife 6, 1–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lippens J. L., et al., Fourier transform-ion cyclotron resonance mass spectrometry as a platform for characterizing multimeric membrane protein complexes. J. Am. Soc. Mass Spectrom. 29, 183–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta K., et al., Identifying key membrane protein lipid interactions using mass spectrometry. Nat. Protoc. 13, 1106–1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liko I., et al., Lipid binding attenuates channel closure of the outer membrane protein OmpF. Proc. Natl. Acad. Sci. U.S.A. 115, 6691–6696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marty M. T., Hoi K. K., Gault J., Robinson C. V., Probing the lipid annular belt by gas-phase dissociation of membrane proteins in nanodiscs. Angew. Chem. Int. Ed. Engl. 55, 550–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruotolo B. T., Benesch J. L. P., Sandercock A. M., Hyung S. J., Robinson C. V., Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Dixit S. M., Polasky D. A., Ruotolo B. T., Collision induced unfolding of isolated proteins in the gas phase: Past, present, and future. Curr. Opin. Chem. Biol. 42, 93–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabuck-Gibbons J. N., Keating J. E., Ruotolo B. T., Collision induced unfolding and dissociation differentiates ATP-competitive from allosteric protein tyrosine kinase inhibitors. Int. J. Mass Spectrom. 427, 151–156 (2018). [Google Scholar]
- 39.Hopper J. T. S., Oldham N. J., Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: The effect of ligand binding on conformational stability. J. Am. Soc. Mass Spectrom. 20, 1851–1858 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Tian Y., Han L., Buckner A. C., Ruotolo B. T., Collision induced unfolding of intact antibodies: Rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal. Chem. 87, 11509–11515 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Hernandez-Alba O., Wagner-Rousset E., Beck A., Cianférani S. C., Native mass spectrometry, ion mobility, and collision-induced unfolding for conformational characterization of IgG4 monoclonal antibodies. Anal. Chem. 90, 8865–8872 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Cong X., Liu W., Laganowsky A., Characterization of membrane protein-lipid interactions by mass spectrometry ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 28, 579–586 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Laganowsky A., et al., Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landreh M., et al., Integrating mass spectrometry with MD simulations reveals the role of lipids in Na+/H+ antiporters. Nat. Commun. 8, 13993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fantin S. M., et al., Collision induced unfolding classifies ligands bound to the integral membrane translocator protein. Anal. Chem. 91, 15469–15476 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Fantin S. M., Huang H., Sanders C. R., Ruotolo B. T., Collision-induced unfolding differentiates functional variants of the KCNQ1 voltage sensor domain. J. Am. Soc. Mass Spectrom. 31, 2348–2355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyung S. J., Robinson C. V., Ruotolo B. T., Gas-phase unfolding and disassembly reveals stability differences in ligand-bound multiprotein complexes. Chem. Biol. 16, 382–390 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Byrne D. P., et al., cAMP-dependent protein kinase (PKA) complexes probed by complementary differential scanning fluorimetry and ion mobility-mass spectrometry. Biochem. J. 473, 3159–3175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchison J. M., et al., Bicelles rich in both Sphingolipids and cholesterol and their use in studies of membrane proteins. J. Am. Chem. Soc. 142, 12715–12729 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlebach J. P., et al., Reversible folding of human peripheral myelin protein 22, a tetraspan membrane protein. Biochemistry 52, 3229–3241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson K., Langridge D., Dixit S. M., Ruotolo B. T., An improved calibration approach for traveling wave ion mobility spectrometry: Robust, high-precision collision cross sections. Anal. Chem. 93, 3542–3550, 10.1021/acs.analchem.0c04948 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Mittendorf K. F., Kroncke B. M., Meiler J., Sanders C. R., The homology model of PMP22 suggests mutations resulting in peripheral neuropathy disrupt transmembrane helix packing. Biochemistry 53, 6139–6141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allison T. M., Landreh M., Benesch J. L. P., Robinson C. V., Low charge and reduced mobility of membrane protein complexes has implications for calibration of collision cross section measurements. Anal. Chem. 88, 5879–5884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marklund E. G., Degiacomi M. T., Robinson C. V., Baldwin A. J., Benesch J. L. P., Collision cross sections for structural proteomics. Structure 23, 791–799 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Haynes S. E., et al., Variable-velocity traveling-wave ion mobility separation enhancing peak capacity for data-independent acquisition proteomics. Anal. Chem. 89, 5669–5672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polasky D. A., Dixit S. M., Fantin S. M., Ruotolo B. T., CIUSuite 2: Next-generation software for the analysis of gas-phase protein unfolding data. Anal. Chem. 91, 3147–3155 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Tobler A. R., et al., Transport of Trembler-J mutant peripheral myelin protein 22 is blocked in the intermediate compartment and affects the transport of the wild-type protein by direct interaction. J. Neurosci. 