Public health officials have raised concerns about the use of nonsteroidal anti-inflammatory drugs (NSAIDs) for treating symptoms of coronavirus disease 2019 (COVID-19). NSAIDs inhibit the enzymes cyclooxygenase-1 (COX-1) and cyclooxygenase-2 (COX-2), which are critical for the generation of prostaglandins—lipid molecules with diverse roles in homeostasis and inflammation.
KEYWORDS: SARS-CoV-2, COVID-19, NSAIDs, antibody response
ABSTRACT
Identifying drugs that regulate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and its symptoms has been a pressing area of investigation during the coronavirus disease 2019 (COVID-19) pandemic. Nonsteroidal anti-inflammatory drugs (NSAIDs), which are frequently used for the relief of pain and inflammation, could modulate both SARS-CoV-2 infection and the host response to the virus. NSAIDs inhibit the enzymes cyclooxygenase-1 (COX-1) and cyclooxygenase-2 (COX-2), which mediate the production of prostaglandins (PGs). Since PGs play diverse biological roles in homeostasis and inflammatory responses, inhibiting PG production with NSAIDs could affect COVID-19 pathogenesis in multiple ways, including (i) altering susceptibility to infection by modifying expression of angiotensin-converting enzyme 2 (ACE2), the cell entry receptor for SARS-CoV-2; (ii) regulating replication of SARS-CoV-2 in host cells; and (iii) modulating the immune response to SARS-CoV-2. Here, we investigate these potential roles. We demonstrate that SARS-CoV-2 infection upregulates COX-2 in diverse human cell culture and mouse systems. However, suppression of COX-2 by two commonly used NSAIDs, ibuprofen and meloxicam, had no effect on ACE2 expression, viral entry, or viral replication. In contrast, in a mouse model of SARS-CoV-2 infection, NSAID treatment reduced production of proinflammatory cytokines and impaired the humoral immune response to SARS-CoV-2, as demonstrated by reduced neutralizing antibody titers. Our findings indicate that NSAID treatment may influence COVID-19 outcomes by dampening the inflammatory response and production of protective antibodies rather than modifying susceptibility to infection or viral replication.
IMPORTANCE Public health officials have raised concerns about the use of nonsteroidal anti-inflammatory drugs (NSAIDs) for treating symptoms of coronavirus disease 2019 (COVID-19). NSAIDs inhibit the enzymes cyclooxygenase-1 (COX-1) and cyclooxygenase-2 (COX-2), which are critical for the generation of prostaglandins—lipid molecules with diverse roles in homeostasis and inflammation. Inhibition of prostaglandin production by NSAIDs could therefore have multiple effects on COVID-19 pathogenesis. Here, we demonstrate that NSAID treatment reduced both the antibody and proinflammatory cytokine response to SARS-CoV-2 infection. The ability of NSAIDs to modulate the immune response to SARS-CoV-2 infection has important implications for COVID-19 pathogenesis in patients. Whether this occurs in humans and whether it is beneficial or detrimental to the host remains an important area of future investigation. This also raises the possibility that NSAIDs may alter the immune response to SARS-CoV-2 vaccination.
INTRODUCTION
During the ongoing coronavirus disease 2019 (COVID-19) pandemic, a common concern has been whether widely used anti-inflammatory medications affect the risk of infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, or disease severity. Used ubiquitously for the relief of pain and inflammation, nonsteroidal anti-inflammatory drugs (NSAIDs) have been one such target of concern, with the health minister of France and the medical director of the National Health Service of England recommending the use of acetaminophen over NSAIDs for treating COVID-19 symptoms (1, 2).
NSAIDs function by inhibiting the cyclooxygenase (COX) isoforms COX-1 and COX-2. COX-1 is constitutively expressed in most cells, while COX-2 expression is induced by inflammatory stimuli (3). COX-1 and COX-2 metabolize arachidonic acid into prostaglandin H2, which can then be converted to several different bioactive prostaglandins (PGs), including PGD2, PGE2, PGF2a, and PGI2 (3). PGs signal through specific receptors to perform diverse roles, such as regulating immune responses and gastrointestinal barrier integrity (3). Several potential hypotheses have linked NSAID use and COVID-19 pathogenesis. First, it has been suggested that NSAID use may upregulate angiotensin-converting enzyme 2 (ACE2), the cell entry receptor for SARS-CoV-2, and increase the risk of infection (4, 5). Second, NSAIDs may directly affect SARS-CoV-2 replication, since COX signaling has been shown to regulate replication of other viruses, including mouse coronavirus (6). Third, given their anti-inflammatory properties, NSAIDs may impair the immune response to SARS-CoV-2 and delay disease resolution or, alternatively, dampen the cytokine storm associated with severe disease (1). Therefore, given the widespread use of NSAIDs, evaluation of the interaction between NSAIDs and SARS-CoV-2 is warranted.
NSAIDs may modulate multiple stages of the SARS-CoV-2 life cycle. As described above, one potential mechanism is that NSAIDs could lead to ACE2 upregulation and thus increase susceptibility to SARS-CoV-2. Ibuprofen treatment of diabetic rats was found to increase ACE2 expression in the heart (7). In addition, inhibition of the PGE2 receptor EP4 in human and mouse intestinal organoids increases ACE2 expression (8), suggesting that NSAID inhibition of COX/PGE2 signaling could similarly lead to ACE2 upregulation. NSAIDs could also affect a later stage of the SARS-CoV-2 life cycle. For porcine sapovirus, feline calicivirus, murine norovirus, and mouse coronavirus, COX inhibition impairs viral replication (6, 9, 10). COX inhibition was found to impair mouse coronavirus infection at a postbinding step early in the replication cycle, potentially entry or initial genome replication (6). Furthermore, SARS-CoV, the closest relative of SARS-CoV-2 among human coronaviruses and cause of the 2002-2003 epidemic (11), stimulates COX-2 expression via its spike and nucleocapsid proteins (12, 13), indicating the potential relevance of this pathway for SARS-CoV-2.
NSAIDs could also regulate the immune response to SARS-CoV-2 in multiple ways that ameliorate or exacerbate COVID-19. While mounting an immune response is necessary for clearing SARS-CoV-2 infection and establishing immunological memory to combat reinfection, it has also been appreciated that hyperinflammatory responses underlie the pathology of severe COVID-19 (14). Studies using immunomodulatory agents to treat COVID-19 suggest that immunostimulation is helpful early in the disease course, whereas immunosuppression may be more beneficial later (14). For example, dexamethasone treatment decreases mortality in COVID-19 patients on respiratory support but is potentially harmful for those with milder disease, suggesting that late-stage disease is mediated by hyperinflammation and therefore benefits from immunosuppression (15). Disease severity and mortality in COVID-19 patients is associated with elevated levels of proinflammatory cytokines, including interleukin-1β (IL-1β), IL-6, interferon gamma (IFN-γ), and tumor necrosis factor alpha (TNF-α), as well as chemokines such as CCL2, CCL4, CXCL9, and CXCL10 (16, 17). Since PGs can regulate and amplify the production of these cytokines (18, 19), NSAIDs could potentially mitigate the hyperinflammatory pathology of COVID-19. However, PGs can also be immunosuppressive in certain contexts, such that NSAIDs may instead promote immune responses. For instance, PGD2 and PGE2 have been shown to impair both innate and adaptive immunity to influenza A virus, with PGD2 having a similar impact on SARS-CoV (20, 21). In addition, PGD2 signaling prevents excessive inflammasome activation during murine coronavirus-induced encephalitis (22). Therefore, reducing PGD2 and PGE2 levels with NSAID treatment could improve the induction of antiviral immunity and yet also promote hyperinflammatory responses. Conversely, NSAID treatment could have detrimental effects on resolution of infection by inhibiting production of PGI2, which is antiviral in respiratory syncytial virus infection (23). Furthermore, NSAIDs may inhibit antibody production to SARS-CoV-2, which has been observed for other viruses, but the effect of this on disease severity is unclear as antibodies can be protective or pathogenic (24, 25). Altogether, given the complex and sometimes conflicting roles of PGs, it is difficult to predict the overall effect of NSAIDs on the immune response to SARS-CoV-2 and, ultimately, disease outcome.
We therefore systematically assessed the effect of NSAIDs on SARS-CoV-2 infection and the immune response to SARS-CoV-2. We found that SARS-CoV-2 infection induced COX-2 expression in human cells and mice. However, suppression of COX-2 by two commonly used NSAIDs, ibuprofen and meloxicam, had no effect on ACE2 expression, viral entry, or viral replication. In a mouse model of SARS-CoV-2 infection, NSAID treatment impaired the production of proinflammatory cytokines and neutralizing antibodies but did not affect weight loss, viral burden, or activation of innate and adaptive immune cells in the lung. These results indicate that NSAID use in humans may affect COVID-19 pathogenesis by mitigating the inflammatory response and the production of protective antibodies rather than by directly influencing viral replication.
RESULTS
SARS-CoV-2 infection induces PTGS2 expression in human cells and mice.
