In the present study, we experimentally infected pythons and boas with reptarenavirus via either intraperitoneal injection or tracheal instillation. The aims were to experimentally induce boid inclusion body disease (BIBD) and to develop an animal model for studying disease transmission and pathogenesis.
KEYWORDS: experimental infection, immune response, reptarenavirus
ABSTRACT
Boid inclusion body disease (BIBD) causes losses in captive snake populations globally. BIBD is associated with the formation of cytoplasmic inclusion bodies (IBs), which mainly comprise reptarenavirus nucleoprotein (NP). In 2017, BIBD was reproduced by cardiac injection of boas and pythons with reptarenaviruses, thus demonstrating a causative link between reptarenavirus infection and the disease. Here, we report experimental infections of Python regius (n = 16) and Boa constrictor (n = 16) with three reptarenavirus isolates. First, we used pythons (n = 8) to test two virus delivery routes: intraperitoneal injection and tracheal instillation. Viral RNAs but no IBs were detected in brains and lungs at 2 weeks postinoculation. Next, we inoculated pythons (n = 8) via the trachea. During the 4 months following infection, snakes showed transient central nervous system (CNS) signs but lacked detectable IBs at the time of euthanasia. One of the snakes developed severe CNS signs; we succeeded in reisolating the virus from the brain of this individual and could demonstrate viral antigen in neurons. In a third attempt, we tested cohousing, vaccination, and sequential infection with multiple reptarenavirus isolates on boas (n = 16). At 10 months postinoculation, all but one snake tested positive for viral RNA in lung, brain, and/or blood, but none exhibited the characteristic IBs. Three of the four vaccinated snakes seemed to sustain challenge with the same reptarenavirus; however, neither of the two snakes rechallenged with different reptarenaviruses remained uninfected. Comparison of the antibody responses in experimentally versus naturally reptarenavirus-infected animals indicated differences in the responses.
IMPORTANCE In the present study, we experimentally infected pythons and boas with reptarenavirus via either intraperitoneal injection or tracheal instillation. The aims were to experimentally induce boid inclusion body disease (BIBD) and to develop an animal model for studying disease transmission and pathogenesis. Both virus delivery routes resulted in infection, and infection via the trachea could reflect the natural route of infection. In the experimentally infected snakes, we did not find evidence of inclusion body (IB) formation, characteristic of BIBD, in pythons or boas. Most of the boas (11/12) remained reptarenavirus infected after 10 months, which suggests that they developed a persistent infection that could eventually have led to BIBD. We demonstrated that vaccination using recombinant protein or an inactivated virus preparation prevented infection by a homologous virus in three of four snakes. Comparison of the antibody responses of experimentally and naturally reptarenavirus-infected snakes revealed differences that merit further studies.
INTRODUCTION
There are descriptions of a plague called boid inclusion body disease (BIBD) in captive snake populations since the 1970s (1). The disease mostly affects members of the families Boidae and Pythonidae and may lead to the eradication of entire snake collections (1, 2). BIBD manifests itself in a variety of clinical conditions, such as neurological signs, including regurgitation, head tremors, and loss of coordination, and abnormal skin shedding, secondary bacterial infections, and neoplastic diseases (1, 2). From the early days of BIBD research, and as the name implies, a hallmark of the disease is the formation of cytoplasmic ultrastructurally electron-dense and histologically eosinophilic inclusion bodies (IBs) in almost all cell types (3, 4). The standard antemortem diagnosis of BIBD relies on IB detection in blood smears or tissue biopsy specimens (1, 5, 6). Even before the causative agent of BIBD was identified, the IBs were found to consist mainly of a 68-kDa protein, unknown at the time (4). In 2012 and 2013, the findings of three independent groups linked BIBD with arenavirus infection (7–9). Furthermore, several groups could demonstrate that the “68-kDa protein” actually represents the arenavirus nucleoprotein (NP) (5, 7, 9). Although BIBD, as defined by the presence of IBs, is always connected to reptarenavirus infection, increasing evidence indicates that reptarenavirus infection does not readily induce IB formation (6, 10, 11). However, throughout this report and in line with the original name of the disease, we consider the presence of IBs a pathognomonic hallmark of BIBD and debate on the disease definition in the Discussion.
The identification of arenaviruses in snakes led to the establishment of two new genera, Mammarenavirus (previously known as arenaviruses) and Reptarenavirus (BIBD-associated arenaviruses), within the family Arenaviridae (12). Independently, two groups then made the observation that snakes with BIBD most often, if not always, carry several reptarenavirus L and S segments (13, 14). These studies dramatically expanded the number of fully sequenced reptarenavirus L segments from 4 to approximately 150 (13, 14). Currently, the L segments of close to 30 reptarenavirus species are known based on the ICTV (International Committee on Taxonomy of Viruses) species demarcation criteria (species sharing <76% nucleotide identity) (12). The high genetic diversity makes nucleic acid-based approaches to BIBD diagnosis challenging, and thus, the detection of reptarenavirus antigen (NP) serves as an alternative (6). Additionally, reptarenavirus infection appears not to readily induce detectable IBs (6, 11, 15), which suggests that BIBD pathogenesis may involve additional factors. For example, vertical transmission of coinfecting reptarenavirus L and S segments with the concurrent presence of IBs was demonstrated (16), and thus, congenital, perinatal, or neonatal infection could be a prerequisite for IB formation. In addition, Haartman Institute snake virus 1 (HISV-1) was identified in a snake with BIBD (14), which led to the establishment of the third arenavirus genus, Hartmanivirus (17, 18). This was followed by the observation that snakes with BIBD fairly often also carry hartmaniviruses; however, so far, hartmanivirus infection has not been linked to BIBD (11, 19, 20).
At present, the family Arenaviridae comprises four genera: Mammarenavirus, Reptarenavirus, Hartmanivirus, and Antennavirus (18). The genome of all except antennaviruses is a bisegmented negative-sense RNA (21). The L segment of mammarenaviruses and reptarenaviruses encodes an RNA-dependent RNA polymerase (RdRp) and a zinc finger matrix Z protein (ZP), while the S segment encodes the glycoprotein (GP) precursor (GPC) and NP (22). The L segment of hartmaniviruses lacks the open reading frame (ORF) for ZP (19).
The literature describes at least three attempts to reproduce BIBD in vivo. In 1994, Schumacher and coworkers injected two 3-month-old Burmese pythons (Python molurus bivittatus) with cell supernatants of cultured primary kidney cells of a Boa constrictor snake with BIBD (3). Both animals developed central nervous system (CNS) signs, leading to the death of the first animal at 6 weeks postinoculation and euthanasia of the second after 10 weeks (3). Pathological examination revealed nonsuppurative, lymphocyte-dominated encephalitis with neuronal degeneration in both animals. IBs were found only in the second animal and only in neurons in the brain and pituitary gland but not in other organs. The authors’ attempts to reisolate and identify the causative agent were unsuccessful (3). In 2000, Wozniak and coworkers infected four boa constrictors intraperitoneally with a filtered liver homogenate from a BIBD-positive donor and observed IBs in hepatocytes at 10 weeks postinfection (4). They succeeded in isolating the IBs and in generating a monoclonal antibody against the 68-kDa protein (most likely reptarenavirus NP, in retrospect) but could not characterize the causative agent (4). At the time of the studies by Schumacher et al. and Wozniak et al., BIBD was suspected to be caused by an unknown retrovirus. In 2017, Stenglein and coworkers reported that they had reproduced BIBD in Python regius and B. constrictor by cardiac injection of purified reptarenavirus (10). The authors diagnosed classical BIBD, as defined by IB formation, in boas but did not observe IBs in pythons (10). Furthermore, while the boas remained clinically healthy for 2 years after infection, the pythons developed severe CNS signs within 2 months (10). These findings highlight the complexity of BIBD pathogenesis and provide further evidence that the disease outcome might vary not only between viruses but also between snake species.
Here, we report the results of a series of experimental infections of ball pythons (P. regius) and common boas (B. constrictor). When we initiated the experimental infections, in 2013, our primary aim was to demonstrate the etiologic relationship between reptarenavirus infection and BIBD. We tested two different routes, intracoelomic and tracheal, for inoculation of the snakes with purified cell culture-grown reptarenaviruses. We also studied the possibility of vaccinating the snakes against reptarenavirus infection and potential transmission during cohousing. During the third set of experimental infections, we learned that snakes with BIBD are often coinfected with several reptarenavirus species (14), and we decided to attempt to inoculate snakes with multiple reptarenaviruses in both co- and superinfection setups. We subjected all snakes to a full postmortem examination and used reverse transcription-PCR (RT-PCR) to detect viral RNA, immunohistochemistry (IHC) (anti-reptarenavirus NP) to detect viral antigen, enzyme-linked immunosorbent assays (ELISAs) to detect antireptarenavirus antibodies in snakes, and vesicular stomatitis viruses (VSVs) pseudotyped with reptarenavirus glycoproteins to detect neutralizing antibodies (NAbs) in the snakes.
RESULTS
Selection of the infection route.
