Kaposi’s sarcoma-associated herpesvirus (KSHV) causes Kaposi’s sarcoma (KS). The contribution of KSHV microRNAs (miRNAs) to oncogenesis is not fully understood.
KEYWORDS: Kaposi's sarcoma-associated herpesvirus, endothelial cells, extracellular acidification, glycolysis, microRNA, oncogenesis, oxygen consumption, tubule formation, wound healing
ABSTRACT
Kaposi’s sarcoma (KS) results from the transformation of Kaposi's sarcoma-associated herpesvirus (KSHV)-infected endothelial cells. The contribution of the KSHV microRNAs (miRNAs) to the process of oncogenesis in endothelial cells has not been fully elucidated. To better understand the contributions of individual miRNAs to oncogenesis-related cellular phenotypes, we used KSHV miRNA knockout mutants, each lacking one of the 12 miRNA genes. An additional mutant lacked all miRNAs. Since KSHV infection causes a variety of phenotypic changes in endothelial cells, we tested the mutants for their ability to effect such changes in telomerase-immortalized vein endothelial (TIVE) cells infected with each of the mutant viruses. Wild type- and mutant-infected as well as uninfected cells were evaluated for perturbations to proliferation, migration, tubule formation, and glycolysis. We found broad variation between the different viruses in these aspects. With respect to proliferation rate, ΔmiR-K12-3, ΔmiR-K12-8, and ΔmiR-K12-11 showed significant impairment. Cells infected with ΔmiR-K12-11 had reduced migration. In tubule formation, the ΔmiR-K12-5, -6, and -7 viruses were deficient. At the same time, cells infected with the ΔmiR-K12-10 virus showed dysregulated glycolysis. By combining these observations with previously published KSHV miRNA targetome lists from ribonomics data, we were able to functionally validate a number of new miRNA targets in specific pathways. As proof of concept, miR-K12-3 was shown to target cathepsin D, a strong promoter of apoptosis. Taken together, the results demonstrate that KSHV miRNAs play different roles in inducing the phenotypic changes that are characteristic of transformed cells.
IMPORTANCE Kaposi’s sarcoma-associated herpesvirus (KSHV) causes Kaposi’s sarcoma (KS). The contribution of KSHV microRNAs (miRNAs) to oncogenesis is not fully understood. This is particularly true for human endothelial cells, the cell type from which KS tumors are derived. Here, we used a panel of KSHV miRNA knockout viruses to shed light on the roles of individual miRNAs in the process of transformation. Latently infected endothelial cells were studied for phenotypic changes related to cancer, including proliferation, migration, angiogenesis, glycolysis, and apoptosis. The mutant-infected cell lines displayed a wide range of phenotypes in these selected measures of oncogenesis, which differed from those of wild-type-infected cells and from each other. These results indicate that KSHV miRNAs contribute to different aspects of oncogenesis and that each one has a unique role to play.
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi’s sarcoma (KS) (1), an endothelial cell tumor, as well as primary effusion lymphoma (PEL) (2) and multicentric Castleman’s disease (MCD) (3), two B-cell proliferation disorders. As a gammaherpesvirus, KSHV undergoes a latent phase of replication that has been tied to oncogenesis. A limited subset of genes is expressed during latency, including the viral microRNAs (miRNAs) (4). miRNAs are short RNAs, 22 nucleotides on average, and function as important posttranscriptional regulators of gene expression. They modulate the translation of target mRNAs by binding to imperfectly complementary sequences, which are often, but not always, located within the 3′ untranslated regions (UTRs) of the transcripts (5, 6). Dysregulation of human miRNA expression is often associated with both tumorigenesis and maintenance of tumor growth (7).
There is evidence that the KSHV miRNAs are drivers of tumorigenesis. For example, KSHV miR-K12-11 is the ortholog of the cellular oncomir miR-155, and its KSHV ortholog, miR-K12-11, has been shown to induce splenic B-cell expansion when expressed in human hematopoietic progenitors in nude mice (8, 9). KSHV also drives phenotypic changes in endothelial cells, which are hallmarks of transformation. Infection with the virus has been shown to induce angiogenesis through upregulation of hypoxia-induced factor 1α (HIF1α) (10). KSHV infection induces the Warburg effect, a shift from aerobic to anaerobic glycolysis, in latently infected endothelial cells (11). In fact, later experiments demonstrated that transducing lymphatic endothelial cells (LECs) with a lentivirus expressing the KSHV miRNA cluster was sufficient to recapitulate this phenotype (12). It was also found that KSHV can induce greater invasiveness in oral fibroblasts (13).
We sought to characterize the contributions of individual KSHV miRNAs to the previously noted phenotypic changes that cells undergo upon infection with KSHV. To this end, we utilized a panel of miRNA knockout mutants in the BAC16 background that were generated previously (14). Telomerase immortalized vein endothelial (TIVE) cells were infected separately with each of the mutant viruses and allowed to attain latency. Cell proliferation, migration, tubule formation as a proxy for angiogenesis, and the Warburg effect were evaluated for each of the mutant-infected cell lines and compared to WT-infected cells. We found broadly varying effects of specific miRNA mutant viruses on endothelial cell phenotype. These results indicate that different KSHV miRNAs target specific pathways in the KSHV-induced transformation of endothelial cells. Furthermore, this approach was combined with data mining of previously published ribonomics data for miRNA targetome analysis and our quick cross-linking sequencing and analysis of hybrids (qCLASH) data set to identify and functionally validate and link specific KSHV miRNAs to pathways involved in KSHV-related tumorigenesis (15–17).
RESULTS
Generating latently infected TIVE cells with BAC16-derived KSHV miRNA deletion mutants.