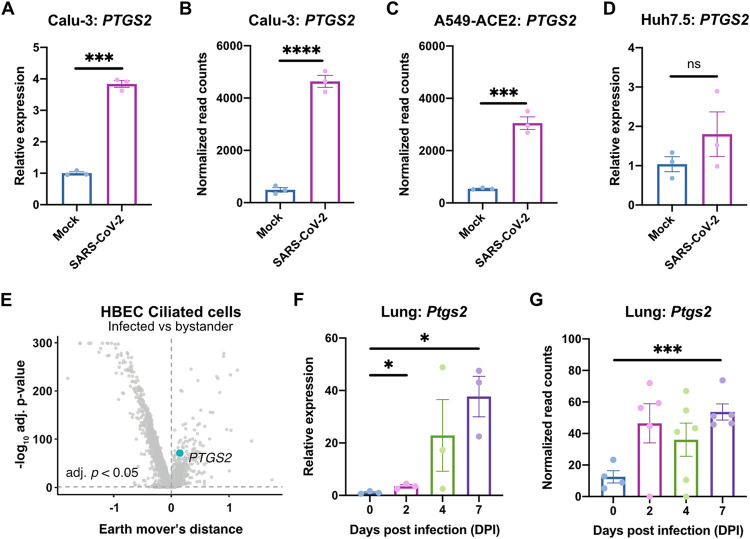
To determine the role of the COX-2 pathway in SARS-CoV-2 infection, we evaluated induction of PTGS2 (encoding COX-2) in human cells and mice. We found that SARS-CoV-2 infection of human lung cancer cell line Calu-3 led to significant upregulation of PTGS2 (Fig. 1A). This is consistent with RNA sequencing (RNA-seq) data sets of SARS-CoV-2-infected Calu-3 cells and ACE2-overexpressing A549 cells, another lung cancer cell line (Fig. 1B and C) (26). However, infection of human liver cancer cell line Huh7.5 did not lead to significant PTGS2 induction, demonstrating cell type specificity of PTGS2 induction by SARS-CoV-2 (Fig. 1D).
FIG 1.
SARS-CoV-2 infection induces PTGS2 expression in human cells and mice. (A) Calu-3 cells were infected with SARS-CoV-2 at an MOI of 0.05. PTGS2 expression was measured at 2 dpi, normalized to ACTB. (B and C) PTGS2 expression in Calu-3 (B) and ACE2-overexpressing A549 (A549-ACE2) (C) cells following SARS-CoV-2 infection. The data are from GSE147507 (26). (D) Huh7.5 cells were infected with SARS-CoV-2 at an MOI of 0.05. PTGS2 expression was measured at 2 dpi, normalized to ACTB. (E) HBECs were cultured at an air-liquid interface and then infected at the apical surface with 104 PFU of SARS-CoV-2. Cells were collected at 1, 2, and 3 dpi for single-cell RNA sequencing (scRNA-seq) (28). A volcano plot of differentially expressed genes in infected versus bystander ciliated cells pooled from all time points is shown. PTGS2 is highlighted. (F) K18-hACE2 mice were infected intranasally with 1.2 × 106 PFU of SARS-CoV-2. Ptgs2 expression in the lung was measured at 0, 2, 4, and 7 dpi. (G) Ptgs2 expression in the lung of K18-hACE2 mice following intranasal SARS-CoV-2 infection. The data are from GSE154104 (36). All data points in this figure are presented as means ± the standard errors of the mean (SEM). Data were analyzed by Welch’s two-tailed, unpaired t test (A, D, and F); Student two-tailed, unpaired t test (B, C, and G); and two-sided Mann-Whitney U test with continuity and Benjamini-Hochberg correction (E). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Data in panels A and D are representative of two independent experiments with three replicates per condition.
We next assessed whether SARS-CoV-2 induces PTGS2 in a more physiologically relevant cell culture system. We cultured primary human bronchial epithelial cells (HBECs) for 28 days at an air-liquid interface, which supports pseudostratified mucociliated differentiation providing an in vitro model of airway epithelium (27). We infected HBECs with SARS-CoV-2 at the apical surface of the culture and then performed single-cell RNA sequencing at 1, 2, and 3 days postinfection (dpi) (28). As we previously reported that ciliated cells in air-liquid interface cultures are the major target of infection (28), we looked for PTGS2 induction in this cell type. Aggregating ciliated cells across the three time points, we found that infected ciliated cells expressed higher levels of PTGS2 compared to uninfected bystander ciliated cells (Fig. 1E), indicating that PTGS2 is also induced by SARS-CoV-2 in a cell-intrinsic manner in ciliated cells, a physiologically relevant target cell.
To determine the relevance of these findings in vivo, we utilized transgenic mice expressing human ACE2 driven by the epithelial cell keratin 18 promoter (K18-hACE2) (29). As SARS-CoV-2 does not efficiently interact with mouse ACE2 (4), human ACE2-expressing mice are required to support SARS-CoV-2 infection (30–35). K18-hACE2 mice were initially developed as a model of SARS-CoV infection and have recently been demonstrated as a model of severe SARS-CoV-2 infection in the lung (29, 36). We found that intranasal infection of K18-hACE2 mice with SARS-CoV-2 led to significant upregulation of Ptgs2 in the lung at multiple time points postinfection (Fig. 1F), consistent with recent SARS-CoV-2-infected K18-hACE2 lung RNA-seq data (Fig. 1G) (36). Taken together, these results demonstrate that SARS-CoV-2 infection induces PTGS2 in diverse in vitro and in vivo airway and lung systems, across multiple independent studies. These findings therefore suggest that COX-2 signaling may be a relevant pathway for regulating SARS-CoV-2 infection or the immune response to the virus.
NSAID treatment does not affect ACE2 expression in human cells and mice.
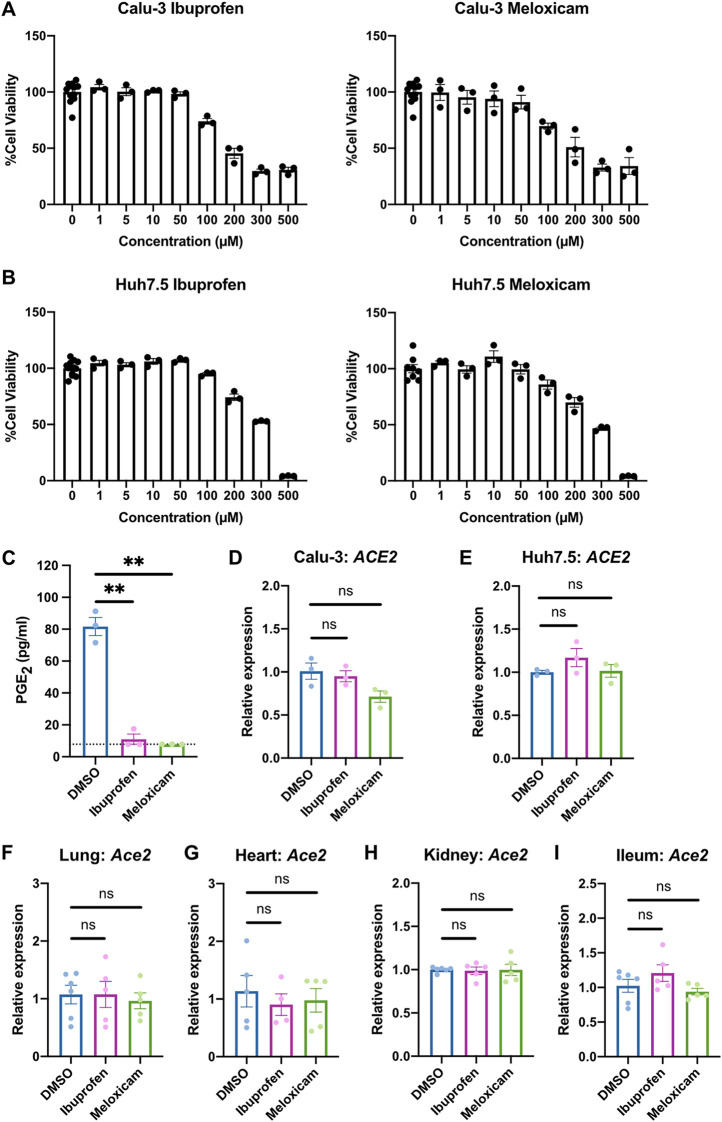
We next explored whether inhibition of COX-2 could affect viral infection by regulating ACE2 expression, as has been reported in studies of diabetic rats and intestinal organoids (7, 8). We utilized two NSAIDs, the nonselective COX-1/COX-2 inhibitor ibuprofen and the selective COX-2 inhibitor meloxicam, which are commonly used clinically. We determined the maximum nontoxic doses of ibuprofen and meloxicam on Calu-3 and Huh7.5 cells (Fig. 2A and B) and demonstrated that both drugs are bioactive in Calu-3 cells, as measured by reduced PGE2 levels (Fig. 2C). Treatment of Calu-3 or Huh7.5 cells with ibuprofen or meloxicam did not significantly affect ACE2 expression (Fig. 2D and E). To test whether NSAID treatment affects Ace2 expression in diverse tissues in vivo, we treated C57BL/6 mice with therapeutic doses of ibuprofen and meloxicam (37–40), which did not lead to changes in Ace2 expression in the lung, heart, kidney, or ileum (Fig. 2F to I). These data indicate that inhibition of the COX-2 pathway by NSAIDs does not affect ACE2 expression in multiple cell and tissue types in vitro or in vivo.
FIG 2.
