The major barrier toward the eradication of HIV-1 infection is the presence of a small reservoir of latently infected cells, which include CD4+ T cells and macrophages that escape immune-mediated clearance and the effects of antiretroviral therapy. There remain crucial gaps in our understanding of the molecular mechanisms that lead to transcriptionally silent or latent HIV-1 infection of macrophages.
KEYWORDS: Escherichia coli, HIV-1, Neisseria gonorrhoeae, interferon regulatory factors, latency, macrophages, Toll-like receptors
ABSTRACT
Macrophages are infected by human immunodeficiency virus type 1 (HIV-1) in vivo and contribute to both viral spread and pathogenesis. Recent human and animal studies suggest that HIV-1-infected macrophages serve as a reservoir that contributes to HIV-1 persistence during antiretroviral therapy. The ability of macrophages to serve as persistent viral reservoirs is likely influenced by the local tissue microenvironment, including interactions with pathogenic and commensal microbes. Here, we show that the sexually transmitted pathogen Neisseria gonorrhoeae (gonococcus [GC]) and the gut-associated microbe Escherichia coli, which encode ligands for both Toll-like receptor 2 (TLR2) and TLR4, repressed HIV-1 replication in macrophages and thereby induced a state reminiscent of viral latency. This repression was mediated by signaling through TLR4 and the adaptor protein Toll/interleukin 1 (IL-1) receptor domain-containing adapter-inducing beta interferon (IFN) (TRIF) and was associated with increased production of type I interferons. Inhibiting TLR4 signaling, blocking type 1 interferon, or knocking down TRIF reversed lipopolysaccharide (LPS)- and gonococcus-mediated repression of HIV-1. Finally, the repression of HIV-1 in macrophages was associated with the recruitment of interferon regulatory factor 8 (IRF8) to the interferon-stimulated response element (ISRE) downstream of the HIV-1 5′ long terminal repeat (LTR). Our data indicate that IRF8 is responsible for repression of HIV-1 replication in macrophages in response to TRIF-dependent signaling during N. gonorrhoeae and E. coli coinfection. These findings highlight the potential role of macrophages as HIV-1 reservoirs, as well as the roles of the tissue microenvironment and coinfections as modulators of HIV-1 persistence.
IMPORTANCE The major barrier toward the eradication of HIV-1 infection is the presence of a small reservoir of latently infected cells, which include CD4+ T cells and macrophages that escape immune-mediated clearance and the effects of antiretroviral therapy. There remain crucial gaps in our understanding of the molecular mechanisms that lead to transcriptionally silent or latent HIV-1 infection of macrophages. The significance of our research is in identifying microenvironmental factors, such as commensal and pathogenic microbes, that can contribute to the establishment and maintenance of latent HIV-1 infection in macrophages. It is hoped that identifying key processes contributing to HIV-1 persistence in macrophages may ultimately lead to novel therapeutics to eliminate latent HIV-1 reservoirs in vivo.
INTRODUCTION
Macrophages are among the immune cells located within the gastrointestinal and genitourinary mucosae thought to play a role in human immunodeficiency virus type 1 (HIV-1) sexual transmission and pathogenesis (1–3). A number of studies examining either HIV-1 infection of human vaginocervical or gastrointestinal tissue explants or rhesus macaque simian immunodeficiency virus (SIVmac) infection in rhesus macaque animal models have shown that macrophages are among the first cells infected during mucosal transmission (2, 4, 5). Macrophages can be productively infected with HIV-1 and are thought to be a source of virus persistence in vivo (6). Given their role in transmission, pathogenesis, and viral persistence, it is important to understand how the local mucosal microenvironment and cellular signaling pathways modulate interactions between macrophages and HIV-1.
Sexually transmitted infections (STIs) have been shown to be cofactors that enhance HIV-1 transmission (7). Neisseria gonorrhoeae (gonococcus [GC]) is a nonulcerative STI that is thought to augment mucosal transmission of HIV-1, both by inducing inflammation and by directly activating virus infection and replication (8–13). The role of GC in HIV-1 persistence is less well understood. Several studies have implicated GC-encoded pathogen-associated molecular patterns (PAMPs) as mediators of both inflammation and HIV-1 activation in target cells such as macrophages; however, the interactions between GC and macrophages are complex. GC encodes PAMPs capable of engaging Toll-like receptors (TLRs), including TLR2, TLR4, and TLR9 (14, 15). While the effects of coinfection with live GC on HIV-1 replication in macrophages have not been reported, purified lipooligosaccharide (LOS), as well as Escherichia coli lipopolysaccharide (LPS), have been shown to repress virus replication through the production of type 1 interferons (IFNs) (16, 17). In the case of LPS, repression is due to undefined effects at the level of gene expression. Although it is not entirely clear how TLR2 signaling affects HIV-1 expression in macrophages, studies have shown that purified TLR2 ligands activate virus replication in macrophages (18) and in latently infected T cells (19).
Here, we demonstrate that coinfection with GC and E. coli repress HIV-1 expression in macrophages. To investigate the underlying mechanism(s) responsible for this repression, we examined the individual effects of TLR2 and TLR4 signaling on HIV-1 expression in macrophages. TLR2 signaling activated HIV-1 expression in macrophages, whereas TLR4 signaling repressed virus expression. Importantly, TLR4 signaling overcame the activation effects of TLR2 signaling in macrophages. The TLR4-mediated repression of HIV-1 in macrophages coinfected with GC or E. coli was dependent on signaling through Toll/interleukin 1 (IL-1) receptor domain-containing adapter-inducing IFN-β (TRIF) and required type 1 IFN production. Finally, we showed that TLR4 signaling leads to the late-phase recruitment of IRF8 to the interferon-stimulated response element (ISRE) downstream of the HIV-1 5′ LTR in infected macrophages. Taken together, our data suggest that TRIF-mediated signaling represses HIV-1 replication in response to GC or E. coli coinfection in an IRF8-dependent manner and shifts macrophages from a state of robust HIV-1 expression to a state of persistent low-level/latent infection.
RESULTS
HIV-1 gene expression in MDMs is enhanced or repressed in a TLR-specific manner.
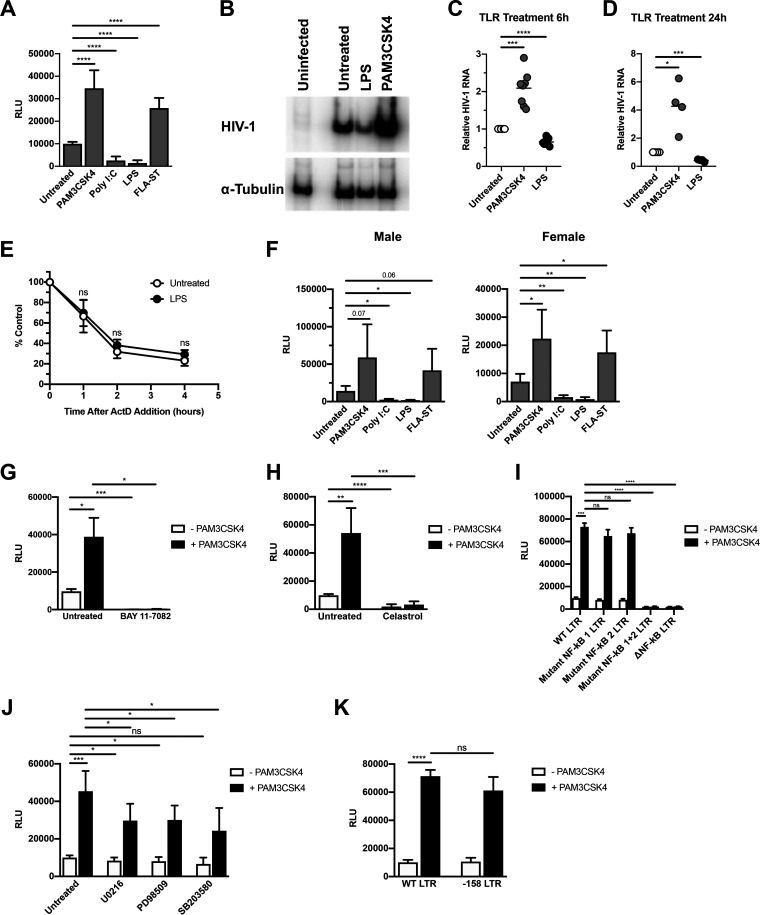
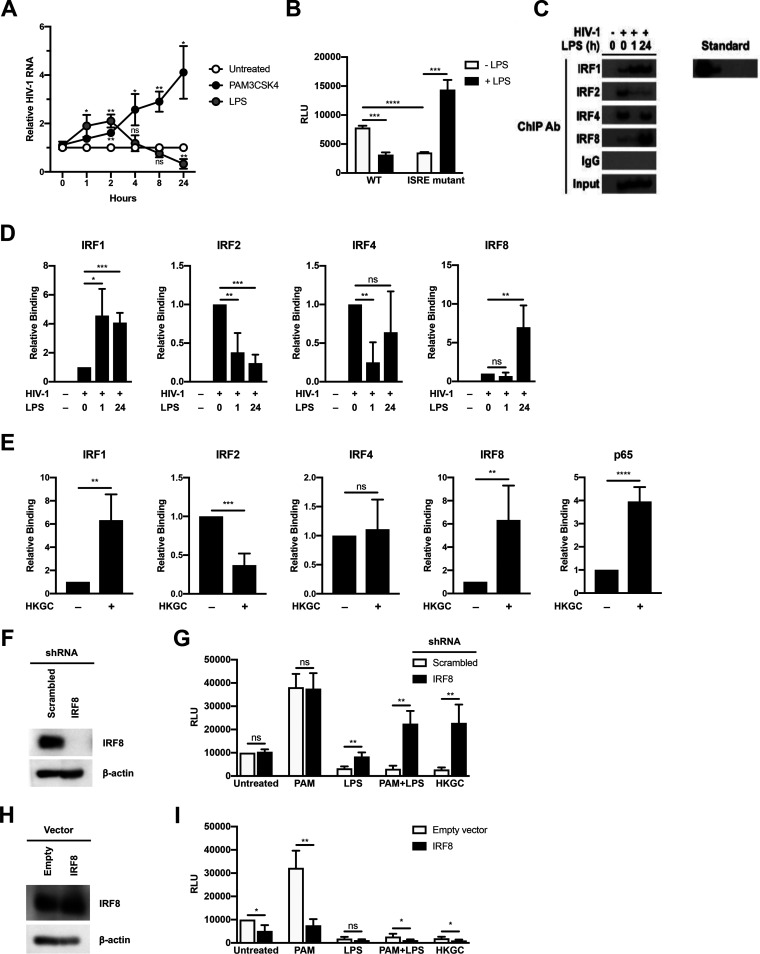
To determine how purified TLR ligands affected HIV-1 gene expression, monocyte-derived macrophages (MDMs) were infected with a single-round infectious HIV-1 reporter virus, and then treated with ligands for TLR2, TLR3, TLR4, or TLR5. Ligands that activated TLR2 or TLR5 enhanced HIV-1 replication, whereas ligands for TLR3 or TLR4 repressed HIV-1 expression (Fig. 1A). The effects of TLR ligands on HIV-1 replication occurred at the level of transcription, as treatment with the TLR2/1 ligand PAM3CSK4 led to an increase in HIV-1 mRNA accumulation, whereas treatment with the TLR4 ligand LPS led to a decrease in HIV-1 transcript levels (Fig. 1B to D). TLR treatment had no effect on viral RNA stability, as viral RNA from LPS-treated MDMs had a similar decay rate to that from untreated MDMs (Fig. 1E). Recent studies have demonstrated that myeloid cells from males and females have different susceptibilities to HIV-1 infection, largely due to differential levels of innate immune responses and steroid hormones (20–22). We therefore sought to determine whether there was a sex-based difference in the response to TLR ligand treatment in MDMs. We found that TLR stimulation had similar effects on HIV-1 expression in MDMs from both male and female donors (Fig. 1F). These results indicate that MyD88-dependent signaling enhances HIV-1 transcription, whereas TRIF-dependent signaling inhibits HIV-1 transcription in MDMs.
FIG 1.