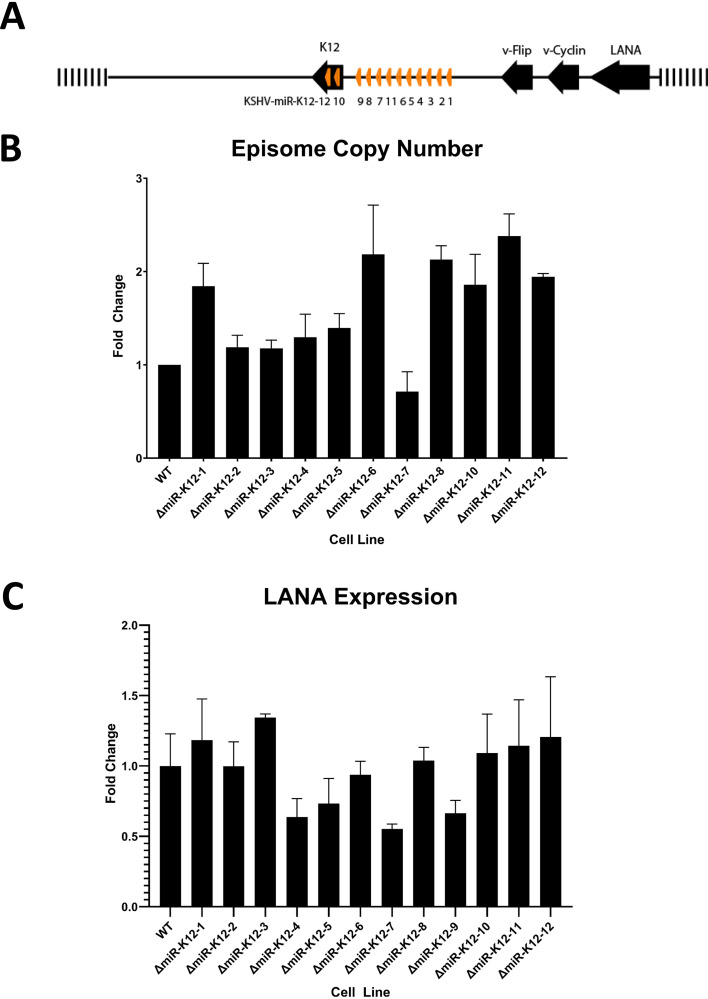
The location of the 12 KSHV miRNA genes in the latency region is shown in Fig. 1A. The generation and quality control of 13 mutant viruses that each have one of the 12 miRNA genes deleted or all 12 (Δall) have previously been described. These mutants exist in latently infected iSLK cells that can be induced to generate high-titer cell-free virus (14). TIVE cells were infected with all 13 mutants and WT BAC16 with approximately 1,000 genome equivalents and selected with hygromycin B until 100% green fluorescent protein (GFP) positive to generate 14 different cell lines. For one mutant, ΔmiR-K12-9, several attempts were required before a successful infection could be established. In the end, the amount of virus had to be doubled in order for a small proportion of cells to become infected with this mutant. Cells were expanded and cryopreserved prior to the phenotype analysis described below. It is important to note that human telomerase-immortalized vein endothelial cells are not transformed, since they do not form tumors in nude mice (18). TIVE cells, like primary endothelial cells, have slow growth, are dependent on endothelial cell growth factors, and have retained the ability to form tubules on Matrigel, an endothelial cell-specific phenotype. The advantage of using TIVE cells over human umbilical vein endothelial cells (HUVEC) for this study, which required large cell numbers for many assays, is that HUVEC have donor-to-donor variations that complicate the monitoring of miRNA effects in the context of viral infection.
FIG 1.
Generation and quality control of viral miRNA mutant- and WT-infected TIVE cells. (A) KSHV miRNA locus showing the cluster of 10 miRNAs and 2 in the K12 open reading frame. The -Δall mutant refers to a virus that has the entire cluster deleted plus the two in K12 changed without affecting the K12 ORF. (B) qPCR analysis of episome copy number. Genomic DNA from infected TIVE cells was quantified using LANA-specific primers and compared to a standard curve. (C) RNA-seq-based quantification of LANA expression levels normalized to GAPDH.
Prior to phenotypic characterization of these mutant virus-infected TIVE cells, we performed quantitative PCR (qPCR) for viral episome copy number. This analysis was performed at three different time points with freshly thawed cells over a 1-year period. As shown in Fig. 1B, the maximum fold change between the wild type (WT) and all mutants was approximately 2-fold. Given that KSHV episome segregation in culture is heterogeneous with respect to every cell division (19, 20), differences of less than 2-fold suggest that, overall, cells harbor similar episome numbers. Second, to ensure comparable gene expression of the KLAR region, we analyzed a transcriptome sequencing (RNA-seq) data set from mutant- and wild-type-infected TIVE cells (to be reported elsewhere) for expression levels of the latency-associated nuclear antigen (LANA). As shown in Fig. 1C, the maximum fold change in expression level between the wild type and all analyzed mutants again is less than 2-fold. Since it was previously shown that latent gene expression differs from cell to cell within clonal cultures (21), these results further support equal levels of viral gene expression in these cell lines. Different biological replicates of assays were performed with different frozen stocks of the same cell lines.
Several KSHV miRNA knockout viruses, including ΔmiR-K12-11, show significant defects in proliferation.
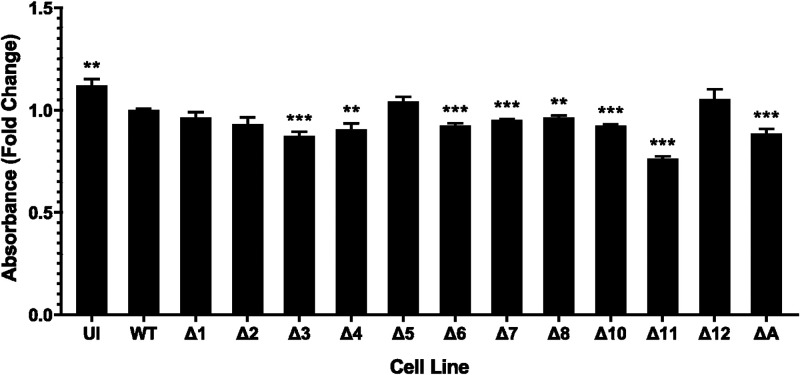
Cell proliferation assays were performed to measure cell growth rate. This type of colorimetric assay detects the bioreduction of a tetrazolium compound, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS), by cells. Although we had observed that WT KSHV-infected cells grew faster and needed to be subcultivated more frequently than uninfected cells, the results of this assay indicated that uninfected cells proliferated at a greater rate than WT-infected cells. We can only surmise that this is related to the metabolic disturbances caused when cells are infected with KSHV (11). However, this should not negate the usefulness of the cell proliferation assay when comparing the different mutant-infected cell lines with WT BAC16. The absorbance was measured for each cell line and compared with that of WT-infected cells. We observed that cells infected with the ΔmiR-K12-11 virus had the most profound defect in proliferative ability. The ΔmiR-K12-3, -4, -6, -7, -8, and -10 cell lines had a modest but significantly slower growth in this metric as well (Fig. 2). Since cells multiply more slowly when these miRNAs are absent, this suggests that these miRNAs play a role in the KSHV-induced enhancement of cell proliferation. Indeed, the findings for ΔmiR-K12-11 cells were expected based on previous work (9). miR-K12-4 was previously identified as capable of rescuing a proliferative phenotype in Δcluster-infected rat mesenchymal precursor cells (22). miR-K12-10 and miR-K12-11 both downregulate the transforming growth factor β (TGFβ) pathway, thereby encouraging cell survival (23, 24). To our knowledge, miR-K12-6 and miR-K12-8 have not previously been identified as potential modulators of cell proliferation or cell survival enhancement.
FIG 2.
KSHV miRNA deletion mutants show changes to cell proliferation. Shown are cell proliferation assay results of TIVE cells infected with viral mutants. Results are averages from three independent experiments consisting of four replicates each. UI, uninfected. Error bars show standard errors. P values versus WT by 2-tailed Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Cells infected with ΔmiR mutants, including ΔmiR-K12-7 and ΔmiR-K12-11, show reduced migration speed.