NSAID treatment does not affect ACE2 expression in human cells and mice. (A and B) Calu-3 (A) and Huh7.5 (B) cells were treated with different concentrations of ibuprofen or meloxicam for 48 h. Cell viability was measured and calculated as a percentage of no treatment. (C) Calu-3 cells were treated with DMSO, 50 μM ibuprofen, or 50 μM meloxicam for 48 h. The levels of prostaglandin E2 (PGE2) were measured in the supernatant. The dotted line represents the limit of detection. (D and E) Calu-3 (D) and Huh7.5 (E) cells were treated with DMSO, 50 μM ibuprofen, or 50 μM meloxicam for 24 h. ACE2 expression was measured and normalized to ACTB. (F to I) C57BL/6 mice were treated intraperitoneally with DMSO, 30 mg/kg ibuprofen, or 1 mg/kg meloxicam daily for 4 days. Ace2 expression was measured in the lung (F), heart (G), kidney (H), and ileum (I), normalized to Actb. All data points in this figure are presented as means ± the SEM. Data were analyzed by Welch’s two-tailed, unpaired t test (C to I). **, P < 0.01; ns, not significant. Data in panels A to E are representative of two independent experiments with three replicates per condition; data in panels F to I are pooled from two independent experiments with a total of four to six mice per condition.
NSAID treatment does not affect SARS-CoV-2 entry or replication in vitro.
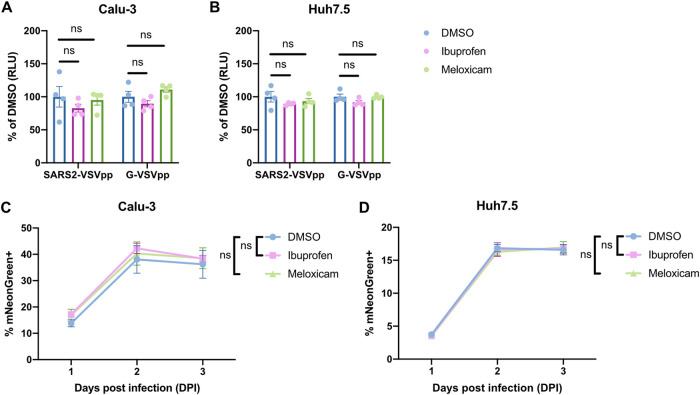
To assess whether NSAID treatment functionally affects SARS-CoV-2 entry, we used a vesicular stomatitis virus (VSV) core expressing Renilla luciferase pseudotyped with the SARS-CoV-2 spike protein (SARS2-VSVpp). We used VSV glycoprotein (G) pseudovirus (G-VSVpp) as a control (41, 42). Quantification of luciferase activity showed that pretreatment of Calu-3 or Huh7.5 cells with ibuprofen or meloxicam did not significantly affect SARS2-VSVpp or G-VSVpp entry (Fig. 3A and B), confirming that NSAID inhibition of COX-2 does not impact susceptibility to infection.
FIG 3.
NSAID treatment does not affect SARS-CoV-2 entry or replication in vitro. (A and B) Calu-3 (A) and Huh7.5 (B) cells were pretreated with DMSO, 50 μM ibuprofen, or 50 μM meloxicam for 24 h and then infected with SARS2-VSVpp or G-VSVpp expressing Renilla luciferase. Luminescence was measured at 24 h postinfection (hpi) and normalized to DMSO for each infection. (C and D) Calu-3 (C) and Huh7.5 (D) cells were pretreated with DMSO, 50 μM ibuprofen, or 50 μM meloxicam for 24 h and then infected with mNeonGreen reporter replication-competent SARS-CoV-2 (icSARS-CoV-2-mNG) at an MOI of 1. The frequency of infected cells was measured by mNeonGreen expression at 1, 2, and 3 dpi. All data points in this figure are presented as means ± the SEM. Data were analyzed by Student two-tailed, unpaired t test (A and B) and two-way ANOVA (C and D). ns, not significant. Data in panels A and B are representative of two independent experiments with four replicates per condition; data in panels C and D are representative of two independent experiments with five replicates per condition.
Next, we studied whether COX-2 inhibition affects SARS-CoV-2 replication. Viruses from several different families have been shown to induce COX-2 signaling in host cells, which can have either proviral or antiviral functions (9, 43, 44). To this end, we utilized a replication-competent SARS-CoV-2 expressing a mNeonGreen reporter (icSARS-CoV-2-mNG) to study the effect of COX-2 inhibition by NSAIDs on viral replication (45). We assessed icSARS-CoV-2-mNG replication in Calu-3 cells, which upregulate PTGS2 in response to SARS-CoV-2 infection (Fig. 1A and B), and Huh7.5 cells, which do not (Fig. 1D). By quantifying the percentage of mNeonGreen-expressing cells, we found that treatment of Calu-3 or Huh7.5 cells with ibuprofen or meloxicam did not impact icSARS-CoV-2-mNG replication (Fig. 3C and D). These results indicate that SARS-CoV-2 induction of the COX-2 pathway in Calu-3 human lung cells does not regulate viral replication.
NSAID treatment does not affect SARS-CoV-2-induced weight loss or lung viral burden in mice.
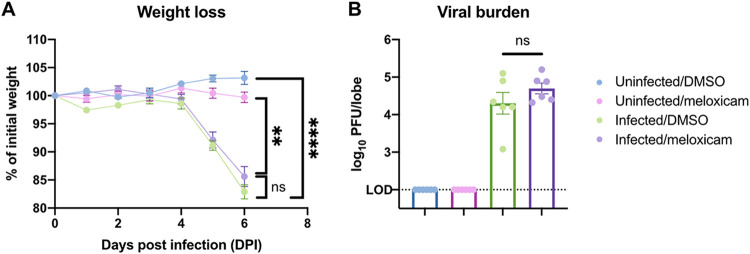
Since NSAIDs do not directly affect SARS-CoV-2 entry or replication in vitro, we next investigated whether they regulate SARS-CoV-2 replication and immunity in vivo. We treated K18-hACE2 mice with meloxicam 1 day prior to SARS-CoV-2 infection and continued daily meloxicam treatment throughout the course of the infection. Dimethyl sulfoxide (DMSO) control- and meloxicam-treated K18-hACE2 mice experienced similar weight loss beginning at 5 dpi (Fig. 4A). In addition, viral lung burden at 6 dpi was similar between infected DMSO- and meloxicam-treated mice (Fig. 4B), suggesting that meloxicam does not affect viral replication in vitro or in vivo.
FIG 4.
NSAID treatment does not affect SARS-CoV-2-induced weight loss or lung viral burden in mice. (A and B) K18-hACE2 mice were treated intraperitoneally with DMSO or 1 mg/kg meloxicam daily for 7 days starting 1 day prior to infection. K18-hACE2 mice were infected intranasally with 103 PFU of SARS-CoV-2 or left uninfected and monitored daily. (A) Weight change expressed as a percentage of initial weight. (B) Viral burden in the lungs at 6 dpi measured by plaque assay. All data points in this figure are presented as means ± the SEM. Data were analyzed by two-way ANOVA (A) and Student two-tailed, unpaired t test (B). **, P < 0.01; ****, P < 0.0001; ns, not significant. Data in panels A and B are pooled from two independent experiments with a total of six mice per condition.
NSAID treatment does not affect innate or adaptive immune cell activation in the lungs of SARS-CoV-2-infected mice.
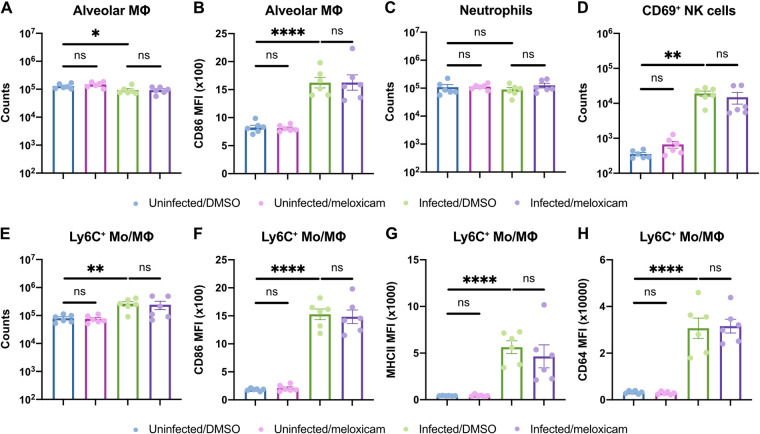
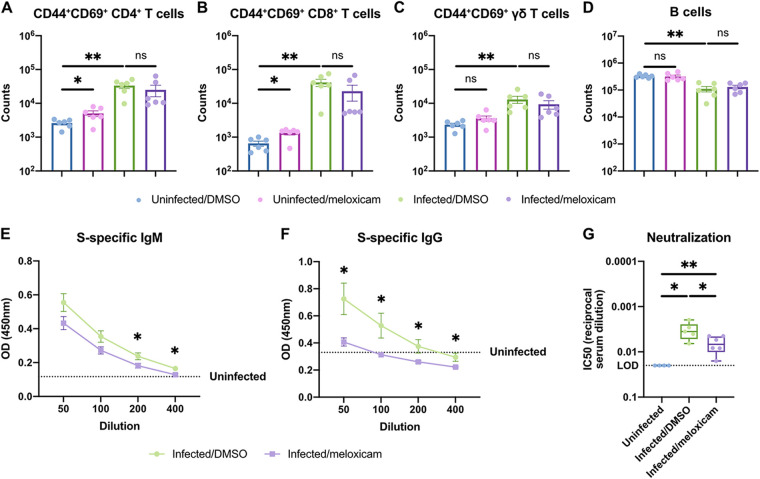
As NSAIDs regulate inflammation, we next assessed whether NSAIDs perturb the host immune response to SARS-CoV-2 in mice. First, we characterized the abundance and activation status of innate and adaptive immune cell types in the lung at 6 dpi (gating strategy is shown in Fig. S1A and B in the supplemental material). SARS-CoV-2 infection led to a slight decrease in alveolar macrophage abundance and upregulation of activation marker CD86. However, this was not modified by meloxicam treatment (Fig. 5A and B). Neutrophil counts were not significantly altered by infection or meloxicam treatment at this time point (Fig. 5C). Activated CD69+ NK cells increased with infection in both DMSO and meloxicam-treated mice (Fig. 5D). Ly6C+ monocyte/macrophage numbers and expression of activation markers CD86, MHCII, and CD64 all increased following SARS-CoV-2 infection but were not affected by meloxicam treatment (Fig. 5E to H). Furthermore, activated (CD44+ CD69+) T cell populations, including CD4+ T cells, CD8+ T cells, and γδ T cells, increased with infection but were not significantly affected by concomitant meloxicam treatment (Fig. 6A to C). In contrast, B cells were decreased in the lungs of infected mice, independent of meloxicam treatment (Fig. 6D). Together, these results indicate that meloxicam treatment does not affect the overall numbers and activation status of innate or adaptive immune cells in the lung during SARS-CoV-2 infection.