Treatment with purified Toll-like receptor (TLR) ligands alters HIV-1 replication at the level of transcription. (A) Monocyte-derived macrophages (MDMs) were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were treated with the TLR2 ligand PAM3CSK4 (100 ng/ml), the TLR3 ligand poly(I·C) (25 μg/ml), the TLR4 ligand lipopolysaccharide (LPS) (100 ng/ml), or the TLR5 ligand FLA-ST (100 ng/ml) for 18 h. The cells were then lysed and assayed for luciferase activity. Bars represent the mean (± standard deviation [SD]) of 11 donors, each donor tested in triplicate. (B to D) MDMs were infected as described above. At 48 h after infection, cells were treated with PAM3CSK4 (100 ng/ml) or LPS (100 ng/ml) for 6 h (B and C) or 24 h (D). Cells were then lysed and assayed for viral RNA accumulation by reverse transcription-PCR (RT-PCR). Shown are data from one representative donor at 6 h (B) and composite data from eight donors at 6 h (C) and four donors at 24 h (D). (E) MDMs were infected as in panel A. At 48 h after infection, cells were treated with the TLR4 ligand LPS (100 ng/ml) for 4 h. Cells were then treated with actinomycin D (10 μg/ml) to inhibit transcription. Total cytoplasmic RNA was prepared from the treated cultures at the indicated time points following actinomycin D treatment and analyzed by reverse transcription-quantitative PCR (RT-qPCR) for the expression of HIV-1 RNA. The data are the means (± SD) from four donors. (F) MDMs were infected and treated with TLR ligands as in (A). The cells were then lysed and assayed for luciferase activity. Bars represent the mean (± SD) of five male donors and five female donors; each donor was tested in triplicate. Although virus replication was decreased overall in MDMs from female donors compared to MDMs from male donors, it was activated by treatment with PAM3CSK4 and FLA-ST and repressed by poly(I·C) and LPS, in a manner similar to that seen in MDMs from male donors. (G to H) MDMs were infected as in panel A, and, 48 h after infection, cells were treated with PAM3CSK4 (100 ng/ml) in the presence or absence of 10 μM BAY 11-7082 (G) or 10 μM celastrol (H) for 18 h. The cells were then lysed and assayed for luciferase activity. The data are the mean (± SD) of three donors (BAY 11-7082) or six donors (celastrol); each donor was tested in triplicate. (I) HEK293-TLR2CFPTLR1YFP cells were transfected with HIV-1 long terminal repeat (LTR)-luciferase reporter constructs with intact NF-κB, mutated NF-κB, or deleted NF-κB binding sites. Following transfection, cells were treated with PAM3CSK4 (100 ng/ml) for 18 h and then harvested and assayed for luciferase activity. Data represent the mean (± SD) of three independent experiments, each performed in triplicate. (J) MDMs were infected as in (A), and 48 h after infection, cells were treated with PAM3CSK4 (100 ng/ml) in the presence or absence of U0126 (10 μM), PD98059 (50 μM), or SB203580 (10 μM) for 18 h. Cells were then lysed and assayed for luciferase activity. The data are the mean (± SD) of six donors; each donor was tested in triplicate. (K) HEK293-TLR2CFPTLR1YFP cells were transfected with HIV-1 LTR-luciferase reporter constructs with intact AP-1 sites (wild-type [WT] LTR) or deleted AP-1 binding sites (−158 LTR). Following transfection, cells were treated with PAM3CSK4 (100 ng/ml) for 18 h and then harvested and assayed for luciferase activity. Data are the mean (± SD) of three independent experiments, each performed in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
MyD88-dependent TLR signaling leads to the activation of both NF-κB and AP-1 transcription factors, among others (23). The HIV-1 LTR contains binding sites for both NF-κB and AP-1. The two NF-κB sites are thought to be essential for HIV-1 transcription (24, 25), whereas the AP-1 sites, while not essential, are thought to enhance HIV-1 transcription (26, 27). Previous studies demonstrated that treatment of HIV-infected MDMs with the TLR2/TLR1 ligand PAM3CSK4 led to an increased association of the p65 subunit of NF-κB and the c-fos subunit of AP-1 with the 5′ LTR, which in turn correlated with increased virus replication (18); however, the contributions made by each pathway to TLR2-mediated activation have not been previously characterized. To determine the roles of NF-κB and AP-1 in TLR2-activated HIV replication in MDMs, HIV-1- infected cells were treated with either BAY 11-7082, an inhibitor of IκB kinase (28); celastrol, a small-molecule inhibitor of the IκB kinase complex (29); or inhibitors that disrupt AP-1 signaling. As shown in Fig. 1G and H, BAY 11-7082 and celastrol treatment completely ablated TLR2/1-enhanced HIV-1 expression. Similarly, the use of an LTR-based reporter construct with mutations in the NF-κB binding sites did not result in increased gene expression in response to TLR2 signaling (Fig. 1I). Treatment of HIV-infected macrophages with inhibitors of kinases upstream of AP-1 activation, such as MEK1/2 (U0126, PD98509), and p38 (SB203580), resulted in modest, but reproducible, decreases in TLR2-mediated activation of HIV-1 (Fig. 1J). Similarly, LTR reporter constructs lacking AP-1 binding sites were activated in response to TLR2 signaling at levels similar to that of the wild-type (WT) construct, further demonstrating the nonessential role of AP-1 in TLR2-mediated HIV-1 activation (Fig. 1K). Although the regulation of HIV-1 transcription through multiple transcription factor binding sites in and adjacent to the 5′ LTR is complex, these data suggest that, in MDMs, TLR2-activated HIV-1 expression is mediated primarily through NF-κB, with a minor contribution from AP-1 signaling.
Coinfection with Neisseria gonorrhoeae or Escherichia coli represses HIV-1 replication in MDMs.
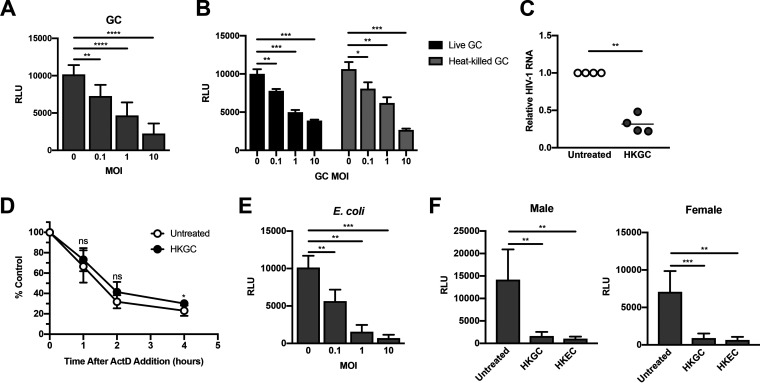
Our preliminary studies using purified TLR ligands in isolation suggested that different TLR signaling cascades had diverse effects on HIV-1 replication. Since most pathogens encode multiple TLR ligands, we sought to determine the effects of intact pathogens on HIV-1 replication. We incubated HIV-infected MDMs with N. gonorrhoeae (gonococcus [GC]), which expresses ligands for TLR2, TLR4, and TLR9. We found that increasing amounts of GC led to a dose-dependent decrease in HIV-1 replication in MDMs (Fig. 2A). Bacterial replication was not required for these effects, as heat-killed GC led to repression of HIV-1 replication in MDMs (Fig. 2B). GC-mediated repression occurred at the level of viral transcription (Fig. 2C) and did not decrease viral RNA stability (Fig. 2D). In addition, repression of HIV-1 replication is not specific for GC, but may be a generalized response to Gram-negative bacteria, as coinfection with E. coli also repressed HIV-1 replication in MDMs in a manner similar to GC (Fig. 2E). Similar to what we observed with purified LPS, the biological sex of the donors had no effect on GC- or E. coli-mediated HIV-1 repression in MDMs (Fig. 2F). Despite the presence of both activating (TLR2) and repressing (TLR4) TLR ligands, both GC and E. coli mediated repression of HIV-1 replication in macrophages. This finding raised several possibilities, as follows: (i) the dominance of TLR4 signaling over TLR2 signaling in MDMs; (ii) different expression levels of TLR2-, TLR4-, and TLR4-associated molecules such as CD14 and MD-2 on MDMs; (iii) different cytokine profiles produced in response to GC or E. coli; and/or (iv) variable expression of signaling molecules downstream of TLRs. These scenarios were further explored.
FIG 2.
Treatment with intact Neisseria gonorrhoeae and Escherichia coli represses HIV-1 replication at the level of transcription. (A) MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were cultured overnight with increasing amounts of gonococcus (GC). Cells were then lysed and assayed for luciferase activity. Bars represent mean (± SD) of seven donors; each donor was tested in triplicate. (B) MDMs were infected as in panel A and, 48 h after infection, were cultured with increasing amounts of live or heat-killed (56°C treatment) GC overnight. The cells were then lysed, and luciferase activity was measured. The data are the mean (± SD) of three donors; each donor was tested in triplicate. (C) MDMs were infected as described above and, 48 h after infection, were treated with heat-killed GC (HKGC) at a multiplicity of infection (MOI) of 10 for 24 h. Cells were then lysed and assayed for viral RNA accumulation by RT-qPCR. Shown are data from four donors. (D) MDMs were infected as in panel A and, 48 h after infection, cells were treated with heat-killed GC (MOI = 10) for 4 h. Cells were then treated with actinomycin D (10 μg/ml) to inhibit transcription. Total cytoplasmic RNA was prepared from the treated cultures at the indicated time points following actinomycin D treatment and analyzed by RT-qPCR for the expression of HIV-1 RNA. The data are the means (± SD) from four donors. (E) MDMs were infected as in panel A and, 48 h after infection, the cells were cultured overnight with increasing amounts of E. coli. Cells were then lysed and assayed for luciferase activity. Bars represent mean (± SD) of four donors, with each donor tested in triplicate. (F) MDMs were infected as in panel A and, 48 h after infection, cells were treated with heat-killed GC (MOI = 10) or heat-killed E. coli (HKEC) (MOI = 10) for 18 h. The cells were then lysed and assayed for luciferase activity. Bars represent the mean (± SD) of five male donors and five female donors; each donor was tested in triplicate. Although virus replication was decreased overall in MDMs from female donors compared to that in MDMs from male donors, it was repressed by HKGC and HKEC in a manner similar to that seen in MDMs from male donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
TLR4 signaling is dominant in MDMs.
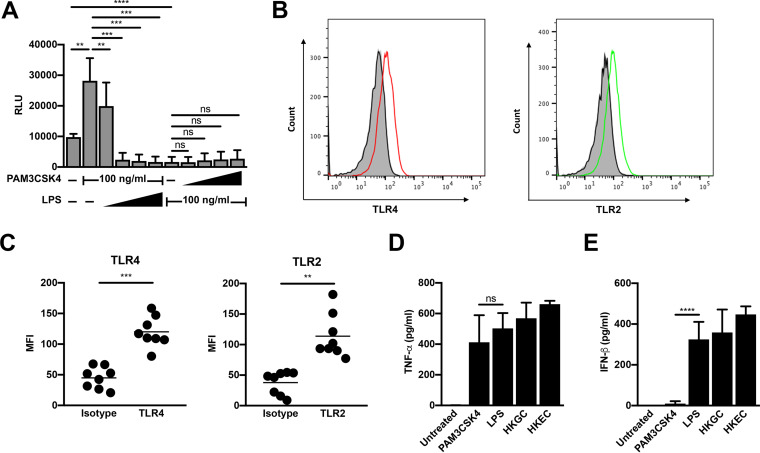
To determine whether certain TLR pathways are dominant in MDMs, we performed cotreatments of HIV-infected MDMs with the TLR2 ligand PAM3CSK4 and the TLR4 ligand LPS. We found that increasing the concentration of LPS against a fixed concentration of PAM3CSK4 led to a reversal of TLR2-mediated activation of HIV-1 and, eventually, to repression of HIV-1 replication (Fig. 3A). Conversely, increasing the concentration of PAM3CSK4 against a fixed concentration of LPS did not reverse LPS-mediated repression of HIV-1 (Fig. 3A). Flow cytometry was used to determine that the different responses of MDMs were likely not due to receptor expression, as MDMs express both TLR2 and TLR4 (Fig. 3B and C). In addition, MDMs produced both tumor necrosis factor alpha (TNF-α) and beta interferon (IFN-β) in response to LPS treatment, GC coinfection, and E. coli coinfection. Whereas treatment of MDMs with LPS resulted in a similar cytokine profile to that of coinfection, treatment of MDMs with the TLR2/1-ligand PAM3CSK4 resulted in the production of TNF-α, but not in appreciable levels of IFN-β (Fig. 3D and E). Taken together, our data suggest that TLR4 signaling, which negatively regulates LTR-driven gene expression, is dominant in MDMs.
