Significance
Insulin and IGF-1 receptors share many downstream signaling pathways but have unique biological effects. Here, using global phosphoproteomics, we demonstrate that there are important differences in phosphorylation-mediated signaling between IR and IGF1R in both the basal and ligand-stimulated states involving multiple pathways of cellular regulation. Thus, mTORC1 and PIP3/AKT signaling, which are important in metabolism, are preferentially regulated by IR, while Rho GTPases, mitosis, and cell cycle proteins, which are involved in control of cellular growth, are preferentially regulated by IGF1R. These differences can be mapped to effects of both the extracellular and intracellular domains of these receptors. Thus, despite their high homology, IR and IGF1R preferentially regulate distinct networks of phosphorylation, contributing to the unique effects of these hormones.
Keywords: insulin signaling, protein phosphorylation, cellular signaling, IGF-1 signaling, kinases
Abstract
Insulin and insulin-like growth factor 1 (IGF-1) receptors share many downstream signaling pathways but have unique biological effects. To define the molecular signals contributing to these distinct activities, we performed global phosphoproteomics on cells expressing either insulin receptor (IR), IGF-1 receptor (IGF1R), or chimeric IR-IGF1R receptors. We show that IR preferentially stimulates phosphorylations associated with mammalian target of rapamycin complex 1 (mTORC1) and Akt pathways, whereas IGF1R preferentially stimulates phosphorylations on proteins associated with the Ras homolog family of guanosine triphosphate hydrolases (Rho GTPases), and cell cycle progression. There were also major differences in the phosphoproteome between cells expressing IR versus IGF1R in the unstimulated state, including phosphorylation of proteins involved in membrane trafficking, chromatin remodeling, and cell cycle. In cells expressing chimeric IR-IGF1R receptors, these differences in signaling could be mapped to contributions of both the extra- and intracellular domains of these receptors. Thus, despite their high homology, IR and IGF1R preferentially regulate distinct networks of phosphorylation in both the basal and stimulated states, allowing for the unique effects of these hormones on organismal function.
Insulin and insulin-like growth factor 1 (IGF-1) act through their cognate receptors to regulate a wide variety of biological processes. The insulin and IGF-1 receptors (IR and IGF1R) are highly homologous and share many overlapping signaling pathways (1). Activation of both receptors results in stimulation of two major canonical pathways of signaling: the PI 3-kinase/Akt pathway, which is linked to most metabolic actions of these hormones, and the Ras/MAP kinase pathway, which is linked to regulation of cell and organismal growth and differentiation (2–5). Perturbations in these pathways can lead to insulin resistance, as manifested by glucose intolerance, dyslipidemia, and increased risk of cardiovascular disease or alterations in growth at both the cellular and organismal level (1, 3). Due to the high degree of homology between these receptors and shared downstream signaling, defining differential molecular signatures has been challenging. This is further complicated by the fact that although insulin and IGF-1 preferentially bind to their own receptors, both ligands can also bind to the alternate receptor with lower affinity (6, 7). Many studies have tried to identify the differences between IR and IGF1R which account for their differences in vivo (2, 8). Some of the differential effects between insulin and IGF-1 have been ascribed to differences in the relative concentration of the hormones, the presence of IGF-1 binding proteins, the relative expression level of receptors in different tissues and the affinity of the two ligands for the two different receptors (1, 9).
Some insight has come from the use of chimeric receptors. For example, activation of chimeric receptors containing the extracellular domains (ECDs) of the neurotrophin receptor (TrkC) fused to the intracellular domains (ICDs) of the insulin or IGF-1 receptors have suggested that the IGF1R intracellular domain (IGF1R-ICD) is more important for mitogenic response, while the IR-ICD is more strongly related to metabolic effects (10, 11). Similar conclusions have been reached using chemical and antibody-based inhibitors of these receptors (12). More recently, using chimeric receptors containing the ECD of IR fused to the ICD of IGF1R and vice versa, we showed that both the intracellular and extracellular domains of IR and IGF1R contribute to their effects on gene expression and that this is in part due to differences in their intracellular juxtamembrane regions and ability to engage IRS proteins versus Shc as substrates of the receptor (2).
In addition to these ligand-stimulated and tyrosine-kinase-dependent events, previous studies from our laboratory have demonstrated a class of ligand and tyrosine-kinase-independent effects mediated by the unoccupied IR and IGF1R. Thus, cells lacking both IR and IGF1R are resistant to apoptosis (13) and show altered expression of a network of imprinted genes and miRNAs associated with changes in DNA methylation (14). These findings suggest that these receptors have receptor-dependent but ligand-independent signaling events, but how these are mediated is unknown.
Intracellular signal transduction by membrane receptors such as IR and IGF1R is primarily mediated by the reversible phosphorylation of networks of signaling molecules. While IR and IGF1R are tyrosine kinases with a limited number of primary substrates, these in turn activate downstream kinases that lead to much broader networks of phosphorylation on serine and threonine residues. The resultant phosphorylated proteins may have altered activity levels, tertiary structure, and/or subcellular localization. To define the molecular differences in signaling between IR and IGF1R that create their unique physiological actions and to determine to what extent these differences are contributed by the ECDs vs. ICDs of these receptors, we performed global phosphoproteomic analysis of preadipocytes in which both endogenous receptors had been genetically inactivated to create double knock out (DKO) cells after which the cells were reconstituted with IR, IGF1R, and chimeric receptors consisting of the ECD of IR and the ICD of IGF1R (named hereafter as IR_IGF1R) and vice versa (i.e., IGF1R_IR) and studied both with and without ligand stimulation to understand signaling by the unoccupied receptor. We show that despite many similarities in signaling, there are major differences in the phosphoproteome mediated by activated IR versus IGF1R which contribute to their specific patterns of action. In addition, there are important and previously unrecognized differences in the basal phosphoproteome in cells expressing IR versus IGF1R. Using chimeric receptors we show that these differences in both the basal and stimulated states are contributed to by both the ICD and ECD of these receptors. Together, these unique signaling networks lead to the important differential effects of insulin and IGF-1 at the postreceptor level.
Results
Mass Spectrometry–Based Phosphoproteomic Characterization of IR, IGF1R, and Chimeric Receptors Consisting of Their Extracellular and Intracellular Domains.
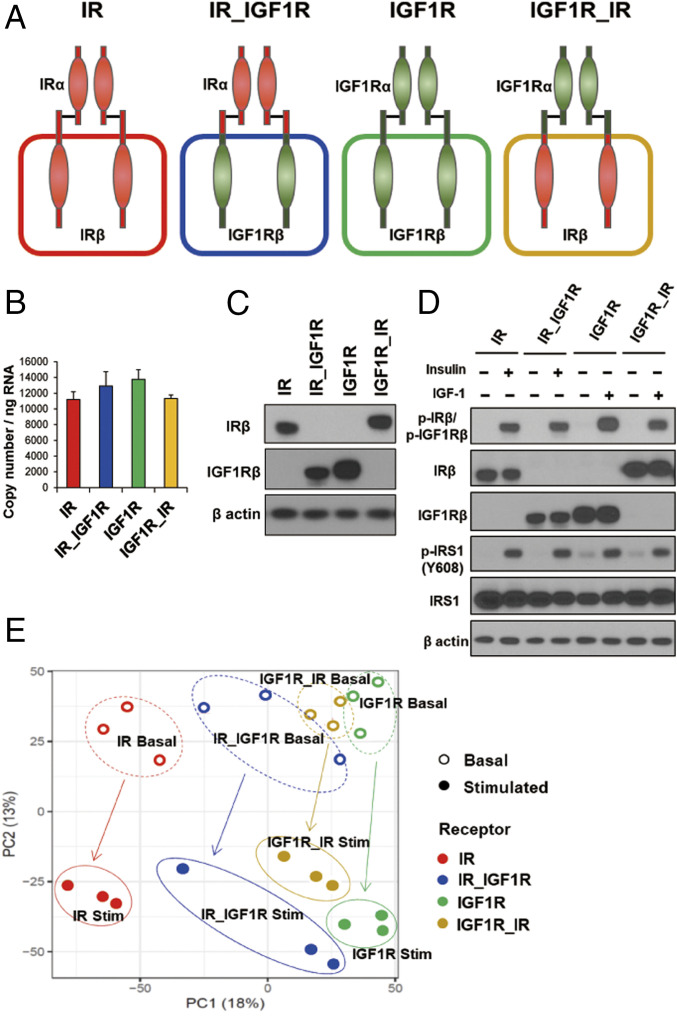
To identify the differences between insulin and IGF-1 signaling via their cognate receptors, we generated brown preadipocytes in which both endogenous IR and IGF1R had been genetically inactivated using Cre-lox recombination and then reconstituted these DKO cells with either wild-type mouse IR (A isoform), mouse IGF1R, or one of two chimeric receptors, termed IR_IGF1R and IGF1R_IR. IR_IGF1R consists of the IR-ECD fused to the IGF1R transmembrane and ICD and the IGF1R_IR consists of the ECD of IGF1R fused to the transmembrane and ICD of IR (2, 13) (Fig. 1A). While DKO cells showed no detectable levels of mRNA and proteins for either IR or IGF1R (SI Appendix, Fig. S1 A and B), cells expressing only IR, only IGF1R, or the two chimeric receptors had similar expression at both the mRNA (Fig. 1B) and protein levels (Fig. 1C and SI Appendix, Fig. S1 C and D). Insulin stimulation of cells expressing IR or IR_IGF1R and IGF-1 stimulation of cells expressing IGF1R and IGF1R_IR led to robust and similar levels of receptor autophosphorylation as measured by an antibody that detects the major autophosphorylation site of both receptors, as well as phosphorylation of tyrosine 608 on IRS1, the major endogenous substrate of these receptors in these cells (Fig. 1D).