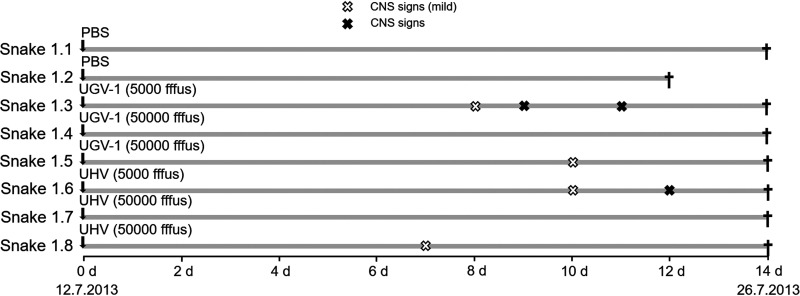
In the first experimental infection involving eight juvenile, ∼2-month-old ball pythons (P. regius), we tested whether the route of administration would affect the course of infection. Cell culture adaptation is for many viruses known to cause virus attenuation. Thus, we decided to use two virus preparations, University of Helsinki virus (UHV) (containing UHV-1 and aurora borealis virus 1 [ABV-1]), which has been propagated in tissue cultures for >8 years, and University of Giessen virus 1 (UGV-1), after a single passage. The viruses were purified by ultracentrifugation, diluted in phosphate-buffered saline (PBS), and used to inoculate each of three snakes (Table 1), two via the coelomic cavity (5,000 and 50,000 fluorescent focus-forming units [FFFU]) and the third via the respiratory route, by instillation into the trachea (50,000 FFFU), to best mimic a possible natural route of infection. Some animals exhibited slight lethargy, but none showed clinical signs during the following 2 weeks (Fig. 1). At the time of euthanasia, at 2 weeks postinoculation, RT-PCR confirmed UGV-1 infection of the brain regardless of the route of infection, whereas for UHV, only tracheal instillation resulted in the detection of viral RNA and only in the lung (Table 1). None of the snakes showed IB formation in blood or tissues, and there was no evidence of viral NP expression in any tissue, including the brain.
TABLE 1.
First experimental infection of ∼2-month-old pythons (P. regius)
| Animal | Inoculation route(s) | Virus, dose (FFFU) | Sample collection time point (day postinoculation) | Result(s)a |
|||
|---|---|---|---|---|---|---|---|
| RT-PCR (brain, lung, blood) | Blood smear | Histology | IHC | ||||
| 1.1 | Trachea and coelomic cavity | Mock (PBS) | 14 | Neg, Neg, Neg | Neg | Neg | Neg |
| 1.2 | Trachea and coelomic cavity | Mock (PBS) | 12 | Neg, Neg, Neg | Neg | Neg | Neg |
| 1.3 | Coelomic cavity | UGV-1, 5,000 | 14 | UGV-1, Neg, Neg | Neg | Neg | Neg |
| 1.4 | Coelomic cavity | UGV-1, 50,000 | 14 | UGV-1, Neg, Neg | Neg | Neg | Neg |
| 1.5 | Trachea | UGV-1, 50,000 | 14 | UGV-1, UGV-1, Neg | Neg | Neg | Neg |
| 1.6 | Coelomic cavity | UHV (UHV-1 and ABV-1), 5,000 | 14 | Neg, Neg, Neg | Neg | Neg | Neg |
| 1.7 | Coelomic cavity | UHV (UHV-1 and ABV-1), 50,000 | 14 | Neg, Neg, Neg | Neg | Neg | Neg |
| 1.8 | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 14 | Neg, UHV-1 and ABV-1, ABV-1 | Neg | Neg | Neg |
Neg, negative.
FIG 1.
Schematic representation of the first experimental infection timeline. The experiment included eight ball pythons, which were monitored for 14 days postinoculation. The vertical arrows indicate inoculation. White (mild tremor) and black (tremor) X marks in the infection timeline mark the observed CNS signs. The black crosses indicate euthanasia.
Experimental infection of a group of ball pythons.
After demonstrating inoculation via the trachea to be effective in the initial trial, we decided to employ tracheal inoculation in the subsequent experiments because it likely reflects a natural route of infection. For the experiment, we inoculated four juvenile pythons (animals 2.3 to 2.6) at the age of ∼2 months with UHV, two (animals 2.7 and 2.8) with UGV-1, and two control animals (animals 2.1 and 2.2) with PBS (Table 2 and Fig. 2). We monitored the snakes daily for signs of disease, and at 19 days postinfection (dpi), animals 2.3, 2.6, 2.7, and 2.8 showed mild head tremor (Fig. 2). By mild head tremor, we refer to mild right-left and/or up-down shaking of the head when the head of the snake is elevated from the ground, e.g., when aiming at prey. Animal 2.6 also exhibited abnormal tail postures. At 22 dpi, two of these snakes (animals 2.3 and 2.7) were euthanized as scheduled, together with one control snake (animal 2.2). Both animals 2.3 and 2.7 were found to be infected; animal 2.3 was RT-PCR positive for both inoculated viruses (UHV-1 and ABV-1) but only in the lung, whereas animal 2.6 exhibited viral RNA in brain and lungs. Neither animal showed IB formation or viral antigen expression (Table 2).
TABLE 2.
Second experimental infection
| Animal | Species | Age category (age [mo]) | Inoculation route | Virus, dose (FFFU) | Sample collection time point (day postinoculation) | Result(s)a |
|||
|---|---|---|---|---|---|---|---|---|---|
| RT-PCR (brain, lung, blood) | Blood smear | Histology | IHC | ||||||
| 2.1 | P. regius | Juvenile (∼2) | Trachea | Mock (PBS) | 118 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.2 | P. regius | Juvenile (∼2) | Trachea | Mock (PBS) | 22 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.3 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 22 | Neg, UHV-1 and ABV-1, Neg | Neg | Neg | Neg |
| 2.4 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 69 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.5 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 30 | ABV-1, ABV-1, ABV-1 | Neg | Neg | Pos |
| 2.6 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 117 | Neg, ABV-1, Neg | Neg | Neg | Neg |
| 2.7 | P. regius | Juvenile (∼2) | Trachea | UGV-1, 50,000 | 22 | UGV-1, UGV-1, UGV-1 | Neg | Neg | Neg |
| 2.8 | P. regius | Juvenile (∼2) | Trachea | UGV-1, 50,000 | 117 | Neg, UGV-1, Neg | Neg | Neg | Neg |
| 2.9 | B. constrictor | Juvenile (∼4) | None | Cohousing with animal 2.4 | 117 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.10 | B. constrictor | Juvenile (∼4) | None | Cohousing with animal 2.8 | 117 | Neg, Neg, Neg | Neg | Neg | Neg |
Pos, positive.
FIG 2.
Schematic representation of the second experimental infection timeline. The experiment included eight ball pythons and two common boas, which were monitored up to 118 days postinoculation. The vertical arrows indicate inoculation. The observed CNS signs are marked by white (mild tremor) and black (tremor) X marks, and the cohousing of snakes is indicated by shading of the infection timeline. The black crosses indicate euthanasia.
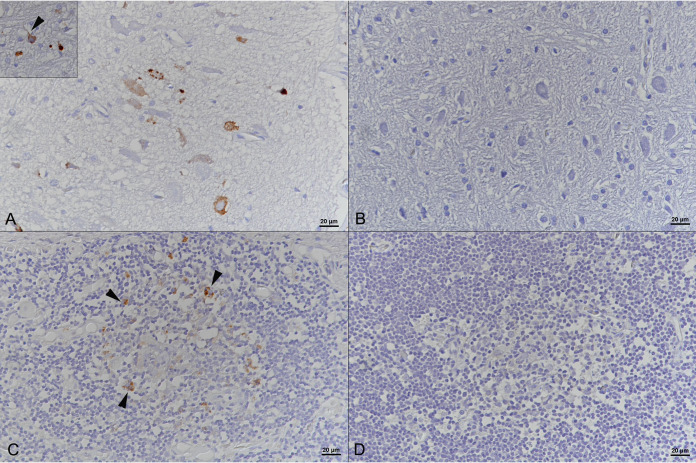
At 25 dpi, we observed neurological signs in animal 2.5 (body balance and coordination problems). At 29 dpi, during feeding, animal 2.5 showed tremor and lethargy and had severe difficulties in swallowing its feed (a frozen-thawed mouse); the snake had to be euthanized the following day since the clinical signs had worsened (Fig. 2). We found virus by RT-PCR in both lungs and brain and could purify the virus from I/1Ki cells inoculated with a brain homogenate. We could not detect IBs in blood cells, but the animal exhibited reptarenavirus NP expression in neurons in the brain and in cells with the morphology of macrophages and/or dendritic cells in spleen and thymus (Fig. 3 and Table 2). At 34 dpi, animal 2.8 showed CNS signs (body balance and coordination problems), and at 37 dpi, animal 2.4 showed head tremors; in both snakes, the clinical signs subsided during the following days (Fig. 2). At 43 dpi, having received 16 juvenile common boas, we decided to investigate whether cohousing with experimentally infected ball pythons would result in virus transmission across the two species. We placed one boa each in the box of one python (animal 2.9 with animal 2.4, and animal 2.10 with animal 2.8). At 54 dpi, animal 2.8 again showed CNS signs (tremors and disorientation). The next day, animal 2.4 also showed similar signs. However, the clinical signs of animal 2.8 improved during the following days (Fig. 2). At 61 dpi, animal 2.6, which had shown mild CNS signs early after inoculation (day 19), had diarrhea but was otherwise in good condition. At 69 dpi, we sacrificed animal 2.4 since the mild CNS signs had by then persisted for 2 weeks. The animal did not exhibit IBs or viral antigen expression, and we did not find reptarenavirus RNA in the tissues studied. The boa (animal 2.9) that had been cohoused with this animal was moved to the box of animal 2.6.
FIG 3.
Brain and spleen of Python regius euthanized 22 days after intratracheal instillation of UHV (animal 2.5). (A and C) Immunohistochemistry (IHC) showing reptarenavirus nucleoprotein in the cytoplasm of neurons (inset, arrowhead) in the brain (A) and in macrophages/dendritic cells (arrowheads) in the spleen (C). (B and D) Negative-control slides. Shown is IHC employing a broadly cross-reactive rabbit anti-pan-reptarenavirus antiserum (39) and hematoxylin counterstaining.