FIG 3.
TLR4 signaling is dominant in MDMs. (A) MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were treated with a fixed concentration of PAM3CSK4 (100 ng/ml) and increasing concentrations of LPS (1 to 1,000 ng/ml, as indicated) or a fixed concentration of LPS (100 ng/ml) and increasing concentrations of PAM3CSK4 (1 to 1,000 ng/ml, as indicated) for 18 h. Cells were then lysed and assayed for luciferase activity. The data are the mean (± SD) of six donors; each donor was tested in triplicate. (B and C) At 8 days postisolation, MDMs were stained with antibodies against TLR2 or TLR4 or relevant isotype controls. Receptor expression was assessed by flow cytometry. Histograms from one representative donor are shown in panel B. Gray, unstained cells; black line, isotype control; red line, TLR4; green line, TLR2. Mean fluorescent intensity (MFI) ± SD from eight donors is depicted in panel C. (D and E) MDMs were treated with the TLR2 ligand PAM3CSK4 (100 ng/ml), the TLR4 ligand LPS (100 ng/ml), heat-killed GC (MOI = 10), or heat-killed E. coli (MOI = 10) for 18 h. Cell supernatant was harvested, filtered through a 0.2-μm filter, and analyzed by enzyme-limited immunosorbent assay (ELISA) for tumor necrosis factor alpha (TNF-α) (D) and beta interferon (IFN-β) (E) production. Data represent mean (± SD) of seven donors (four donors for heat-killed E. coli). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
LPS- and GC-mediated repression of HIV-1 in MDMs is dependent on TRIF-mediated type I IFN production.
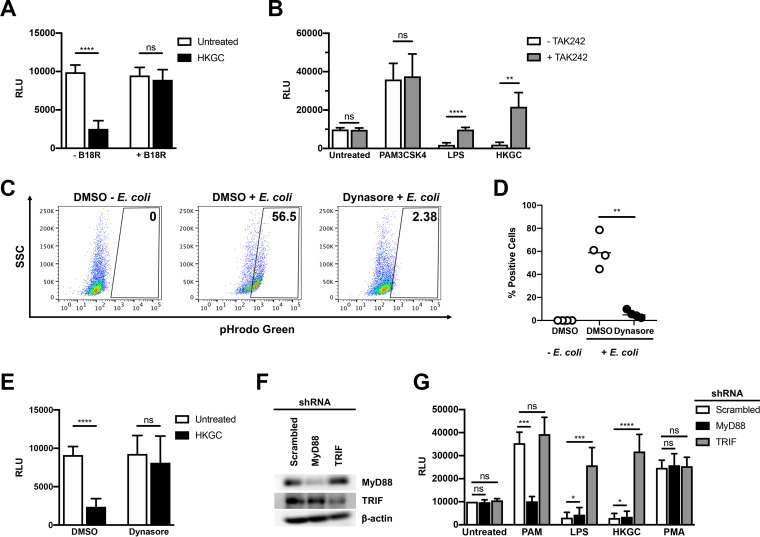
Since LPS and GC both induce type I IFN production, whereas the TLR2 ligand PAM3CSK4 does not, we wished to determine whether GC-stimulated production of IFN-α/β contributes to repression of HIV-1 in MDMs. We found that treatment of HIV-infected MDMs with the vaccinia virus-encoded soluble type I IFN receptor B18R reversed GC-mediated inhibition of HIV-1 replication, suggesting that TLR4-mediated IFN production is required for HIV-1 repression by GC (Fig. 4A). Since both purified TLR4 ligands and GC, which encodes ligands for TLR2, TLR4, and TLR9, repress HIV-1 replication in MDMs, we predicted that downstream effector molecules of TLR4 signaling would contribute to the repression of HIV-1 replication in MDMs. First, we confirmed that TLR4 signaling was responsible for GC-mediated HIV-1 repression in MDMs. Treatment with the TLR4-specific inhibitor TAK242 reversed the LPS- and GC-dependent repression of HIV-1 in MDMs (Fig. 4B). Treatment with TAK242 had no effect on TLR2-mediated activation of HIV-1 replication in MDMs, consistent with reports that TAK242 is specific for TLR4 (30).
FIG 4.
LPS- and GC-mediated repression of HIV-1 replication in MDMs requires TLR4, TRIF, and type I IFNs. (A) MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, cells were treated with GC (MOI = 10) in the absence (white bars) or presence (black bars) of B18R (100 ng/ml) for 18 h. The cells were then lysed and assayed for luciferase activity. The data are the mean (± SD) of seven donors; each donor was tested in triplicate. (B) MDMs were infected as in panel A and, 48 h after infection, were treated with PAM3CSK4 (100 ng/ml), LPS (100 ng/ml), or heat-killed GC (MOI = 10) in the absence (white bars) or presence (gray bars) of TAK242 (1 μg/ml) for 18 h. The cells were then lysed and assayed for luciferase activity. The data are the mean (± SD) of six donors; each donor was tested in triplicate. (C and D) MDMs were incubated with dimethyl sulfoxide (DMSO) or the dynamin inhibitor Dynasore (80 μM) for 15 min at 37°C. The cells were washed with phosphate-buffered saline (PBS) and incubated with pHrodo green E. coli (1 mg/ml) for 2 h at 37°C. Endocytosis/phagocytosis was measured by flow cytometry. Shown are data from one representative donor (C) and composite data from four donors (D). (E) MDMs were infected as in panel A and, 48 h after infection, were treated with vehicle control (white bars) or with Dynasore (80 μM); black bars for 15 min prior to treatment with heat-killed GC (MOI = 10) for 18 h. The cells were then lysed and assayed for luciferase activity. The data are the mean (± SD) of six donors, each donor tested in triplicate. (F and G) MDMs were transfected with a control scrambled shRNA, shRNA targeting MyD88, or shRNA targeting TRIF. Knockdown of protein expression was detected by Western blot analysis (F). Transfected MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were treated with PAM3CSK4 (100 ng/ml), LPS (100 ng/ml), heat-killed GC (MOI = 10), or phorbol myristate acetate (PMA) (10 nM) for 18 h. The cells were then lysed and assayed for luciferase activity (G). The data are the mean (± SD) of six donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
It has been shown that TLR4, which can utilize both MyD88 and TRIF adaptor proteins, initiates different signaling pathways dependent upon its cellular location. Cell-surface TLR4 engagement leads to MyD88-dependent signaling, whereas endosomal TLR4 engagement leads to TRIF-dependent signaling (31). To examine whether TRIF-dependent signaling is responsible for HIV-1 repression, we blocked dynamin-dependent endocytosis of TLR4 with Dynasore, which prevents TRIF-dependent signaling while leaving MyD88-dependent signaling intact. As shown, blocking endocytosis-mediated TLR4 internalization (Fig. 4C and D) reversed GC-mediated repression of HIV-1 in MDMs (Fig. 4E). Given the ability of GC to signal through both TLR2-MyD88 and TLR4-TRIF, one might expect the inhibition of type I IFN signaling by B18R or the inhibition of endocytosis by Dynasore to lead to augmented viral gene expression through intact MyD88 signaling. However, we did not observe this, likely due to incomplete inhibition of either IFN signaling or endocytosis.
To confirm the role of MyD88 in TLR2-mediated HIV-1 activation and TRIF in TLR4-mediated HIV-1 repression, we used short hairpin RNAs (shRNAs) to knock down the two molecules in HIV-infected MDMs (Fig. 4F). Knockdown of MyD88 led to a loss of TLR2-mediated HIV-1 activation but had no effect on LPS- or GC-mediated HIV-1 repression (Fig. 4G). In contrast, knockdown of TRIF had no effect on TLR2-mediated HIV-1 activation, but reversed LPS- and GC-mediated repression of HIV-1 replication (Fig. 4G). Knockdown of either MyD88 or TRIF had no effect on the activation of HIV-1 by the phorbol ester phorbol myristate acetate (PMA), which signals directly through protein kinase C, independently of TLRs (Fig. 4G). These data suggest that the TLR4-TRIF-type I IFN axis in MDMs leads to GC- and E. coli-mediated repression of HIV-1 replication.
TLR4 signaling leads to differential IRF recruitment to the HIV-1 LTR.
Since type I IFN production is critical for GC- and E. coli-mediated HIV-1 repression in MDMs, we examined the role of interferon-stimulated genes (ISGs) in HIV-1 regulation. Previous studies have shown that ISGs are temporally regulated in macrophages in response to innate immune sensors and type I IFN signaling (32, 33). To determine whether the repressive effects of LPS were due to early- or late-phase ISGs, HIV-1-infected MDMs were treated with the TLR2 ligand PAM3CSK4 or the TLR4 ligand LPS, and total cytoplasmic RNA was extracted at various times posttreatment. Treatment of HIV-infected MDMs with the TLR2 ligand PAM3CSK led to a continuous increase in HIV-1 RNA levels (Fig. 5A). In contrast, treatment of HIV-infected MDMs with the TLR4 ligand LPS led to an initial short-lived increase in HIV-1 RNA levels; however, levels steadily declined thereafter (Fig. 5A), indicating that HIV-1 transcription displays a biphasic response to TLR4 stimulation in MDMs. This suggests that late-phase proteins induced by type I IFNs are responsible for TLR4-mediated decreases in HIV-1 transcription. It is known that HIV-1 contains an interferon-stimulated response element (ISRE) in the Gag-leader sequence (GLS), immediately downstream from the 5′ LTR. Because type I IFN is required for LPS- and GC-mediated repression of HIV-1 in MDMs, we assessed the role of the ISRE in this process using transient-transfection assays with mutated LTR-reporter constructs in HEK293 cells expressing TLR4, MD-2, and CD14. We found that LPS treatment repressed LTR-driven reporter-gene expression in cells expressing WT ISRE elements, but not in cells transfected with an LTR-luciferase construct containing a mutated ISRE (Fig. 5B). This suggests that transcription factor engagement of the ISRE governs TLR4-mediated HIV-1 repression.
FIG 5.