Fig. 1.
Phosphoproteomic analysis of cells expressing IR, IGF1R, and the chimeric receptors IR_IGF1R and IGF1R_IR. (A) Schematic representation of the experimental strategy applied to analyze the phosphoproteome of DKO preadipocytes reconstituted with IR, IGF1R, and chimeric receptors IR_IGF1R and IGF1R_IR. (B) Relative mRNA levels of recombinant receptors of IR, IGF1R, IR_IGF1R, and IGF1R_IR as determined by qPCR using cDNA standards for quantitation. Data are means ± SEM copy number per nanogram of total RNA, n = 3. (C) Immunoblotting of IR and IGF1R using antibodies specific for IRβ (#3025, Cell Signaling) and IGF1Rβ (#3027, Cell Signaling) in lysates from cells expressing IR, IGF1R, IR_IGF1R, and IGF1R_IR receptor cDNAs. (D) Immunoblotting to determine phosphorylated and total receptor protein levels and IRS-1Y608 phosphorylation in lysates from IR and chimeric receptor IR_IGF1R stimulated with 10 nM insulin or IGF1R and chimeric receptor IGF1R_IR stimulated with 10 nM IGF1 for 15 min. (E) PCA of the phosphosites identified by LC-MS/MS from three independent lines of DKO preadipocytes reconstituted with IR, IGF1R, and chimeric receptors IR_IGF1R and IGF1R_IR in the basal and hormone-stimulated states. Cells were serum starved for 5 h with DMEM containing 0.1% BSA before insulin and IGF1 stimulation.
All four cell lines were then subjected to a global phosphoproteomic analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (15), both in the absence and presence of appropriate ligand stimulation for the ECD (insulin or IGF-1 at 10 nM for 15 min), each performed as three independent biological replicates. Across all cells and conditions, an average of 26,076 phosphosites were identified and quantified in each cell line. Principal component analysis (PCA) and T-distributed stochastic neighbor embedding (t-SNE) indicated clear separation of the phosphoproteome in these four receptors (Fig. 1E and SI Appendix, Fig. S1E) with the different receptors reflected primarily by principal component 1 and the effects of ligand stimulation by principal component 2 (Fig. 1E). The greatest separation occurred between IR and IGF1R in both the basal and stimulated states, with the two chimeric receptors having intermediate positions driven more by the ECD than the ICD.
Differential Phosphorylated Patterns Regulated by IR and IGF1R.
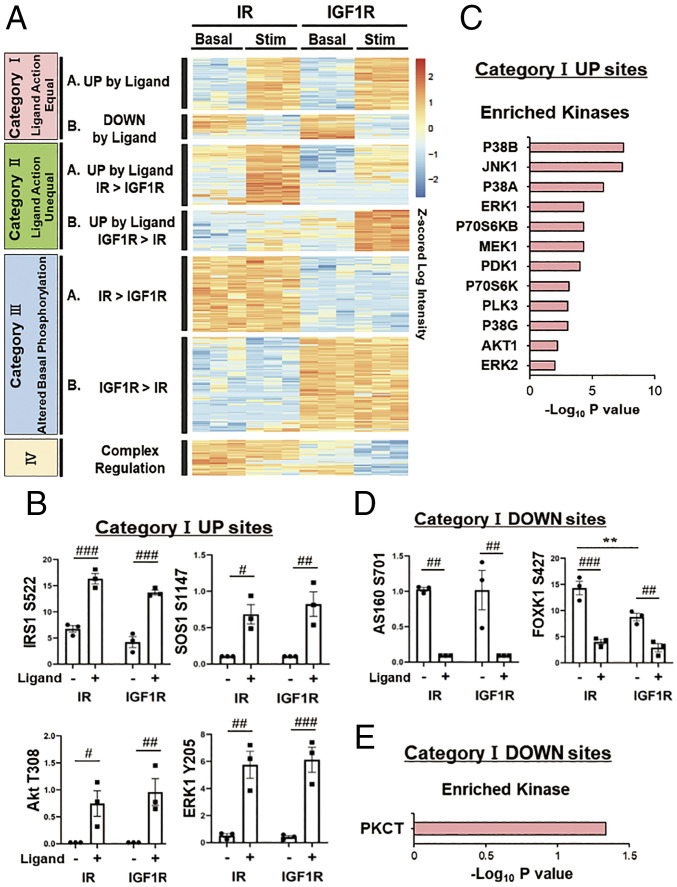
To identify the specific differences in signaling between IR and IGF1R, we analyzed changes in the phosphoproteome between IR and IGF1R in the basal and ligand-stimulated states using heatmap analysis with hierarchical clustering. A total of 3,208 phosphosites (12.3% of the total identified sites) were significantly different between the two receptors in either the basal or stimulated states at an false discovery rate (FDR) of <0.05. These formed seven major clusters based on changes caused by ligand stimulation and differences in phosphorylation in the basal, i.e., unstimulated state (Fig. 2A). Specifically, four of these differential clusters related to ligand action and could be divided into two subclusters: one in which the stimulated IR and stimulated IGF1R produced equal up-regulation or equal down-regulation designated as category IA and category IB, respectively. These contained 446 and 200 phosphosites each. The second subcluster consisted of two ligand up-regulated clusters, where one receptor showed significantly more effect than the other. These were designated as category IIA for sites stimulated by IR > IGF1R and category IIB for sites stimulated by IGF1R > IR and contained 489 and 340 sites, respectively. Finally, and somewhat unexpectedly, there were also two large clusters which were not ligand regulated but differed significantly between IR and IGF1R in the basal state. These were designated category III sites and subdivided into those which showed significantly greater phosphorylation in cells expressing IR than cells expressing IGF1R (category IIIA) or vice versa, i.e., greater in cells expressing IGF1R than cells expressing IR (category IIIB). These two clusters contained 636 and 798 phosphosites, respectively. Finally, there were some category IV sites (n = 299); these showed a more complex pattern due to a combination of receptor and ligand stimulation effects.
Fig. 2.
Differential phosphorylated protein patterns regulated by IR and IGF1R in the basal and stimulated states. (A) Heatmap showing the hierarchical clustering of the phosphopeptides in IR- and IGF1R-expressing cells in the basal and ligand-stimulated states. IR was stimulated with 10 nM insulin, and IGF1R was stimulated with 10 nM IGF1, both for 15 min. Values are Z-scores of log2-transformed intensity values. (B) Analysis of some important phosphosites in category IA, i.e., the equally up-regulated by ligand action for IR and IGF1R. Data are means ± SEM of phosphosite intensity values for three independent samples. #P < 0.05, ##P < 0.01, ###P < 0.001 basal vs. ligand, two-way ANOVA. (C) Kinase enrichment analysis in the cluster of category IA. Plots are –log10 transforms of the enrichment P value. (D) Analysis of some important phosphosites in category IB, i.e., sites equally down-regulated by ligand for both IR and IGF1R. Data are means ± SEM of phosphosite intensity values. ##P < 0.01, ###P < 0.001 basal vs. ligand, **P < 0.01 IR vs. IGF1R, two-way ANOVA. (E) Kinase enrichment analysis for phosphosites in category IB. Plots are –log10 transforms of enrichment P value.