At 83 to 85 dpi, animal 2.6 showed lethargy, and the cohoused boa (animal 2.9) displayed abnormal tail postures. In addition, animal 2.8 was lethargic at 85 dpi. The clinical signs of all three snakes improved during the following days, but from 98 dpi onward, animal 2.6 was again lethargic. At 100 dpi, animal 2.8 showed similar lethargy, but again, both snakes improved during the following days. At 109 dpi, when feeding, both snakes again showed CNS signs (mild tremor) and refused to feed. The boa (animal 2.9) cohoused with a python (animal 2.6) showed similar signs and difficulties in eating. At 117 dpi, we decided to euthanize these three animals (2.6, 2.8, and 2.9) as well as the boa (animal 2.10) that had shared the box with the python (animal 2.8). At 118 dpi, we sacrificed the remaining control snake, animal 2.1. All except the control animal (2.1) and the cohoused boas were found to be reptarenavirus positive by RT-PCR in the lung, and animal 2.7 also carried the virus in the brain. The histological analysis did not reveal IB formation in any of these animals; reptarenavirus antigen expression was also not detected (Table 2).
Vaccination and experimental infection challenge of common boas.
Since the first two rounds of experimental infections had been unsuccessful in terms of replicating IB formation, we finally decided to attempt inoculation of common boas since the virus isolates originate from this species. We also decided to attempt vaccination prior to inoculations and used purified, detergent-inactivated UHV (three animals, 3.3 to 3.5) or recombinant UHV-1 NP (23) (snake 3.6). We gave the first vaccinations the day after the boas had arrived (−74 dpi), i.e., the start day for the python-boa cohousing experiment. Around 2 weeks (−61 dpi) and 4 weeks (−48 dpi) later, we boosted animals 3.3 to 3.6 with the same antigens (Table 3 and Fig. 4).
TABLE 3.
Third experimental infection
| Animal | Species | Age category (age [mo]) | Inoculation route | Virus, dose (FFFU) | Sample collection time (day postinoculation) | Result(s) |
|||
|---|---|---|---|---|---|---|---|---|---|
| RT-PCR (brain, lung, blood) | Blood smear | Histology | IHC | ||||||
| 2.1 | P. regius | Juvenile (∼2) | Trachea | Mock (PBS) | 118 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.2 | P. regius | Juvenile (∼2) | Trachea | Mock (PBS) | 22 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.3 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 22 | Neg, UHV-1 and ABV-1, Neg | Neg | Neg | Neg |
| 2.4 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 69 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.5 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 30 | ABV-1, ABV-1, ABV-1 | Neg | Neg | Pos |
| 2.6 | P. regius | Juvenile (∼2) | Trachea | UHV (UHV-1 and ABV-1), 50,000 | 117 | Neg, ABV-1, Neg | Neg | Neg | Neg |
| 2.7 | P. regius | Juvenile (∼2) | Trachea | UGV-1, 50,000 | 22 | UGV-1, UGV-1, UGV-1 | Neg | Neg | Neg |
| 2.8 | P. regius | Juvenile (∼2) | Trachea | UGV-1, 50,000 | 117 | Neg, UGV-1, Neg | Neg | Neg | Neg |
| 2.9 | B. constrictor | Juvenile (∼4) | None | Cohousing with animal 2.4 | 117 | Neg, Neg, Neg | Neg | Neg | Neg |
| 2.10 | B. constrictor | Juvenile (∼4) | None | Cohousing with animal 2.8 | 117 | Neg, Neg, Neg | Neg | Neg | Neg |
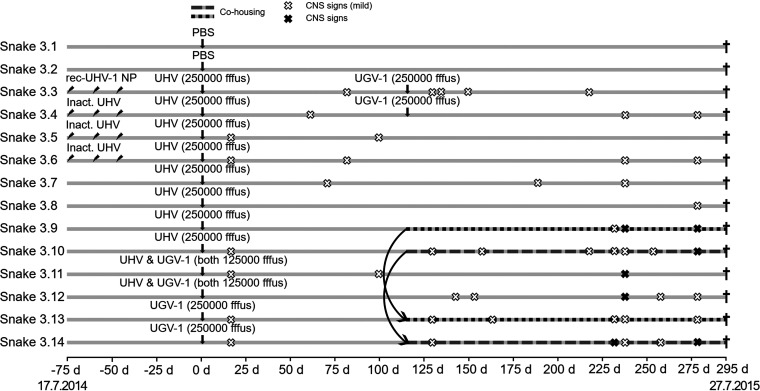
FIG 4.
Schematic representation of the third experimental infection timeline. The experiment included 14 common boas, which were monitored up to 295 days (∼10 months) postinoculation. The inclined arrows indicate immunization, and the vertical arrows indicate inoculation time points. White (mild tremor) and black (tremor) X marks indicate the observed CNS signs, and the cohousing of snakes is indicated by shading of the infection timeline. The black crosses indicate euthanasia.
Since we had successfully infected the pythons but had been unsuccessful in inducing IB formation, we decided to increase the amount of input virus and chose to use an infectious dose that was 5-fold higher than the one used previously. We also wanted to attempt coinfection of some snakes with UHV and UGV-1 but at this point were unaware that our UHV preparation was indeed a mix of ABV-1 and UHV-1. We inoculated the vaccinated boas (animals 3.3 to 3.6) and four nonvaccinated boas (animals 3.7 to 3.10) with UHV (250,000 FFFU/snake), two boas (animals 3.11 and 3.12) with a mix of UHV and UGV-1 (125,000 FFFU/snake each), and two boas (animals 3.13 and 3.14) with UGV-1 (250,000 FFFU/snake) (Table 3). At 14 dpi, we observed mild tremors in animals 3.5, 3.6, 3.10, 3.11, 3.13, and 3.14, but the signs waned in the following days. Afterward, mild CNS signs were observed occasionally: at 57 dpi (animal 3.4, stargazing), 59 dpi (animal 3.4, tremor), 68 dpi (animal 3.7, tremor), and 79 dpi (animals 3.3 and 3.6, tremor) (Fig. 4).
After the discovery that snakes with BIBD often carry several reptarenavirus L and S segments (14), we decided to superinfect some snakes by reinoculation at 115 dpi: animals 3.3 and 3.4 (originally inoculated with UHV) received 250,000 FFFU/snake of UGV-1. At this point, we had also determined using next-generation sequencing (NGS) that our UHV preparation actually contains two viruses (ABV-1 and UHV-1) and decided to retry virus transmission during cohousing. Therefore, at 115 dpi, we placed animal 3.9 (UHV inoculated) in the box of animal 3.13 (UGV-1 inoculated), and animal 3.10 (UHV inoculated) in the box of animal 3.14 (UGV-1 inoculated). We continued monitoring the snakes, and at the following time points, we observed intermittent clinical signs that affected all animals at some point: 127 dpi (animals 3.3, 3.10, 3.13, and 3.14, mild tremors), 132 dpi (3.3, mild tremor), 140 dpi (3.12, mild tremor and disorientation), 149 dpi (3.3, disorientation), 152 dpi (3.12, stargazing), 155 dpi (3.10, tremors and disorientation), 161 dpi (3.13, mild tremor), 186 dpi (3.7, disorientation), 215 dpi (3.10, mild tremor; 3.3, disorientation), 229 dpi (3.9, 3.10, 3.13, and 3.14, mild tremors; 3.14, lethargy), 235 dpi (3.4, 3.6, 3.7, 3.9 to 3.11, 3.13, and 3.14, mild tremor; 3.9, 3.11, and 3.12, lethargy), 251 dpi (3.10 and 3.11, mild tremors; 3.4, 3.8, 3.10, and 3.12, lethargy), 261 dpi (3.12, tremor), 265 dpi (3.12, tremor; 3.14, stargazing), and 276 dpi (3.4, 3.6, 3.8 to 3.10, and 3.12 to 3.14, lethargy) (Fig. 4).
At 294 dpi, we euthanized animals 3.1 to 3.8, and at 295 dpi, we euthanized animals 3.9 to 3.14, as scheduled. Using RT-PCR, we confirmed that all of the animals except snake 3.4 were infected. They carried viral RNA in one or more of the tissues studied; five animals (3.4, 3.8, and 3.10 to 3.12) were also found to be viremic. None of the animals exhibited IBs or reptarenavirus antigen in any tissue or the blood (Table 3).
Immune response against reptarenavirus NP in experimentally and naturally infected snakes.