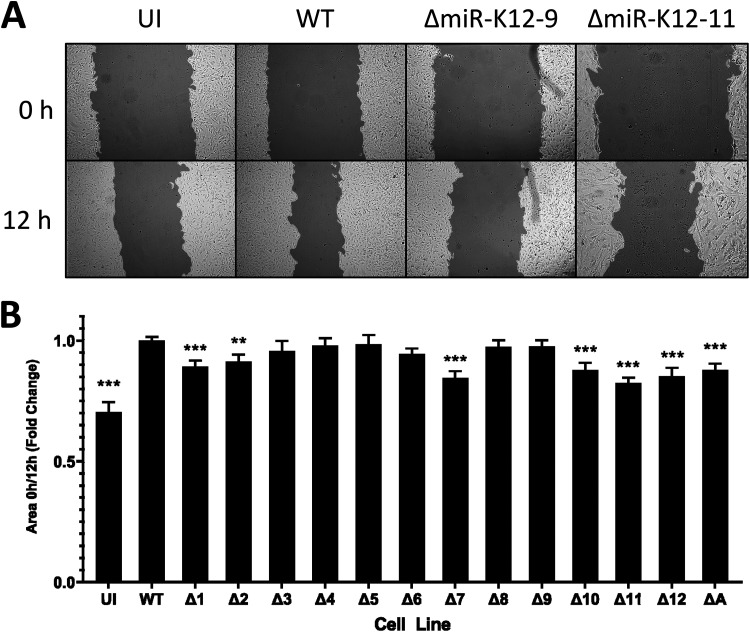
Wound-healing assays are designed to measure the rate at which cells can migrate. The assay involves growing cells to confluence and then scratching the monolayer with a pipette tip to create a uniform “wound.” Endothelial cells, being the component cells of blood vessels, will naturally migrate to patch any injury. In cancer, the cell migration rate is enhanced, contributing to the invasiveness of tumors. Wound-healing assays were performed on each of the mutant-infected cell lines as well as WT-infected and uninfected cells. Photographs were taken of the cells at 0 and 12 h after scratching. The ratio between the scratch area at 0 h and at 12 h was determined using TScratch (25). Infection with WT KSHV increased the migration rate of TIVE cells (Fig. 3A and B). We found that ΔmiR-K12-7- and ΔmiR-K12-11-infected TIVE cells as well as Δall-infected cells moved into the cell-free area more slowly than WT-infected cells. ΔmiR-K12-1-, -10-, and -12-infected cells also showed a deficiency in this respect, although to a lesser extent (Fig. 3B). miR-K12-11 was previously shown to indirectly downregulate dual-specificity phosphatase 1 (DUSP1), an inhibitor of mitogen-activated protein kinase (MAPK) signaling. The restoration of MAPK signaling has a number of downstream effects, including the release of promigratory factors (26). This may explain why the absence of miR-K12-11 leads to a reduction in the migration speed of infected cells. Another interesting result was that ΔmiR-K12-9-infected cells, although they did not show a significant difference in migration speed compared with WT-infected cells, were found to be considerably larger than all of the other cell lines tested (Fig. 3A). As a consequence of their greater size, ΔmiR-K12-9-infected cells had to be seeded at a lower density to produce a flat, even monolayer. This slight difference in protocol may somewhat complicate the comparison of WT- and ΔmiR-K12-9-infected cells.
FIG 3.
ΔmiR-K12-11-infected TIVE cells and several other mutants show reduced migration speed. (A) Representative images of uninfected (UI) or wild-type (WT)-, ΔmiR-K12-9-, or ΔmiR-K12-11-infected TIVE cells at 0 and 12 h after a scratch was made in the cell monolayer. (B) Quantification of wound-healing assays. The ratio of area covered by cells at 0 h postscratching compared to the cell-covered area at 12 h postscratching was determined. Data are averages from 3 independent experiments consisting of 3 to 6 replicates per treatment, normalized to the WT. Error bars show standard errors. P values versus WT by 2-tailed Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Multiple miRNAs strongly affect tubule formation, a surrogate marker for angiogenesis.
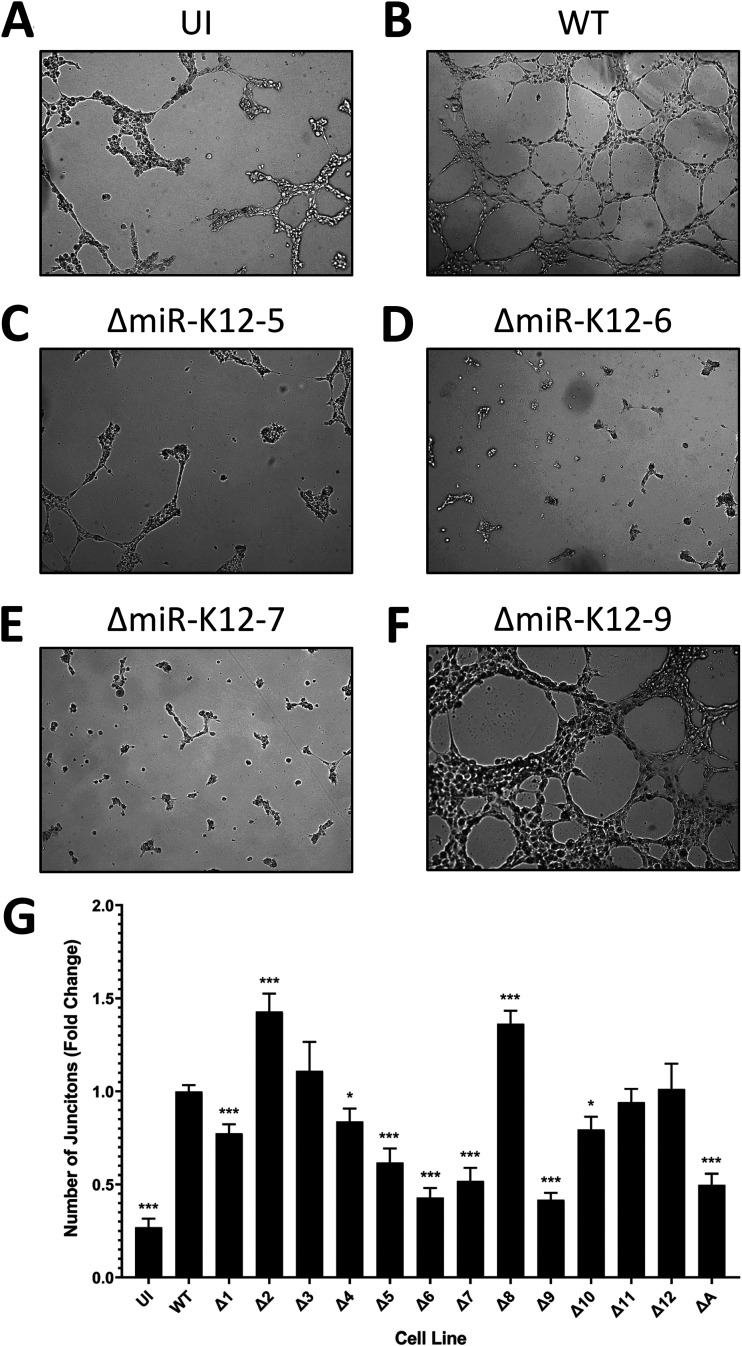
To evaluate the angiogenic abilities of each mutant virus, the virus-infected cell lines were used in tubule formation assays. TIVE cells are blood endothelial cells, so they can form vessels in culture as long as they are provided with an appropriate extracellular matrix. The cells were plated on Matrigel, which serves as the matrix, and photographed at 6 and 12 h after plating. The number of junctions between three or more tubules was counted. It was observed that cells infected with WT KSHV formed more junctions than uninfected cells (Fig. 4A and B). This was not unexpected, as it has already been shown that KSHV enhances angiogenesis (27, 28). Cells infected with ΔmiR-K12-5, -6, and -7 showed a significant defect in the ability to form tubules (Fig. 4C, D, and E). They formed fewer junctions than even uninfected cells. ΔmiR-K12-9-infected cells had a lower number of junctions than WT-infected cells, but, perhaps due to the larger size of these cells, it looked as though ΔmiR-K12-9-infected cells formed a larger network of tubules than the other cell lines. This may mean that the ability to form tubules was not altered but rather that fewer junctions could be found in the same-sized microscope field (Fig. 4F). Cells infected with ΔmiR-K12-1, -10, and Δall also formed fewer junctions than WT-infected cells. In contrast, the cells infected with ΔmiR-K12-2 and -8 actually showed a significant increase in the number of junctions that formed (Fig. 4G). This suggests that these two miRNAs actually suppress angiogenesis, perhaps serving as a necessary counterbalance to the proangiogenic miRNAs. Overall, there is a strong indication that miR-K12-5, -6, and -7 are indispensable to KSHV-induced angiogenesis. miR-K12-6, in particular, has already been strongly implicated in angiogenesis. It was found that miR-K12-6-3p targets SH3 domain binding glutamate-rich protein (SH3BGR), resulting in enhanced cell migration and angiogenesis (29). The same researchers also noted that miR-K12-6-5p targets the metastasis suppressor CD82. Downregulation of CD82 resulted in greater invasiveness and angiogenic ability in KSHV-infected primary endothelial cells (30). Others showed that miR-K12-6-5p targets breakpoint cluster region protein (BCR), leading to the derepression of ras-related C3 botulinum toxin substrate 1 (Rac1) and the subsequent enhancement of angiogenesis (31). To our knowledge, miR-K12-7 has not previously been identified to be specifically involved in targeting angiogenesis.