Category IA phosphosites (i.e., those equally up-regulated by ligand with both IR and IGF1R) represent 446 phosphosites on 249 proteins. These include many sites well known to be regulated by both IR and IGF1R, such as phosphorylation of IRS-1S522, AKTT308, and ERK1Y205 (Fig. 2B). Kinase enrichment analysis of experimentally defined kinase phosphosites showed enrichment of several kinases known to be involved in insulin and IGF-1 signaling, including p38 MAP kinase, JNK-1, ERK, PDK1, S6, and Akt1 (Fig. 2C) (P < 0.05). Category IB contained 200 phosphosites on 126 proteins which were equally down-regulated by IR and IGF1R and included AS160S701 (isoform 2 of AS160, UniProtKB: Q8BYJ6-2) and FOXK1S427 (Fig. 2D). Kinase enrichment analysis in this small cluster showed only one potentially involved kinase, PKC-θ, although other novel PKCs are known to share overlapping specificity (Fig. 2E). Together with tyrosine kinases, protein phosphatases regulate the phosphorylation state of many important signaling molecules involved in cell growth and metabolism. We therefore also analyzed enriched potential phosphatases for categories IA and IB phosphosites (SI Appendix, Fig. S2 A and B). Category IA sites showed enrichment of receptor type protein tyrosine phosphatases (PTPs) (PTPRE and PTPRF), nonreceptor type PTPs (PTPN2, PTPN7, and PTPN14), and MAPK phosphatases (DUSP1, DUSP4, DUSP5, and DUSP9).
Differential Phosphorylated Patterns by IR and IGF1R in Ligand Action.
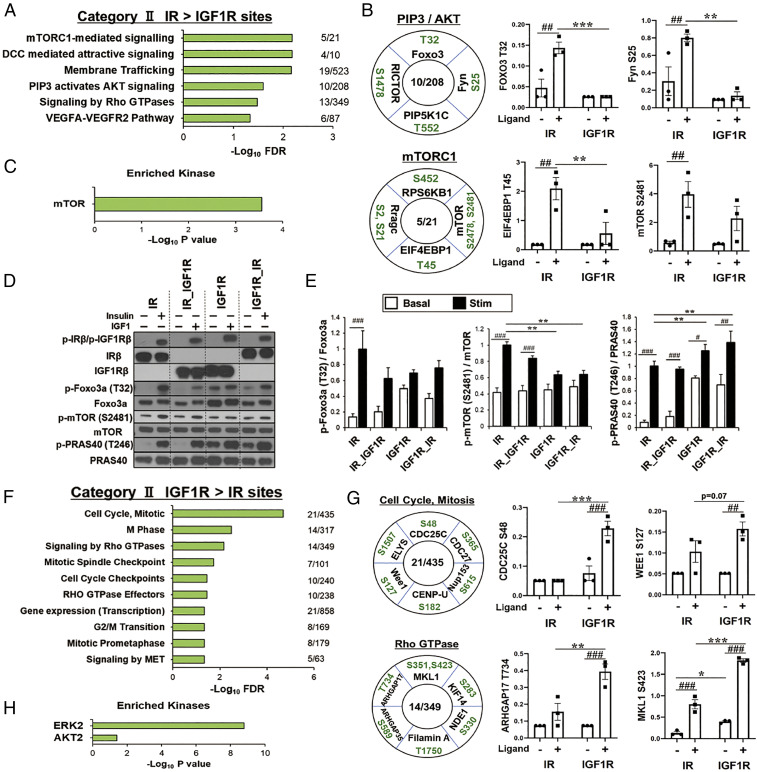
To understand the differences in downstream signaling between IR and IGF1R by ligand action, we next analyzed the two clusters with unequal ligand action (categories IIA and IIB). There were 489 phosphosites on 274 proteins in category IIA, i.e., phosphosites significantly up-regulated by IR > IGF1R following ligand stimulation. Pathway analysis of proteins of this cluster showed six significantly enriched pathways often linked to insulin action, including mammalian target of rapamycin complex 1 (mTORC1) signaling, membrane trafficking, PIP3-activated AKT signaling, and signaling by the Ras homolog family of guanosine triphosphate hydrolases (Rho GTPases) (Fig. 3A). Examples of proteins and phosphosites in these pathways are shown and quantified in Fig. 3B and SI Appendix, Fig. S3A. Proteins involved in mTORC1 whose phosphorylation was up-regulated in IR > IGF1R included core components of mTORC1, such as mTORS2481, and major proteins downstream of mTORC1 (RPS6KB1S452 and EIF4EBP1T45). Not surprisingly, this cluster also included increased phosphorylation of proteins involved in PIP3 and AKT signaling, including FOXO3T32, RICTORS1478, PIP5K1CT552, and FynS25. Kinase enrichment analysis of the phosphosites in this cluster revealed strong enrichment for mTOR, as a major driver of this pathway (Fig. 3C). Many of these changes were confirmed by immunoblotting using phosphosite-specific antibodies (Fig. 3 D and E and SI Appendix, Fig. S3 B–D). Indeed, the phosphorylation of mTORS2481, which only tended to be higher in IR-expressing cells by phosphoproteomics, was significantly higher by IR when assessed by immunoblotting. Increased phosphorylation of EIF4EBP1T37/46 was also higher in IR expressing cells. Interestingly, these occurred despite equal ligand-stimulated phosphorylation of Akt Thr-308 by IR and IGF1R (Fig. 2B and SI Appendix, Fig. S3 C and D). Phosphatase analysis of category IIA sites showed enrichment for two nonreceptor type PTPs, PTPN11 and PTPN13 (SI Appendix, Fig. S3E). PTPN13 has been shown to be involved in multiple signaling pathways including dephosphorylation of IRS-1 and inhibition of PI3K (16).
Fig. 3.
Differential phosphorylated patterns by IR and IGF1R in the ligand action. (A) REACTOME pathway enrichment analysis of phosphosites in category IIA, i.e., those up-regulated by ligand for IR > IGF1R. Plots are –log10 transforms of enrichment FDR value. (B) Representation of some important phosphosites in selected pathways and quantification of exemplary phosphosites in the cluster of category IIA. Numbers in the center of each circle indicate number of proteins regulated within each pathway. Data are means ± SEM of phosphosite intensity values. ##P < 0.01 basal vs. ligand, **P < 0.01, ***P < 0.001 IR vs. IGF1R, two-way ANOVA. (C) Kinase enrichment analysis for phosphosites in category IIA. Plots are –log10 transforms of enrichment P value. (D and E) Validation of phosphoproteomics by immunoblotting in lysates from IR, IR_IGF1R, IGF1R, and IGF1R_IR cells. Representative immunoblot analysis (D) and quantification of phosphorylation of p-Foxo3aT32, p-mTORS2481, and p-PRAS40T246 (E). Data are means ± SEM (n = 3 per group). #P < 0.05, ##P < 0.01, ###P < 0.001 basal vs. ligand, **P < 0.01, two-way ANOVA. (F) REACTOME pathways enrichment analysis of phosphosites up-regulated by ligand for IGF1R > IR (category IIB). Plots are –log10 transforms of enrichment FDR value. (G) Representation of some important phosphosites on selected pathways and quantification of exemplary phosphosites in the cluster of category IIB. Data are means ± SEM of phosphosites intensity values. ##P < 0.01, ###P < 0.001 basal vs. ligand, *P < 0.05, **P < 0.01, ***P < 0.001 IR vs. IGF1R, two-way ANOVA. (H) Kinase enrichment analysis of phosphosites in category IIB. Plots are –log10 transforms of enrichment P value.
By comparison, category IIB consisted of 340 phosphosites on 248 proteins that were up-regulated by ligand in IGF1R–expressing cells significantly greater than in IR-expressing cells. Pathway analysis showed that this cluster was significantly enriched for proteins related to cell cycle, mitosis, and signaling by Rho GTPases (Fig. 3F). Examples of proteins and phosphosites in these pathways with some phosphosites quantified are shown in Fig. 3G and SI Appendix, Fig. S3F. Proteins involved in cell cycle and mitosis whose phosphorylation was up-regulated in ligand-stimulated IGF1R > IR included well-recognized substrates such as CDC25CS48 and CDC27S365, which play key roles in the regulation of cell division, as well as lesser known proteins such as CENP-US182, which plays a role in assembly of kinetochore proteins and mitotic progression; ELYSS1507, a protein required for the assembly of a functional nuclear pore complex (NPC) (17); and Wee1S127, which acts as a negative regulator of entry into mitosis (18). Activated IGF1R was also more effective than activated IR to increase phosphorylation of proteins involved in signaling by Rho GTPases, including multiple Rho GAPs (ARHGAP17T734 and ARHGAP35S589), which promote GTP hydrolysis and thus put Rho GTPases in an inactive state, as well as MKL1S351,S423,T822, which controls expression of genes regulating myogenic differentiation and cytoskeletal organization (19); filamin AT1750, which regulates actin remodeling; NDE1S330, which plays an essential role in microtubule organization (20); and KIF14S283, which is involved in chromosome segregation and cytokinesis (21). Kinase enrichment analysis of phosphosites in this cluster revealed these to be downstream of ERK2 and AKT2 (Fig. 3H). Phosphatase analysis showed phosphosites enriched for dephosphorylation by the protein phosphatases PHLPP1 and PHLPP2 (SI Appendix, Fig. S3G), which have been shown to be important negative regulators of Akt and protein kinase C (PKC) isoforms (22). We also calculated changes in phosphorylation as fold stimulation, i.e., the stimulated level divided by basal level of phosphosites in Fig. 3 B and G and SI Appendix, Fig. S3 A and F (SI Appendix, Fig. S3 H and I). In most cases, the differences seen between IR and IGF1R were maintained, although in a few they were not, due to changes in basal phosphorylation. In agreement with the phosphoproteomics, Western blotting revealed that phosphorylation of PRAS40, a core component of mTORC1, at Thr-246 was increased by IGF1R > IR (Fig. 3 D and E). However, this was due primarily to increases in basal phosphorylation. Thus, when calculated as fold change, i.e., stimulated level divided by basal level, IR had a much stronger effect to stimulate phosphorylation of PRAS40 than IGF1R (SI Appendix, Fig. S3B). To what extent absolute levels of phosphorylation versus stimulation ratio play a role in final biological effect remains to be determined.