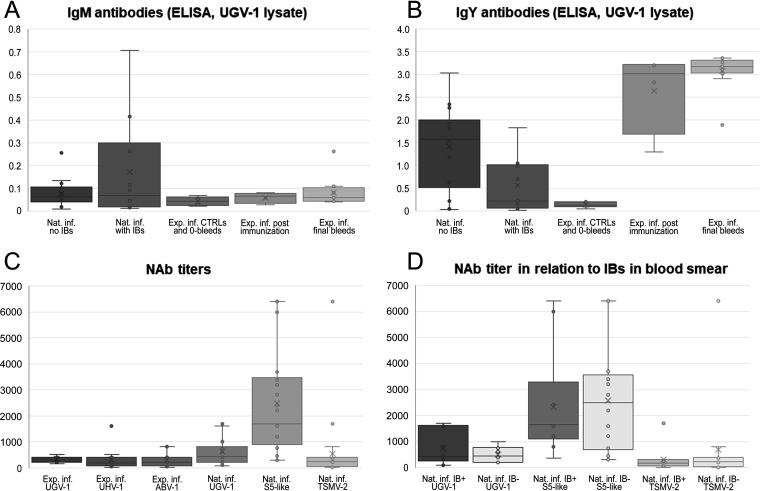
Unlike Stenglein and colleagues (10), we did not succeed in inducing IB formation, the hallmark of BIBD (1, 3–7, 9), by experimental reptarenaviruses infection in pythons or boas. We recently learned that reptarenavirus-infected snakes with IBs in blood cells have lower levels of antireptarenavirus antibodies than reptarenavirus-infected snakes without IBs (11). Thus, we compared the antibody responses of the experimentally infected snakes to the responses in naturally reptarenavirus-infected boas (the latter using a panel of 24 plasma samples available from a previous study [11]) by employing tools developed previously (11, 23–25). We used an ELISA with purified UGV-1 as the antigen to determine the levels of IgM and IgY antibodies against reptarenavirus NP (Table 4). None of the python sera produced a signal in the ELISA, most likely indicating a lack of cross-reactivity of our anti-boa immunoglobulin reagents to python immunoglobulins rather than a lack of antibodies. The ELISA results show that the uninfected boas that served as control snakes did not have anti-reptarenavirus NP antibodies, confirming that the snakes had not been in contact with reptarenaviruses prior to vaccination and/or inoculation (Table 4 and Fig. 5A). This result also indicates that the vaccinations with both the inactivated UHV preparation and recombinant UHV-1 NP had induced the formation of anti-NP antibodies (Table 4 and Fig. 5A and B). We took serum samples from the vaccinated snakes on day 74 after the initial vaccination, which is the likely reason for the presence of IgY but not IgM class anti-NP antibodies. At the end of the experiment, ∼10 months after virus challenge, all snakes had IgY class anti-NP antibodies (Table 4 and Fig. 5B). Their anti-NP IgY levels were slightly higher than those of the naturally infected snakes, some of which were also anti-NP IgM positive (Table 4 and Fig. 5A). We then compared the results of snakes without IB to those of snakes with IB (i.e., confirmed BIBD) and observed that the latter had lower levels of anti-NP antibodies (Table 4 and Fig. 5A and B), a finding reported previously (11).
TABLE 4.
Antibody responses in experimentally compared to naturally reptarenavirus-infected boasa
| Animal | Virus(es) used for inoculation or immunization | ELISA titer (UGV-1 lysate) |
Blood smear score | Neut. titer |
RT-PCR result(s) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| IgY | IgM | UGV-1 | UHV-1 | ABV-1 | TSMV-2 | S5-like | ||||
| 3.1 0-bleed | 0.151 | 0.052 | 0 | 0 | 0 | 0 | ND | ND | Neg | |
| 3.1. final | 0.201 | 0.058 | 0 | 0 | 0 | 0 | ND | ND | Neg | |
| 3.2 0-bleed | 0.199 | 0.067 | 0 | 0 | 0 | 0 | ND | ND | Neg | |
| 3.2 final | 0.109 | 0.026 | 0 | 0 | 0 | 0 | ND | ND | Neg | |
| 3.3 postimm. | Immunized with recombinant UHV-1 NP | 3.227 | 0.069 | 0 | ND | ND | ND | ND | ND | Neg |
| 3.3 final | UHV-1, ABV-1, later UGV-1 | 3.311 | 0.109 | 0 | 250 | 150 | 75 | ND | ND | UGV-1 |
| 3.4 postimm. | Immunized with inactivated UHV (UHV-1 and ABV-1) | 2.838 | 0.059 | 0 | ND | ND | ND | ND | ND | Neg |
| 3.4 final | UHV-1, ABV-1, later UGV-1 | 3.092 | 0.109 | 0 | 150 | 100 | 75 | ND | ND | UGV-1 |
| 3.5 postimm. | Immunized with inactivated UHV (UHV-1 and ABV-1) | 1.303 | 0.080 | 0 | ND | ND | ND | ND | ND | Neg |
| 3.5 final | UHV-1 and ABV-1 | 1.901 | 0.045 | 0 | 500 | 500 | 200 | ND | ND | UHV-1, ABV-1 |
| 3.6 postimm. | Immunized with inactivated UHV (UHV-1 and ABV-1) | 3.212 | 0.026 | 0 | ND | ND | ND | ND | ND | Neg |
| 3.6 final | UHV-1 and ABV-1 | 3.103 | 0.041 | 0 | 200 | 400 | 50 | ND | ND | Neg |
| 3.7 final | UHV-1 and ABV-1 | 3.018 | 0.081 | 0 | 200 | 150 | 400 | ND | ND | ABV-1 |
| 3.8 final | UHV-1 and ABV-1 | 3.177 | 0.066 | 0 | 400 | 0 | 800 | ND | ND | ABV-1 |
| 3.9 final | UHV-1 and ABV-1 | 3.177 | 0.042 | 0 | 400 | 200 | 800 | ND | ND | ABV-1 |
| 3.10 final | UHV-1 and ABV-1 | 2.910 | 0.039 | 0 | 400 | 200 | 50 | ND | ND | ABV-1 |
| 3.11 final | UHV-1, ABV-1, and UGV-1 | 3.267 | 0.059 | 0 | 400 | 50 | 0 | ND | ND | UGV-1 |
| 3.12 final | UHV-1, ABV-1, and UGV-1 | 3.319 | 0.060 | 0 | 400 | 400 | 400 | ND | ND | UGV-1 |
| 3.13 final | UGV-1 | 3.372 | 0.262 | 0 | 400 | 50 | 400 | ND | ND | UGV-1 |
| 3.14 final | UGV-1 | 3.370 | 0.049 | 0 | 400 | 1,600 | 200 | ND | ND | UGV-1 |
| Nat. inf. 1 | 3.042 | 0.061 | 0 | 200 | 0 | 6,400 | UGV, TSMV-2 | |||
| Nat. inf. 2 | 1.839 | 0.085 | 0 | 1,000 | 50 | 3,200 | TSMV-2 | |||
| Nat. inf. 3 | 1.186 | 0.047 | 0 | 500 | 400 | 3,400 | S5, TSMV-2 | |||
| Nat. inf. 4 | 0.045 | 0.008 | 0 | 700 | 6,400 | 6,400 | Neg | |||
| Nat. inf. 5 | 0.150 | 0.089 | 2 | 90 | 100 | 800 | UGV, S5, TSMV-2 | |||
| Nat. inf. 6 | 0.621 | 0.045 | 0 | 800 | 350 | 1,600 | S5 | |||
| Nat. inf. 7 | 0.222 | 0.116 | 2 | 450 | 50 | 350 | UGV, S5 | |||
| Nat. inf. 8 | 1.914 | 0.026 | 0 | 750 | 400 | 3,500 | Neg | |||
| Nat. inf. 9 | 0.161 | 0.021 | 1 | 400 | 300 | 1,600 | UGV, S5 | |||
| Nat. inf. 10 | 2.348 | 0.122 | 0 | 400 | 0 | 2,200 | S5 | |||
| Nat. inf. 11 | 2.270 | 0.257 | 0 | 600 | 800 | 750 | Neg | |||
| Nat. inf. 12 | 0.054 | 0.018 | 3 | 800 | 1,700 | 2,400 | UGV, S5 | |||
| Nat. inf. 13 | 1.546 | 0.101 | 0 | 200 | 0 | 300 | UGV, TSMV-2 | |||
| Nat. inf. 14 | 0.085 | 0.012 | 2 | 1,700 | 50 | 1,200 | UGV, S5, TSMV-2 | |||
| Nat. inf. 15 | 1.828 | 0.262 | 1 | 1,700 | 300 | 1,200 | UGV | |||
| Nat. inf. 16 | 1.588 | 0.132 | 0 | 800 | 75 | 3,700 | UGV, S5, TSMV-2 | |||
| Nat. inf. 17 | 1.049 | 0.416 | 3 | 1,600 | 0 | 6,400 | UGV, S5, TSMV-2 | |||
| Nat. inf. 18 | 0.699 | 0.015 | 1 | 300 | 250 | 6,000 | UGV, S5, TSMV-2 | |||
| Nat. inf. 19 | 0.217 | 0.017 | 0 | 250 | 250 | 2,800 | S5, TSMV-2 | |||
| Nat. inf. 20 | 0.034 | 0.060 | 0 | 200 | 200 | 300 | S5, TSMV-2 | |||
| Nat. inf. 21 | 1.581 | 0.078 | 0 | 400 | 400 | 450 | S5, TSMV-2 | |||
| Nat. inf. 22 | 0.020 | 0.045 | 1 | 400 | 250 | 1,700 | UGV, S5, TSMV-2 | |||
| Nat. inf. 23 | 1.813 | 0.054 | 0 | 200 | 200 | 1,200 | UGV, S5, TSMV-2 | |||
| Nat. inf. 24 | 0.983 | 0.706 | 1 | 75 | 100 | 1,700 | S5, TSMV-2 | |||
Abbreviations: 0-bleed, blood sample collected before initiation of the experimental work; final, blood sample collected during necropsy; postimm., blood sample collected following immunizations and prior to virus challenge; Nat. inf., naturally infected animal; ND, not determined. Blood smears were scored on a scale of 0 to 3, where a score of 0 is no IBs, 1 is small and/or rare IBs, 2 is medium-sized and/or occasional IBs, and 3 is large and/or numerous IBs.
FIG 5.