FIG 4.
Several miRNA knockouts show a marked reduction in tubule formation, a proxy for angiogenesis. (A to F) Representative images of uninfected (UI) cells or cells infected with wild-type (WT), ΔmiR-K12-5, ΔmiR-K12-6, ΔmiR-K12-7, or ΔmiR-K12-9 virus. Magnification is ×50. (G) Quantification of tubule formation assays. The number of junctions between three or more tubules was counted for each image. Data are averages from 3 independent experiments consisting of 12 replicates per treatment, normalized to the WT. Error bars show standard errors. P values versus the WT by 2-tailed Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
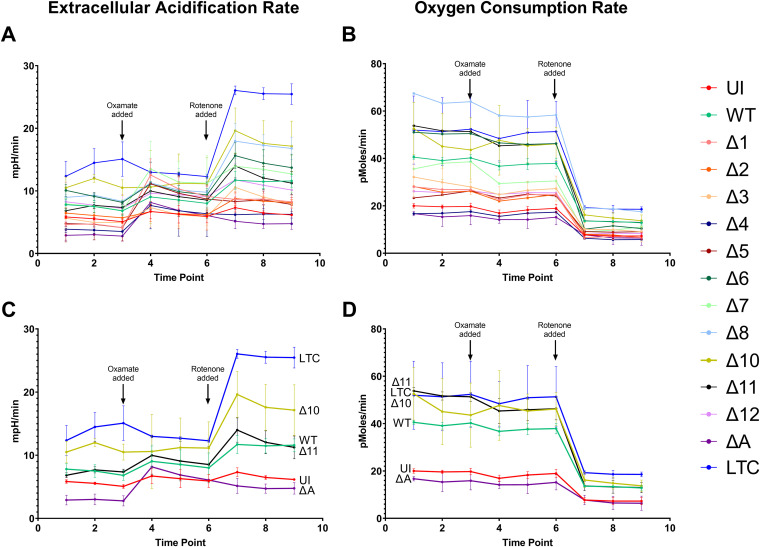
ΔmiR-K12-10-infected cells display highly dysregulated glycolysis.
The Warburg effect is a phenomenon in cancer cells that is characterized by an increased rate of glycolysis in conjunction with increased use of lactic acid fermentation as a source of ATP (32). Cells experiencing the Warburg effect do not rely on oxidative phosphorylation as their main source of ATP, so they require less oxygen than normal cells and would be expected to have a lower oxygen consumption rate (OCR). Another characteristic of these cells is the accumulation of lactic acid, which leads to an increase in the extracellular acidification rate (ECAR). KSHV infection is known to induce the Warburg effect in endothelial cells. It has also been shown that the miRNA cluster alone is sufficient to produce this effect (11). To assess whether or not the various KSHV mutant-infected cell lines were capable of inducing the Warburg effect, an Agilent Seahorse XF96 machine was used to simultaneously measure the OCR and ECAR of living cells. At certain time points, oxamate, an inhibitor of glycolysis, and rotenone, an inhibitor of oxidative phosphorylation, were added. Interrupting these two metabolic pathways was intended to give a clearer picture of which cell lines were relying more heavily on which pathways. In addition to uninfected and WT-infected cells, we used an additional cell line in this assay. Telomerase immortalized vein endothelial long-term culture (TIVE-LTC) cells are both KSHV positive and fully transformed (18). As a transformed cell line, we expected that TIVE-LTC cells would show the strongest induction of the Warburg effect and, therefore, would make an appropriate positive control.
Much to our surprise, the TIVE-LTC cells had a much higher OCR than uninfected TIVE cells. This is the opposite of what was anticipated based on results obtained by others (11). WT-infected cells had an OCR that was intermediate between these two extremes. Cells infected with the virus from which all miRNAs were deleted had an OCR similar to that of uninfected cells. The cells infected with the ΔmiR-K12-10 virus had OCR readings on par with that of TIVE-LTC cells (Fig. 5A).
FIG 5.
miR-K12-10 may be important for KSHV-induced changes to glycolysis. Shown are the extracellular acidification rate and oxygen consumption rate of uninfected, WT-infected, or mutant-infected cell lines. Panels A and B show results for all cell lines, while panels C and D show the results only for the more divergent cell lines. The time points at which oxamate, an inhibitor of glycolysis, and rotenone, an inhibitor of oxidative phosphorylation, were added are indicated. Results are averages from 8 replicates of each cell line. Error bars show standard errors. Results from some mutants with intermediate phenotypes were omitted for clarity. (A and C) Extracellular acidification rate. (B and D) Oxygen consumption rate.
The ECAR results were more in line with expectations. TIVE-LTC cells had the highest ECAR, while uninfected and Δall-infected cells had the lowest. WT-infected cells had an intermediate phenotype, while ΔmiR-K12-10-infected cells had an ECAR more similar to that of TIVE-LTC cells (Fig. 5B).
miR-K12-3 blocks apoptosis by strongly suppressing cathepsin D.