LPS- and GC-mediated repression of HIV-1 in MDMs is associated with changes in interferon regulatory factor (IRF) recruitment to the interferon-stimulated response element (ISRE). (A) MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were treated with PAM3CSK4 (100 ng/ml) or LPS (100 ng/ml). At various time points after TLR stimulation, cells were harvested, lysed, and total cytoplasmic RNA was extracted. Viral RNA accumulation was assessed by RT-PCR. The data are the mean (± SD) of four donors. (B) HEK293-TLR4CFP/MD-2/CD14 cells were transfected with HIV-1 LTR/Gag-leader sequence (GLS)-luciferase reporter constructs with an intact ISRE or mutated ISRE binding site. Following transfection, cells were treated with LPS (100 ng/ml) for 18 h and then harvested and assayed for luciferase activity. Data are the mean (± SD) of three independent experiments, each performed in triplicate. (C and D) MDMs were infected with a single-round replication-defective HIV-GFP reporter virus and, 48 h after infection, cells were treated with LPS (100 ng/ml). At either 1 or 24 h after LPS treatment, cells were fixed with formaldehyde, lysed, sonicated, and subjected to immunoprecipitation with antibodies against IRF1, IRF2, IRF4, IRF8, or rabbit IgG (isotype control). Association with the HIV-1 ISRE was assessed by PCR using HIV-1 specific primers. Data from one representative donor (C). Composite data representing the mean (± SD) from five donors (D). (E) MDMs were infected as in panel C and, 48 h after infection, were treated with heat-killed GC (MOI = 10). At 24 hours after GC treatment, cells were fixed with formaldehyde, lysed, sonicated, and subjected to immunoprecipitation with antibodies against IRF1, IRF2, IRF4, IRF8, NF-κB p65, or rabbit IgG (isotype control). Association with the HIV-1 ISRE was assessed by PCR using HIV-1-specific primers. Composite data from five donors are shown. (F and G) MDMs transfected with a controlled scrambled shRNA (white bars) or with shRNA targeting IRF8 (black bars). Knockdown of protein expression was detected by Western blot (F). Transfected MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were treated with PAM3CSK4 (100 ng/ml), LPS (100 ng/ml), a combination of PAM3CSK4 and LPS (each at 100 ng/ml), or GC (MOI = 10) for 18 h. Cells were then lysed and assayed for luciferase activity (G). The experiment was performed using cells from five different donors. (H and I) MDMs transfected with an empty vector (white bars) or a vector encoding IRF8 (black bars). IRF8 protein expression was detected by Western blot analysis (H). Transfected MDMs were infected with a single-round, replication-defective HIV-luciferase reporter virus and, 48 h after infection, were treated with PAM3CSK4 (100 ng/ml), LPS (100 ng/ml), a combination of PAM3CSK4 and LPS (each at 100 ng/ml), or GC (MOI = 10) for 18 h. Cells were then lysed and assayed for luciferase activity (I). The experiment was performed using cells from four different donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Previous studies have shown that IRF1 and IRF2 both bind to this ISRE in vitro and that IRF1 and IRF2 expression are associated with enhanced HIV-1 transcription (34). Two other IRFs, IRF4 and IRF8, are also expressed in macrophages (35) and have been shown to increase in response to type I IFN signaling and other signals (36, 37). Interestingly, IRF8 has been implicated in maintaining HIV-1 latency in infected monocytic cell lines (34, 38, 39), and IRF4 has been implicated in negative regulation of TLR signaling (40). We therefore investigated whether various IRFs are recruited to the HIV-1 ISRE in response to LPS and GC treatment in MDMs. Using chromatin immunoprecipitation analysis, we found that IRF1, IRF2, IRF4, and IRF8 all are able to associate with the 5′ LTR and GLS containing the ISRE in HIV-infected MDMs (Fig. 5C and D). Early after treatment with LPS, the levels of IRF1 associated with this region of the viral promoter increased, whereas the levels of IRF2 and IRF4 decreased. By 24 h posttreatment with LPS, the levels of IRF4 and IRF8 associated with this region increased. It is of particular note that the levels of IRF8 recruitment increased to well above those seen in unstimulated MDMs (Fig. 5D). A similar pattern of IRF recruitment to the 5′ LTR and GLS occurred in GC-treated MDMs (Fig. 5E), suggesting that repression of HIV-1 transcription in response to LPS and GC treatment is due to enhanced IRF8 recruitment to the 5′ LTR and GLS. To confirm the central role of IRF8 in TLR4-mediated repression of HIV-1 expression in MDMs, we used shRNA to knockdown IRF8 expression in MDMs (Fig. 5F). Reducing IRF8 expression reversed TLR4-mediated HIV-1 repression in response to treatment with LPS or GC. Knockdown of IRF8 led to activation of HIV-1 expression in cells treated with a combination of PAM3CSK4 and LPS or GC, similar to that seen with treatment with PAM3CSK4 alone (Fig. 5G). In contrast, overexpression of IRF8 in MDMs led to decreased HIV-1 expression in untreated MDMs and reversed the activation of HIV-1 expression in PAM3CSK4-treated MDMs, but it had no effect on LPS-mediated repression in MDMs (Fig. 5H and I). There was a small, but significant, enhancement of HIV-1 repression in MDMs treated with a combination of PAM3CSK4 and LPS or with GC.
Treatment with LPS or GC induces persistent low-level/latent HIV-1 infection in MDMs.
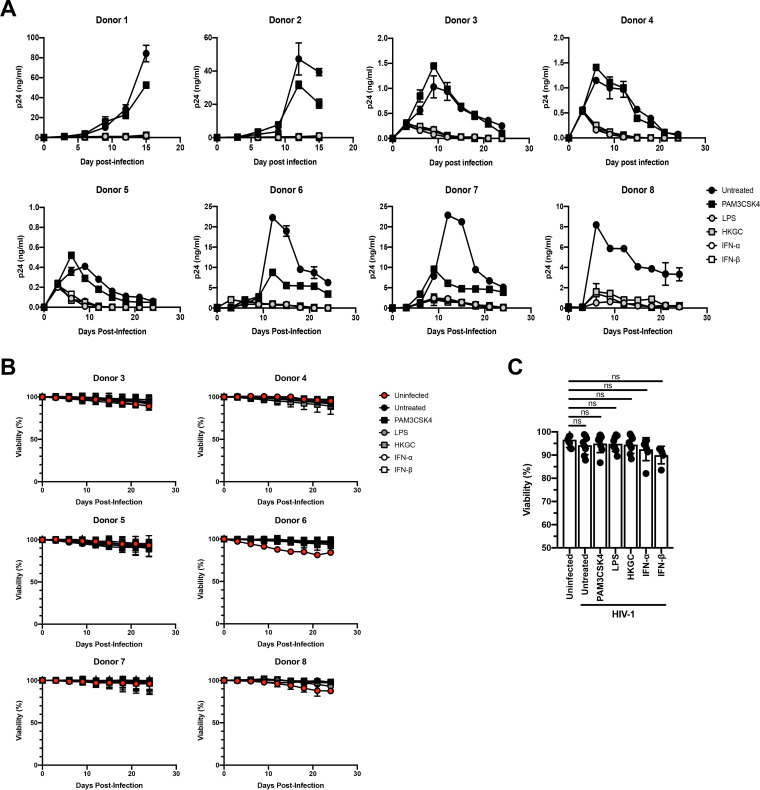
Recent studies in animals and human tissues demonstrate that HIV-1 can form persistent low-level or latent infections in macrophages (41–44). Our data suggest that engagement of the TLR4-TRIF-type I IFN axis in macrophages can repress virus replication and we wished to determine whether signaling through this axis could contribute to the establishment of persistent low-level or latent HIV-1 infection in macrophages. To this end, HIV-1-infected MDMs were treated a single time with the TLR2 ligand PAM3CSK4, the TLR4 ligand LPS, heat-killed GC, IFN-α, or IFN-β at day 3 postinfection. As shown in Fig. 6A, while there was a range of virus replication in the various donors, we found that treatment with a single dose of LPS, heat-killed GC, IFN-α, or IFN-β consistently led to a prominent, sustained decrease in HIV-1 replication in MDMs, whereas treatment with PAM3CSK4 led to a transient increase in HIV-1 replication followed by a slight decrease in replication. Importantly, these treatments did not significantly alter cellular viability (Fig. 6B and C). These data suggest that engagement of the TLR4-TRIF-type I IFN axis can promote low-level persistent/latent HIV-1 infection in MDMs.
FIG 6.
Treatment with LPS, GC, or type I IFNs induces a low-level persistent/latent infection in MDMs. MDMs were infected with replication-competent HIV-1Ba-L. At day 3 postinfection, MDMs from donors 1 to 5 were treated with a single dose of PAM3CSK4 (100 ng/ml), LPS (100 ng/ml), GC (MOI = 10), IFN-α (1,000 U/ml), or IFN-β (1,000 U/ml). MDMs from donors 6 and 7 were treated with PAM3CSK4, LPS, GC, or IFN-β. MDMs from donor 8 were treated with LPS, GC, or IFN-β. Cell-free supernatants were harvested every 3 days, and virus production was monitored by p24 enzyme-linked immunosorbent assay (ELISA). Data from eight independent donors, tested in triplicate (donors 1 to 5) or duplicate (donors 6 to 8), are shown. (B) Cell viability was monitored by measuring lactate dehydrogenase (LDH) in cell-free supernatants every 3 days during culture for donors 3 to 8 using a commercial LDH assay. (C) Cell viability was determined at the end of culture for donors 1 to 8 by measuring total LDH in cell lysates using a commercial LDH assay. Data from eight independent donors, tested in triplicate (donors 1 to 5) or duplicate (donors 6 to 8), are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
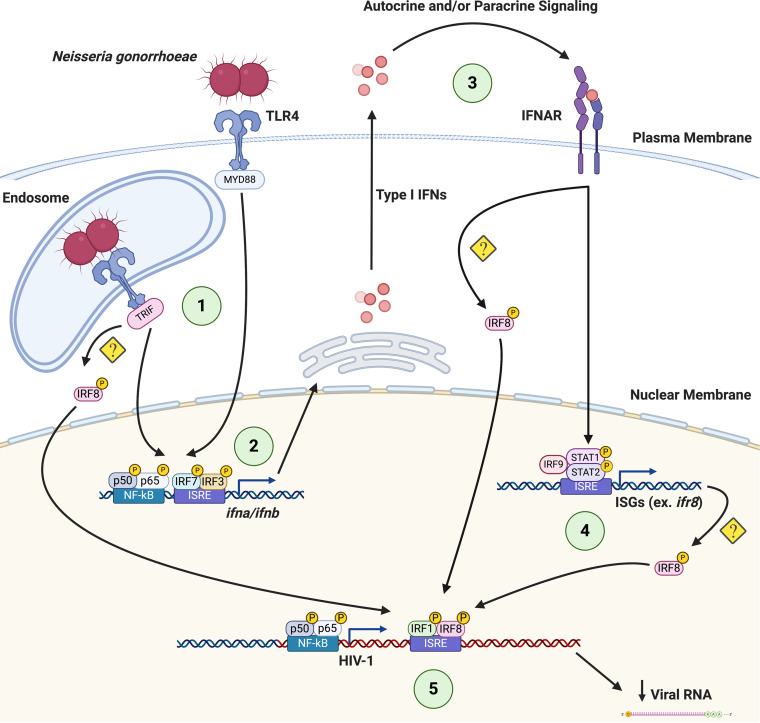
Taken together, these findings suggest that both LPS and GC activate TLR4-mediated TRIF signaling in MDMs, resulting in the production of type I IFNs. In turn, type I IFNs work in an autocrine or paracrine fashion to induce the expression of IRF8, which then binds to the ISRE present in the GLS of HIV-1 to repress viral transcription (Fig. 7).
FIG 7.
Coinfection with GC or E. coli represses HIV-1 replication by altering IRF recruitment to the HIV-1 GLS. (1) Upon engagement of TLR4 by GC (or E. coli) at the cell surface and in the endosome, signaling pathways are initiated that lead to the activation and nuclear translocation of transcription factors such as NF- κB, IRF3, and IRF7. (2) NF-κB, IRF3, and IRF7 are recruited to the IFN-α and/or IFN-β promoters to drive type I IFN expression. (3) Type I IFNs act through paracrine or autocrine signaling to drive the expression of ISGs, including IRFs (4). (5) During the late phase of the response, IRF8 is recruited to the HIV-1 GLS in a TLR4-TRIF-type I IFN-dependent manner, leading to the repression of HIV-1 transcription. Figure created with Biorender.com.
DISCUSSION
In these studies, we provide evidence that the interaction between commensal and pathogenic bacteria can repress HIV-1 replication in macrophages by altering the recruitment of transcription factors to the HIV-1 GLS, thereby inducing a state reminiscent of proviral latency. We further demonstrate that TLR2 ligands trigger MyD88-mediated signaling that increases virus expression via the activation of NF-κB, whereas TLR4 ligands trigger TRIF-dependent production of type I IFNs. Type I IFN signaling, in turn, is associated with the recruitment of IRF8 to the ISRE located in the GLS and a shift to low-level or latent HIV-1 infection.