An Integrated Protein Phosphorylation Map of IR vs. IGF1R in Ligand Action.
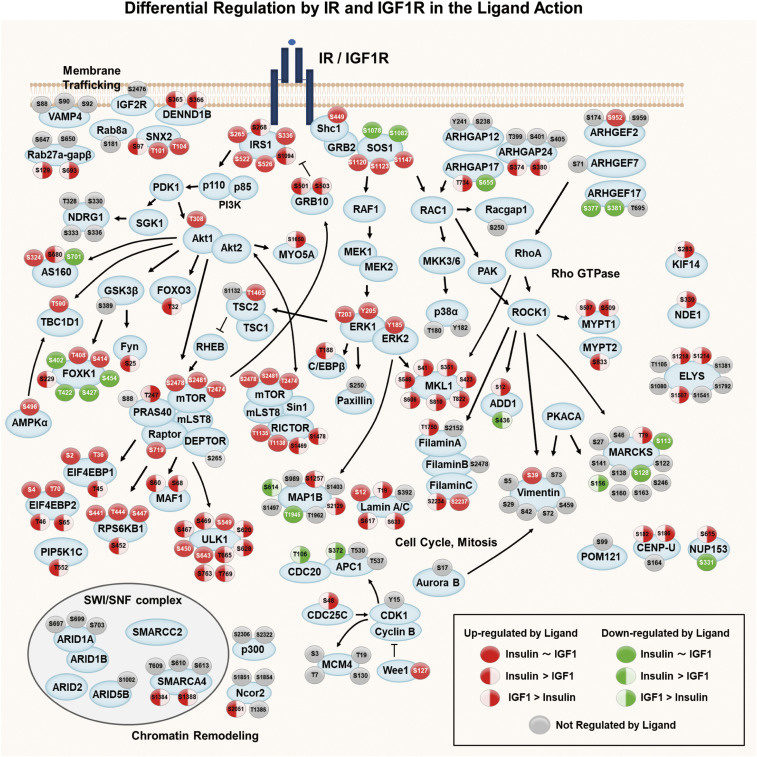
To visualize the differential signaling events between IR and IGF1R, as well as their points of overlap, we combined category IA, IB, IIA, and IIB phosphosites to generate an integrated signaling map showing phosphorylations mediated by IR and/or IGF1R following ligand stimulation (Fig. 4). It is clear that a larger fraction of phosphosites were up-regulated by ligand stimulation than were down-regulated. Differences of action on different sites in a single protein are also apparent. For example, of the six phosphoserine sites on IRS1, a major substrate in the insulin/IGF-1 signaling networks, four (IRS1S265,S336,S522,S526) were equally stimulated by both ligand-activated receptors, whereas two phosphosites (IRS1S268, S1094) were preferentially stimulated by IR compared to IGF1R. On the other hand, Shc1S449, AktT308, ERK1T203,Y205, and ERK2Y185 were stimulated equally by activated IR and IGF1R, as was vimentinS39, a site previously reported to be downstream of Akt1 (23). Three phosphorylation sites on mTOR were identified by phosphoproteomics as increased by ligand stimulation and were not significantly different between IR and IGF1R, although phosphorylation of mTORS2481 was significantly higher in IR when assessed by immunoblotting (Fig. 3E). Whether this difference reflects effects of conformational changes or is due to other modifications remains to be determined. Interestingly, many phosphosites associated with mTORC1 signaling (4EBP1T45, 4EBP2T46,S65, ULK1S467,S469, S763,T769, and MAF1S60,S68), many phosphosites related to PIP3-activated AKT signaling (FOXO3T32, FynS25, and FOXK1S229), and sites on proteins involved in membrane trafficking (DENND1BS365,S366 and SNX2S97) were more highly up-regulated by IR than IGF1R.
Fig. 4.
Integrated map showing differential signaling networks stimulated by the ligand-activated IR vs. IGF1R. Signaling map showing phosphosites regulated by ligand for IR and/or IGF1R identified by phosphoproteomics. Red-filled circles represent sites for which phosphorylation was increased and green-filled circles, sites for which phosphorylation was decreased by ligand (P < 0.05). Circles with solid fill represent sites equally regulated by ligand for both receptors. Sites with red or green only on the Left represent sites regulated by IR greater than IGF1R. Sites with green or red fill only on the Right represent sites for which regulation was greater by IGF1R than IR. Arrows indicate known protein–protein interactions and phosphorylation/dephosphorylation events curated from databases of experimentally defined kinase–substrate relationships (PhosphositePlus) and the literature.
On the other hand, many phosphosites on proteins associated with cell cycle, mitosis, and signaling by Rho GTPases were up-regulated to significantly higher levels by ligand-stimulated IGF1R versus IR, including CDC25CS48, Lamin A/CT19,S633, CENP-US182,S186, and NUP153S615 (all proteins involved in cell cycle and mitosis) and ARHGAP17T734, MYPT1S597, filamin AT1750, MKL1S41,S351,S423,S548,S606,S810,T822, KIF14S283, and NDE1S330 (all proteins are associated with signaling by Rho GTPases). Finally, among phosphosites down-regulated by ligand stimulation, MAP1BS614 was more down-regulated by IR than IGF1R, while APC1S372, CDC20T106, and MARCKSS156 were more down-regulated by IGF1R than IR.
Differential Phosphorylated Patterns by IR and IGF1R in the Basal State.
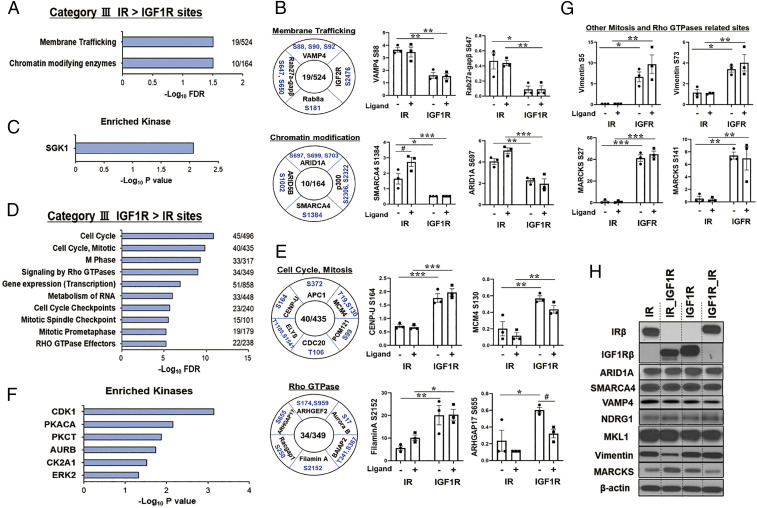
Unexpected, but even more striking than changes in ligand stimulation, were changes in phosphorylation in the basal state, i.e., in cells expressing unoccupied IR and IGF1R. These were designated as categories IIIA and IIIB. The former included 636 phosphosites on 280 proteins for which basal phosphorylation was significantly higher in cells expressing IR versus cells expressing IGF1R. Enrichment analysis of these proteins revealed two overrepresented REACTOME pathways: membrane trafficking and chromatin modifying enzymes (Fig. 5A). Some representative phosphosites in these pathways are shown and quantified in Fig. 5B and include increased basal and stimulated phosphorylation of proteins involved in membrane trafficking, such as VAMP4S88,S90,S92, IGF-2RS2476, Rab27a-gapβS647,S650, and Rab8aS181. VAMP4 is particularly important, since VAMP4 regulates GLUT4 and CD36 translocation, which are involved in uptake of glucose and fatty acids (24). There was also increased phosphorylation of proteins involved in chromatin modification, including ARID1AS697,S699,S703, ARID5BS1002, SMARCA4S1384, and p300S2306,2322. ARID1A, ARID5B, and SMARCA4 involved in chromatin remodeling and critical in metabolic reprogramming and adaptation to nutritional signals (25). p300 is a transcriptional coactivator and histone acetyltransferase and is typically recruited to transcriptional enhancers and regulates gene expression by acetylating chromatin (26). Kinase enrichment analysis of this cluster showed significant enrichment of sites for the serine/threonine-protein kinase SGK1 (Fig. 5C). Several sites on NDRG1 (NDRG1T328,S330,S333,S336) were up-regulated in the basal state only in IR-expressing cells. NDRG1 is a downstream protein of SGK1 and has a role for attenuating ErbB signaling (27).