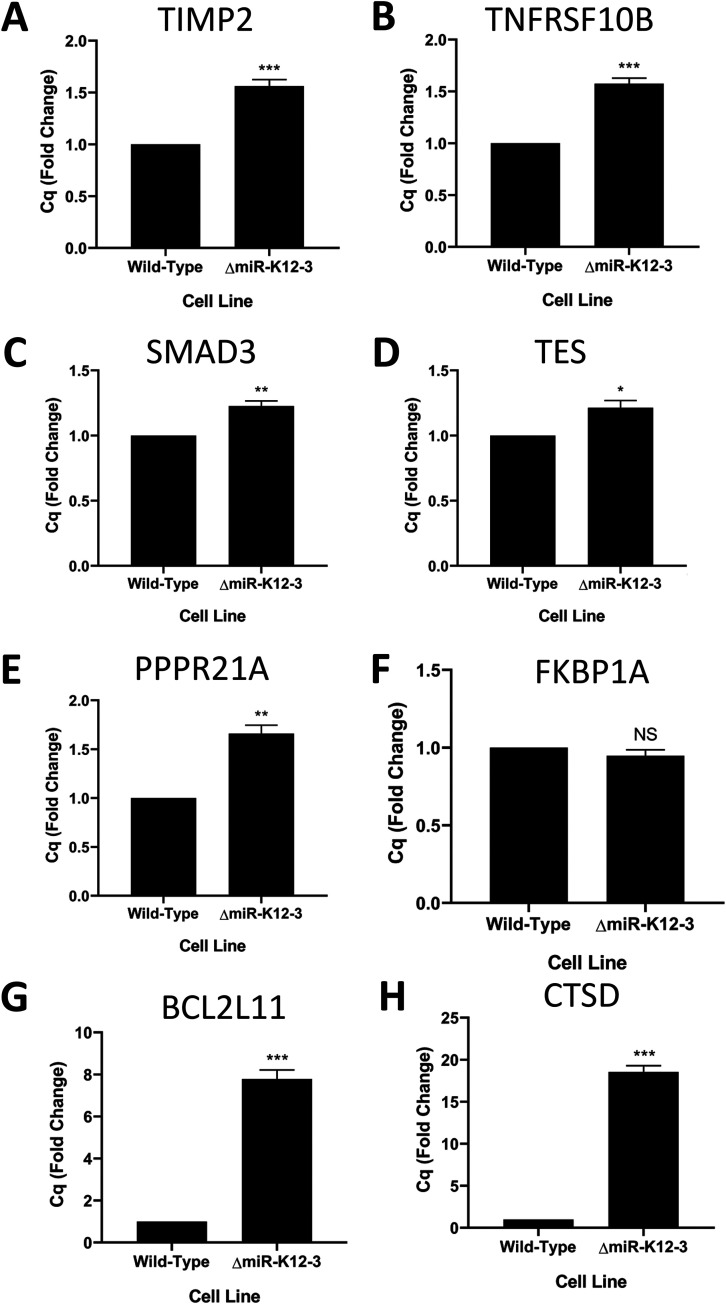
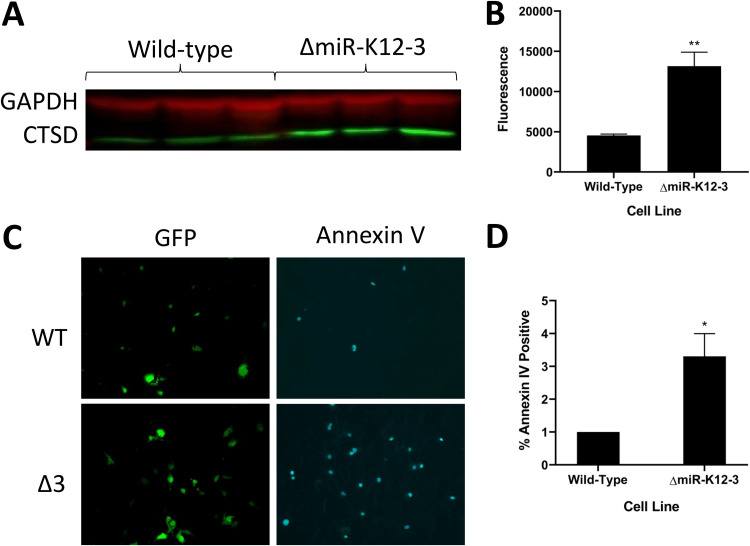
The long-term goal of these studies is to explain the observed phenotypes by linking specific miRNAs to host genes and their networks within the context of viral infection. Table 1 summarizes the observed findings for the ΔmiR-K12-3 mutant in the proliferation, migration, angiogenesis, and glycolysis assays. Recently, we performed quick cross-linking sequencing and analysis of hybrids (qCLASH) in TIVE-LTC cells, which identified several thousand high-confidence miRNA targets (17). In a first step to understand how specific miRNAs other than miR-K12-11 contribute to proliferation, we searched for miR-K12-3 targets within the qCLASH data set and found a number of genes that can play roles in regulating proliferation or apoptosis; we note that the cell proliferation assay does not distinguish between inhibitors of cell growth or apoptosis (Table 1). These were TIMP2 (TIMP metallopeptidase inhibitor 2), TNFRSF10B (TNF receptor superfamily member 10b), SMAD3 (SMAD family member 3), TES (testin LIM domain protein), PPPR21A, FKBP1A (FKBP prolyl isomerase 1A), BCL2L11 (BCL2 like 11), and CTSD (cathepsin D). Performing quantitative reverse transcription-PCR (qRT-PCR) assays in ΔmiR-K12-3- versus WT-infected cells revealed that seven of eight genes were significantly derepressed in the absence of miR-K12-3 (Fig. 6). The CTSD mRNA, a strong inducer of apoptosis, was upregulated more than 15-fold in the absence of mir-K12-3 (Fig. 6H). Although not previously known to be a KSHV miRNA target, the CTSD transcript contains three binding sites for miR-K12-3 identified by qCLASH (17). CTSD has been shown to be negatively regulated by vIL6, thereby leading to the suppression of apoptosis in PEL cells (33). This suggests that the presence of CTSD is particularly detrimental to KSHV latency. Next, we measured the expression of CTSD protein by Western blotting in WT- and ΔmiR-K12-3-infected TIVE cells. CTSD expression was significantly lower in WT-infected cells at both the mRNA (Fig. 6H) and the protein level (Fig. 7A and B), validating CTSD as an miR-K12-3 target in TIVE cells. To further follow up on the idea that miR-K12-3 is an inhibitor of apoptosis, we performed an annexin V detection assay. When comparing the proportion of annexin V-positive cells in WT- and ΔmiR-K12-3-infected TIVE cells, we observed that the ΔmiR-K12-3-infected cells had a significantly greater proportion of cells stained positive for annexin V (Fig. 7C and D). This suggests that miR-K12-3 is at least partly responsible for the inhibition of apoptosis seen in KSHV-infected TIVE cells. These results also validate the interaction that was found by qCLASH (17).
TABLE 1.
Summary of phenotypes observed for mutant-infected TIVE cellsa
| Virus | Example target gene(s) | Phenotype |
|||
|---|---|---|---|---|---|
| Proliferation | Migration | Angiogenesis | ECAR/OCR | ||
| ΔmiR-K12-1 | ST13, ANXA5, NHLRC2, LASS2 | Normal | Greatly reduced | Greatly reduced | Greatly reduced/greatly reduced |
| ΔmiR-K12-2 | PIP5K1A, XRCC6, DDX3X, VIM | Normal | Slightly reduced | Greatly increased | Greatly reduced/greatly reduced |
| ΔmiR-K12-3 | TES, PPP2R1A, TIMP2, BCL2L11, TNFRSF10B, CTSD, SMAD3 | Greatly reduced | Normal | Normal | Greatly reduced/greatly reduced |
| ΔmiR-K12-4 | TPD52, CHD1, SENP7, DDOST, HIPK2, STAT1 | Greatly reduced | Normal | Normal | Greatly reduced/greatly reduced |
| ΔmiR-K12-5 | LARP1 | Normal | Normal | Greatly reduced | Greatly reduced/greatly reduced |
| ΔmiR-K12-6 | GSTP1, VIM, YOD1, TRAF7, TPT1, MTA2 | Greatly reduced | Normal | Greatly reduced | Normal/normal |
| ΔmiR-K12-7 | PKM, APEX1, MKRN1 | Greatly reduced | Greatly reduced | Greatly reduced | Normal/greatly reduced |
| ΔmiR-K12-8 | FOSL2, VCL, SP1, RB1 | Moderately reduced | Normal | Greatly increased | Normal/greatly increased |
| ΔmiR-K12-9 | ACIN1 | NA | Normal | Greatly reduced | NA |
| ΔmiR-K12-10 | PPP1R10, PCNA, PDHX | Greatly reduced | Greatly reduced | Slightly reduced | Greatly increased/greatly increased |
| ΔmiR-K12-11 | NOP58, MAP3K2, CDC42EP3, EIF2A, ANAPC7, EEA1 | Greatly reduced | Greatly reduced | Normal | Greatly reduced/normal |
| ΔmiR-K12-12 | TGM2, MEF2D | Normal | Greatly reduced | Normal | Normal/greatly reduced |
| ΔmiR-All | Greatly reduced | Greatly reduced | Greatly reduced | Greatly reduced/greatly reduced | |
Example target genes were previously identified by ribonomics methods but have yet to be validated, with the exception of the miR-K12-3 targets.