Antibody responses in experimentally versus naturally infected common boas. (A) Box plot of IgM class antibodies against reptarenavirus NP (using a concentrated UGV-1 lysate as the antigen). The boxes from left to right represent naturally infected snakes without IBs, naturally infected snakes with IBs, the 0-bleeds (blood samples collected prior to immunization or inoculation), samples collected following immunization, and samples collected from the experimentally infected snakes at the time of euthanasia. The y axis represents optical density at 450 nm (OD450) values as the ELISA readout. (B) Box plot of IgY class antibodies against reptarenavirus NP (using a concentrated UGV-1 lysate as the antigen). The boxes from left to right represent naturally infected snakes without IBs, naturally infected snakes with IBs, the 0-bleeds collected prior to immunization or inoculation, samples collected following immunization, and samples collected from the experimentally infected snakes at the time of euthanasia. The y axis represents OD450 values as the ELISA readout. (C) Box plot of neutralizing antibody (NAb) titers as studied using VSV pseudotypes with reptarenavirus glycoproteins. The boxes from left to right represent neutralizing antibodies against UGV-1 in experimentally infected snakes, neutralizing antibodies against UHV-1 in experimentally infected snakes, neutralizing antibodies against ABV-1 in experimentally infected snakes, neutralizing antibodies against UGV-1 in naturally infected snakes, neutralizing antibodies against S5-like glycoproteins in naturally infected snakes, and neutralizing antibodies against TSMV-2 in naturally infected snakes. The y axis represents the last dilution producing a 50% reduction in the number of fluorescent foci. (D) Box plot of NAb titers as studied using VSV pseudotypes with reptarenavirus glycoproteins in naturally infected snakes with and without IBs. The boxes from left to right represent neutralizing antibodies against UGV-1 in snakes with IBs, neutralizing antibodies against UGV-1 in snakes without IBs, neutralizing antibodies against S5-like glycoproteins in snakes with IBs, neutralizing antibodies against S5-like glycoproteins in snakes without IBs, neutralizing antibodies against TSMV-2 in snakes with IBs, and neutralizing antibodies against TSMV-2 in snakes without IBs. The y axis represents the last dilution producing a 50% reduction in the number of fluorescent foci.
Reptarenavirus NAbs in experimentally and naturally infected snakes.
As the analysis of anti-reptarenavirus NP antibodies indicated potential differences in the immune responses of experimentally versus naturally infected snakes, we wanted to compare the NAb responses in both groups of snakes. Using a fluorescent replication-defective recombinant vesicular stomatitis virus (rVSV-ΔG*eGFP) system, we had previously generated single-round infectious particles pseudotyped with reptarenaviral GPCs (ABV-1, UHV-1, UGV-1, S5-like, and tavallinen suomalainen mies virus 2 [TSMV-2]) (24), which we employed to determine the 50% focus reduction neutralization test (FRNT50) titers for the sera. We studied the boa sera from the third experimental infection against ABV-1, UHV-1, and UGV-1 GP-bearing pseudotypes and used UGV-1, S5-like, and TSMV-2 GP-bearing pseudotypes for the sera of naturally infected snakes from our previous study (11). The latter were selected based on the results of RT-PCRs targeting the respective S segments (11). The results indicate differences in the neutralizing titers of the experimentally infected snakes (Table 4 and Fig. 5C and D). Four snakes (animals 3.3, 3.4, 3.10, and 3.11) showed the highest FRNT50 titers against UGV-1, two (animals 3.6 and 3.14) showed the highest titers against UHV-1, and three (animals 3.7, 3.8, and 3.9) showed the highest titers against ABV-1. Snake 3.5 showed equal FRNT50 titers for UGV-1 and UHV-1, snake 3.12 showed equal titers for all studied viruses, and snake 3.13 showed equal titers for UGV-1 and ABV-1. Two of the snakes, animals 3.8 (inoculated with UHV) and 3.10 (inoculated with both UHV and UGV-1), did not show NAbs against UHV-1 and ABV-1, respectively, but animal 3.9 had a good NAb response against UGV-1. Animal 3.12 had the highest recorded neutralizing titer against UHV-1, reaching a value of 1,600. Two snakes, animals 3.1 and 3.2, mounted the weakest NAb responses with the highest titer reaching 250, while for other experimentally infected snakes, the highest titers were ≥400.
The naturally infected snakes showed much higher NAb titers; for most animals, the highest titer was clearly above 1,000, and for some, we recorded neutralizing titers as high as 6,400 (Table 4 and Fig. 5C). We further observed that a high neutralizing titer against a given virus did not provide neutralization against other reptarenaviruses (see, e.g., Nat. inf. 1, 10, 13, and 17 in Table 4), which suggests that the level of cross-neutralization might be low.
DISCUSSION
BIBD has remained an enigmatic disease for decades. Before identifying reptarenaviruses as the likely causative agents, at least two studies had reproduced the disease under experimental conditions using cell culture-isolated causative agents (3, 4). In the 1994 report by Schumacher and coauthors, one of the two infected Burmese pythons (Python bivittatus) developed severe CNS signs and died at 6 weeks postinoculation, and the second was euthanized at 10 weeks postinoculation due to severe CNS signs (3). The authors observed IBs in the brain of one of the snakes; however, they failed to reisolate the infectious agent (3). Retrospectively, it is possible that reisolation per se was successful but that the authors merely failed to detect the causative agent since reptarenaviruses do not induce a cytopathic effect in cell culture. In the second study, by Wozniak and colleagues, the authors inoculated four juvenile common boas with a liver homogenate from a boa with BIBD and included two equal-sized control groups: noninoculated and inoculated with a liver homogenate from a healthy boa (4). By 10 weeks postinoculation, all four snakes inoculated with the liver homogenate from a BIBD-positive snake had developed IBs in the liver, but none developed clinical disease during the 1-year surveillance period (4). After the identification of reptarenaviruses as the most likely etiological agents of BIBD, Stenglein and colleagues performed an experimental infection on two ball pythons and common boas each (10). The pythons developed CNS signs within 2 months postinoculation but did not show IB formation, and only the brain samples tested positive for the viral NP, i.e., the main IB component (10). The authors monitored the experimentally infected boas for 2 years, and in addition to IB formation in several tissues, the animals showed virus secretion via feces and urates but remained clinically healthy (10).
Based on the studies by Schumacher et al. and Wozniak et al. (3, 4), and following anecdotal evidence of breeders employing pythons as sentinels of BIBD since they can rapidly develop CNS signs, we initially made an attempt to experimentally infect juvenile ball pythons. The experimental inoculation of the first set of pythons (n = 8) revealed that both tracheal instillation and intraperitoneal injection result in virus replication in multiple tissues. We considered inoculation via the trachea to better mimic the natural infection route and thus used this approach in the subsequent experiments. In the second experimental infection of ball pythons, one individual developed severe CNS signs, and we could reisolate the virus from the brain of this snake; however, none of the snakes demonstrated IBs, even at 4 months postinoculation. For the third experimental infection, 16 common boa siblings were available, and we cohoused two of these boas with pythons that had been experimentally infected prior to the initiation of this experiment; however, we could not confirm horizontal transmission. Considering the results of Stenglein and coworkers, transmission from pythons to boas during cohousing can be considered unlikely, since the authors did not find reptarenavirus RNA in python excreta (10). We immunized four boas prior to virus inoculations and used a larger amount of virus for the tracheal instillations. While the experiment was ongoing, we learned that snakes with BIBD often harbor several reptarenaviruses and that our “UHV inoculum” actually contained UHV-1 and ABV-1 at an ∼1:1 ratio. On top of the coinfection with two distinct reptarenaviruses, we then superinfected some of the snakes with a genetically distinct reptarenavirus (UHV-inoculated snakes reinoculated with UGV-1) ∼3.5 months after the initial inoculation. We also attempted cohousing to demonstrate horizontal transmission, but none of the snakes developed IBs during the 10-month surveillance period, even though some boas were RT-PCR positive for multiple reptarenaviruses. Some of the snakes, including the boas cohoused with pythons, showed transient CNS signs and anorexia, even though they were eventually reptarenavirus RT-PCR and IB negative. It is possible that the behavioral changes were stress induced rather than reptarenavirus infection induced. Theoretically, the signs could also be linked to an infection by an unknown agent. Our findings in pythons were similar to those of Stenglein and colleagues (10); however, unlike other studies (3, 4, 10), we did not detect IB formation in the boas. On the other hand, our findings in the pythons, i.e., clinical evidence that they are more prone to developing CNS signs than boas upon infection, concur with those made in previous studies (3, 4, 10).
More specific diagnostic tools, such as RT-PCR and immunohistology, have become available after identifying reptarenaviruses as the causative agents of BIBD (7–10, 13, 16, 26). These tools have allowed the identification of reptarenavirus-infected snakes that do not demonstrate the presence of IBs (6, 10, 11, 26), the morphological hallmark of BIBD. Furthermore, increasing evidence, including the data presented in this study, suggests that reptarenavirus infection may have a dramatically different outcome depending on the host species (10, 26). We apply the term “BIBD” for the disease because it affects and is diagnosed mainly in boid snakes and also to distinguish it from IBD (inflammatory bowel disease), a common condition in humans and several companion animal species. Recent evidence suggests that reptarenavirus infection affects multiple snake species but is frequently not associated with IB formation in nonboids (26–28). We thus suggest that the term BIBD be reserved for disease that manifests with IB formation as a result of reptarenavirus infection. Because reptarenavirus infection in nonboids is often associated with CNS signs, we propose such disease to instead be referred to as, e.g., reptarenavirus-associated neurological syndrome.