FIG 5.
NSAID treatment does not affect innate immune cell activation in the lungs of SARS-CoV-2-infected mice. (A to H) K18-hACE2 mice were treated intraperitoneally with DMSO or 1 mg/kg meloxicam daily for 7 days starting 1 day prior to infection. K18-hACE2 mice were infected intranasally with 103 PFU of SARS-CoV-2 or left uninfected. Flow cytometric analysis of the lungs at 6 dpi for alveolar macrophage (MΦ) counts (A) and expression of CD86 (B), neutrophil counts (C), activated CD69+ natural killer (NK) cell counts (D), and Ly6C+ monocyte/macrophage (Mo/MΦ) counts (E) and expression of CD86 (F), MHCII (G), and CD64 (H) results are shown. All data points in this figure are presented as means ± the SEM. Data were analyzed by two-tailed Mann-Whitney test (A to H). *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant. Data in panels A to H are pooled from two independent experiments with a total of six mice per condition.
FIG 6.
NSAID treatment impairs systemic neutralizing antibody responses but not adaptive immune cell activation in the lungs of SARS-CoV-2-infected mice. (A to G) K18-hACE2 mice were treated intraperitoneally with DMSO or 1 mg/kg meloxicam daily for 7 days starting 1 day prior to infection. K18-hACE2 mice were infected intranasally with 103 PFU of SARS-CoV-2 or left uninfected. Flow cytometric analysis of the lungs at 6 dpi for activated CD44+CD69+ CD4+ T cells (A), CD44+ CD69+ CD8+ T cells (B), CD44+ CD69+ γδ T cells (C), and B cells (D) was performed. (E and F) Spike (S)-specific IgM (E) and IgG (F) titers in the serum at 6 dpi. (G) Neutralizing antibody titers in the serum at 6 dpi measured by SARS2-VSVpp pseudovirus neutralization assay. Data points in panels A to F are presented as means ± the SEM. Data points in panel G are presented as boxplots. Data were analyzed by two-tailed Mann-Whitney test (A to D, G) and Student two-tailed, unpaired t test (E and F). *, P < 0.05; **, P < 0.01; ns, not significant. Data in panels A to G are pooled from two independent experiments with a total of four to six mice per condition.
NSAID treatment impairs systemic neutralizing antibody responses to SARS-CoV-2.
While meloxicam did not alter the abundance of immune cell populations in the lung, NSAID treatment might regulate the function of these cell types. To assess the effect of meloxicam treatment on antibody production in infected mice, we measured the serum levels of spike-specific IgM and IgG antibodies at 6 dpi. DMSO-treated mice demonstrated detectable levels of both spike-specific IgM and IgG antibodies, which were significantly reduced in meloxicam-treated mice (Fig. 6E and F). Corroborating the difference in antibody titers, serum from DMSO-treated mice exhibited greater neutralization capacity compared to serum from meloxicam-treated mice (Fig. 6G).
NSAID treatment dampens the induction of proinflammatory cytokines that are upregulated by SARS-CoV-2 infection in mice.
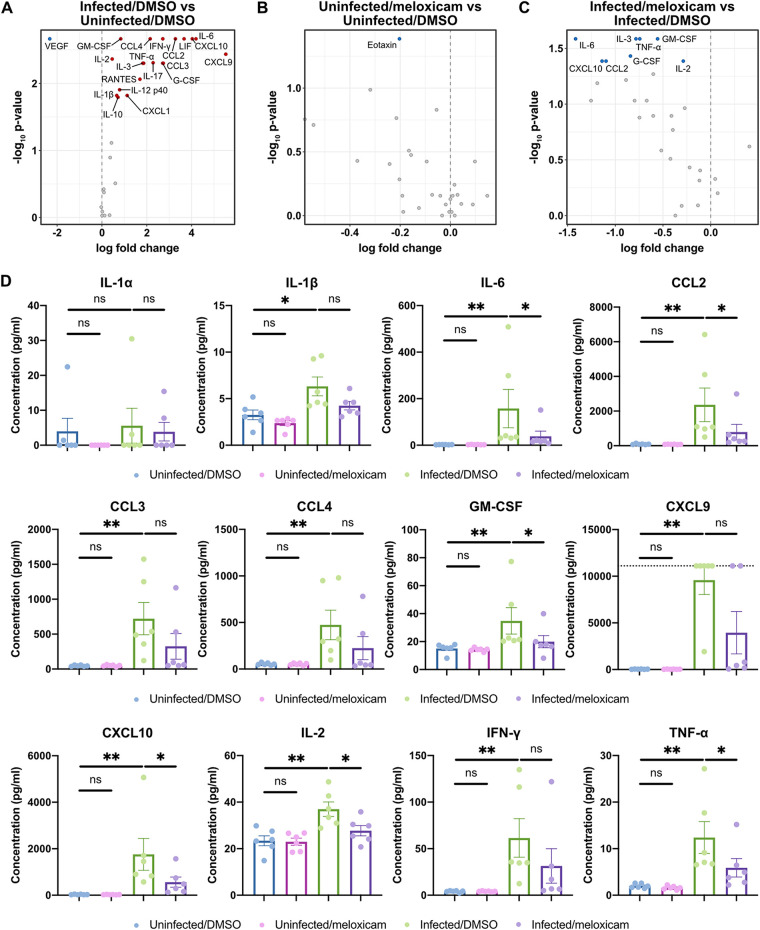
Finally, we studied whether NSAID treatment modulates the cytokine response to SARS-CoV-2 infection. SARS-CoV-2 infection led to increased production of proinflammatory cytokines (IL-1β, IL-6, IFN-γ, TNF-α, and granulocyte-macrophage colony-stimulating factor [GM-CSF]); T cell growth factors (IL-2); and chemokines (CCL2, CCL4, CXCL9, and CXCL10) that have been associated with disease severity and mortality in COVID-19 patients (Fig. 7A) (16, 17). Among uninfected mice, meloxicam treatment led to minimal changes in cytokine production (Fig. 7B; see also Fig. S2 in the supplemental material). Among infected mice, meloxicam treatment decreased the production of a subset of cytokines upregulated by infection, including IL-6, CCL2, GM-CSF, CXCL10, IL-2, and TNF-α, while others were unaffected (Fig. 7C and D; see also Fig. S2). Together, these results demonstrate that NSAID treatment partially dampens the cytokine response to SARS-CoV-2 infection.
FIG 7.
NSAID treatment dampens the induction of proinflammatory cytokines that are upregulated by SARS-CoV-2 infection in mice. (A to D) K18-hACE2 mice were treated intraperitoneally with DMSO or 1 mg/kg meloxicam daily for 7 days starting 1 day prior to infection. K18-hACE2 mice were infected intranasally with 103 PFU of SARS-CoV-2 or left uninfected. Cytokine levels were measured in lung homogenates at 6 dpi. (A to C) Volcano plots detailing the differential abundance of cytokines in lung homogenates from infected versus uninfected mice treated with DMSO (A), uninfected mice treated with meloxicam versus DMSO (B), and infected mice treated with meloxicam versus DMSO (C). Significantly upregulated (red) and downregulated (blue) cytokines are labeled. (D) Levels of proinflammatory cytokines in lung homogenates from uninfected mice treated with DMSO, uninfected mice treated with meloxicam, infected mice treated with DMSO, and infected mice treated with meloxicam. The dotted line represents the upper limit of quantification. Data points in panel D are presented as means ± the SEM. Data were analyzed by two-tailed Mann–Whitney test (A to D). *, P < 0.05; **, P < 0.01; ns, not significant. Data in panels A to D are pooled from two independent experiments with a total of six mice per condition. Additional cytokine data are shown in Fig. S2 in the supplemental material.
DISCUSSION
Given the concerns about NSAID use in patients with COVID-19, we studied whether NSAIDs affect SARS-CoV-2 infection and the immune response to the virus. We found that SARS-CoV-2 infection induced PTGS2 upregulation in diverse systems, including Calu-3 and A549 lung cancer cell lines, primary HBEC air-liquid interface cultures, and the lungs of K18-hACE2 mice. Inhibition of COX-2 with the commonly used NSAIDs ibuprofen and meloxicam did not affect ACE2 expression in multiple cell and tissue types in vitro or in vivo, nor did it affect SARS-CoV-2 entry or replication. In K18-hACE2 mice infected with SARS-CoV-2, NSAID treatment impaired the production of neutralizing antibodies and proinflammatory cytokines but did not influence weight loss, viral burden, or activation state of innate and adaptive immune cells in the lung. Our findings therefore rule out a direct effect of NSAIDs on SARS-CoV-2 infection but indicate that NSAIDs could modulate COVID-19 severity by dampening production of neutralizing antibodies and inflammatory cytokines.