FIG 6.
Multiple genes related to proliferation and apoptosis are derepressed in the absence of miR-K12-3. (A to G) mRNA expression levels of the indicated genes measured by qRT-PCR. Data are averages from 3 biological replicates, normalized to the WT. Error bars show standard errors. P values versus the WT by 2-tailed Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
FIG 7.
Cells infected with ΔmiR-K12-3 show higher cathepsin D expression and increased apoptosis compared to WT-infected cells. (A) Representative image of a Western blot comparing CTSD protein levels in WT-infected and ΔmiR-K12-3-infected TIVE cells. Three biological replicates are shown for each condition. (B) Quantification of CTSD protein levels. Western blots using fluorescently tagged secondary antibodies were photographed on a Li-Cor instrument and fluorescence was quantified. The graph shows the average from three replicates, normalized to GAPDH. Error bars show standard errors. (C) Representative images of WT- and ΔmiR-K12-3-infected TIVE cells with Alexa Fluor 647-labeled annexin V. GFP indicates normal cells. (D) Quantification of annexin V micrographs. The percentage of annexin V-positive cells was calculated for each microscope field. Results are averages from four independent experiments consisting of three replicates per treatment. Error bars represent standard errors. P values versus WT by 2-tailed Student’s t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
DISCUSSION
The phenotypic changes that occur in endothelial cells latently infected with KSHV are characteristic of the process of oncogenesis. Since the KSHV miRNAs are expressed during latency, they likely play important roles in the observed changes. To evaluate what contributions individual KSHV miRNAs make to changes in cellular phenotype, we infected endothelial cells with a panel of miRNA knockout viruses. We monitored differences in cell proliferation, migration, angiogenesis, and glycolysis between cells infected with WT and mutant viruses. As a general observation, it was noted that the deletion of different miRNAs could, in some cases, result in the same or similar phenotypes. This is indicative of the importance of redundancy in miRNA targeting. We have previously shown that different viral miRNAs can target the same gene or separate genes in the same pathway (17, 34). However, simultaneous deletion of all of the miRNAs in the Δall virus did not have much of an additive effect. The reason for this is unclear, but it may be an unforeseen consequence of making a 1.8-kb deletion within the KLAR miRNA cluster region of the virus. This deletion likely impacts the expression ratio of several latency-associated genes, impacting the host cellular transcriptome.
Cells infected with the mutant lacking miR-K12-11 showed a pronounced deficit in proliferative ability, indicating that this miRNA is needed in both lymphoid and endothelial cells to sustain the high levels of cell proliferation seen in WT KSHV infection. This agrees well with the work of others showing that ectopic expression of miR-K12-11 can enhance B-cell proliferation (9, 35). It will be interesting to see whether miR-K12-11 targets are similar or different between B cells and endothelial cells. Two miRNAs whose absence had a modest impact on cell proliferation, miR-K12-6 and miR-K12-8, are not currently well understood in this context and may be a subject for future study. Through recent ribonomics analysis of the KSHV miRNA targetome, we identified RB1, a negative regulator of the cell cycle, to be a target of miR-K12-8. We also found that miR-K12-6 may disrupt TGF-β signaling and thereby promote cell survival by targeting SMAD4 (17). These potential targets will need to be validated in the future.
Wound-healing assays demonstrated that miR-K12-7 and miR-K12-11 are important drivers of cell migration, a surrogate for metastasis. While miR-K12-11 was previously shown to be indirectly involved in the enhancement of cell migration (26), the relationship of miR-K12-7 to this process will need to be further elucidated. One putative target of miR-K12-7 identified through qCLASH is cytoplasmic FMR1 interacting protein 1 (CYFIP1), which is thought to have a role in suppressing tumor invasion (36). This interaction will require verification.
Three of the KSHV miRNAs, miR-K12-5, -6, and -7, were found to be significant factors in virally induced angiogenesis. Others have identified miR-K12-6 as a component of this process (29–31), but miR-K12-5 and -7 need to be studied further. It is difficult to draw any solid conclusions from the OCR/ECAR data other than that miR-K12-10 should be thoroughly characterized for its role in changes to cellular metabolism.
The deletion of miR-K12-9 had an unexpectedly profound impact on the character of infected cells. It was observed that cells infected with this virus were unusually large and had extremely slow growth. Indeed, the difficulty of working with these cells made it so two of the four phenotypic assays could not be completed with them. The ΔmiR-K12-9 virus expresses neither miR-K12-9 nor miR-K12-9*. miR-K12-9* is known to downregulate RTA, the KSHV gene responsible for initiating lytic replication of the virus, thereby contributing to the maintenance of latency (37). In the absence of miR-K12-9/9*, KSHV-infected cells may undergo more frequent lytic reactivation. Lysis of cells due to lytic replication may have contributed to the perceived slow growth of the ΔmiR-K12-9-infected cell line; however, we did not observe an increased number of dead cells compared to other cell lines. Interestingly, miR-K12-9 is the most polymorphic miRNA in PEL cells, indicating miR-K12-9 is dispensable in some circumstances (38). A number of single-nucleotide polymorphisms (SNPs) have been found in miR-K12-9 pre-miRNA sequences from a variety of cell lines and patients, with the greatest variation of all KSHV miRNAs (39). Certain SNPs in the miR-K12-9 gene correlate with specific clinical diagnoses: patients with MCD, in which KSHV is more lytic than in KS, have divergent miR-K12-9 sequences (40). SNPs within the pre-miRNA sequence can impact miRNA processing by Drosha and Dicer, leading to changes in mature miRNA expression. As a result of SNPs, the PEL-derived BC-3 cell line does not express mature miR-K12-9 (41). Despite this, in our specific endothelial cell system, miR-K12-9 appears to be virtually required for normal growth. Therefore, further investigation of miR-K12-9 is warranted.
As a first attempt to go from observed phenotypes to identifying miRNA targets conveying this biology, we followed miR-K12-3, which showed significant changes in cell proliferation and glycolysis. We first identified a number of genes in our ribonomics data set that might be linked to these processes and, as a proof of concept, demonstrated CTSD, a strong promoter of apoptosis (42), as a novel miR-K12-3 target. CTSD, whose mRNA contains 3 miR-K12-3 binding sites within its open reading frame (ORF), is downregulated by miR-K12-3 at the mRNA and protein level. Moreover, comparing TIVE cells in the presence and absence of miR-K12-3 showed significant differences in annexin V staining. This novel finding underlines the importance for inhibition of apoptosis in latently infected cells, which has also been reported by others through different miRNA-independent mechanisms.