Fig. 5.
Proteins differentially phosphorylated in cells expressing IR versus IGF1R in the unstimulated (basal) state. (A) REACTOME pathway enrichment analysis of phosphosites in the IR > IGF1R cluster (category IIIA). Plots are –log10 transforms of enrichment FDR value. (B) Representation of some important phosphosites in selected pathways, and quantification of exemplary phosphosites in the cluster of category IIIA. Numbers in the center indicate number of proteins regulated within each pathway. Data are means ± SEM of phosphosites intensity values. #P < 0.05 basal vs. ligand, *P < 0.05, **P < 0.01, ***P < 0.001 IR vs. IGF1R, two-way ANOVA. (C) Kinase enrichment analysis of phosphosites in category IIIA. Plots are –log10 transforms of enrichment P value. (D) REACTOME pathway enrichment analysis of phosphosites in the IGF1R basal > IR basal cluster (category IIIB). Plots are –log10 transforms of enrichment FDR value. (E) Representation of some important phosphosites on selected pathways, and quantification of exemplary phosphosites in category IIIB. Data are means ± SEM of phosphosites intensity values. #P < 0.05 basal vs. ligand, *P < 0.05, **P < 0.01, ***P < 0.001 IR vs. IGF1R, two-way ANOVA. (F) Kinase enrichment analysis of phosphosites in category IIIB. Plots are –log10 transforms of enrichment P value. (G) Quantification of exemplary phosphosites of vimentin and MARCKS which are secondary downstream targets of Rho GTPases signaling and cell cycle and mitosis. *P < 0.05, **P < 0.01, ***P < 0.001 IR vs. IGF1R, two-way ANOVA. (H) Immunoblotting of total protein levels for ARID1A, SMARCA4, VAMP4, NDRG1, MKL1, vimentin, and MARCKS in lysates from cells expressing IR, IR_IGF1R, IGF1R, and IGF1R_IR.
Finally, 798 phosphosites on 497 proteins showed significantly increased basal phosphorylation in cells expressing IGF1R versus IR (category IIIB). Enrichment analysis revealed that many of these fell into 10 pathways, most related to cell cycle, mitosis, and Rho GTPase signaling (Fig. 5D). This is similar to the pathways identified to be ligand-regulated by IGF1R > IR (category IIB). Quantitation of some representative phosphosites is shown in Fig. 5E and SI Appendix, Fig. S4A. Proteins involved in cell cycle and mitosis whose basal phosphorylation was up-regulated in cells expressing IGF1R included APC1S372 and CDC20T106; MCM4T19,S130, which is essential for the initiation of eukaryotic genome replication (28); POM121S99, an essential component of the nuclear pore complex (29); and CENP-US164 and ELYST1105,S1541 involved in mitotic progression and nuclear pore function. There was also increased phosphorylation of proteins involved in signaling by Rho GTPases, including Aurora B kinaseS17, which participates in alignment and segregation of chromosomes during mitosis; Racgap1S250, a GTPase-activating protein for RhoA required for cytokinesis; the Rho GTPase-activating protein and exchange factor ARHGAP17S655 and ARHGEF2S174,S959; and filamin AS2152. Kinase enrichment analysis indicated that this cluster included potential targets of CDK1, PKA-CA, PKC-θ, AUR-B, CK2A1, and ERK2 (Fig. 5F), kinases closely linked to cell cycle and mitosis. There was also multisite phosphorylation of vimentin (S5, S29, S42, S72, S73, and S459) and MARCKS (S27, S46, S113, S122, S128, S138, S141, S156, S160, S163, and S246), two other proteins associated with mitosis and Rho GTPase signaling (30, 31), that were increased to a greater extent in cells expressing IGF1R than cells expressing IR (Fig. 5G). In most cases, these differences were not due to changes in total protein level (Fig. 5H and SI Appendix, Fig. S4 B–D). Thus, immunoblotting of ARID1A, SMARCA4, VAMP4, NDRG1, FOXK1, MKL1, vimentin, and Lamin A/C showed no differences in total protein between IR- and IGF1R–expressing cells, indicating that altered phosphorylation of these proteins was due to changes in signaling not protein abundance. The one exception was MARCKS protein, which was higher in cells expressing IGF1R than in IR, suggesting that the higher multisite phosphorylation of MARCKS protein in IGF1R–expressing cells was due, at least in part, to a higher level of protein.
An Integrated Protein Phosphorylation Map of IR vs. IGF1R in the Basal State.
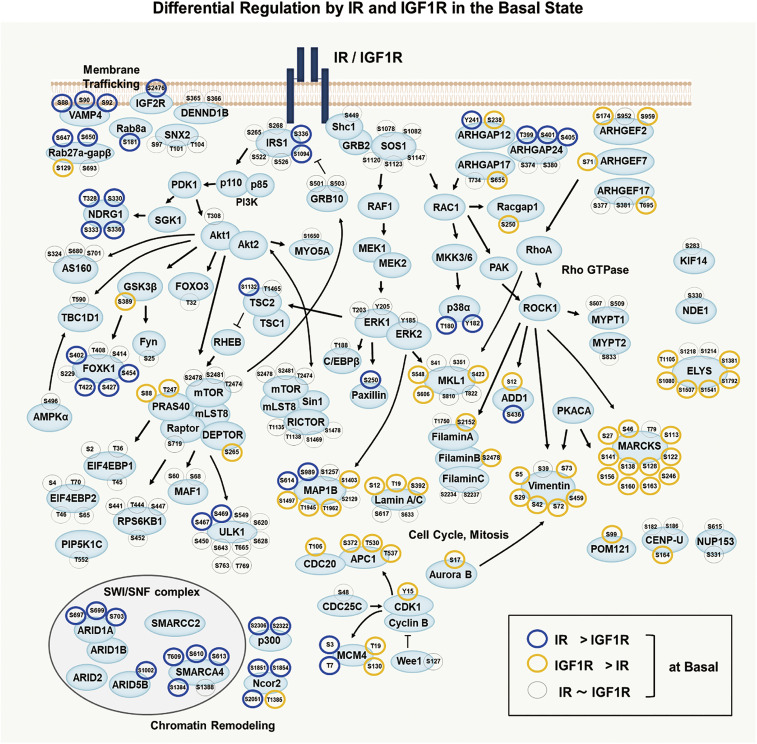
An integrated signaling map of differences in basal phosphorylation between IR- and IGF1R–expressing cells, indicating important differences in signaling by the unoccupied IR and IGF1R, is shown in Fig. 6. Consistent with the pathway analysis, many phosphosites up-regulated by unoccupied IR greater than unoccupied IGF1R were on proteins related to membrane trafficking (VAMP4S88,S90,S92, IGF-2RS2476, Rab27a-gapβS647,S650, and Rab8aS181) or chromatin modification (ARID1AS697,S699,S703, SMARCA4T609,S610,S613, p300S2306,S2322, and Ncor2S1851,S1854,S2051). These could reflect a “priming” effect of unoccupied IR itself on glucose transport and gene expression. In addition, multiple phosphosites on NDRG1 (T328, S330, S333, and S336) that are downstream of SGK-1 were also up-regulated in the basal state in IR-expressing cells. On the other hand, many phosphosites of proteins related to Rho GTPase signaling, including multiple Rho GEFs (ARHGEF2S174,S959, ARHGEF7S71,S959, ARHGEF17T695) and Rho GAPs (ARHGAP12S238 and ARHGAP17S655, S1288), MKL1S423,S548,S606, filamin AS2152, and Racgap1S250, and many proteins related to cell cycle and mitosis, including vimentin, Aurora B kinaseS17, CDK1Y15, APC1S372,T530,T537, CDC20T106, and Lamin A/CS12,T19,S392 were up-regulated in cells expressing IGF1R greater than IR in the basal state. Thus, even in the unoccupied state, IR is preferentially generating signals involved in metabolic effects, whereas IGF1R is priming pathways involved in mitogenic control. A summary of all the categories in the heatmap (Fig. 2A) and their enriched pathways and kinases are shown in SI Appendix, Table S1.
Fig. 6.
Integrated map showing altered signaling in cells expressing IR vs. IGF1R in the basal state. Diagram of intracellular signaling networks regulated by IR and IGF1R in the basal state as identified by phosphoproteomics. Each of the indicated phosphosite was analyzed by two-way ANOVA. Blue and yellow halos represent sites increased in cells expressing IR versus IGF1R, respectively, in the basal (nonstimulated) state, which were significantly different at the P < 0.05 level. Arrows indicate protein–protein interactions and phosphorylation/dephosphorylation events curated from databases of experimentally defined kinase–substrate relationships (PhosphositePlus) and the literature.