An important question arising from our findings is how SARS-CoV-2 infection induces COX-2 expression. One possibility is that the pattern recognition receptor retinoic acid inducible gene-I (RIG-I), which can recognize double-stranded RNA generated during viral genome replication and transcription (46), may drive this response. Indeed, COX-2 induction by influenza A virus is RIG-I-dependent (47), and we showed here that Huh7.5 cells, which are defective in RIG-I signaling (48), do not upregulate PTGS2 in response to SARS-CoV-2. Alternatively, SARS-CoV-2 proteins may mediate the induction of COX-2 through their complex effects on host cells. In the case of SARS-CoV, transfection of plasmids encoding either the spike or the nucleocapsid genes is sufficient to stimulate COX-2 expression (12, 13). SARS-CoV spike protein induces COX-2 expression through both calcium-dependent PKCα/ERK/NF-κB and calcium-independent PI3K/PKCε/JNK/CREB pathways (13), while the nucleocapsid protein directly binds to the COX-2 promoter to regulate its expression (12). Any of these potential mechanisms are consistent with our HBEC scRNA-seq results demonstrating that SARS-CoV-2 increases PTGS2 expression in a cell-intrinsic manner.
One of the effects of NSAID treatment on the immune response to SARS-CoV-2 was impairment of early, neutralizing spike-specific antibodies. These early humoral responses are mediated by short-lived plasmablasts, and their requirement for T cell help is unclear (49). NSAID treatment could therefore act by inhibiting activation (T cell-dependent or T cell-independent), proliferation, differentiation, or antibody-secreting capacity of spike-specific B cells and plasmablasts. B cells have been found to upregulate COX-2 expression following activation with T cell-dependent and T cell-independent stimuli (50, 51), and treatment of purified B cell cultures with NSAIDs reduces IgM and IgG production (52). COX-2 inhibition reduces expression of BLIMP-1 and XBP-1 (53), which are essential transcription factors for plasmablast differentiation, providing a potential mechanism by which NSAIDs impair antibody production. In addition, in mice infected with vaccinia virus, antiviral antibody production is impaired by chronic but not acute COX-2 inhibition (24), indicating that the duration of NSAID treatment may regulate the impact on antibody production. Given that K18-hACE2 mice succumb to lethal SARS-CoV-2 disease within 7 days (36, 54), we could not assess the effect of NSAID treatment on long-term antibody production. This could be explored in nonlethal models of SARS-CoV-2 infection, as well as in the setting of vaccination.
Understanding the effect of NSAID treatment on cytokine production is also critical, as cytokines may be protective early in COVID-19 but potentially pathological at later stages, thus informing the timing of immunomodulatory drugs like NSAIDs. We observed that NSAID treatment decreased the production of a subset of cytokines that were induced by infection, including IL-6, CCL2, GM-CSF, CXCL10, IL-2, and TNF-α. This is consistent with prior reports of COX-2 inhibitors decreasing the production of these cytokines in various inflammatory settings (55–59). Mechanistically, NSAIDs may reduce production of these cytokines through inhibition of the PGE2/NF-κB positive-feedback loop, in which NF-κB and COX-2 can reciprocally activate their respective signaling pathways and amplify inflammatory responses (18, 19, 60).
However, it is less clear whether this dampened cytokine response is beneficial, detrimental, or neutral in the setting of COVID-19 given the many roles that cytokines can play in controlling infection or driving immunopathology. GM-CSF is a myelopoietic growth factor, as well as proinflammatory cytokine, with pathogenic GM-CSF-producing Th1 cells being reported in patients with severe COVID-19 (61). Reduction of GM-CSF production by meloxicam could therefore indicate a beneficial effect of NSAIDs on restraining hyperinflammatory responses. IL-2, IFN-γ, TNF-α can be coproduced by polyfunctional T cells, which play important roles in the control of viral infections, and yet these cytokines can also promote lethal cytokine shock and tissue damage (62–64). In addition, while IL-6 is correlated with disease severity in COVID-19, clinical trials blocking IL-6 signaling have not shown clear evidence of benefit for patients (65). While we observed decreased cytokine responses with NSAID treatment in K18-hACE2 mice, these changes did not translate into differences in weight loss or viral burden, suggesting that these features of the disease may involve other cytokines or pathology at other sites (e.g., the brain) not affected by NSAID treatment (36, 54). The timing, duration, and dosing of NSAID treatment may also matter for COVID-19 pathogenesis, potentially with early treatment impacting the initiation of antiviral immune responses and later treatment suppressing immune-driven pathology (14). In the present study, we treated K18-hACE2 mice continuously throughout the course of infection, but it would be interesting to explore in future work whether limiting NSAID treatment to particular phases of disease has differential effects on pathogenesis.
Here, we demonstrated that SARS-CoV-2 infection induces COX-2 expression in cell lines, primary airway epithelial cells, and mice. Inhibition of COX-2 by NSAIDs did not affect viral entry or replication in vitro or in vivo. However, NSAID treatment impaired the production of proinflammatory cytokines and neutralizing antibodies in response to SARS-CoV-2 infection in mice. NSAIDs could therefore have complex effects on the host response to SARS-CoV-2. While studies thus far have not observed worse clinical outcomes in COVID-19 patients taking NSAIDs (66–71), evaluation of the breadth, potency, and durability of the humoral immune response is warranted in patients on NSAIDs in response to both natural infection and vaccination.
MATERIALS AND METHODS
Cell lines.
Calu-3 and Huh7.5 were from ATCC. Calu-3 cells were cultured in Eagle minimum essential medium with 10% heat-inactivated fetal bovine serum (FBS), 1% GlutaMAX (Gibco), and 1% penicillin/streptomycin. Huh7.5 cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% heat-inactivated FBS and 1% penicillin/streptomycin. All cell lines tested negative for Mycoplasma spp.
Generation of SARS-CoV-2 stocks.
As previously described (42), SARS-CoV-2 P1 stock was generated by inoculating Huh7.5 cells with SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources, NR-52281) at a multiplicity of infection (MOI) of 0.01 for 3 days. The P1 stock was then used to inoculate Vero-E6 cells, and after 3 days, the supernatant was harvested and clarified by centrifugation (450 × g for 5 min), filtered through a 0.45-μm filter, and stored in aliquots at –80°C. Virus titer was determined by plaque assay using Vero-E6 cells (42).
To generate icSARS-CoV-2-mNG stocks (45), lyophilized icSARS-CoV-2-mNG was reconstituted in 0.5 ml of deionized water. Then, 50 μl of virus was diluted in 5 ml of medium and added to 107 Vero-E6 cells. After 3 days, the supernatant was harvested and clarified by centrifugation (450 × g for 5 min), filtered through a 0.45-μm filter, and stored in aliquots at –80°C.
All work with SARS-CoV-2 or icSARS-CoV-2-mNG was performed in a biosafety level 3 facility with approval from the office of Environmental Health and Safety and the Institutional Animal Care and Use Committee at Yale University.
Preparation of NSAIDs.
Ibuprofen (I4883) and meloxicam (M3935) were purchased from Sigma-Aldrich. For cell culture experiments, ibuprofen and meloxicam were solubilized in DMSO at a stock concentration of 10 mM and then diluted in medium to make working solutions. For mouse experiments, stock solutions of ibuprofen (300 mg/ml) and meloxicam (10 mg/ml) were prepared in DMSO and then diluted 100-fold in phosphate-buffered saline (PBS) to make working solutions. To determine the maximum nontoxic dose of NSAIDs to use for cell culture experiments, cells were treated with different concentrations of ibuprofen or meloxicam for 48 h, and cell viability was measured by a CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer’s instructions.
Mice.
C57BL/6J and K18-hACE2 [B6.Cg-Tg(K18-ACE2)2Prlmn/J (29)] were purchased from Jackson Laboratory. Mice were bred in-house using mating trios to enable utilization of littermates for experiments. Mice of both sexes between 6 and 8 weeks old were used for this study. C57BL/6J and K18-hACE2 mice were anesthetized using 30% (vol/vol) isoflurane diluted in propylene glycol (30% isoflurane) and administered 30 mg/kg ibuprofen, 1 mg/kg meloxicam, or an equivalent amount of DMSO intraperitoneally in a volume of 10 ml/kg daily for 4 or 7 days as indicated in the figure legends. K18-hACE2 mice were anesthetized using 30% isoflurane and administered 1.2 × 106 PFU or 103 PFU of SARS-CoV-2 intranasally as indicated in the figure legends. Mice were monitored daily for weight and survival. Animal use and care was approved in agreement with the Yale Animal Resource Center and Institutional Animal Care and Use Committee (no. 2018-20198) according to the standards set by the Animal Welfare Act.
Analysis of RNA-seq data.
We utilized RNA-seq data from recent published studies to assess the impact of SARS-CoV-2 infection on PTGS2 expression. From GSE147507 (26), we reanalyzed the raw count data from Calu-3 and A549-ACE2 cells, comparing SARS-CoV-2 infection to matched mock-treated controls. We performed differential expression analysis using the Wald test from DESeq2 (72), using a Benjamini-Hochberg adjusted P < 0.05 as the cutoff for statistical significance. For visualization of PTGS2 expression, the DESeq2-normalized counts were exported and plotted in GraphPad Prism. Statistical significance was assessed using a Student two-tailed, unpaired t test.