Taken together, this study has laid the groundwork in characterizing which KSHV miRNAs are responsible for the oncogenesis-related phenotypic changes observed in human endothelial cells latently infected with KSHV. Moreover, comparing miRNA targetome data from us and others in combination with RNA-seq studies of both cell lines and primary tumors will allow us to further decipher how specific pathways are affected by KSHV miRNAs.
MATERIALS AND METHODS
Viruses.
The KSHV mutant viruses used in this study were developed previously (14). Briefly, mutations were made to KSHV in the BAC16 background. Each single miRNA deletion mutant contains only a small deletion which prevents the selected miRNA from forming a hairpin. This prevents the expression of both the leading and passenger strands from the locus. For the ΔmiR-K12-10 and ΔmiR-K12-12 viruses, both of which are located within the open reading frame of kaposin, synonymous mutations were introduced to prevent hairpin formation but keep the coding sequence of kaposin intact. The Δall virus has the entire miRNA cluster region of the genome deleted (miR-K12-1 through miR-K12-9 and miR-K12-11) and has mutations in the miR-K12-10 and miR-K12-12 genes.
Establishment of mutant-infected TIVE cell lines.
TIVE cells (18) were grown in Medium 199 (number 10-060-CV; Corning) supplemented with 20% fetal bovine serum (FBS), 60 μg/ml ECGF (number E2759-5X15MG; Millipore Sigma), and 1% penicillin-streptomycin (P/S). Cells were seeded at high density in 6-well plates and infected separately with each iSLK-derived, mutant, or wild-type virus at approximately 1,000 genome copies per cell. For the ΔmiR-K12-9 virus, which could not establish an infection under these conditions, even after several attempts, cells were infected with approximately 2,000 genome copies per cell in the presence of Polybrene. Seventy-two hours after infection, the medium was replaced with medium containing 50 μg/ml hygromycin (number 30-240-CR; Corning) to selectively kill any noninfected cells. Subsequently, cells were maintained in medium containing 100 μg/ml hygromycin. After selection for several weeks, 100% GFP-positive lines of these 13 mutants plus the wild type were cryopreserved. All cells used in later experiments came from this initial infection and have been quality controlled by qPCR for viral episome copy number and by RNA-seq for expression levels of the major latency-associated antigen, LANA. Cells were maintained and used in experiments for approximately 6 months, at which time they were discarded and new cells stored from the initial infection were thawed and utilized for subsequent assays.
Other cell culture.
TIVE-LTC cells are the result of a fusion between KSHV-infected TIVE cells and the SLK/Caki-1 cell line (43). The TIVE-LTC cells created in this event remain infected with KSHV in the absence of selection. They readily form tumors in mice and continue to express major endothelial cell markers (18). TIVE-LTC cells were maintained in Dulbecco’s modified Eagle medium (DMEM) DMEM with 10% FBS and 1% P/S.
Cell proliferation assay.
Ninety-six-well plates were seeded at a density of 5,000 cells per well and grown for 72 h in the aforementioned medium, which contains phenol red. At that time, without replacing the medium, cell proliferation was measured using a CellTiter 96 aqueous nonradioactive cell proliferation assay (number G5421; Promega) according to the manufacturer’s instructions.
Wound-healing assay.
Cells were seeded at a density of 2 × 105 cells per ml in 48-well plates for a total of 5 × 104 cells per well. The ΔmiR-K12-9-infected cells, which were much larger than those of other cell lines, were seeded at a density of 1 × 105 cells per ml. Approximately 24 h later, each monolayer was scratched with a 1,000-μl pipet tip. Photographs of each scratch were taken at 0 h and 12 h after scratching. The ratio between the scratch area at 0 h and at 12 h was determined using TScratch (105).
Tubule formation assay.
BD Matrigel (number 356234; BD) was diluted to 4 mg/ml protein, and 100 μl of this solution was distributed to each well of a 48-well plate. The Matrigel was allowed to solidify at 37°C for 30 min before adding 5 × 104 cells per well. Photographs were taken of two fields per well at 6 and 12 h after plating. The number of junctions between three or more tubules was counted for each field.
Cell metabolism assay.
Agilent XF96 extracellular flux assay plates were seeded at a density of 2.5 × 104 cells per well. The corresponding sensor cartridge was hydrated according to the manufacturer’s instructions. Unbuffered DMEM was prepared by reconstituting DMEM powder without glucose, l-glutamine, phenol red, sodium pyruvate, and sodium bicarbonate (number D5030; Sigma) in distilled, deionized water. Glucose (number 41095-5000; Acros Organics), sodium pyruvate (number 25-000-CI; Cellgro), and l-glutamine (number 25030; Gibco) were added to final concentrations of 10 mM, 1 mM, and 2 mM, respectively. Phenol red (number AC611785000, discontinued; Acros Organics) was added to an approximate final concentration of 15 mg/liter. The pH of the medium was then adjusted to 7.4. Twenty-four hours after plating, the regular cell culture medium was replaced with unbuffered DMEM, and the cells were placed in a CO2-free incubator for 1 h. The oxygen consumption rate and extracellular acidification rate of the cells were determined on an Agilent Seahorse XF96 machine. Oxamate was added to a final concentration of 1 μM at time point 3, and rotenone was added to a final concentration of 100 mM at time point 6.
Apoptosis assay.
The GFP-certified apoptosis/necrosis detection kit (number ENZ-51002-25; Enzo) was used by following the protocol for fluorescence/confocal microscopy. The annexin V-EnzoGold apoptosis detection reagent was replaced with Alexa Fluor 647 annexin V conjugate (number A23204; Thermofisher), which can be detected through the Cy5 channel, so that the assay was in line with the capabilities of our fluorescence microscope. GFP-positive and Cy5-positive cells were counted using ImageJ software, and the ratio of Cy5-positive to GFP-positive cells was determined.
Western blotting.
Protein levels were determined by Western blotting using a rabbit monoclonal antibody against CTSD (number 12517-R010; Sino Biological) at a 1:500 dilution and a fluorescence-labeled secondary antibody, IRDye 800CW goat anti-rabbit IgG (number 12517-R010; Li-Cor), at a 1:10,000 dilution. Fluorescence was measured on a Li-Cor instrument.
qRT-PCR.
RNA was extracted from cells using RNA-Bee (number CS-501B; Bio-Connect) according to the manufacturer’s instructions and stored at –80°C. Superscript IV reverse transcriptase (number 18090010; Invitrogen) was used with oligo(dT) to prepare cDNA. qPCR was performed using FastStart essential DNA green master mix (number 06402712001; Roche) and a Roche LightCycler 96 instrument.
qPCR.
DNA was extracted from cells using DNAzol (number 10503027; Invitrogen) according to the manufacturer’s instructions. qPCR was performed using Roche products as described above.
RNA-seq.
Total RNA was extracted from cells with RNA-Bee and treated with the NEBNext rRNA depletion kit (human/mouse/rat) (number E6350; NEB). RNA-seq libraries were prepared using the NEBNext Ultra II directional RNA library prep kit (number E7765; NEB) with NEBNext multiplex oligonucleotides for Illumina (96 unique dual-index primer pairs) (number E6440; NEB) (Table 2). Libraries were sequenced on an Illumina NovaSeq 6000.