A number of studies have shown that IRFs play an important role in the regulation of HIV-1 replication. There is an ISRE located downstream from the 5′ LTR in the GLS that is essential for efficient viral replication (26, 45). This ISRE is typically bound by IRF1 and/or IRF2, leading to activation of virus transcription (34, 46) through the recruitment of transcriptional coactivators, such as the histone acetyltransferase (HAT) p300/CBP (47). IRF1 and IRF2 are ubiquitously expressed in cells, although they can be upregulated by type I IFNs (36) and, in the case of IRF1, by TLR signaling (48, 49) and HIV-1 infection (45, 50), illustrating how HIV-1 can coopt the antiviral IFN response to augment its own replication. Once associated with the ISRE, IRF1 can cooperatively bind to both NF-κB at the HIV-1 LTR and the viral transactivator Tat at the HIV-1 TAR loop to augment viral transcription/elongation (34, 51). Our studies demonstrate that both IRF1 and IRF2 associate with the HIV-1 ISRE in unstimulated MDMs (Fig. 5). Upon stimulation with TLR4 ligands, IRF1 recruitment to the HIV-1 ISRE is enhanced (Fig. 5), consistent with the prevailing theory that TLR-MyD88 signaling can activate IRF1 (52). This is accompanied by a concomitant decrease of IRF2 binding. These data suggest that IRF1 binding to the ISRE as either monomers or homodimers activates HIV-1 expression, whereas IRF2 binding to the ISRE as monomers, homodimers, or heterodimers with IRF1 represses HIV-1 expression. Unfortunately, chromatin immunoprecipitation (ChIP) analysis of HIV-infected MDMs using current tools does not permit differentiating between the association of various homodimers and heterodimers with the ISRE at a population level.
We demonstrate that at late time points after TLR4 engagement, IRF8 is recruited to the GLS downstream from the 5′ LTR (Fig. 5) and that this is associated with decreased HIV-1 transcription (Fig. 1). Macrophages express high basal levels of IRF8, although its expression can be further enhanced in response to type I IFNs (36, 37) or TLR signaling (53, 54). IRF8 has been shown to bind to IRF1, in addition to other transcription factors, and to serve as either a transcriptional activator or a transcriptional inhibitor of other genes in a context-dependent manner (55–57). Previous studies have shown that IRF8 can repress HIV-1 expression (34, 38, 39). In fact, the interaction between IRF8 and IRF1 has been shown to repress HIV-1 transcription in Jurkat cells (34). This may be due to IRF8-mediated disruptions of the IRF1-Tat interaction and/or the IRF1-NF-κB interaction (51) that increase viral replication. Based on our data, we propose that changes in the IRF binding pattern to the ISRE in response to TLR signaling have profound effects on HIV-1 replication. In unstimulated HIV-infected macrophages, the ISRE is most likely occupied by IRF1/IRF2 heterodimers that allow for a low level of virus replication (Fig. 5D). Early after stimulation of TLR4 with LPS, there is a switch to IRF1 homodimers present at the ISRE that allow for high levels of virus replication due to cooperative binding between IRF1, NF-κB, and HIV-1 Tat (Fig. 5D). At late time points after TLR4 stimulation with LPS or GC, during the IFN feedback phase of the response, the ISRE is occupied by IRF1/IRF8 heterodimers (Fig. 5D). These IRF1/IRF8 heterodimers likely block the cooperative interaction(s) between IRF1, NF-κB, and Tat, thereby repressing HIV-1 replication. Although we also demonstrate that there is a transient decrease in IRF4 recruitment to the HIV-1 ISRE following treatment with LPS, the biological significance of this finding is uncertain. Prior studies have provided evidence for an LPS/TLR4-mediated repression of HIV-1 expression through the induction of type I IFNs and other mechanisms (16, 17, 58–62). Our data extend these findings and demonstrate that LPS treatment, as well as infection with the sexually transmitted pathogen GC or the gut-associated microbe E. coli, represses HIV-1 expression in MDMs through the TLR4-mediated, TRIF-dependent production of type I IFNs and the subsequent recruitment of IRF8 to the HIV-1 ISRE (Fig. 7). The exact mechanism whereby IRF8 is recruited to the HIV-1 GLS is not certain. This process may involve direct activation of IRF8 by TLR4 or the type I IFN receptor (IFNAR), or increased expression of IRF8 downstream of type I IFN signaling.
Importantly, our data suggest that the microbial environment can influence the state of HIV-1 replication and the establishment of latency in human macrophages as part of the viral reservoir in infected individuals under antiretroviral therapy (ART) regimens. Macrophages can be productively infected with HIV-1 in vivo, and viral replication can be modulated by copathogens through their interactions with innate immune receptors such as TLRs (18, 63). We demonstrate that productive infection of macrophages can be altered by TLR signaling in response to purified ligands and bacterial coinfection, with TLR2- and TLR5-mediated signaling activating HIV-1 and TLR3- and TLR4-mediated signaling repressing HIV-1 replication in MDMs (Fig. 1).
In addition to their role in HIV-1 production, macrophages also contribute to HIV-1 persistence in vivo. Although CD4+ memory T cells are thought to constitute the majority of the HIV-1 reservoir, several studies have demonstrated that tissue-resident macrophages in the lymph nodes (64–66), gastrointestinal tract (5, 67), genitourinary tract (2, 42, 68), liver (69–71), and lung (72–74), as well as perivascular macrophages and microglial cells in the brain (41, 75–80), can serve as tissue reservoirs for HIV-1. In simian-human immunodeficiency virus (SHIV)-infected rhesus macaques, in vivo viral replication was sustained by tissue macrophages after depletion of CD4+ T cells (81). Moreover, HIV-1 persistence in macrophages was confirmed in HIV-1-infected humanized myeloid-only mice in which viral rebound was observed in a subset of the animals following treatment interruption (43). These studies demonstrate that macrophages have the capacity to serve as bona fide HIV-1 reservoirs in vivo. Our findings that pathogenic and commensal bacteria, through engagement of TLRs, can influence HIV-1 replication in macrophages have potential clinical significance. For example, sexually transmitted infections (STIs) that induce robust type I interferon production, such as GC or HSV-2, may repress virus replication in genitourinary tract macrophages that harbor HIV-1 provirus and contribute to viral escape from the immune system and from ART.
The major obstacle to the eradication of HIV-1 is the presence of a persistent viral reservoir that can resurface upon discontinuation of ART. The potential contribution of HIV-1 in tissue macrophages to virus rebound with the cessation of ART is not entirely understood, but recent primate studies suggest that the functional macrophage reservoir can contribute to viral rebound upon treatment cessation (82–84). Our data demonstrate that interactions between macrophages and pathogenic or commensal microorganisms within the genitourinary and gastrointestinal tracts, such as GC and E. coli, may alter the ability of macrophages to serve as reservoirs for viral persistence in the host. Our findings are consistent with independent studies that demonstrate that repeated stimulation of M1-polarized MDMs with proinflammatory cytokines (TNF-α) and/or type II IFNs (interferon-γ) induce a state akin to HIV-1 latency (85). In addition, the oral pathogen Porphyromonas gingivalis has been shown to influence the establishment and maintenance of persistent HIV-1 infection in MDMs (86). Finally, studies have demonstrated that a subset of HIV-1-infected macrophages enter a state of viral latency characterized by altered metabolic signatures (87) and apoptotic mechanisms (88). Taken together, these studies demonstrate that coinfection, inflammatory stimuli, and metabolic alterations can influence the establishment and maintenance of the HIV-1 reservoir in macrophages. As an example, gastrointestinal macrophages constitute a major cellular reservoir for HIV-1 (5, 89–91) and are frequently exposed to microbes and microbial products either through luminal sampling (92) or microbial translocation, the latter of which is increased in HIV-positive individuals (93). Our data suggest that interactions such as those between intestinal macrophages and gut-associated microbes may have clinical significance for the establishment and maintenance of the latent HIV-1 reservoir.
Our results demonstrating that Neisseria gonorrhoeae and E. coli repress HIV-1 replication in macrophages by altering transcription factor recruitment to the HIV-1 GLS and induce a state of viral latency confirm the need for further in vitro, ex vivo, and in vivo studies regarding the effects of sexually transmitted pathogens and commensal microbes on HIV-1 persistence.
MATERIALS AND METHODS
Ethics statement.
This research has been determined to be exempt by the Institutional Review Boards of the Boston University Medical Center and University of Utah Health, since it does not meet the definition of human subject research.
Cell isolation and culture.
Primary human CD14+ monocytes were isolated from the peripheral blood mononuclear cells of healthy donors using anti-CD14 magnetic beads (Miltenyi Biotec) per the manufacturer's instructions. Primary monocyte-derived macrophages (MDMs) were generated by culturing CD14+ monocytes in the presence of 10% human AB serum and 10% fetal bovine serum (FBS) for 6 days. Following differentiation, MDMs were cultured in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.29 mg/ml l-glutamine. The genetic sex of a subset of the donors was determined by PCR amplification of the SRY gene located on the Y chromosome. PM1 cells were cultured in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.29 mg/ml l-glutamine. 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.29 mg/ml l-glutamine. MAGI-CCR5 cells were cultured in DMEM supplemented with 10% FBS, 100U/ml penicillin, 100 μg/ml streptomycin, 0.29 mg/ml l-glutamine, 500 μg/ml G418, 1 μg/ml puromycin, and 0.1 μg/ml hygromycin B. HEK293-TLR2CFPTLR1YFP cells and HEK293-TLR4CFP/MD-2/CD14 cells were cultured in DMEM supplemented with 10% FBS, 10 μg/ml ciprofloxacin, 0.29 mg/ml l-glutamine, and 500 μg/ml G418.
Bacterial culture.
Neisseria gonorrhoeae (GC) strain FA1090B was a generous gift from Caroline Genco. GC was cultured from a glycerol stock on GC agar plates supplemented with IsoVitalex enrichment supplement (Becton, Dickinson) in a humidified 37°C incubator with 5% CO2. E. coli strain DH5α was purchased from New England Biolabs and was cultured from a glycerol stock on LB agar plates at 37°C. Where indicated, bacteria were heat inactivated (heat killed) by incubation at 56°C for 2 h. Heat inactivation was monitored by culture on GC or LB agar plates as described above.
Flow cytometry.
TLR expression on viable MDMs was assessed 8 days after isolation using antibodies against TLR2 (clone TL2.1) and TLR4 (clone HTA125) (both from eBioscience) and eFluor 450 fixable viability dye (eBioscience). MDMs were stained in plates, washed with phosphate-buffered saline (PBS), fixed using Cytofix (BD Biosciences), and then detached after incubation in PBS supplemented with 20 mM EDTA for 1 h at 4°C. Flow cytometric data were acquired using a Becton-Dickenson FACScan II or LSRFortessa instruments, and data were analyzed using FlowJo software.
TLR ligands, interferons, and chemical inhibitors.
PAM3CSK4, FSL-1, Salmonella enterica subsp. enterica serovar Typhimurium flagellin (FLA-ST), poly(I·C), and E. coli K-12 LPS were obtained from InvivoGen. TLR ligands were reconstituted in endotoxin-free H2O. IFN-α and IFN-β were purchased from PBL InterferonSource. B18R was purchased from Abcam. BAY 11-7082, celastrol, U0126, PD95809, and SB203580 were purchased from Sigma and reconstituted in dimethyl sulfoxide (DMSO). Dynasore was purchased from Tocris Bioscience and was reconstituted in DMSO.
Virus production.
Single-round replication-defective HIV-1 reporter viruses were generated by packaging a luciferase-expressing reporter virus, BruΔEnvLuc2, or an enhanced green fluorescent protein (GFP)-expressing reporter virus, BruΔEnvEGFP3, with the envelope glycoproteins from VSV (VSV-G). In these constructs, reporter gene expression is under the control of the 5′ LTR. Reporter virus stocks were generated by transfecting HEK293T cells using the calcium phosphate method as described previously (18). Replication competent HIV-1Ba-L was generated by infection of PM1 cells as described previously (18). Virus titers were determined using MAGI-CCR5 cells, and p24gag content was determined by enzyme-limited immunosorbent assay (ELISA) as described previously (18).
Virus infections.
To assess viral replication, macrophages (2.5 × 105 cells/well in 24-well plates) were incubated with VSV-G-pseudotyped HIV-luciferase reporter virus at a multiplicity of infection (MOI) of 0.1 for 4 h at 37°C. Cells were washed four to five times with PBS to remove unbound virus, and then cultured in growth medium. Following 48 h of culture, cells were treated with TLR ligands or vehicle, as indicated in the text and figure legends. After 18 h, the cells were washed twice with PBS and lysed in PBS-0.02% Triton X-100. Luciferase activity was measured using BrightGlo luciferase reagent (Promega) and an MSII luminometer.