For analysis of HBEC air-liquid interface cultures infected with SARS-CoV-2, we utilized a previously generated catalog of differentially expressed genes that our group recently described in a preprint study (28). The differential expression table is publicly available (https://github.com/vandijklab/HBEC_SARS-CoV-2_scRNA-seq). Here, we specifically investigated PTGS2 expression in ciliated cells, comparing infected cells to bystander cells (cells aggregated across the 1, 2, and 3 dpi time points). The cutoff for statistical significance was set at adjusted P < 0.05, and the results were visualized as a volcano plot in R.
From GSE154104 (36), we reanalyzed the raw count data from the lungs of K18-hACE2 mice infected with SARS-CoV-2, performing pairwise comparisons of mice at 2 dpi, 4 dpi, and 7 to 0 dpi controls (prior to infection). For visualization of Ptgs2 expression, the DESeq2-normalized counts were exported and plotted in GraphPad Prism. Statistical significance was assessed using a Student two-tailed, unpaired t test.
PGE2 ELISA.
Levels of PGE2 in cell culture supernatants were measured using the Prostaglandin E2 enzyme-linked immunosorbent assay (ELISA) kit (Cayman Chemical) according to the manufacturer’s instructions. Absorbance was measured at 410 nm on a microplate reader (Molecular Devices), and PGE2 concentrations were calculated using a standard curve.
Quantitative PCR.
Cells or tissues were lysed in TRIzol (Life Technologies), and total RNA was extracted using the Direct-zol RNA Miniprep Plus kit (Zymo Research) according to the manufacturer’s instructions. cDNA synthesis was performed using random hexamers and ImProm-II reverse transcriptase (Promega). qPCR was performed with Power SYBR Green (Thermo Fisher) and run on the QuantStudio3 (Applied Biosystems). Target mRNA levels were normalized to those of ACTB or Actb. The qPCR primer sequences were as follows: ACTB (human), GAGCACAGAGCCTCGCCTTT (forward) and ATCATCATCCATGGTGAGCTGG (reverse); PTGS2 (human), AGAAAACTGCTCAACACCGGAA (forward) and GCACTGTGTTTGGAGTGGGT (reverse); ACE2 (human), GGGATCAGAGATCGGAAGAAGAAAA (forward) and AAGGAGGTCTGAACATCATCAGTG (reverse); Actb (mouse), ACTGTCGAGTCGCGTCCA (forward) and ATCCATGGCGAACTGGTGG (reverse); Ptgs2 (mouse), CTCCCATGGGTGTGAAGGGAAA (forward) and TGGGGGTCAGGGATGAACTC (reverse); and Ace2 (mouse), ACCTTCGCAGAGATCAAGCC (forward) and CCAGTGGGGCTGATGTAGGA (reverse).
Pseudovirus production.
VSV-based pseudotyped viruses were produced as previously described (41, 42). Vector pCAGGS containing the SARS-related coronavirus 2, Wuhan-Hu-1 Spike glycoprotein gene, NR-52310, was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH. 293T cells were transfected with the pCAGGS vector expressing the SARS-CoV-2 spike glycoprotein and then incubated with replication-deficient VSV expressing Renilla luciferase for 1 h at 37°C (41). The virus inoculum was then removed, and the cells were washed with PBS before adding media with anti-VSV-G clone I4 to neutralize residual inoculum. No antibody was added to cells expressing VSV-G. Supernatant containing pseudoviruses was collected 24 h postinoculation, clarified by centrifugation, and stored in aliquots at –80°C.
Pseudovirus entry and neutralization assays.
We plated 3 × 104 Calu-3 or 1 × 104 Huh7.5 cells in a 100-μl volume in each well of a 96-well plate. The following day, the medium was replaced with 50 μM ibuprofen, 50 μM meloxicam, or an equivalent amount of DMSO. One day later, 10 μl of SARS-CoV-2 spike protein-pseudotyped (SARS2-VSVpp) or VSV glycoprotein-typed virus was added. The luciferase activity was measured at 24 hpi using the Renilla luciferase assay system (Promega). Each well of cells was lysed with 50 μl of lysis buffer, and 15 μl of cell lysate was then mixed with 15 μl of luciferase assay reagent. The luminescence was measured on a microplate reader (BioTek Synergy).
For neutralization assays, 2 × 104 Huh7.5 cells were plated in a 100-μl volume in each well of a 96-well plate. The following day, serial dilutions of serum were incubated with SARS2-VSVpp pseudovirus for 1 h at 37°C. The growth medium was then aspirated from the cells and replaced with 50 μl of the serum/virus mixture. Luciferase activity was measured at 24 hpi as detailed above. Half-maximal inhibitory concentrations (IC50s) were calculated as previously described (73).
icSARS-CoV-2-mNG assay.
We plated 6.5 × 103 Calu-3 or 2.5 × 103 Huh7.5 cells in 20 μl of phenol red-free medium containing 50 μM ibuprofen, 50 μM meloxicam, or an equivalent amount of DMSO in each well of a black-walled, clear-bottom 384-well plate. The following day, icSARS-CoV-2-mNG was added at an MOI of 1 in a 5-μl volume. The frequency of infected cells was measured by mNeonGreen expression at 1, 2, and 3 dpi by high content imaging (BioTek Cytation 5) configured with brightfield and GFP cubes. The total cell numbers were quantified by Gen5 software for brightfield images. Object analysis was used to determine the number of mNeonGreen-positive cells. The percentage of infection was calculated as the ratio of the number of mNeonGreen-positive cells to the total number of cells in brightfield.
Measurement of lung viral burden and cytokines.
Lungs were perfused with 3 ml of sterile PBS. The left lobe was collected and homogenized in 1 ml of DMEM supplemented with 2% heat-inactivated FBS and 1% antibiotic-antimycotic. The viral burden was measured in lung homogenates by plaque assay on Vero-E6 cells as previously described (42). To measure cytokines, lung homogenates were incubated with a final concentration of 1% Triton X-100 for 1 h at room temperature to inactivate SARS-CoV-2. Cytokine analysis was performed by Eve Technologies using their Mouse Cytokine Array/Chemokine Array 31-Plex (MD31) platform.
Flow cytometry analysis of lung immune cells.
Lungs were perfused with 3 ml of sterile PBS. The right inferior lobe was collected and digested with 0.5 mg/ml collagenase IV (Sigma-Aldrich) and 100 U/ml DNase I (Sigma-Aldrich) in complete RPMI for 45 min at 37°C. Single-cell suspensions of digested lung tissue were preincubated with Fc block (clone 2.4G2) for 5 min at room temperature before staining. The cells were stained with the following antibodies or viability dyes for 30 min at 4°C: PE anti-CD64 (clone X54-5/7.1), PE/Cy7 anti-Ly6C (clone HK1.4), PerCP/Cy5.5 anti-CD45.2 (clone 104), APC anti-CD86 (clone GL1), AF700 anti-CD19 (clone 6D5), DAPI (4′,6′-diamidino-2-phenylindole; Thermo Fisher), BV510 anti-I-A/I-E (clone M5/114.15.2), BV605 anti-CD11c (clone N418), Live/Dead Fixable Green (Thermo Fisher), PE anti-TCRγ/δ (clone GL3), PE/Cy7 anti-CD69 (clone H1.2F3), PerCP/Cy5.5 anti-CD8 (clone 53.6-7), APC anti-CD45.2 (clone 104), APC/Cy7 anti-TCRβ (clone H57-597), AF700 anti-CD4 (clone RM4-5), PB anti-Ly6G (clone 1A8), BV510 anti-NK1.1 (clone PK136), and BV605 anti-CD44 (clone IM7). After being washed, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature to inactivate SARS-CoV-2. Samples were acquired on a CytoFLEX S (Beckman Coulter) and analyzed using FlowJo software (BD).
Spike-specific ELISAs.
Serum was incubated with a final concentration of 0.5% Triton X-100 and 0.5 mg/ml RNase A to inactivate any potential SARS-CoV-2. SARS-CoV-2 stabilized spike glycoprotein (BEI Resources, NR-53524) was coated at a concentration of 2 μg/ml in carbonate buffer on 96-well MaxiSorp plates (Thermo Fisher) overnight at 4°C. Plates were blocked with 1% BSA in PBS for 1 h at room temperature. Serum samples were serially diluted in 1% BSA in PBS and incubated in plates for 2 h at room temperature. Antibody isotypes were detected with anti-mouse IgM-HRP or anti-mouse IgG Fc-HRP (Southern Biotech) by incubation for 1 h at room temperature. The plates were developed with TMB stabilized chromogen (Thermo Fisher), stopped with 3 N hydrochloric acid, and read at 450 nm on a microplate reader.
Statistical analysis.
Data analysis was performed using GraphPad Prism 8 unless otherwise indicated. Data were analyzed using Welch’s two-tailed, unpaired t test; Student two-tailed, unpaired t test; two-tailed Mann-Whitney test; or two-way ANOVA, as indicated. P < 0.05 was considered statistically significant.
Data availability.
All previously published data are available as described above. Cytokine data generated in this study are available in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Benhur Lee, Pei-Yong Shi, the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), Paulina Pawlica, Joan Steitz, Michael Diamond, Adam Bailey, Emma Winkler, and BEI Resources for providing critical reagents and expertise. We thank all members of the Wilen and Eisenbarth labs for helpful discussions. We thank Yale Environmental Health and Safety for providing necessary training and support for SARS-CoV-2 research.