19, 2027–2036 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fortun J., et al., Alterations in degradative pathways and protein aggregation in a neuropathy model based on PMP22 overexpression. Neurobiol. Dis. 22, 153–164 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Liu N., Yamauchi J., Shooter E. M., Recessive, but not dominant, mutations in peripheral myelin protein 22 gene show unique patterns of aggregation and intracellular trafficking. Neurobiol. Dis. 17, 300–309 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Tobler A. R., Liu N., Mueller L., Shooter E. M., Differential aggregation of the Trembler and Trembler J mutants of peripheral myelin protein 22. Proc. Natl. Acad. Sci. U.S.A. 99, 483–488 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nodera H., Nishimura M., Logigian E. L., Herrmann D. N., Kaji R., HNPP due to a novel missense mutation of the PMP22 gene. Neurology 60, 1863–1864 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Kumar C. V., Swetha R. G., Anbarasu A., Ramaiah S., Computational analysis reveals the association of threonine 118 methionine mutation in PMP22 resulting in CMT-1A. Adv. Bioinforma. 2014, 502618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bello M., Torres M. J., Méndez-Tenorio A., Correa-Basurto J., Conformational changes associated with L16P and T118M mutations in the membrane-embedded PMP22 protein, consequential in CMT-1A. J. Biomol. Struct. Dyn. 35, 2880–2894 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Sakakura M., Hadziselimovic A., Wang Z., Schey K. L., Sanders C. R., Structural basis for the Trembler-J phenotype of Charcot-Marie-Tooth disease. Structure 19, 1160–1169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders C. R., Ismail-Beigi F., McEnery M. W., Mutations of peripheral Myelin protein 22 result in defective trafficking through mechanisms which may be common to diseases involving tetraspan membrane proteins. Biochemistry 40, 9453–9459 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Fortun J., et al., The formation of peripheral myelin protein 22 aggregates is hindered by the enhancement of autophagy and expression of cytoplasmic chaperones. Neurobiol. Dis. 25, 252–265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isaacs A. M., et al., Identification of a new Pmp22 mouse mutant and trafficking analysis of a Pmp22 allelic series suggesting that protein aggregates may be protective in Pmp22-associated peripheral neuropathy. Mol. Cell. Neurosci. 21, 114–125 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Duarte J. M., Biyani N., Baskaran K., Capitani G., An analysis of oligomerization interfaces in transmembrane proteins. BMC Struct. Biol. 13, 21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H., et al., Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arkhipov A., et al., Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlebach J. P., et al., Topologically diverse human membrane proteins partition to liquid-disordered domains in phase-separated lipid vesicles. Biochemistry 55, 985–988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lorent J. H., Levental I., Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 192, 23–32 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Gopalakrishnan G., et al., Lipidome and proteome map of myelin membranes. J. Neurosci. Res. 91, 321–334 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Marinko J. T., Kenworthy A. K., Sanders C. R., Peripheral myelin protein 22 preferentially partitions into ordered phase membrane domains. Proc. Natl. Acad. Sci. U.S.A. 117, 14168–14177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marinko J. T., Carter B. D., Sanders C. R., Direct relationship between increased expression and mistrafficking of the Charcot-Marie-Tooth-associated protein PMP22. J. Biol. Chem. 295, 11963–11970 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryan M. C., Shooter E. M., Notterpek L., Aggresome formation in neuropathy models based on peripheral myelin protein 22 mutations. Neurobiol. Dis. 10, 109–118 (2002). [DOI] [PubMed] [Google Scholar]
- 77.Notterpek L., Ryan M. C., Tobler A. R., Shooter E. M., PMP22 accumulation in aggresomes: Implications for CMT1A pathology. Neurobiol. Dis. 6, 450–460 (1999). [DOI] [PubMed] [Google Scholar]
- 78.Lee S., et al., Elevated peripheral myelin protein 22, reduced mitotic potential, and proteasome impairment in dermal fibroblasts from charcot-marie-tooth disease type 1A patients. Am. J. Pathol. 188, 728–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw IM-MS data files have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.c.5271845.v1).