The lack of IB formation in the experimentally infected boas is peculiar. The different outcome of our experimental infection than that in the study by Stenglein and colleagues could rather conveniently be explained by the use of different viruses. However, there are several counterarguments to this claim. We used viruses that have been isolated from snakes with BIBD, i.e., snakes displaying IBs in blood and tissues. We have also demonstrated that these viruses induce IB formation in cell culture (9, 19, 23, 29, 30). Furthermore, the evidence suggests that the UGV-like (or S6-like according to Stenglein et al.) S segment is most commonly associated with the presence of IBs in infected snakes (11, 13, 16). Based on the above-described results, it seems unlikely that our selection of viruses would have affected the outcome. Another possibility is that the host’s genotype would have affected the outcome; however, studying this hypothesis would be very challenging if not impossible to demonstrate in light of the currently limited knowledge on the Boa constrictor genome. It would theoretically be possible that reptarenavirus infection alone is not sufficient to induce BIBD and that a coinfection with some unidentified virus, e.g., a retrovirus, would be required for the IB phenotype. For this explanation to be true, all of the previous experimental infections would have needed to have such a coinfecting agent, which seems rather unlikely.
In our previous study, we found low anti-NP antibody responses in snakes with BIBD (11); thus, we decided to study the antibody responses of the experimentally infected snakes. We observed that the experimentally infected boas had mounted a strong antibody response against NP while lacking IBs. Also considering the findings of our previous study (11), the results might suggest an association of low anti-NP antibody responses and IB development. In this study, we found lower NAb titers in the experimentally infected snakes than in the naturally reptarenavirus-infected snakes. Although the NAb titers against different viruses may not be directly comparable for technical reasons, the NAb titers against UGV-1 concur with the above-described data. In fact, based on the Mann-Whitney test, the differences in NAb titers (these could be compared only for UGV-1) were not statistically significant. However, the naturally versus experimentally infected animals showed a statistically significant difference in the amount of anti-NP IgY antibodies determined by ELISAs. Viremia in snakes with BIBD regardless of a strong NAb response is interesting, and it could be an indication of the role of snakes as reptarenavirus reservoirs since persistently infected Calomys musculinus (dryland vesper mouse), the primary reservoir host of Junin virus (JUNV), a mammarenavirus, also possesses NAbs (31–33). The same is true for persistently infected hantavirus rodent hosts (34, 35). Studies on JUNV and lymphocytic choriomeningitis virus (LCMV) suggest that mutations to the targets of NAbs could at least partially explain this persistence (31–33, 36). On the other hand, the study by Stenglein and colleagues successfully reproduced BIBD in boas as judged by IB formation. It would be interesting to study the blood samples of this particular infection trial, especially since the authors managed to collect sequential samples from the infected animals. While one might expect that the antibody response followed a similar course, one could also speculate that, e.g., the inoculation route would contribute to potential differences in the responses. The direct cardiac venipuncture for inoculation used by Stenglein and colleagues could have allowed the virus to spread more rapidly, thus delaying the immune response. Also, during the current experiment, snakes were housed at a consistent environmental temperature of 27°C to 29°C, without a temperature gradient. As poikilothermic animals, snakes could alter their behavior as a result of reptarenavirus infection to control the immune response. Such behavioral changes have been described in other reptiles following infection (37). Actually, we observed that some reptarenavirus-infected snakes spent a large amount of time in their water basins, which could be indicative of their desire to control their body temperature.
In our study, vaccination of snakes with either inactivated virus or recombinant NP resulted in strong antibody responses. The subsequent challenge with the same virus (UHV, containing both UHV-1 and ABV-1), 74 days after the initial immunization, was sustained in three of the four vaccinated snakes. One of the vaccinated snakes remained uninfected throughout the experiment, and only one snake became infected with the viruses used for the immunization. Subsequent challenge, 115 days after the initial challenge and 189 days after the first immunization, with a different reptarenavirus (UGV-1) resulted in infection of both challenged animals. Unfortunately, our animal experimentation permits did not allow blood collection via cardiac venipuncture, due to which we obtained only a small amount of blood after completing the immunizations and thus could not analyze NAb titers. It is possible that immunization of snakes with detergent-inactivated reptarenaviruses did not induce a high-enough NAb response to protect against virus challenge. Weakly neutralizing or nonneutralizing antibodies against reptarenavirus GPs could also boost the infection via antibody-dependent enhancement (ADE), i.e., by enabling the virus to enter Fc receptor-expressing cells. ADE could allow infection by viruses bearing the GPs of different reptarenavirus species; e.g., antibodies against UHV-1 GPs could facilitate infection by virions with UGV-1 GPs. Further studies are needed to reveal whether a NAb response can be induced by, e.g., recombinant reptarenavirus GPs and whether the NAb response would actually protect the snakes against virus challenge. However, our results suggest that immunization of snakes with either inactivated reptarenavirus or recombinant NP might result in at least short-term protection against virus challenge. There are currently no vaccines against reptarenaviruses, although an effective vaccine might allow reptarenavirus eradication and enable BIBD-free snake collections. Because UGV-like viruses appear to dominate in BIBD-positive animals (11, 13, 16), future studies should address the possibility of vaccinating snakes using either inactivated UGV or recombinant NP of UGV. They should also determine the virus dose suitable for vaccine challenge experiments.
MATERIALS AND METHODS
Ethics statement.
The experimental infection was approved by the National Animal Experiment Board (Eläinkoelautakunta [ELLA]) of Finland (permit number ESAVI/4690/04.10.07/2013). All animals were euthanized according to schedule 1 procedures to minimize suffering.
Cells, viruses, and purification of viruses.
The continuous B. constrictor kidney cell line I/1Ki, generated and maintained as described previously (9, 23), served for virus production and virus reisolation attempts from tissues and blood of the experimentally infected animals. One isolate used in this study, University of Helsinki virus (UHV), was initially described previously (9), but we later found that it actually comprises two reptarenaviruses, UHV-1 (GenBank accession numbers KR870020.1 and KR870011.1) and aurora borealis virus 1 (ABV-1) (GenBank accession numbers KR870021.1 and KR870010.1), at roughly equal amounts as judged by reads obtained by NGS (14). The other isolate (T10404) used was from snake 5 reported previously (9), which was later named University of Giessen virus 1 (UGV-1) (GenBank accession numbers KR870022.1 and KR870012.1). The propagation, purification, and storage of UHV and UGV-1 preparations used for inoculation were described previously (23). Virus titration was done as described previously for hantaviruses (38).
For the reisolation of virus from infected snakes, 100 μl of EDTA blood was diluted 1:5 in fully supplemented growth medium (Eagle’s minimal essential medium [Sigma] with 10% fetal bovine serum [Gibco], 2 mM l-glutamine [Sigma], 100 IU penicillin, and 100 μg/ml streptomycin [Sigma]). The brain, lung, liver, kidney, and heart (∼10- to 20-mm3-piece) samples were homogenized using a scalpel, and the tissue mash was resuspended in fully supplemented growth medium by up-down pipetting. The unfiltered diluted blood and the tissue homogenates were overlaid on I/1Ki cells (80 to 90% confluent), with each tissue in a separate bottle. Cells were kept with a minimal volume of the inoculum (∼500 μl/25-cm2 bottle) for 1 h at 30°C, after which 3 ml of fully supplemented medium was added and the cells were incubated for 24 h at 30°C, followed by medium exchange and incubation for 10 to 14 days at 30°C. The cells were analyzed for viral antigen expression by Western blotting. The cell culture supernatant collected at 5 and 10 days postinfection (dpi) was clarified by centrifugation (3,000 × g for 5 min) and filtered through a 0.45-μm filter, and the viruses were pelleted by ultracentrifugation (27,000 × g at 5°C for 2 h) through a 1-ml 30% sucrose cushion (in phosphate-buffered saline [PBS], pH 7.4) in an SW41 rotor (Beckman Coulter). The pelleted virus material was resolubilized in PBS and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Animals and infection.
Animals that were BIBD negative by blood smear (16 Python regius animals obtained from a commercial German breeder and 16 Boa constrictor animals from a private Swiss breeder) were used. The animals were housed in aerated plastic boxes (dimensions, 40 by 30 by 19 cm) (Smartstore Classic 15; Orthex Group) held in temperature (between 27°C and 30°C)- and daylight (12 h of light)-controlled cabinets (Ehret GmbH, Germany). The humidity (approximately 60 to 80%) inside housing boxes was maintained by evaporation from a water supply.
For the initial trial (Table 1), involving eight juvenile (∼2 months of age) ball pythons (P. regius) from a single clutch, three snakes were infected with a UHV preparation (containing UHV-1 and ABV-1 but for simplicity referred to as UHV), three were infected with UGV-1, and two remained as controls. The infected animals in both groups were inoculated as follows: one received 5,000 fluorescent focus-forming units (FFFU) intracoelomically, one received 50,000 FFFU intracoelomically, and one received 50,000 FFFU instilled into the trachea (the volume of the inoculum was 500 μl in PBS). The control animals received 500 μl PBS intracoelomically and intratracheally, respectively. The snakes were monitored daily for clinical signs and euthanized at 14 dpi.
In the second experimental infection (Table 2), again involving eight 2-month-old ball pythons from a single clutch, four ball pythons received the UHV preparation, two received UGV-1, and two were administered the equivalent amount of PBS. Virus inocula (50,000 FFFU for both UGV-1 and UHV inoculations, all diluted in 500 μl PBS) were instilled into the trachea. The snakes were monitored daily and fed at 1- to 2-week intervals. At 44 dpi, two juvenile (4 months of age) common boas were included in the experiment: one was cohoused with a UHV-inoculated python (animal 2.4), and the other was cohoused with a UGV-1-inoculated python (animal 2.8). All snakes were monitored daily for any clinical signs. The animals were euthanized as follows: animal 2.1 at 118 dpi; 2.2, 2.3, and 2.7 at 22 dpi; 2.5 at 30 dpi (due to severe CNS signs); 2.4 at 69 dpi; and 2.6 and 2.8 at 117 dpi. The two cohoused boas (animals 2.9 and 2.10) were also euthanized at 117 dpi (73 days after initiation of cohousing), at the scheduled end of the experiment.