HIV-1 transcription.
Total cytoplasmic RNA was isolated from MDMs using the RNeasy minikit (Qiagen). RNA (100 ng) was analyzed by reverse transcription-PCR (RT-PCR) using the OneStep RT-PCR kit (Qiagen). RNA was reverse transcribed and amplified in a total volume of 50 μl containing 2.5 mM MgCl2, 400 μM concentrations of each deoxynucleoside triphosphate, 10 U of RNasin RNase inhibitor (Promega), 5 μCi of α-32P dATP, and 0.6 μM HIV-1-specific primers. RNA samples were reverse transcribed for 30 min at 50°C. After an initial denaturing step at 95°C for 15 min, cDNA products were amplified for 25 cycles, each consisting of a 30-s denaturing step at 94°C, a 45-s annealing step at 65°C, and a 1-min extension step at 72°C. The amplification concluded with a 10-minute extension step at 72°C. Samples were resolved on 5% nondenaturing polyacrylamide gels, visualized by autoradiography, and quantified in a Molecular Dynamics PhosphorImager SI using ImageQuant software. Alternatively, HIV-1 RNA was analyzed using the QuantiTect SYBR green RT-PCR kit (Qiagen) in a LightCycler 480 (Roche). The HIV-1 primers were specific for the R and U5 regions of the LTR and amplify both spliced mRNAs and genomic RNA. The HIV-1 primers were sense primer 5′-GGCTAACTAGGGAACCCACTGC-3′ and antisense primer 5′-CTGCTAGAGATTTTCCACACTGAC-3′. α-Tubulin primers were sense primer 5′-CACCCGTCTTCAGGGCTTCTTGGTTT-3′ and antisense primer 5′-CATTTCACCATCTGGTTGGCTGGCTC-3′. RNA standards corresponding to 500, 50, and 5 ng of RNA from PAM3CSK4-activated MDMs were included in each experiment to ensure that all amplifications were within the linear range of the assay.
HIV-1 RNA stability assays.
MDMs (2 × 106 cells/well in 6-well plates) were incubated with VSV-G-pseudotyped HIV-luciferase reporter virus at an MOI of 0.1 for 4 h at 37°C. Cells were washed 4 to 5 times with PBS to remove unbound virus and cultured in growth medium. Following 48 h of culture, cells were treated with TLR ligands (PAM3CSK4 or LPS at 100 ng/ml) or vehicle for 4 h. Actinomycin D (10 μg/ml) was then added to cells to block de novo RNA synthesis, and total cytoplasmic RNA was isolated at given times as described in the figure legends. Viral RNA was analyzed using the QuantiTect SYBR green RT-PCR kit (Qiagen) in a LightCycler 480 (Roche) with primers specific for the R and U5 regions of the LTR as described above.
Cytokine release assays.
MDMs (2.5 × 105 cells/well) were treated with PAM3CSK4 (100 ng/ml), LPS (100 ng/ml), or GC (MOI of 10) for 24 h. Cell-free culture supernatants were collected and analyzed for TNF-α (eBioscience) or IFN-β (PBL Interferon Source) release by commercially available ELISA following the manufacturer's instructions.
Chromatin immunoprecipitation assays.
MDMs (1.2 × 107) were incubated with VSV-G-pseudotyped HIV-enhanced GFP (EGFP) reporter virus at an MOI of 2 for 4 h at 37°C. Cells were washed 4 to 5 times with PBS to remove unbound virus and cultured in growth medium. Following 48 h of culture, MDMs were treated with TLR ligands for various times, as described in the text. Cells were then fixed in 1% formaldehyde for 10 min at room temperature, quenched with 125 mM glycine, and lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml aprotinin, and 1 μg/ml pepstatin A). Cellular lysates were sonicated using a cup horn (550 sonic dismembrator; Fisher Scientific) at a power setting of 5 with 25 20-s pulses on ice, which fragmented the chromatin to an average length of approximately 1,000 bp. Samples were diluted and immunoprecipitated with antibodies against NF-κB p65, IRF1, IRF2, IRF4, IRF8, rabbit IgG, or goat IgG (all from Santa Cruz Biotechnology). Purified DNA samples from both ChIPs and input controls were resuspended in distilled H2O and analyzed by semiquantitative PCR. PCR mixtures contained 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 1.5 mM MgCl2; 100 pmol of each primer; 200 μM each dATP, dGTP, dCTP, and dTTP; 5 μCi α32P-dATP; and 2.5 units of AmpliTaq Gold (Applied Biosystems) in a 50-μl reaction volume. Following an initial denaturation step at 95°C for 15 min, DNA was amplified for 30 cycles, each consisting of a 30-s denaturing step at 94°C, a 45-s annealing step at 65°C, and a 1-min extension step at 72°C. Samples were electrophoresed on 5% nondenaturing polyacrylamide gels, visualized by autoradiography, and quantified using a Molecular Dynamics PhosphorImager SI using ImageQuant software. Alternatively, purified DNA from ChIPs and input controls were analyzed using the PowerUp SYBR green mastermix (Applied Biosystems) in a LightCycler 480 (Roche). The primers used to amplify specifically the HIV-1 5′ LTR and GLS were 5′-TGGAAGGGCTAATTTACTCCC-3′ (sense) and 5′-CATCTCTCTCCTTCTAGCCTC-3′ (antisense). Control amplifications of a serial dilution of purified genomic DNA from latently infected U1 cells were performed with each primer set to ensure that all amplifications were within the linear range of the reaction. To calculate the relative levels of association with the LTR, PhosphorImager data of the PCR products obtained for immunoprecipitated chromatin samples were normalized against the PCR products obtained for input DNA (% input). Values were normalized across donors and expressed as relative binding.
LTR mutant construction.
The reported plasmid pLTR(Sp1)-luciferase was generated by PCR amplification of pNL4-3 using the sense primer 5′-CGGGGTACCCCGTGGAAGGGCTAATTTGGTCCC-3′ and the antisense primer 5′-CCGCTCGAGCGGCATCTCTCTCCTTCTAGCCTC-3′, digestion with KpnI and XhoI, and ligation into KpnI/XhoI-digested pGL3-Basic (Promega). Mutations to the NF-κB and IRF binding sites in pLTR(Sp1)-luciferase were generated using the QuikChange IIXL site-directed mutagenesis kit (Stratagene). Primers used for site-directed mutagenesis are listed in Table 1. The −158 LTR-luciferase construct was generated by deleting the LTR sequence upstream of position −158 (relative to the start site of transcription) of pNL4-3, which includes the AP-1 binding sites located in the U3 portion of the 5′ LTR, digestion of the resulting fragment with KpnI and XhoI, and ligation into KpnI/XhoI-digested pGL3-Basic (Promega).
TABLE 1.
Primers used for PCR-based mutagenesis
| Primer name | Sequence |
|---|---|
| Forward mutI NF-κB | GGACTTTCCGCTGTCTACTTTCCAGG |
| Reverse mutI NF-κB | CCTGGAAAGTAGACAGCGGAAAGTCC |
| Forward mutII NF-κB | GCTTTCTACAATCTACTTTCCGCTGG |
| Reverse mutII NF-κB | CCAGCGGAAAGTAGATTGTAGAAAGC |
| Forward mutI&II NF-κB | GCTTTCTACAATCTACTTTCCGCTGTCTACTTTCCAGG |
| Reverse mutI&II NF-κB | CCTGGAAAGTAGACAGCGGAAAGTAGATTGTAGAAAGC |
| Forward delNF-κB | GCTGACATCGAGCTTTCTACAAAGGGAGGTGTGGCCTGGGCGGG |
| Reverse delNF-κB | CCCGCCCAGGCCACACCTCCCTTTGTAGAAAGCTCGATGTCAGC |
| Forward mutISRE | GCCCGAACAGGGACTTGCCCGCGCCCGTAAAGCCAGAGGAGATC |
| Reverse mutISRE | GATCTCCTCTGGCTTTACGGGCGCGGGCAAGTCCCTGTTCGGGC |
shRNA knockdown of MyD88, TRIF, and IRF8.
MDMs (1.2 × 107) were transfected with plasmids that encoded either a mixture of three to five shRNAs directed against MyD88, a mixture of three to five shRNAs directed against TRIF, or a mixture of three to five control shRNAs (InvivoGen) and a blasticidin resistance gene using Oligofectamine (Invitrogen) per the manufacturer’s instructions. Transfected cells were selected by culture in the presence of blasticidin for 48 h, and either used in HIV-1 replication assays or lysed for immunoblot analysis to measure MyD88 and TRIF expression using a rabbit monoclonal antibody to MyD88 (Cell Signaling Technology), a rabbit polyclonal antibody to TRIF (Cell Signaling Technology), or a mouse monoclonal antibody to β-actin (Sigma). Similarly, MDMs were transfected with plasmids that encoded either a mixture of three to five shRNAs directed against IRF8 (Sigma) or a mixture of control shRNAs (Sigma) and a puromycin resistance gene using Oligofectamine (Invitrogen) per the manufacturer’s instructions. Transfected cells were selected by culture in the presence of puromycin for 48 h and either used in HIV-1 replication assays or lysed for immunoblot analysis to measure IRF8 expression using a rabbit monoclonal antibody (Cell Signaling Technology).
Overexpression of IRF8.
MDMs (1.2 × 107) were transfected with a plasmid that encoded IRF8 (OriGene) and a neomycin resistance gene using Oligofectamine (Invitrogen) per the manufacturer’s instructions. Transfected cells were selected by culture in the presence of neomycin for 48 h and then used for HIV-1 replication assays or lysed for immunoblot analysis to measure IRF8 expression using a rabbit monoclonal antibody (Cell Signaling Technology).
Endocytosis/phagocytosis assays.
MDMs (5 × 105/well) were treated with Dynasore (80 μM) or DMSO and then incubated with pHrodo green E. coli particles (Thermo Fisher) at 1 mg/ml for 2 h at 37°C. The MDMs were then washed three times with PBS, incubated with eFluor 450 fixable viability dye (eBioscience) for 15 min at 4°C, and analyzed by flow cytometry. Flow cytometric data were acquired using a Becton-Dickenson LSRFortessa instrument, and data were analyzed using FlowJo software.
Viability assays.
Viability of uninfected and HIV-1-infected MDMs was monitored over time (longitudinal) and at the end of culture (endpoint) using the CytoTox-One homogeneous membrane integrity assay (Promega) per the manufacturer’s instructions.
Statistical analysis.
Comparison between experimental samples was performed with a paired two-tailed t test, with P < 0.5 denoting significant differences. Experiments were performed in triplicate using cells from a minimum of four independent donors (unless otherwise indicated) to control for interdonor variability.
ACKNOWLEDGMENTS
We gratefully acknowledge Laura Dickey for her scientific and editorial suggestions. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV IG from NABI and the National Heart, Lung, and Blood Institute (Luiz Barbosa), anti-p24gag monoclonal antibody clone 183-H12-5C from Bruce Chesebro and Kathy Wehrly, and MAGI-CCR5 cells from Julie Overbaugh.
This work was supported by funds obtained from National Institutes of Health (www.nih.gov) grants AI073149, (G.A.V.), AI143567-02 (V.P.), T32-AI07309 (T.M.H.), and T32-AI0764206 (T.M.H.); a Boston University Division of Graduate Medical Sciences Graduate Student Research Fellowship award (T.M.H.); and a Resident Research Award from the Department of Pathology at the University of Utah (T.M.H.).
We have no conflicts of interest.
T.M.H. and G.A.V. designed the study. T.M.H. and G.A.V. developed the methodology. T.M.H. conducted experiments. T.M.H., V.P., and G.A.V. wrote the manuscript. T.M.H., V.P., and G.A.V. acquired funds. T.M.H. and G.A.V. supervised the study.