Domain-Dependent Effects of IR and IGF1R in the Basal and Stimulated States.
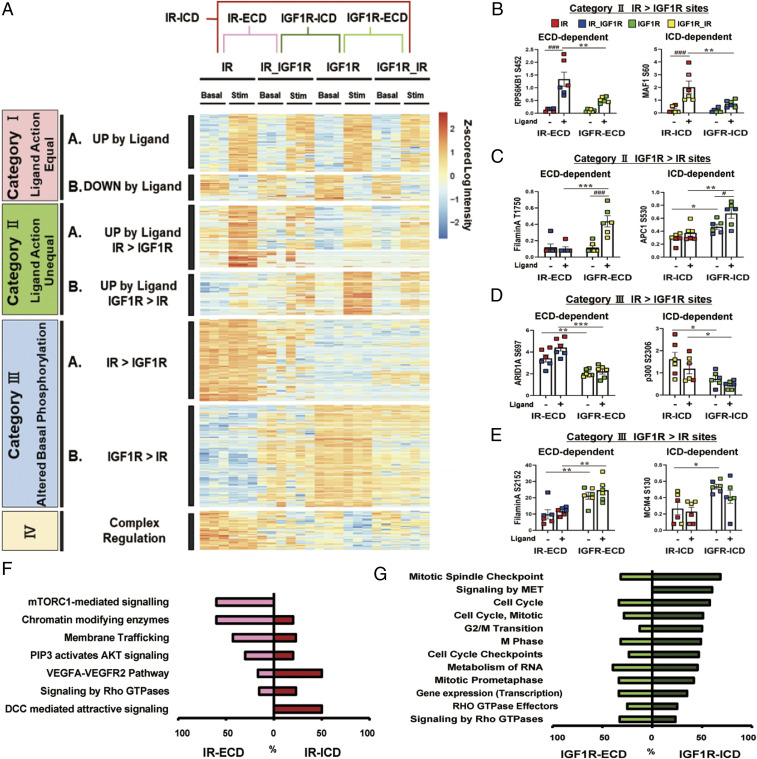
Although the intracellular domain of IR and IGF1R contain the kinase domain involved in signaling, both the extracellular and intracellular domains undergo conformational changes during ligand signaling (32) and have the ability to interact with other proteins in the cell (33, 34). To determine to what extent differences in phosphorylation were determined by effects of the extracellular domains vs. intracellular domains of these receptors, we analyzed phosphoproteome of cells expressing the chimeric receptors exchanging ICD and ECD of the IR and IGF1R. A hierarchical clustering map of the phosphoproteomic data including chimeric receptors IR_IGF1R and IGF1R_IR along with the native receptors is shown in Fig. 7A. For categories IA and IB (i.e., sites equally up- or down-regulated by ligand activation of IR and IGF1R), the changes in phosphorylation following ligand stimulation of the chimeric receptors showed similar changes to the native IR and IGF1R. By contrast, regulation of phosphorylation by categories II and III phosphosites were changed in different ways following exchange of the intracellular and extracellular domains of the two receptors, allowing a further subclassification of the categories II and III to identify the domain dependency of these phosphosites (SI Appendix, Fig. S5). Fig. 7 B–E shows examples of domain-dependent changed phosphosites in categories II and III. In category IIA, up-regulation of RPS6KB1S452 by ligand depended on IR-ECD, while up-regulation of MAF1S60 depended on IR-ICD. In category IIB, up-regulation of Filamin AT1750 by ligand depended on IGF-R-ECD, while up-regulation of APC1S530 depended on IGF1R-ICD. In category IIIA, higher phosphorylation in ARID1AS697 depended on IR-ECD, while higher phosphorylation in p300S2306 depended on IR-ICD. In category IIIB, higher phosphorylation in Filamin AS2152 depended on IGF1R-ECD, while higher phosphorylation in MCM4S130 depended on IGF1R-ICD.
Fig. 7.
Domain-dependent effects of IR and IGF1R in the basal and ligand-stimulated states. (A) A hierarchical clustering heatmap showing sites that were differential between IR vs. IGF1R in the basal or stimulated states (shown in Fig. 2A) to which has been added the phosphoproteomic data of cells expressing chimeric IR_IGF1R and IGF1R_IR receptors. Values are Z-scores of log2-transformed intensity values. (B–E) Quantification of exemplary phosphosites which were ECD dependent or ICD dependent in category IIA, i.e., ligand-stimulated IR > IGF1R (B); in category IIB, i.e., ligand-stimulated IGF1R > IR (C); in category IIIA, i.e., basal IR > IGF1R (D); and in category IIIB, i.e., basal IGF1R > IR (E). Data are means ± SEM of phosphosites intensity values. #P < 0.05, ###P < 0.001 basal vs. ligand, *P < 0.05, **P < 0.01, ***P < 0.001 IR vs. IGF1R, two-way ANOVA. (F) Difference in proportions of IR-ECD-dependent and IR-ICD-dependent proteins in the enriched REACTOME pathways of the categories IIA and IIIA (Figs. 3A and 5A). (G) Difference in proportions of IGF1R-ECD-dependent and IGF1R-ICD-dependent proteins in the enriched REACTOME pathways of categories IIB and IIIB (Figs. 3F and 5D).
Depending on the downstream pathway being regulated, the differentially regulated phosphosites observed in both the basal and stimulated states in IR- vs. IGF1R–expressing cells were driven largely by either the intracellular or extracellular domains of the two receptors (Fig. 7 F and G and SI Appendix, Fig. S5). Thus, phosphorylation differences on proteins in pathways related to mTORC1 signaling, chromatin modifying enzymes, PIP3-activated AKT signaling, and membrane trafficking were mostly dependent on the IR extracellular domain, whereas pathways of DCC-mediated attractive signaling, which is involved in netrin-induced axon attraction and involves the focal adhesion kinase and src family kinases, and VEGFA-VEGFR2 action, were more dependent on IR intracellular domain (Fig. 7F and SI Appendix, Fig. S5 A and C). Importantly, these differences in phosphorylation control led to differences in biological effects of these receptors and their chimeric counterparts. For example, autophagic flux was more markedly inhibited by ligand stimulation of IR than IGF1R, and this is in agreement with the fact that protein phosphorylations downstream of mTORC1, which is a central regulator of autophagy, were more potently regulated by IR versus IGF1R (SI Appendix, Fig. S6 A and B). Even more striking and consistent with the differences in phosphorylation observed with the chimeric receptors, there was a greater inhibition of autophagy by ligand stimulation of receptors containing the IR-ECD than receptors containing the IGF1R-ECD (SI Appendix, Fig. S6 C and D). Similarly, we also identified IGF1R-ECD–dependent and IGF1R-ICD–dependent proteins showing increased phosphorylation in the basal or stimulated states by IGF1R (Fig. 7G and SI Appendix, Fig. S5 B and D). Thus, most of the pathways associated with cell cycle and mitosis depended more on the IGF1R intracellular domain, while phosphorylation of proteins involved in signaling by Rho GTPases depended more on the presence of the IGF1R extracellular domain. Likewise, many phosphosites associated with membrane trafficking and chromatin remodeling were preferentially up-regulated by IR-ECD as compared with IGF1R-ECD, whereas many phosphosites associated with cell cycle were uniquely up-regulated by IGF1R-ICD as compared with IR-ICD. An integrated signaling map showing phosphorylations that could be specifically linked to either the intracellular or extracellular domains of the insulin and IGR-1 receptors for the ligand-dependent effects, i.e., those observed in the stimulated state, is shown in SI Appendix, Fig. S7 and those which could be linked to the ICD or ECD in the basal, unstimulated state are shown in SI Appendix, Fig. S8. Note for example the cluster of ligand-stimulated sites on ULK1 whose phosphorylation was dependent on IR-ECD > IGF1R-ECD versus the cluster of sites on MKL1 where the converse was true (SI Appendix, Fig. S7). Some of these showed similar differences in the basal state and some did not. There were also sites which were differential based on receptor domains in the basal state only, such as the IR-ECD-dependent phosphorylations on VAMP4 (SI Appendix, Fig. S8). Understanding the exact kinase/phosphatase pathways linked to these receptor differential phosphorylations will be important in developing pathway-specific agonists and antagonists.