This study was supported by NIH Medical Scientist Training Program Training Grant T32GM007205 (J.S.C., R.D.C.), NIH/NHLBI F30HL149151 (J.S.C.), NIH/NCI F30CA250249 (R.D.C.), NIH/NIAID K08 AI128043 (C.B.W.), a Burroughs Wellcome Fund Career Award for Medical Scientists (C.B.W.), the Mathers Charitable Foundation (C.B.W.), the Ludwig Family Foundation (C.B.W.), and an Emergent Ventures Fast Grant (C.B.W.).
Jennifer S. Chen: Conceptualization, Formal Analysis, Investigation, Validation, Visualization, Writing – Original Draft; Mia Madel Alfajaro: Methodology, Investigation; Jin Wei: Methodology, Investigation; Ryan D. Chow: Formal Analysis, Visualization; Renata B. Filler: Investigation; Stephanie C. Eisenbarth: Supervision; Craig B. Wilen: Conceptualization, Formal Analysis, Funding Acquisition, Resources, Supervision, Writing – Original Draft. All authors reviewed and edited the manuscript.
Yale University (C.B.W.) has a patent pending related to this work entitled “Compounds and Compositions for Treating, Ameliorating, and/or Preventing SARS-CoV-2 Infection and/or Complications Thereof.” Yale University has committed to rapidly executable nonexclusive royalty-free licenses to intellectual property rights for the purpose of making and distributing products to prevent, diagnose, and treat COVID-19 infection during the pandemic and for a short period thereafter.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Day M. 2020. Covid-19: ibuprofen should not be used for managing symptoms, say doctors and scientists. BMJ 368:m1086. [DOI] [PubMed] [Google Scholar]
- 2.Powis S. 2020. Novel coronavirus: anti-inflammatory medications. Medicines and Healthcare Products Regulatory Agency, London, United Kingdom. [Google Scholar]
- 3.Ricciotti E, FitzGerald GA. 2011. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol 31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang L, Karakiulakis G, Roth M. 2020. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med 8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raaben M, Einerhand AW, Taminiau LJ, van Houdt M, Bouma J, Raatgeep RH, Büller HA, de Haan CA, Rossen JW. 2007. Cyclooxygenase activity is important for efficient replication of mouse hepatitis virus at an early stage of infection. Virol J 4:55. doi: 10.1186/1743-422X-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao W, Wang C, Chen B, Zhang F, Liu Y, Lu Q, Guo H, Yan C, Sun H, Hu G, Yin X. 2015. Ibuprofen attenuates cardiac fibrosis in streptozotocin-induced diabetic rats. Cardiology 131:97–106. doi: 10.1159/000375362. [DOI] [PubMed] [Google Scholar]
- 8.Miyoshi H, VanDussen KL, Malvin NP, Ryu SH, Wang Y, Sonnek NM, Lai C-W, Stappenbeck TS. 2017. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J 36:5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfajaro MM, Choi J-S, Kim D-S, Seo J-Y, Kim J-Y, Park J-G, Soliman M, Baek Y-B, Cho E-H, Kwon J, Kwon H-J, Park S-J, Lee WS, Kang M-I, Hosmillo M, Goodfellow I, Cho K-O. 2017. Activation of COX-2/PGE2 promotes sapovirus replication via the inhibition of nitric oxide production. J Virol 91:e01656-16. doi: 10.1128/JVI.01656-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alfajaro MM, Cho E-H, Park J-G, Kim J-Y, Soliman M, Baek Y-B, Kang M-I, Park S-I, Cho K-O. 2018. Feline calicivirus- and murine norovirus-induced COX-2/PGE2 signaling pathway has proviral effects. PLoS One 13:e0200726. doi: 10.1371/journal.pone.0200726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan X, Hao Q, Mu Y, Timani KA, Ye L, Zhu Y, Wu J. 2006. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-κB and CCAAT/enhancer binding protein. Int J Biochem Cell Biol 38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Liu M, Yang Y, Gu C, Yue Y, Wu KK, Wu J, Zhu Y. 2007. Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. FASEB J 21:1586–1596. doi: 10.1096/fj.06-6589com. [DOI] [PubMed] [Google Scholar]
- 14.Fajgenbaum DC, June CH. 2020. Cytokine storm. N Engl J Med 383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ, RECOVERY Collaborative Group . 2020. Dexamethasone in hospitalized patients with Covid-19: preliminary report. N Engl J Med doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Cruz CD, Farhadian S, Ko AI, Omer SB, Iwasaki A, Yale IMPACT Team . 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. 7821. Nature 584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann ER, Menon M, Knight SB, Konkel JE, Jagger C, Shaw TN, Krishnan S, Rattray M, Ustianowski A, Bakerly ND, Dark P, Lord GM, Simpson A, Felton T, Ho L-P, Trc NR, Feldmann M, CIRCO, Grainger JR, Hussell T. 2020. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci Immunol 5:eabd6197. doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robb CT, Goepp M, Rossi AG, Yao C. 2020. Non-steroidal anti-inflammatory drugs, prostaglandins, and COVID-19. Br J Pharmacol 177:4899–4920. doi: 10.1111/bph.15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao C, Narumiya S. 2019. Prostaglandin-cytokine crosstalk in chronic inflammation. Br J Pharmacol 176:337–354. doi: 10.1111/bph.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Zhao J, Legge K, Perlman S. 2011. Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest 121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, Wong G, Kobinger G, Xing Z, Couture C, Joubert P, Fritz JH, Powell WS, Divangahi M. 2014. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity 40:554–568. doi: 10.1016/j.immuni.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Vijay R, Fehr AR, Janowski AM, Athmer J, Wheeler DL, Grunewald M, Sompallae R, Kurup SP, Meyerholz DK, Sutterwala FS, Narumiya S, Perlman S. 2017. Virus-induced inflammasome activation is suppressed by prostaglandin D2/DP1 signaling. Proc Natl Acad Sci U S A 114:E5444–E5453. doi: 10.1073/pnas.1704099114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto K, Graham BS, Geraci MW, FitzGerald GA, Egan K, Zhou W, Goleniewska K, O’Neal JF, Morrow JD, Durbin RK, Wright PF, Collins RD, Suzutani T, Peebles RS. 2004. Signaling through the prostaglandin I2 receptor IP protects against respiratory syncytial virus-induced illness. J Virol 78:10303–10309. doi: 10.1128/JVI.78.19.10303-10309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernard MP, Bancos S, Chapman TJ, Ryan EP, Treanor JJ, Rose RC, Topham DJ, Phipps RP. 2010. Chronic inhibition of cyclooxygenase-2 attenuates antibody responses against vaccinia infection. Vaccine 28:1363–1372. doi: 10.1016/j.vaccine.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, Lanzavecchia A, Corti D, Virgin HW. 2020. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. 7821. Nature 584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 26.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung C, Wadsworth SJ, Yang SJ, Dorscheid DR. 2020. Structural and functional variations in human bronchial epithelial cells cultured in air-liquid interface using different growth media. Am J Physiol Lung Cell Mol Physiol 318:L1063–L1073. doi: 10.1152/ajplung.00190.2019. [DOI] [PubMed] [Google Scholar]
- 28.Ravindra NG, Alfajaro MM, Gasque V, Habet V, Wei J, Filler RB, Huston NC, Wan H, Szigeti-Buck K, Wang B, Wang G, Montgomery RR, Eisenbarth SC, Williams A, Pyle AM, Iwasaki A, Horvath TL, Foxman EF, Pierce RW, van Dijk D, Wilen CB. 2020. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium. bioRxiv doi: 10.1101/2020.05.06.081695. [DOI] [PMC free article] [PubMed]
- 29.McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 31.Jiang R-D, Liu M-Q, Chen Y, Shan C, Zhou Y-W, Shen X-R, Li Q, Zhang L, Zhu Y, Si H-R, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang H-J, Baric RS, Zhou P, Yang X-L, Shi Z-L. 2020. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler SP, Fremont DH, Greco S, McCray PB, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS. 2020. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Zhuang Z, Zheng J, Li K, Wong RL-Y, Liu D, Huang J, He J, Zhu A, Zhao J, Li X, Xi Y, Chen R, Alshukairi AN, Chen Z, Zhang Z, Chen C, Huang X, Li F, Lai X, Chen D, Wen L, Zhuo J, Zhang Y, Wang Y, Huang S, Dai J, Shi Y, Zheng K, Leidinger MR, Chen J, Li Y, Zhong N, Meyerholz DK, McCray PB, Perlman S, Zhao J. 2020. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182:734–743. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, Ring A, Wilen CB, Iwasaki A. 2020. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun S-H, Chen Q, Gu H-J, Yang G, Wang Y-X, Huang X-Y, Liu S-S, Zhang N-N, Li X-F, Xiong R, Guo Y, Deng Y-Q, Huang W-J, Liu Q, Liu Q-M, Shen Y-L, Zhou Y, Yang X, Zhao T-Y, Fan C-F, Zhou Y-S, Qin C-F, Wang Y-C. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28:124–133. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang L-I, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1470–1479. doi: 10.1038/s41590-020-0794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park MK, Kang SH, Son JY, Lee MK, Ju JS, Bae YC, Ahn DK. 2019. Co-administered low doses of ibuprofen and dexamethasone produce synergistic antinociceptive effects on neuropathic mechanical allodynia in rats. J Pain Res 12:2959–2968. doi: 10.2147/JPR.S222095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tubbs JT, Kissling GE, Travlos GS, Goulding DR, Clark JA, King-Herbert AP, Blankenship-Paris TL. 2011. Effects of buprenorphine, meloxicam, and flunixin meglumine as postoperative analgesia in mice. J Am Assoc Lab Anim Sci 50:185–191. [PMC free article] [PubMed] [Google Scholar]