For the third experimental infection (Table 3), we received a clutch of 16 common boas, of which three were immunized with purified UHV inactivated by the addition of Triton X-100 (initially to yield 1% [vol/vol], followed by dilution to a final concentration of 0.2% [vol/vol] with PBS) (animals 3.4, 3.5, and 3.6) and one was inoculated with recombinant UHV nucleoprotein (NP) (described in reference 23) (animal 3.3). Briefly, at day 0, the animals were subcutaneously administered either approximately 10,000,000 FFFU of detergent-inactivated UHV or 0.1 mg of recombinant UHV NP emulsified with an equal volume of Freund’s incomplete adjuvant (Thermo Fisher Scientific); the total volume per individual was 125 μl. Thirteen and 26 days after the initial administration, boosters with a similar dose were administered. Seventy-four days after the initial immunizations, eight boas (animals 3.3 to 3.10, including the vaccinated ones [animals 3.3 to 3.6]) received 250,000 FFFU of UHV, two boas (animals 3.11 and 3.12) received 125,000 FFFU of both UHV and UGV-1, and two boas (animals 3.13 and 3.14) received 250,000 FFFU of UGV-1, by tracheal instillation. At 116 dpi, after finding out that snakes with BIBD often carry multiple reptarenavirus L and S segments, two vaccinated snakes were superinfected by administering 250,000 FFFU of UGV-1 (animals 3.3 and 3.4). To study horizontal transmission, the two snakes initially inoculated with UGV-1 (animals 3.13 and 3.14) were placed into boxes with UHV-inoculated snakes for cohousing (animals 3.9 and 3.10, respectively) to mimic the introduction of new snakes into a collection and to study if horizontal transmission contributed to the development of BIBD. The snakes were monitored daily for any clinical signs and fed at 1- to 3-week intervals.
Prior to virus inoculation, a blood sample had been collected from the tail vein of each animal. Snakes were euthanized by decapitation after sedation by exposure to CO2. A blood sample was collected, animals were necropsied, and organ samples were collected immediately into TRIzol (Life Technologies) (for RT-PCR) and paraformaldehyde (PFA) (4% solution in PBS) (for histology and immunohistology) and fresh frozen at −70°C for virus isolation and further analyses.
SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblotting were done as described previously (9, 23). Antibodies against UHV NP, described previously (23), were used for detection in immunoblots. The visualization of immunoblots probed (at a 1:10,000 dilution) with goat anti-rabbit IR800Dye (Li-Cor Biosciences) or goat anti-rabbit Alexa Fluor 680 (Invitrogen) was done using the Odyssey infrared imaging system (Li-Cor Biosciences).
Reverse transcription-PCR and Sanger sequencing.
RNA isolation from tissue and blood samples was done as described previously (16). Reverse transcription-PCRs (RT-PCRs) for UHV-1, ABV-1, and UGV-1 L and/or S segments were done initially using the primers and protocol described previously (16), and the RT-PCR products were analyzed by standard agarose gel electrophoresis, visualized by using GelRed nucleic acid stain (Biotium), and subjected to Sanger sequencing (core facility of the Haartman Institute, University of Helsinki, Finland). The extracted RNAs were later reanalyzed using a one-step TaqMan assay with the following primers and probes targeting the S segment, according to the TaqMan Fast virus 1-step master mix (Thermo Scientific) product guidelines: UGV-1 probe (6-carboxyfluorescein [FAM]–CTCGACAAGCGTGGGCGGAGG–black hole quencher 1 [BHQ-1]), UGV-1-fwd (5′-CAAGAAAAACCACACTGCACA-3′), UGV-rev (5′-AACCTGTTGTGTTCAGTAGT-3′), UHV-1 probe (FAM–TCCTCTGCCGCAAAAGACTATGTCACAG–BHQ-1), UHV-1-fwd (5′-ACAAACTGAATAAGACTGCTGCATT-3′), UHV-1-rev (5′-AGGGCTATACACACATAGTTGGATG-3′), ABV probe (FAM–CATGAATTCTTCATCGACATCAGAAACCG–BHQ-1), ABV-1-fwd (5′-CCGTACTGCACAACTGATGATG-3′), and ABV-1-rev (5′-AGCAACACAGGAGTAACCTGTCAC-3′). The cycling conditions were (i) reverse transcription for 5 min at 50°C and (ii) reverse transcriptase inactivation for 20 s at 95°C, 40 cycles of denaturation for 3 s at 95°C, and annealing-extension for 30 s at 60°C.
Histology and immunohistochemistry.
For histology and immunohistochemistry (IHC), samples of brain, lung, liver, kidney, pancreas, spleen, small intestine, and heart were fixed in PFA for 48 h and routinely embedded in paraffin wax. Sections (3 to 4 μm) were prepared and stained with hematoxylin-eosin (HE). For all RT-PCR-positive animals, consecutive sections were prepared and subjected to IHC for viral NP, employing the recently described broadly cross-reactive rabbit anti-pan-reptarenavirus antiserum (39), according to a previously described protocol (9, 23).
Focus reduction neutralization test using replication-incompetent vesicular stomatitis virus pseudotyped with reptarenavirus glycoproteins.
The production of single-cycle replication, GP-deficient, recombinant vesicular stomatitis virus (VSV) expressing enhanced green fluorescent protein (scrVSVΔG-eGFP) pseudotyped with different reptarenavirus GPs was done as described previously (24). Each pseudotyped scrVSVΔG-eGFP batch was titrated with a 10-fold dilution series on a 96-well plate of clean I/1Ki cells as described previously (24), and the dilution yielding 50 to 150 fluorescent cells was selected for a focus reduction neutralization test (FRNT). To demonstrate neutralizing antibodies (NAbs) against UHV-1, UGV-1, and ABV-1, EDTA plasma was prepared from the blood samples (11). A 2-fold dilution series of the plasma (1:50 to 1:6,400) was incubated with the different pseudotyped VSVs (50 to 150 FFFU) at 30°C for 60 min, with a plasma-pseudovirus mixture volume of 200 μl. The plasma-pseudovirus mixtures were then laid onto 80 to 90% confluent I/1Ki cells grown on 96-well plates (Viewplate-96 black, optically clear bottom, tissue culture treated, sterile, 96-well with lid; PerkinElmer), at 50 μl/well. After 2 h of incubation at 30°C, the virus-plasma mixture was replaced with fresh fully supplemented medium (see “Cells, viruses, and purification of viruses” above), and the plate was incubated for 16 to 24 h at 30°C. Infected cells were enumerated using fluorescence microscopy. All experiments were performed in triplicate. Plasma samples of 24 naturally reptarenavirus-infected snakes from a previous study (11) were analyzed using VSVs pseudotyped with S5-like, tavallinen suomalainen mies virus 2 (TSMV-2), and UGV-1 GPs. These reptarenavirus S segments had been found in the collection, and the snakes had been analyzed by RT-PCR for their presence at the time of sampling (11). The neutralizing titer was determined as the plasma dilution that induced at least a 50% reduction in the number of fluorescent foci.
ACKNOWLEDGMENTS
This work was supported by grants from the Jenny and Antti Wihuri Foundation (to J.H.) and the Academy of Finland (grant numbers 308613 and 314119 to J.H.).