Discussion
Physiologically, in both humans and mice, insulin is a major regulator of glucose and lipid metabolism, whereas IGF-1 is a major regulator of growth and development (4, 10, 11, 35, 36). These ligands bind to two highly homologous receptors (IR and IGF1R) that show many similarities in structure and share many overlapping downstream signaling pathways (1, 9). Separation of the actions of these hormones and receptors is further complicated by the fact that at high concentrations these ligands can bind to each other’s receptors and show overlapping effects (9, 37, 38), and by the fact that most cells express both IR and IGF1R and these can form hybrid IR/IGF1R receptors, which can also bind both ligands and have a mixture of signaling effects (36). Several studies have shown differences in the ability of IR and IGF1R to engage their immediate substrate proteins, such as IRS-1, IRS-2, and Shc (2, 39, 40); however, these tyrosine kinases have a limited number of primary substrates, and most final effects are mediated by a much broader network of serine and threonine phosphorylations leading to altered activity, structure, and/or subcellular localization of these proteins.
To determine differences in signaling between IR and IGF1R that could account for their unique physiological actions, we performed global phosphoproteomic analysis of preadipocytes in which we had inactivated genes for production of both endogenous receptors, then reconstituted these cells with cDNAs for IR only, IGF1R only, or chimeric IR_IGF1R and IGF1R_IR receptors. Analysis of phosphoproteomic data from these cells revealed many similarly regulated sites, but more importantly a broad network of differentially regulated phosphorylations regulated by basal and ligand-stimulated IR and IGF1R. Not surprisingly, sites that were similarly up- or down-regulated by ligands of these two highly homologous receptors (categories IA and IB) included many of the most studied phosphosites on IRS1, SOS1, and Akt1. Kinase enrichment analysis reveals these to be targets of many well-known kinases downstream of both IR and IGF1R, including p38, JNK1, ERK, PDK1, S6, and Akt1. More interesting are the two clusters of ligand-regulated phosphosites which show preferential activation by IR (category IIA) or IGF1R (category IIB). These include many phosphosites associated with mTOR signaling, PI 3 kinase-AKT signaling, and membrane trafficking that were preferentially up-regulated by IR, whereas many phosphosites on proteins associated with regulation of Rho GTPases and cell cycle were uniquely up-regulated by IGF1R.
More important is the observation that over 1,400 phosphosites were differentially phosphorylated in cells expressing IR vs. IGF1R even in the basal, unstimulated state (categories IIIA and IIIB), indicating the existence of major differences in signaling by these receptors in the unoccupied state. Among these, phosphosites associated with membrane trafficking and chromatin remodeling were preferentially up-regulated by IR, whereas phosphosites on proteins associated with Rho GTPases, cell cycle, and mitosis were uniquely up-regulated by IGF1R. The differential phosphorylations of these proteins at the basal state were, for the most part, not due to changes in total protein levels. On the other hand, these differences in the phosphoproteome under the basal condition could lead to functional differences in cells expressing these two receptors. Thus, we have previously shown that cells lacking both IR and IGF1R have altered sensitivity to apoptosis and altered imprinted gene expression and that some of these effects can be reversed by reexpression of IR or IGF1R even in the absence of ligand stimulation (13, 14). Similar effects have been observed by other types of unoccupied (unliganded) receptors, such as regulation of osteoclast apoptosis by unoccupied αvβ3 integrin (41), regulation of neuronal apoptosis by the unliganded neurotrophin receptor (42), and inhibitory signals from unoccupied GPCR (43). These and other studies indicate that IR and IGF1R have multiple signaling states, including the unoccupied and occupied by ligand states.
Thus, despite their highly homologous structure, their use of overlapping signaling networks, and their ability to bind both insulin and IGF-1, IR and IGF1R regulate phosphorylation of unique networks of proteins in both the basal and stimulated states. Consistent with their in vivo biology, proteins regulated by IR are more involved in control of metabolism, while those regulated by IGF1R are more involved in control of mitogenesis (4, 44). These data suggest that differences in relative levels of these receptors in tissue contribute to their differential postreceptor effects even in the unoccupied state.
Previous studies have suggested that differences in regulation of both intracellular signaling and downstream actions on gene expression are mainly dependent on differences in the intracellular domains of IR and IGF1R (2, 45), although some small differences in these actions have been suggested to depend on differences in extracellular domains (2). When the data of differential regulation of phosphorylation by IR and IGF1R were compared to the phosphoproteome of cells expressing IR and IGF1R chimeric receptors, i.e., receptors with the ECD of IR fused to the ICD of IGF1R and vice versa, we found a large number of domain-dependent effects of IR and IGF1R. Thus, phosphorylation events which were higher in both the basal and stimulated states in cells expressing IR > IGF1R in categories IIA and IIIA, such as phosphorylations in the mTORC1 signaling pathway, were mostly dependent on the presence of the IR extracellular domain, rather than intracellular domain. This resulted in parallel changes in downstream effects such as autophagic flux which was inhibited to a greater extent in cell receptors containing receptors with the IR-ECD, i.e., IR_IGF1R and IR. A previous report showed that mTORC1 is an upstream kinase of RagCS21, and this phosphorylation is essential for mTORC1 activation (46). We found that category IIA includes RagCS21, and the phosphorylation of RagCS21 is up-regulated by insulin in IR-expressing cells. This suggests changes in the lysosome state between insulin and IGF-1 stimulation which may contribute to preferential mTORC1 activation by IR. Future studies addressing the spatiotemporal distribution of signaling complexes in this system should shed more light into this matter. Similarly, phosphorylations in pathways of membrane trafficking and on chromatin modifying enzymes depended more on IR-ECD than IR-ICD. On the other hand, among phosphorylations in the IGF1R > IR clusters (categories IIB and IIIB), many of pathways associated with cell cycle and mitosis depended more on the IGF1R-ICD. Likewise, cell cycle and mitosis through the Shc-Grb2-Sos-Ras-Raf-MAPK pathway depended on IGF1R-ICD, while the pathway of signaling by Rho GTPases depended more on IGF1R-ECD. These data are consistent with our previous study which showed that cells containing either normal or chimeric receptors with IGF1R-ICD have higher levels of proliferative potential compared to IR-ICD (2). Taken together, these findings indicate that the extracellular domains of both IR and IGF1R have important roles in control of both basal and stimulated signaling, which are in addition to specificity created by the intracellular domains of the receptors. For practical reasons, these phosphoproteomic data have only been able to compare insulin and IGF-1 action at a single time point and with a single ligand concentration (10 nM and 15 min after ligand stimulation) and in cells expressing somewhat higher than normal levels of receptors. It is possible that there are differences between insulin and IGF-1 in the kinetics and dose–response for activation in these phosphorylation networks, which might show additional differences contributing to their unique actions, since growth responses usually require longer stimulation with higher concentrations of ligand than metabolic responses.
In summary, using a comprehensive global phosphoproteomics approach, we have demonstrated that IR and IGF1R have distinct patterns of signaling in both the basal and ligand-stimulated states. Thus, many phosphosites associated with mTOR signaling were preferentially up-regulated by ligand-stimulated IR, whereas many phosphosites associated with Rho GTPases and cell cycle were uniquely up-regulated by ligand-stimulated IGF1R. Interestingly, there were also many differences in the phosphoproteome between cells expressing IR and IGF1R in the basal state. Thus, many phosphosites on proteins associated with membrane trafficking and chromatin remodeling were higher in IR-expressing cells even in the unstimulated state, whereas many phosphosites associated with Rho GTPases and cell cycle were uniquely higher in cells expressing IGF1R, indicating important and previously unrecognized differences in signaling by the unoccupied IR and IGF1R. Using the cells expressing chimeric receptors, we were able to uncover roles for both the intracellular and extracellular domains of IR and IGF1R in these differential networks of phosphorylation. Together, these data provide insights into the unique roles of IR and IGF1R in biology.
Methods
Materials.
Antibodies for immunoblotting are in SI Appendix, SI Methods. Human insulin was purchased from Sigma and human IGF-1 from Preprotech. Plasmids of IR, IGF1R, and chimeric IR_IGF1R and IGF1R_IR were generated as described previously (2). Detailed methods are in SI Appendix, SI Methods.
Brown Preadipocytes Isolation and Culture.
IR/IGF1R double knockout brown preadipocytes were obtained as previously described (13). Detailed methods are in SI Appendix, SI Methods.
Quantification of mRNA Levels.
Total RNA was isolated from cells using TRIzol reagent (Thermo Fisher Scientific). cDNA was synthesized from 500 ng total RNA using the cDNA Reverse Transcription kit (Applied Biosystems). Quantitative PCR amplification was conducted with the C1000 Thermal Cycler (BioRad, catalog CFX384) using iQ SybrGreen Supermix (Bio-Rad) according to the protocol supplied by the manufacturer. The sequences of primers are as follows: mouse IR, 5′-AAA-TGCAGGAACTCTCGGAAGCCT-3′ and 5′-ACCTTCGAGGATTTGGCAGACCTT-3′; and mouse IGF1R, 5′-ATCGCGATTTCTGCGCCAACA-3′ and 5′-TTCTTCTCTTCATCGCCG-CAGACT-3′.
Insulin and IGF-1 Signaling.