REFERENCES
- 1.Miller CJ, Shattock RJ. 2003. Target cells in vaginal HIV transmission. Microbes Infect 5:59–67. doi: 10.1016/s1286-4579(02)00056-4. [DOI] [PubMed] [Google Scholar]
- 2.Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, Sankaran-Walters S, Dandekar S, Clapham PR, Smythies LE, Smith PD. 2009. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J Virol 83:3258–3267. doi: 10.1128/JVI.01796-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElrath MJ, Smythe K, Randolph-Habecker J, Melton KR, Goodpaster TA, Hughes SM, Mack M, Sato A, Diaz G, Steinbach G, Novak RM, Curlin ME, Curlin M, Lord JD, Maenza J, Duerr A, Frahm N, Hladik F, NIAID HIV Vaccine Trials Network. 2013. Comprehensive assessment of HIV target cells in the distal human gut suggests increasing HIV susceptibility toward the anus. J Acquir Immune Defic Syndr 63:263–271. doi: 10.1097/QAI.0b013e3182898392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen R, Richter HE, Smith PD. 2011. Early HIV-1 target cells in human vaginal and ectocervical mucosa. Am J Reprod Immunol 65:261–267. doi: 10.1111/j.1600-0897.2010.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zalar A, Figueroa MI, Ruibal-Ares B, Bare P, Cahn P, de Bracco MM, Belmonte L. 2010. Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antiviral Res 87:269–271. doi: 10.1016/j.antiviral.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Wong ME, Jaworowski A, Hearps AC. 2019. The HIV reservoir in monocytes and macrophages. Front Immunol 10:1435. doi: 10.3389/fimmu.2019.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galvin SR, Cohen MS. 2004. The role of sexually transmitted diseases in HIV transmission. Nat Rev Microbiol 2:33–42. doi: 10.1038/nrmicro794. [DOI] [PubMed] [Google Scholar]
- 8.Malott RJ, Keller BO, Gaudet RG, McCaw SE, Lai CC, Dobson-Belaire WN, Hobbs JL, St Michael F, Cox AD, Moraes TF, Gray-Owen SD. 2013. Neisseria gonorrhoeae-derived heptose elicits an innate immune response and drives HIV-1 expression. Proc Natl Acad Sci U S A 110:10234–10239. doi: 10.1073/pnas.1303738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvis GA, Chang TL. 2012. Modulation of HIV transmission by Neisseria gonorrhoeae: molecular and immunological aspects. Curr HIV Res 10:211–217. doi: 10.2174/157016212800618138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira VH, Nazli A, Khan G, Mian MF, Ashkar AA, Gray-Owen S, Kaul R, Kaushic C. 2011. Endometrial epithelial cell responses to coinfecting viral and bacterial pathogens in the genital tract can activate the HIV-1 LTR in an NFκB-and AP-1-dependent manner. J Infect Dis 204:299–308. doi: 10.1093/infdis/jir260. [DOI] [PubMed] [Google Scholar]
- 11.Ding J, Rapista A, Teleshova N, Mosoyan G, Jarvis GA, Klotman ME, Chang TL. 2010. Neisseria gonorrhoeae enhances HIV-1 infection of primary resting CD4+ T cells through TLR2 activation. J Immunol 184:2814–2824. doi: 10.4049/jimmunol.0902125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Li G, Bafica A, Pantelic M, Zhang P, Broxmeyer H, Liu Y, Wetzler L, He JJ, Chen T. 2005. Neisseria gonorrhoeae enhances infection of dendritic cells by HIV type 1. J Immunol 174:7995–8002. doi: 10.4049/jimmunol.174.12.7995. [DOI] [PubMed] [Google Scholar]
- 13.Sanyal A, Shen C, Ding M, Reinhart TA, Chen Y, Sankapal S, Gupta P. 2019. Neisseria gonorrhoeae uses cellular proteins CXCL10 and IL8 to enhance HIV-1 transmission across cervical mucosa. Am J Reprod Immunol 81:e13111. doi: 10.1111/aji.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogensen TH, Paludan SR, Kilian M, Ostergaard L. 2006. Live Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis activate the inflammatory response through Toll-like receptors 2, 4, and 9 in species-specific patterns. J Leukoc Biol 80:267–277. doi: 10.1189/jlb.1105626. [DOI] [PubMed] [Google Scholar]
- 15.Dobson-Belaire WN, Rebbapragada A, Malott RJ, Yue FY, Kovacs C, Kaul R, Ostrowski MA, Gray-Owen SD. 2010. Neisseria gonorrhoeae effectively blocks HIV-1 replication by eliciting a potent TLR9-dependent interferon-alpha response from plasmacytoid dendritic cells. Cell Microbiol 12:1703–1717. doi: 10.1111/j.1462-5822.2010.01502.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Mosoian A, Li-Yun Chang T, Zerhouni-Layachi B, Snyder A, Jarvis GA, Klotman ME. 2006. Gonococcal lipooligosaccharide suppresses HIV infection in human primary macrophages through induction of innate immunity. J Infect Dis 194:751–759. doi: 10.1086/506360. [DOI] [PubMed] [Google Scholar]
- 17.Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. 1989. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med 169:1137–1151. doi: 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanley TM, Viglianti GA. 2011. Nuclear receptor signaling inhibits HIV-1 replication in macrophages through multiple trans-repression mechanisms. J Virol 85:10834–10850. doi: 10.1128/JVI.00789-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novis CL, Archin NM, Buzon MJ, Verdin E, Round JL, Lichterfeld M, Margolis DM, Planelles V, Bosque A. 2013. Reactivation of latent HIV-1 in central memory CD4+ T cells through TLR-1/2 stimulation. Retrovirology 10:119. doi: 10.1186/1742-4690-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szaniawski MA, Spivak AM, Bosque A, Planelles V. 2019. Sex influences SAMHD1 activity and susceptibility to human immunodeficiency virus-1 in primary human macrophages. J Infect Dis 219:777–785. doi: 10.1093/infdis/jiy583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler S, Altfeld M. 2016. Sex differences in HIV-1-mediated immunopathology. Curr Opin HIV AIDS 11:209–215. doi: 10.1097/COH.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Garcia M, Biswas N, Patel MV, Barr FD, Crist SG, Ochsenbauer C, Fahey JV, Wira CR. 2013. Estradiol reduces susceptibility of CD4+ T cells and macrophages to HIV-infection. PLoS One 8:e62069. doi: 10.1371/journal.pone.0062069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai T, Akira S. 2006. TLR signaling. Cell Death Differ 13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 24.Nabel G, Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 25.Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ. 1993. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J 12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Lint C, Amella CA, Emiliani S, John M, Jie T, Verdin E. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol 71:6113–6127. doi: 10.1128/JVI.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canonne-Hergaux F, Aunis D, Schaeffer E. 1995. Interactions of the transcription factor AP-1 with the long terminal repeat of different human immunodeficiency virus type 1 strains in Jurkat, glial, and neuronal cells. J Virol 69:6634–6642. doi: 10.1128/JVI.69.11.6634-6642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. 1997. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem 272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Koo TH, Yoon H, Jung HS, Jin HZ, Lee K, Hong YS, Lee JJ. 2006. Inhibition of NF-κB activation through targeting IκB kinase by celastrol, a quinone methide triterpenoid. Biochem Pharmacol 72:1311–1321. doi: 10.1016/j.bcp.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Ono Y, Maejima Y, Saito M, Sakamoto K, Horita S, Shimomura K, Inoue S, Kotani J. 2020. TAK-242, a specific inhibitor of Toll-like receptor 4 signalling, prevents endotoxemia-induced skeletal muscle wasting in mice. Sci Rep 10:694. doi: 10.1038/s41598-020-57714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol 9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasr N, Alshehri AA, Wright TK, Shahid M, Heiner BM, Harman AN, Botting RA, Helbig KJ, Beard MR, Suzuki K, Kelleher AD, Hertzog P, Cunningham AL. 2017. Mechanism of interferon-stimulated gene induction in HIV-1-infected macrophages. J Virol 91. doi: 10.1128/JVI.00744-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheikh F, Dickensheets H, Gamero AM, Vogel SN, Donnelly RP. 2014. An essential role for IFN-β in the induction of IFN-stimulated gene expression by LPS in macrophages. J Leukoc Biol 96:591–600. doi: 10.1189/jlb.2A0414-191R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sgarbanti M, Borsetti A, Moscufo N, Bellocchi MC, Ridolfi B, Nappi F, Marsili G, Marziali G, Coccia EM, Ensoli B, Battistini A. 2002. Modulation of human immunodeficiency virus 1 replication by interferon regulatory factors. J Exp Med 195:1359–1370. doi: 10.1084/jem.20010753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto M, Kato T, Hotta C, Nishiyama A, Kurotaki D, Yoshinari M, Takami M, Ichino M, Nakazawa M, Matsuyama T, Kamijo R, Kitagawa S, Ozato K, Tamura T. 2011. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PLoS One 6:e25812. doi: 10.1371/journal.pone.0025812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barber SA, Fultz MJ, Salkowski CA, Vogel SN. 1995. Differential expression of interferon regulatory factor 1 (IRF-1), IRF-2, and interferon consensus sequence binding protein genes in lipopolysaccharide (LPS)-responsive and LPS-hyporesponsive macrophages. Infect Immun 63:601–608. doi: 10.1128/IAI.63.2.601-608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta M, Shin DM, Ramakrishna L, Goussetis DJ, Platanias LC, Xiong H, Morse HC, 3rd, Ozato K. 2015. IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes. Nat Commun 6:6379. doi: 10.1038/ncomms7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornton AM, Buller RM, DeVico AL, Wang IM, Ozato K. 1996. Inhibition of human immunodeficiency virus type 1 and vaccinia virus infection by a dominant negative factor of the interferon regulatory factor family expressed in monocytic cells. Proc Natl Acad Sci U S A 93:383–387. doi: 10.1073/pnas.93.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munier S, Delcroix-Genete D, Carthagena L, Gumez A, Hazan U. 2005. Characterization of two candidate genes, NCoA3 and IRF8, potentially involved in the control of HIV-1 latency. Retrovirology 2:73. doi: 10.1186/1742-4690-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. 2005. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci U S A 102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abreu C, Shirk EN, Queen SE, Beck SE, Mangus LM, Pate KAM, Mankowski JL, Gama L, Clements JE. 2019. Brain macrophages harbor latent, infectious simian immunodeficiency virus. Aids 33(Suppl 2):S181–S188. doi: 10.1097/QAD.0000000000002269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganor Y, Real F, Sennepin A, Dutertre CA, Prevedel L, Xu L, Tudor D, Charmeteau B, Couedel-Courteille A, Marion S, Zenak AR, Jourdain JP, Zhou Z, Schmitt A, Capron C, Eugenin EA, Cheynier R, Revol M, Cristofari S, Hosmalin A, Bomsel M. 2019. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol 4:633–644. doi: 10.1038/s41564-018-0335-z. [DOI] [PubMed] [Google Scholar]
- 43.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, Garcia JV. 2017. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 23:638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith PD, Meng G, Salazar-Gonzalez JF, Shaw GM. 2003. Macrophage HIV-1 infection and the gastrointestinal tract reservoir. J Leukoc Biol 74:642–649. doi: 10.1189/jlb.0503219. [DOI] [PubMed] [Google Scholar]
- 45.Harman AN, Lai J, Turville S, Samarajiwa S, Gray L, Marsden V, Mercier SK, Mercier S, Jones K, Nasr N, Rustagi A, Cumming H, Donaghy H, Mak J, Gale M, Churchill M, Hertzog P, Cunningham AL. 2011. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 118:298–308. doi: 10.1182/blood-2010-07-297721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsili G, Borsetti A, Sgarbanti M, Remoli AL, Ridolfi B, Stellacci E, Ensoli B, Battistini A. 2003. On the role of interferon regulatory factors in HIV-1 replication. Ann N Y Acad Sci 1010:29–42. doi: 10.1196/annals.1299.005. [DOI] [PubMed] [Google Scholar]