- 39.Santos ARS, Vedana EMA, De Freitas GAG. 1998. Antinociceptive effect of meloxicam, in neurogenic and inflammatory nociceptive models in mice. Inflammation Res 47:302–307. doi: 10.1007/s000110050333. [DOI] [PubMed] [Google Scholar]
- 40.Laird JMA, Herrero JF, Garcia de la Rubia P, Cervero F. 1997. Analgesic activity of the novel COX-2 preferring NSAID, meloxicam in mono-arthritic rats: central and peripheral components. Inflamm Res 46:203–210. doi: 10.1007/s000110050174. [DOI] [PubMed] [Google Scholar]
- 41.Avanzato VA, Oguntuyo KY, Escalera-Zamudio M, Gutierrez B, Golden M, Pond SLK, Pryce R, Walter TS, Seow J, Doores KJ, Pybus OG, Munster VJ, Lee B, Bowden TA. 2019. A structural basis for antibody-mediated neutralization of Nipah virus reveals a site of vulnerability at the fusion glycoprotein apex. Proc Natl Acad Sci U S A 116:25057–25067. doi: 10.1073/pnas.1912503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang S-M, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen H, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB. 2021. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184:76–91. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sander WJ, O’Neill HG, Pohl CH. 2017. Prostaglandin E2 as a modulator of viral infections. Front Physiol 8:89. doi: 10.3389/fphys.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luczak M, Gumulka W, Szmigielski S, Korbecki M. 1975. Inhibition of multiplication of parainfluenza 3 virus in prostaglandin-treated WISH cells. Arch Virol 49:377–380. doi: 10.1007/BF01318248. [DOI] [PubMed] [Google Scholar]
- 45.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi P-Y. 2020. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kindler E, Thiel V, Weber F. 2016. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv Virus Res 96:219–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudek SE, Nitzsche K, Ludwig S, Ehrhardt C. 2016. Influenza A viruses suppress cyclooxygenase-2 expression by affecting its mRNA stability. 1. Sci Rep 6:27275. doi: 10.1038/srep27275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sumpter R, Loo Y-M, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fink K. 2012. Origin and function of circulating plasmablasts during acute viral infections. Front Immunol 3:78. doi: 10.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan EP, Pollock SJ, Pollack SJ, Murant TI, Bernstein SH, Felgar RE, Phipps RP. 2005. Activated human B lymphocytes express cyclooxygenase-2 and cyclooxygenase inhibitors attenuate antibody production. J Immunol 174:2619–2626. doi: 10.4049/jimmunol.174.5.2619. [DOI] [PubMed] [Google Scholar]
- 51.Bernard MP, Phipps RP. 2007. CpG oligodeoxynucleotides induce cyclooxygenase-2 in human B lymphocytes: implications for adjuvant activity and antibody production. Clin Immunol 125:138–148. doi: 10.1016/j.clim.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bancos S, Bernard MP, Topham DJ, Phipps RP. 2009. Ibuprofen and other widely used non-steroidal anti-inflammatory drugs inhibit antibody production in human cells. Cell Immunol 258:18–28. doi: 10.1016/j.cellimm.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernard MP, Phipps RP. 2010. Inhibition of cyclooxygenase-2 impairs the expression of essential plasma cell transcription factors and human B-lymphocyte differentiation. Immunology 129:87–96. doi: 10.1111/j.1365-2567.2009.03152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng J, Wong L-YR, Li K, Verma AK, Ortiz M, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Perlman S. 2020. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tipton DA, Flynn JC, Stein SH, Dabbous MK. 2003. Cyclooxygenase-2 inhibitors decrease interleukin-1β-stimulated prostaglandin E2 and IL-6 production by human gingival fibroblasts. J Periodontol 74:1754–1763. doi: 10.1902/jop.2003.74.12.1754. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Q, Li N, Chen M, Zheng J, Qian Z, Wang X, Huang C, Xu S, Shi G. 2013. Cyclooxygenase-2 promotes hepatocellular apoptosis by interacting with TNF-α and IL-6 in the pathogenesis of nonalcoholic steatohepatitis in rats. Dig Dis Sci 58:2895–2902. doi: 10.1007/s10620-013-2823-6. [DOI] [PubMed] [Google Scholar]
- 57.Kaur J, Sanyal SN. 2011. Diclofenac, a selective COX-2 inhibitor, inhibits DMH-induced colon tumorigenesis through suppression of MCP-1, MIP-1α, and VEGF. Mol Carcinog 50:707–718. doi: 10.1002/mc.20736. [DOI] [PubMed] [Google Scholar]
- 58.Lisboa FA, Bradley MJ, Hueman MT, Schobel SA, Gaucher BJ, Styrmisdottir EL, Potter BK, Forsberg JA, Elster EA. 2017. Nonsteroidal anti-inflammatory drugs may affect cytokine response and benefit healing of combat-related extremity wounds. Surgery 161:1164–1173. doi: 10.1016/j.surg.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 59.Anderson GD, Hauser SD, McGarity KL, Bremer ME, Isakson PC, Gregory SA. 1996. Selective inhibition of cyclooxygenase (COX)-2 reverses inflammation and expression of COX-2 and interleukin 6 in rat adjuvant arthritis. J Clin Invest 97:2672–2679. doi: 10.1172/JCI118717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aoki T, Frȍsen J, Fukuda M, Bando K, Shioi G, Tsuji K, Ollikainen E, Nozaki K, Laakkonen J, Narumiya S. 2017. Prostaglandin E2–EP2–NF-κB signaling in macrophages as a potential therapeutic target for intracranial aneurysms. Sci Signal 10:eaah6037. doi: 10.1126/scisignal.aah6037. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, Sun R, Tian Z, Xu X, Wei H. 2020. Pathogenic T cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci Rev 7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lam JKP, Hui KF, Ning RJ, Xu XQ, Chan KH, Chiang AKS. 2018. Emergence of CD4+ and CD8+ polyfunctional T cell responses against immunodominant lytic and latent EBV antigens in children with primary EBV infection. Front Microbiol 9:416. doi: 10.3389/fmicb.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo SH, Webster RG. 2002. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J Virol 76:1071–1076. doi: 10.1128/jvi.76.3.1071-1076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, Zheng M, Sundaram B, Banoth B, Malireddi RKS, Schreiner P, Neale G, Vogel P, Webby R, Jonsson CB, Kanneganti T-D. 2021. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184:149–168. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang E, Jordan SC. 2020. Tocilizumab for Covid-19: the ongoing search for effective therapies. N Engl J Med 383:2387–2388. doi: 10.1056/NEJMe2032071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imam Z, Odish F, Gill I, O’Connor D, Armstrong J, Vanood A, Ibironke O, Hanna A, Ranski A, Halalau A. 2020. Older age and comorbidity are independent mortality predictors in a large cohort of 1305 COVID-19 patients in Michigan, United States. J Intern Med 288:469–476. doi: 10.1111/joim.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rinott E, Kozer E, Shapira Y, Bar-Haim A, Youngster I. 2020. Ibuprofen use and clinical outcomes in COVID-19 patients. Clin Microbiol Infect 26:1259.e5–1259-e7. doi: 10.1016/j.cmi.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong AY, MacKenna B, Morton C, Schultze A, Walker AJ, Bhaskaran K, Brown J, Rentsch CT, Williamson E, Drysdale H, Croker R, Bacon S, Hulme W, Bates C, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald H, Tomlinson L, Mathur R, Wing K, Forbes H, Parry J, Hester F, Harper S, Evans S, Smeeth L, Douglas I, Goldacre B. 2020. OpenSAFELY: do adults prescribed Non-steroidal anti-inflammatory drugs have an increased risk of death from COVID-19? medRxiv doi: 10.1101/2020.08.12.20171405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abu Esba LC, Alqahtani RA, Thomas A, Shamas N, Alswaidan L, Mardawi G. 2020. Ibuprofen and NSAID use in COVID-19 infected patients is not associated with worse outcomes: a prospective cohort study. Infect Dis Ther doi: 10.1007/s40121-020-00363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bruce E, Barlow-Pay F, Short R, Vilches-Moraga A, Price A, McGovern A, Braude P, Stechman MJ, Moug S, McCarthy K, Hewitt J, Carter B, Myint PK. 2020. Prior routine use of non-steroidal anti-inflammatory drugs (NSAIDs) and important outcomes in hospitalized patients with COVID-19. J Clin Med 9:2586. doi: 10.3390/jcm9082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lund LC, Kristensen KB, Reilev M, Christensen S, Thomsen RW, Christiansen CF, Støvring H, Johansen NB, Brun NC, Hallas J, Pottegård A. 2020. Adverse outcomes and mortality in users of non-steroidal anti-inflammatory drugs who tested positive for SARS-CoV-2: a Danish nationwide cohort study. PLoS Med 17:e1003308. doi: 10.1371/journal.pmed.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All previously published data are available as described above. Cytokine data generated in this study are available in Table S1 in the supplemental material.