Cells were serum starved for 5 h with Dulbecco’s modified Eagle medium (DMEM) containing 0.1% bovine serum albumin (BSA) before hormone stimulation. For all experiments, the ligand-dependent activation of the receptors was performed using the ligand matching the extracellular domain of the receptor. Cells expressing wild-type IR or the chimeric IR_IGF1R receptor were stimulated with 10 nM insulin for 15 min, while cells expressing wild-type IGF1R or the chimeric IGF1R_IR receptor were stimulated with 10 nM IGF-1 for 15 min. After stimulation, cells were washed immediately with ice-cold phosphate-buffered saline (PBS) once before lysis and scraped down in radioimmunoprecipitation assay (RIPA) lysis buffer complemented with 50 mM potassium fluoride (KF), 50 mM β-glycerolphosphate, 2 mM ethylene glycol tetraacetic acid (EGTA) (pH8.0), 1 mM Na3VO4, and 1× protease inhibitor mixture (Biotool).
Immunoblotting.
Detailed methods are described in SI Appendix, SI Methods.
Statistical Analysis.
Data are presented as mean ± SEM. Comparisons between more than two groups were performed using one-way ANOVA followed by Tukey’s multiple comparison test. Comparisons between two groups and two nominal variables were performed using two-way ANOVA followed by Sidak’s correction. In all cases, differences were considered significant when the P value was <0.05.
Phosphoproteome Analysis.
Phosphoproteome analysis was processed as described previously (47). Detailed methods regarding lysis and digestion for phosphoproteomic analysis, phosphopeptide enrichment, LC-MS/MS measurement, and phosphoproteomic data analysis are in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R37DK031036 (to C.R.K.) and the Joslin Diabetes Research Center Grant (P30DK036836). N.J.W.A. was supported by an Excellence Emerging Investigator Grant—Endocrinology and Metabolism (NNF19OC0055001) and by EliteForsk Rejsestipendiat (2016). H.N. was supported by a Sunstar Foundation postdoctoral fellowship. W.C. was supported by NIH Grants K01 DK120740 and P30 DK057521.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019474118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Boucher J., Kleinridders A., Kahn C. R., Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 6, a009191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai W., et al., Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat. Commun. 8, 14892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czech M. P., Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 23, 804–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallagher E. J., LeRoith D., Hyperinsulinaemia in cancer. Nat. Rev. Cancer 20, 629–644 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi C. M., Emanuelli B., Kahn C. R., Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Belfiore A., Frasca F., Pandini G., Sciacca L., Vigneri R., Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 30, 586–623 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Kahn C. R., Freychet P., Roth J., Neville D. M. Jr, Quantitative aspects of the insulin-receptor interaction in liver plasma membranes. J. Biol. Chem. 249, 2249–2257 (1974). [PubMed] [Google Scholar]
- 8.Boucher J., et al., Differential roles of insulin and IGF-1 receptors in adipose tissue development and function. Diabetes 65, 2201–2213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y., et al., How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun. 9, 821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalloo-Hosein H. E., et al., Differential signaling to glycogen synthesis by the intracellular domain of the insulin versus the insulin-like growth factor-1 receptor. Evidence from studies of TrkC-chimeras. J. Biol. Chem. 272, 24325–24332 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Ursø B., et al., Differences in signaling properties of the cytoplasmic domains of the insulin receptor and insulin-like growth factor receptor in 3T3-L1 adipocytes. J. Biol. Chem. 274, 30864–30873 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Cieniewicz A. M., et al., Novel monoclonal antibody is an allosteric insulin receptor antagonist that induces insulin resistance. Diabetes 66, 206–217 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Boucher J., et al., A kinase-independent role for unoccupied insulin and IGF-1 receptors in the control of apoptosis. Sci. Signal. 3, ra87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucher J., et al., Insulin and insulin-like growth factor 1 receptors are required for normal expression of imprinted genes. Proc. Natl. Acad. Sci. U.S.A. 111, 14512–14517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphrey S. J., Azimifar S. B., Mann M., High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Mcheik S., Aptecar L., Coopman P., D’Hondt V., Freiss G., Dual role of the PTPN13 tyrosine phosphatase in cancer. Biomolecules 10, 1659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zierhut C., Funabiki H., Nucleosome functions in spindle assembly and nuclear envelope formation. BioEssays 37, 1074–1085 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt M., et al., Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules 22, 2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parmacek M. S., Myocardin-related transcription factors: Critical coactivators regulating cardiovascular development and adaptation. Circ. Res. 100, 633–644 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Bradshaw N. J., Hennah W., Soares D. C., NDE1 and NDEL1: Twin neurodevelopmental proteins with similar ‘nature’ but different ‘nurture’. Biomol. Concepts 4, 447–464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She Z. Y., Yang W. X., Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J. Cell Sci. 130, 2097–2110 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Tovell H., Newton A. C., PHLPPing the balance: Restoration of protein kinase C in cancer. Biochem. J. 478, 341–355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Q. S., et al., Vimentin is a novel AKT1 target mediating motility and invasion. Oncogene 30, 457–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwenk R. W., et al., Requirement for distinct vesicle-associated membrane proteins in insulin- and AMP-activated protein kinase (AMPK)-induced translocation of GLUT4 and CD36 in cultured cardiomyocytes. Diabetologia 53, 2209–2219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bögershausen N., Wollnik B., Mutational landscapes and phenotypic spectrum of SWI/SNF-related intellectual disability disorders. Front. Mol. Neurosci. 11, 252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bedford D. C., Brindle P. K., Is histone acetylation the most important physiological function for CBP and p300? Aging (Albany NY) 4, 247–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menezes S. V., Sahni S., Kovacevic Z., Richardson D. R., Interplay of the iron-regulated metastasis suppressor NDRG1 with epidermal growth factor receptor (EGFR) and oncogenic signaling. J. Biol. Chem. 292, 12772–12782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forsburg S. L., Eukaryotic MCM proteins: Beyond replication initiation. Microbiol. Mol. Biol. Rev. 68, 109–131 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funakoshi T., et al., Two distinct human POM121 genes: Requirement for the formation of nuclear pore complexes. FEBS Lett. 581, 4910–4916 (2007). [DOI] [PubMed] [Google Scholar]
- 30.El Amri M., Fitzgerald U., Schlosser G., MARCKS and MARCKS-like proteins in development and regeneration. J. Biomed. Sci. 25, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goto H., et al., Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J. Biol. Chem. 273, 11728–11736 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Gutmann T., Kim K. H., Grzybek M., Walz T., Coskun Ü., Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J. Cell Biol. 217, 1643–1649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakaguchi M., et al., FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism. Nat. Commun. 10, 1582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ussar S., Bezy O., Blüher M., Kahn C. R., Glypican-4 enhances insulin signaling via interaction with the insulin receptor and serves as a novel adipokine. Diabetes 61, 2289–2298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Accili D., et al., Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat. Genet. 12, 106–109 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Puche J. E., Castilla-Cortázar I., Human conditions of insulin-like growth factor-I (IGF-I) deficiency. J. Transl. Med. 10, 224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Combettes-Souverain M., Issad T., Molecular basis of insulin action. Diabetes Metab. 24, 477–489 (1998). [PubMed] [Google Scholar]
- 38.Le Roith D., Butler A. A., Insulin-like growth factors in pediatric health and disease. J. Clin. Endocrinol. Metab. 84, 4355–4361 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Rabiee A., Krüger M., Ardenkjær-Larsen J., Kahn C. R., Emanuelli B., Distinct signalling properties of insulin receptor substrate (IRS)-1 and IRS-2 in mediating insulin/IGF-1 action. Cell. Signal. 47, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddle K., Molecular basis of signaling specificity of insulin and IGF receptors: Neglected corners and recent advances. Front. Endocrinol. (Lausanne) 3, 34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H., Ross F. P., Teitelbaum S. L., Unoccupied alpha(v)beta3 integrin regulates osteoclast apoptosis by transmitting a positive death signal. Mol. Endocrinol. 19, 771–780 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Nikoletopoulou V., et al., Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature 467, 59–63 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Sridharan R., Connelly S. M., Naider F., Dumont M. E., Variable dependence of signaling output on agonist occupancy of Ste2p, a G protein-coupled receptor in yeast. J. Biol. Chem. 291, 24261–24279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dauber A., Rosenfeld R. G., Hirschhorn J. N., Genetic evaluation of short stature. J. Clin. Endocrinol. Metab. 99, 3080–3092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lammers R., Gray A., Schlessinger J., Ullrich A., Differential signalling potential of insulin- and IGF-1-receptor cytoplasmic domains. EMBO J. 8, 1369–1375 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang G., et al., RagC phosphorylation autoregulates mTOR complex 1. EMBO J. 38, e99548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batista T. M., et al., A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metab. 32, 844–859.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.