- 47.Marsili G, Remoli AL, Sgarbanti M, Battistini A. 2004. Role of acetylases and deacetylase inhibitors in IRF-1-mediated HIV-1 long terminal repeat transcription. Ann N Y Acad Sci 1030:636–643. doi: 10.1196/annals.1329.074. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y, Park-Min KH, Yarilina A, Ivashkiv LB. 2008. Regulation of STAT pathways and IRF1 during human dendritic cell maturation by TNF-α and PGE2. J Leukoc Biol 84:1353–1360. doi: 10.1189/jlb.0107040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liljeroos M, Vuolteenaho R, Rounioja S, Henriques-Normark B, Hallman M, Ojaniemi M. 2008. Bacterial ligand of TLR2 signals Stat activation via induction of IRF1/2 and interferon-alpha production. Cell Signal 20:1873–1881. doi: 10.1016/j.cellsig.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 50.Sgarbanti M, Marsili G, Remoli AL, Ridolfi B, Stellacci E, Borsetti A, Ensoli B, Battistini A. 2004. Analysis of the signal transduction pathway leading to human immunodeficiency virus-1-induced interferon regulatory factor-1 upregulation. Ann N Y Acad Sci 1030:187–195. doi: 10.1196/annals.1329.024. [DOI] [PubMed] [Google Scholar]
- 51.Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, Orsatti R, Ilari R, Sernicola L, Stellacci E, Ensoli B, Battistini A. 2008. IRF-1 is required for full NF-κB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J Virol 82:3632–3641. doi: 10.1128/JVI.00599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, Takayanagi H, Ohba Y, Taniguchi T, Honda K. 2006. Evidence for licensing of IFN-γ-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci U S A 103:15136–15141. doi: 10.1073/pnas.0607181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura T, Ozato K. 2002. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res 22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J, Kong HJ, Li H, Huang B, Yang M, Zhu C, Bogunovic M, Zheng F, Mayer L, Ozato K, Unkeless J, Xiong H. 2006. IRF-8/interferon (IFN) consensus sequence-binding protein is involved in Toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-γ signaling pathways. J Biol Chem 281:10073–10080. doi: 10.1074/jbc.M507788200. [DOI] [PubMed] [Google Scholar]
- 55.Bovolenta C, Driggers PH, Marks MS, Medin JA, Politis AD, Vogel SN, Levy DE, Sakaguchi K, Appella E, Coligan JE. 1994. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci U S A 91:5046–5050. doi: 10.1073/pnas.91.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meraro D, Hashmueli S, Koren B, Azriel A, Oumard A, Kirchhoff S, Hauser H, Nagulapalli S, Atchison ML, Levi BZ. 1999. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J Immunol 163:6468–6478. [PubMed] [Google Scholar]
- 57.Langlais D, Barreiro LB, Gros P. 2016. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med 213:585–603. doi: 10.1084/jem.20151764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmed N, Hayashi T, Hasegawa A, Furukawa H, Okamura N, Chida T, Masuda T, Kannagi M. 2010. Suppression of human immunodeficiency virus type 1 replication in macrophages by commensal bacteria preferentially stimulating Toll-like receptor 4. J Gen Virol 91:2804–2813. doi: 10.1099/vir.0.022442-0. [DOI] [PubMed] [Google Scholar]
- 59.Bernstein MS, Tong-Starksen SE, Locksley RM. 1991. Activation of human monocyte–derived macrophages with lipopolysaccharide decreases human immunodeficiency virus replication in vitro at the level of gene expression. J Clin Invest 88:540–545. doi: 10.1172/JCI115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Equils O, Salehi KK, Cornateanu R, Cornataeanu R, Lu D, Singh S, Whittaker K, Baldwin GC. 2006. Repeated lipopolysaccharide (LPS) exposure inhibits HIV replication in primary human macrophages. Microbes Infect 8:2469–2476. doi: 10.1016/j.micinf.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Simard S, Maurais E, Gilbert C, Tremblay MJ. 2008. LPS reduces HIV-1 replication in primary human macrophages partly through an endogenous production of type I interferons. Clin Immunol 127:198–205. doi: 10.1016/j.clim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 62.Nordone SK, Ignacio GA, Su L, Sempowski GD, Golenbock DT, Li L, Dean GA. 2007. Failure of TLR4-driven NF-κB activation to stimulate virus replication in models of HIV type 1 activation. AIDS Res Hum Retroviruses 23:1387–1395. doi: 10.1089/aid.2007.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Appelberg KS, Wallet MA, Taylor JP, Cash MN, Sleasman JW, Goodenow MM. 2017. HIV-1 infection primes macrophages through STAT signaling to promote enhanced inflammation and viral replication. AIDS Res Hum Retroviruses 33:690–702. doi: 10.1089/AID.2016.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Embretson J, Zupancic M, Ribas JL, Burke A, Racz P, Tenner-Racz K, Haase AT. 1993. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 65.Orenstein JM, Fox C, Wahl SM. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 66.DiNapoli SR, Ortiz AM, Wu F, Matsuda K, Twigg HL, 3rd, Hirsch VM, Knox K, Brenchley JM. 2017. Tissue-resident macrophages can contain replication-competent virus in antiretroviral-naive, SIV-infected Asian macaques. JCI Insight 2:e91214. doi: 10.1172/jci.insight.91214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yukl SA, Sinclair E, Somsouk M, Hunt PW, Epling L, Killian M, Girling V, Li P, Havlir DV, Deeks SG, Wong JK, Hatano H. 2014. A comparison of methods for measuring rectal HIV levels suggests that HIV DNA resides in cells other than CD4+ T cells, including myeloid cells. AIDS 28:439–442. doi: 10.1097/QAD.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganor Y, Zhou Z, Bodo J, Tudor D, Leibowitch J, Mathez D, Schmitt A, Vacher-Lavenu MC, Revol M, Bomsel M. 2013. The adult penile urethra is a novel entry site for HIV-1 that preferentially targets resident urethral macrophages. Mucosal Immunol 6:776–786. doi: 10.1038/mi.2012.116. [DOI] [PubMed] [Google Scholar]
- 69.Schmitt MP, Gendrault JL, Schweitzer C, Steffan AM, Beyer C, Royer C, Jaeck D, Pasquali JL, Kirn A, Aubertin AM. 1990. Permissivity of primary cultures of human Kupffer cells for HIV-1. AIDS Res Hum Retroviruses 6:987–991. doi: 10.1089/aid.1990.6.987. [DOI] [PubMed] [Google Scholar]
- 70.Hufert FT, Schmitz J, Schreiber M, Schmitz H, Racz P, von Laer DD. 1993. Human Kupffer cells infected with HIV-1 in vivo. J Acquir Immune Defic Syndr (1988) 6:772–777. [PubMed] [Google Scholar]
- 71.Mosoian A, Zhang L, Hong F, Cunyat F, Rahman A, Bhalla R, Panchal A, Saiman Y, Fiel MI, Florman S, Roayaie S, Schwartz M, Branch A, Stevenson M, Bansal MB. 2017. Frontline Science: HIV infection of Kupffer cells results in an amplified proinflammatory response to LPS. J Leukoc Biol 101:1083–1090. doi: 10.1189/jlb.3HI0516-242R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sierra-Madero JG, Toossi Z, Hom DL, Finegan CK, Hoenig E, Rich EA. 1994. Relationship between load of virus in alveolar macrophages from human immunodeficiency virus type 1-infected persons, production of cytokines, and clinical status. J Infect Dis 169:18–27. doi: 10.1093/infdis/169.1.18. [DOI] [PubMed] [Google Scholar]
- 73.Jambo KC, Banda DH, Kankwatira AM, Sukumar N, Allain TJ, Heyderman RS, Russell DG, Mwandumba HC. 2014. Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol 7:1116–1126. doi: 10.1038/mi.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cribbs SK, Lennox J, Caliendo AM, Brown LA, Guidot DM. 2015. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses 31:64–70. doi: 10.1089/AID.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stoler MH, Eskin TA, Benn S, Angerer RC, Angerer LM. 1986. Human T-cell lymphotropic virus type III infection of the central nervous system. A preliminary in situ analysis. JAMA 256:2360–2364. doi: 10.1001/jama.1986.03380170076022. [DOI] [PubMed] [Google Scholar]
- 76.Wiley CA, Schrier RD, Nelson JA, Lampert PW, Oldstone MB. 1986. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci U S A 83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fischer-Smith T, Croul S, Sverstiuk AE, Capini C, L'Heureux D, Regulier EG, Richardson MW, Amini S, Morgello S, Khalili K, Rappaport J. 2001. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J Neurovirol 7:528–541. doi: 10.1080/135502801753248114. [DOI] [PubMed] [Google Scholar]
- 78.Cosenza MA, Zhao ML, Si Q, Lee SC. 2002. Human brain parenchymal microglia express CD14 and CD45 and are productively infected by HIV-1 in HIV-1 encephalitis. Brain Pathol 12:442–455. doi: 10.1111/j.1750-3639.2002.tb00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ko A, Kang G, Hattler JB, Galadima HI, Zhang J, Li Q, Kim WK. 2019. Macrophages but not astrocytes harbor HIV DNA in the brains of HIV-1-infected aviremic individuals on suppressive antiretroviral therapy. J Neuroimmune Pharmacol 14:110–119. doi: 10.1007/s11481-018-9809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Micci L, Alvarez X, Iriele RI, Ortiz AM, Ryan ES, McGary CS, Deleage C, McAtee BB, He T, Apetrei C, Easley K, Pahwa S, Collman RG, Derdeyn CA, Davenport MP, Estes JD, Silvestri G, Lackner AA, Paiardini M. 2014. CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog 10:e1004467. doi: 10.1371/journal.ppat.1004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A 98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Parrilla DR, Ferreira EA, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Mac Gabhann F, O’Connor SL, Gama L, Clements JE. 2019. Myeloid and CD4 T cells comprise the latent reservoir in antiretroviral therapy-suppressed SIVmac251-infected macaques. mBio 10:e01659-19. doi: 10.1128/mBio.01659-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, Bullock BT, Li M, Metcalf Pate KA, Beck SE, Mangus LM, Mankowski JL, Clements JE, Gama L. 2019. Infectious virus persists in CD4+ T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 93:e00065-19. doi: 10.1128/JVI.00065-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andrade VM, Mavian C, Babic D, Cordeiro T, Sharkey M, Barrios L, Brander C, Martinez-Picado J, Dalmau J, Llano A, Li JZ, Jacobson J, Lavine CL, Seaman MS, Salemi M, Stevenson M. 2020. A minor population of macrophage-tropic HIV-1 variants is identified in recrudescing viremia following analytic treatment interruption. Proc Natl Acad Sci U S A 117:9981–9990. doi: 10.1073/pnas.1917034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graziano F, Aimola G, Forlani G, Turrini F, Accolla RS, Vicenzi E, Poli G. 2018. Reversible human immunodeficiency virus type-1 latency in primary human monocyte-derived macrophages induced by sustained M1 polarization. Sci Rep 8:14249. doi: 10.1038/s41598-018-32451-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agosto LM, Hirnet JB, Michaels DH, Shaik-Dasthagirisaheb YB, Gibson FC, Viglianti G, Henderson AJ. 2016. Porphyromonas gingivalis-mediated signaling through TLR4 mediates persistent HIV infection of primary macrophages. Virology 499:72–81. doi: 10.1016/j.virol.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Castellano P, Prevedel L, Valdebenito S, Eugenin EA. 2019. HIV infection and latency induce a unique metabolic signature in human macrophages. Sci Rep 9:3941. doi: 10.1038/s41598-019-39898-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castellano P, Prevedel L, Eugenin EA. 2017. HIV-infected macrophages and microglia that survive acute infection become viral reservoirs by a mechanism involving Bim. Sci Rep 7:12866. doi: 10.1038/s41598-017-12758-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, Shao W, Hunt PW, Somsouk M, Wylie W, Douek DC, Loeb L, Custer J, Hoh R, Poole L, Deeks SG, Hecht F, Palmer S. 2013. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 110:E4987–96. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moore AC, Bixler SL, Lewis MG, Verthelyi D, Mattapallil JJ. 2012. Mucosal and peripheral Lin− HLA-DR+ CD11c/123− CD13+ CD14− mononuclear cells are preferentially infected during acute simian immunodeficiency virus infection. J Virol 86:1069–1078. doi: 10.1128/JVI.06372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yukl SA, Shergill AK, Ho T, Killian M, Girling V, Epling L, Li P, Wong LK, Crouch P, Deeks SG, Havlir DV, McQuaid K, Sinclair E, Wong JK. 2013. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J Infect Dis 208:1212–1220. doi: 10.1093/infdis/jit308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. 2009. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med 206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]