REFERENCES
- 1.Chang L, Jacobson ER. 2010. Inclusion body disease, a worldwide infectious disease of boid snakes: a review. J Exot Pet Med 19:216–225. doi: 10.1053/j.jepm.2010.07.014. [DOI] [Google Scholar]
- 2.Vancraeynest D, Pasmans F, Martel A, Chiers K, Meulemans G, Mast J, Zwart P, Ducatelle R. 2006. Inclusion body disease in snakes: a review and description of three cases in boa constrictors in Belgium. Vet Rec 158:757–760. doi: 10.1136/vr.158.22.757. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher J, Jacobson ER, Homer BL, Gaskin JM. 1994. Inclusion body disease in boid snakes. J Zoo Wildl Med 25:511–524. [Google Scholar]
- 4.Wozniak E, McBride J, DeNardo D, Tarara R, Wong V, Osburn B. 2000. Isolation and characterization of an antigenically distinct 68-kd protein from nonviral intracytoplasmic inclusions in boa constrictors chronically infected with the inclusion body disease virus (IBDV: Retroviridae). Vet Pathol 37:449–459. doi: 10.1354/vp.37-5-449. [DOI] [PubMed] [Google Scholar]
- 5.Chang LW, Fu A, Wozniak E, Chow M, Duke DG, Green L, Kelley K, Hernandez JA, Jacobson ER. 2013. Immunohistochemical detection of a unique protein within cells of snakes having inclusion body disease, a world-wide disease seen in members of the families Boidae and Pythonidae. PLoS One 8:e82916. doi: 10.1371/journal.pone.0082916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang L, Fu D, Stenglein MD, Hernandez JA, DeRisi JL, Jacobson ER. 2016. Detection and prevalence of boid inclusion body disease in collections of boas and pythons using immunological assays. Vet J 218:13–18. doi: 10.1016/j.tvjl.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, DeRisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3:e00180-12. doi: 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2013. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J Gen Virol 94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 9.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2013. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenglein MD, Sanchez-Migallon Guzman D, Garcia VE, Layton ML, Hoon-Hanks LL, Boback SM, Keel MK, Drazenovich T, Hawkins MG, DeRisi JL. 2017. Differential disease susceptibilities in experimentally reptarenavirus-infected boa constrictors and ball pythons. J Virol 91:e00451-17. doi: 10.1128/JVI.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Windbichler K, Michalopoulou E, Palamides P, Pesch T, Jelinek C, Vapalahti O, Kipar A, Hetzel U, Hepojoki J. 2019. Antibody response in snakes with boid inclusion body disease. PLoS One 14:e0221863. doi: 10.1371/journal.pone.0221863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 13.Stenglein MD, Jacobson ER, Chang LW, Sanders C, Hawkins MG, Guzman DS, Drazenovich T, Dunker F, Kamaka EK, Fisher D, Reavill DR, Meola LF, Levens G, DeRisi JL. 2015. Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog 11:e1004900. doi: 10.1371/journal.ppat.1004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hepojoki J, Salmenpera P, Sironen T, Hetzel U, Korzyukov Y, Kipar A, Vapalahti O. 2015. Arenavirus coinfections are common in snakes with boid inclusion body disease. J Virol 89:8657–8660. doi: 10.1128/JVI.01112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aqrawi T, Stohr AC, Knauf-Witzens T, Krengel A, Heckers KO, Marschang RE. 2015. Identification of snake arenaviruses in live boas and pythons in a zoo in Germany. Tierarztl Prax Ausg K Kleintiere Heimtiere 43:239–247. doi: 10.15654/TPK-140743. [DOI] [PubMed] [Google Scholar]
- 16.Keller S, Hetzel U, Sironen T, Korzyukov Y, Vapalahti O, Kipar A, Hepojoki J. 2017. Co-infecting reptarenaviruses can be vertically transmitted in boa constrictor. PLoS Pathog 13:e1006179. doi: 10.1371/journal.ppat.1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maes P, Alkhovsky SV, Bao Y, Beer M, Birkhead M, Briese T, Buchmeier MJ, Calisher CH, Charrel RN, Choi IR, Clegg CS, de la Torre JC, Delwart E, DeRisi JL, Di Bello PL, Di Serio F, Digiaro M, Dolja VV, Drosten C, Druciarek TZ, Du J, Ebihara H, Elbeaino T, Gergerich RC, Gillis AN, Gonzalez J-PJ, Haenni A-L, Hepojoki J, Hetzel U, Ho T, Hong N, Jain RK, Jansen van Vuren P, Jin Q, Jonson MG, Junglen S, Keller KE, Kemp A, Kipar A, Kondov NO, Koonin EV, Kormelink R, Korzyukov Y, Krupovic M, Lambert AJ, Laney AG, LeBreton M, Lukashevich IS, Marklewitz M, Markotter W, et al. 2018. Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018. Arch Virol 163:2295–2310. doi: 10.1007/s00705-018-3843-5. [DOI] [PubMed] [Google Scholar]
- 18.Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, Briese T, Buchmeier MJ, Burt FJ, Calisher CH, Cháng C, Charrel RN, Choi IR, Clegg JCS, de la Torre JC, de Lamballerie X, Dèng F, Di Serio F, Digiaro M, Drebot MA, Duàn X, Ebihara H, Elbeaino T, Ergünay K, Fulhorst CF, Garrison AR, Gāo GF, Gonzalez J-PJ, Groschup MH, Günther S, Haenni A-L, Hall RA, Hepojoki J, Hewson R, Hú Z, Hughes HR, Jonson MG, Junglen S, Klempa B, Klingström J, Kòu C, Laenen L, Lambert AJ, Langevin SA, Liu D, Lukashevich IS, et al. 2019. Taxonomy of the order Bunyavirales: update 2019. Arch Virol 164:1949–1965. doi: 10.1007/s00705-019-04253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hepojoki J, Hepojoki S, Smura T, Szirovicza L, Dervas E, Prahauser B, Nufer L, Schraner EM, Vapalahti O, Kipar A, Hetzel U. 2018. Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses—the representatives of genus Hartmanivirus, family Arenaviridae. PLoS Pathog 14:e1007415. doi: 10.1371/journal.ppat.1007415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Argenta FF, Hepojoki J, Smura T, Szirovicza L, Hammerschmitt ME, Driemeier D, Kipar A, Hetzel U. 2020. Identification of reptarenaviruses, hartmaniviruses and a novel chuvirus in captive Brazilian native boa constrictors with boid inclusion body disease. J Virol 94:e00001-20. doi: 10.1128/JVI.00001-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, Wang W, Eden JS, Shen JJ, Liu L, Holmes EC, Zhang YZ. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 22.Radoshitzky SR, Buchmeier MJ, Charrel RN, Clegg JCS, Gonzalez JJ, Gunther S, Hepojoki J, Kuhn JH, Lukashevich IS, Romanowski V, Salvato MS, Sironi M, Stenglein MD, de la Torre JC, ICTV Report Consortium. 2019. ICTV virus taxonomy profile: Arenaviridae. J Gen Virol 100:1200–1201. doi: 10.1099/jgv.0.001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hepojoki J, Kipar A, Korzyukov Y, Bell-Sakyi L, Vapalahti O, Hetzel U. 2015. Replication of boid inclusion body disease-associated arenaviruses is temperature sensitive in both boid and mammalian cells. J Virol 89:1119–1128. doi: 10.1128/JVI.03119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korzyukov Y, Iheozor-Ejiofor R, Levanov L, Smura T, Hetzel U, Szirovicza L, de la Torre JC, Martinez-Sobrido L, Kipar A, Vapalahti O, Hepojoki J. 2020. Differences in tissue and species tropism of reptarenavirus species studied by vesicular stomatitis virus pseudotypes. Viruses 12:395. doi: 10.3390/v12040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korzyukov Y, Hetzel U, Kipar A, Vapalahti O, Hepojoki J. 2016. Generation of anti-boa immunoglobulin antibodies for serodiagnostic applications, and their use to detect anti-reptarenavirus antibodies in boa constrictor. PLoS One 11:e0158417. doi: 10.1371/journal.pone.0158417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simard J, Marschang RE, Leineweber C, Hellebuyck T. 2020. Prevalence of inclusion body disease and associated comorbidity in captive collections of boid and pythonid snakes in Belgium. PLoS One 15:e0229667. doi: 10.1371/journal.pone.0229667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyndman TH, Marschang RE, Bruce M, Clark P, Vitali SD. 2019. Reptarenaviruses in apparently healthy snakes in an Australian zoological collection. Aust Vet J 97:93–102. doi: 10.1111/avj.12792. [DOI] [PubMed] [Google Scholar]
- 28.Dietz J, Kolesnik E, Heckers KO, Klingberg M-N, Marschang RE. 2020. Detection of an arenavirus in a group of captive Wagler’s pit vipers (Tropidolaemus wagleri). J Zoo Wildl Med 51:236–240. doi: 10.1638/2018-0179. [DOI] [PubMed] [Google Scholar]
- 29.Szirovicza L, Hetzel U, Kipar A, Martinez-Sobrido L, Vapalahti O, Hepojoki J. 2020. Snake deltavirus utilizes envelope proteins of different viruses to generate infectious particles. mBio 11:e03250-19. doi: 10.1128/mBio.03250-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baggio F, Hetzel U, Nufer L, Kipar A, Hepojoki J. 2020. Arenavirus nucleoprotein localizes to mitochondria. bioRxiv 10.1101/2020.11.06.370825. [DOI] [PMC free article] [PubMed]
- 31.Laguens RM, Lampuri JS, Coto CE, Laguens RP. 1982. Pathology of persistent infection of Calomys musculinus with an attenuated strain Junin virus. Medicina (B Aires) 42:273–278. [PubMed] [Google Scholar]
- 32.Lampuri JS, Vidal MD, Coto CE. 1982. Response of Calomys musculinus to experimental infection with Junin virus. Medicina (B Aires) 42:61–66. [PubMed] [Google Scholar]
- 33.Alche LE, Coulombie FC, Coto CE. 1985. Isolation of Junin virus from blood and peripheral lymphocytes of infected Calomys musculinus. Rev Argent Microbiol 17:177–181. [PubMed] [Google Scholar]
- 34.Arikawa J, Ito M, Yao JS, Kariwa H, Takashima I, Hashimoto N. 1994. Epizootiological studies of hantavirus infection among urban rats in Hokkaido, Japan: evidences for the persistent infection from the sero-epizootiological surveys and antigenic characterizations of hantavirus isolates. J Vet Med Sci 56:27–32. doi: 10.1292/jvms.56.27. [DOI] [PubMed] [Google Scholar]
- 35.Botten J, Mirowsky K, Kusewitt D, Bharadwaj M, Yee J, Ricci R, Feddersen RM, Hjelle B. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc Natl Acad Sci U S A 97:10578–10583. doi: 10.1073/pnas.180197197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciurea A, Klenerman P, Hunziker L, Horvath E, Senn BM, Ochsenbein AF, Hengartner H, Zinkernagel RM. 2000. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc Natl Acad Sci U S A 97:2749–2754. doi: 10.1073/pnas.040558797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmerman LM, Vogel LA, Bowden RM. 2010. Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol 213:661–671. doi: 10.1242/jeb.038315. [DOI] [PubMed] [Google Scholar]
- 38.Hepojoki J, Strandin T, Vaheri A, Lankinen H. 2010. Interactions and oligomerization of hantavirus glycoproteins. J Virol 84:227–242. doi: 10.1128/JVI.00481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Argenta FF, Hepojoki J, Smura T, Szirovicza L, Hammerschmitt M, Driemeier D, Kipar A, Hetzel U. 2020. Identification of reptarenaviruses, hartmaniviruses and a novel chuvirus in captive Brazilian native boa constrictors with boid inclusion body disease. bioRxiv 10.1101/2020.01.02.893420. [DOI] [PMC free article] [PubMed]