TABLE 2.
qPCR primers
| Gene | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| TIMP2 | ATGGACTCTTGCACATCACTAC | GGGATGGATAAACAGGGAAACA |
| TNFRSF10B | AGAGTAGATGGTGCTTGAGAATG | CAGGGTGACAGATAACCAGAAG |
| SMAD3 | GACTCAGAACCCAGAGACAATAC | CTGTGTAAGAAACAGGCCATAGA |
| TES | CCTCCCAAATAGCTGGGATTAC | CATCTGAGGTCAGGAGTTTGAG |
| PPPR21A | ATGGGTCTCTCTCCCATCTT | GTTCACACAGTCCAGGTTAGAG |
| FKBP1A | CCTGATGTTCCACTCCACTTT | AACACACATACGAGGAGAAAGG |
| BCL2L11 | CTGCTGGACACACACATACA | GGGCTGAGGAAACAGAGTAAA |
| CTSD | GACATCCACTATGGCTCGGG | AGCACGTTGTTGACGGAGAT |
RNA-seq data analysis.
The complete process of RNA-seq data analysis will be detailed in a future manuscript. For the quantification of ORF73/LANA, a custom reference library was prepared containing the KSHV ORF73/LANA coding sequence (CDS) (GQ994935; ORF73, NCBI; acquired 17 December 2020) and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript (NM_002046.7; Nuccore; 17 December 2020). Deduplicated reads were then aligned to this library using Bowtie2 (version 2.4.2). For quantification of alignments, SAMtools (version 1.10) was utilized to provide counts of aligned sequence reads for GAPDH and LANA, respectively.
ACKNOWLEDGMENTS
We thank Laurence Morel for the use of the Agilent Seahorse XF96 analyzer.
This work was supported by NCI grants RO1 CA119917 and P01 CA 214091 to R.R., F31 CA180522 to L.G., and UF Informatics Institute Fellowship 00130153 to D.S.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280. doi: 10.1182/blood.V86.4.1276.bloodjournal8641276. [DOI] [PubMed] [Google Scholar]
- 4.Boss IW, Plaisance KB, Renne R. 2009. Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol 17:544–553. doi: 10.1016/j.tim.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent OA, Mendell JT. 2006. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 8.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boss IW, Nadeau PE, Abbott JR, Yang Y, Mergia A, Renne R. 2011. A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rgammanull mice. J Virol 85:9877–9886. doi: 10.1128/JVI.05558-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll PA, Kenerson HL, Yeung RS, Lagunoff M. 2006. Latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J Virol 80:10802–10812. doi: 10.1128/JVI.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado T, Carroll PA, Punjabi AS, Margineantu D, Hockenbery DM, Lagunoff M. 2010. Induction of the Warburg effect by Kaposi's sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc Natl Acad Sci U S A 107:10696–10701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yogev O, Lagos D, Enver T, Boshoff C. 2014. Kaposi's sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS Pathog 10:e1004400. doi: 10.1371/journal.ppat.1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai L, Qin Z, Defee M, Toole BP, Kirkwood KL, Parsons C. 2012. Kaposi sarcoma-associated herpesvirus (KSHV) induces a functional tumor-associated phenotype for oral fibroblasts. Cancer Lett 318:214–220. doi: 10.1016/j.canlet.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain V, Plaisance-Bonstaff K, Sangani R, Lanier C, Dolce A, Hu J, Brulois K, Haecker I, Turner P, Renne R, Krueger B. 2016. A toolbox for herpesvirus miRNA research: construction of a complete set of KSHV miRNA deletion mutants. Viruses 8:54. doi: 10.3390/v8020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R. 2012. Ago HITS-CLIP expands understanding of Kaposi's sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog 8:e1002884. doi: 10.1371/journal.ppat.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gay LA, Sethuraman S, Thomas M, Turner PC, Renne R. 2018. Modified cross-linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi's sarcoma-associated herpesvirus microRNA targets in endothelial cells. J Virol 92:e02138-17. doi: 10.1128/JVI.02138-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An FQ, Folarin HM, Compitello N, Roth J, Gerson SL, McCrae KR, Fakhari FD, Dittmer DP, Renne R. 2006. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J Virol 80:4833–4846. doi: 10.1128/JVI.80.10.4833-4846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu YF, Sugden AU, Fox K, Hayes M, Sugden B. 2017. Kaposi's sarcoma-associated herpesvirus stably clusters its genomes across generations to maintain itself extrachromosomally. J Cell Biol 216:2745–2758. doi: 10.1083/jcb.201702013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison TJ, Kedes DH. 2014. Variable episomal silencing of a recombinant herpesvirus renders its encoded GFP an unreliable marker of infection in primary cells. PLoS One 9:e111502. doi: 10.1371/journal.pone.0111502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darst RP, Haecker I, Pardo CE, Renne R, Kladde MP. 2013. Epigenetic diversity of Kaposi's sarcoma-associated herpesvirus. Nucleic Acids Res 41:2993–3009. doi: 10.1093/nar/gkt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moody R, Zhu Y, Jones T, Cui XD, Bedolla R, Lei XF, Bai ZQ, Huang YF, Gao S-J. 2012. Cellular transformation and tumorigenesis mediated by KSHV microRNAs. International Congress on Oncogeneic Herpesviruses and Associated Diseases, Philadelphia, PA. [Google Scholar]
- 23.Lei X, Zhu Y, Jones T, Bai Z, Huang Y, Gao SJ. 2012. A KSHV microRNA and its variants target TGF-beta pathway to promote cell survival. J Virol 86:11698–11711. doi: 10.1128/JVI.06855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Sun R, Lin X, Liang D, Deng Q, Lan K. 2012. Kaposi's sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J Virol 86:1372–1381. doi: 10.1128/JVI.06245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geback T, Schulz MM, Koumoutsakos P, Detmar M. 2009. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 26.Qin Z, Dai L, Defee M, Findlay VJ, Watson DK, Toole BP, Cameron J, Peruzzi F, Kirkwood K, Parsons C. 2013. Kaposi's sarcoma-associated herpesvirus suppression of DUSP1 facilitates cellular pathogenesis following de novo infection. J Virol 87:621–635. doi: 10.1128/JVI.01441-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma T, Patel H, Babapoor-Farrokhran S, Franklin R, Semenza GL, Sodhi A, Montaner S. 2015. KSHV induces aerobic glycolysis and angiogenesis through HIF-1-dependent upregulation of pyruvate kinase 2 in Kaposi's sarcoma. Angiogenesis 18:477–488. doi: 10.1007/s10456-015-9475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Jia X, Shen C, Zhang M, Xu J, Shang Y, Zhu K, Hu M, Yan Q, Qin D, Lee MS, Zhu J, Lu H, Krueger BJ, Renne R, Gao SJ, Lu C. 2016. A KSHV microRNA enhances viral latency and induces angiogenesis by targeting GRK2 to activate the CXCR2/AKT pathway. Oncotarget 7:32286–32305. doi: 10.18632/oncotarget.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Hu H, He Z, Liang D, Sun R, Lan K. 2016. Fine-tuning of the Kaposi's sarcoma-associated herpesvirus life cycle in neighboring cells through the RTA-JAG1-Notch pathway. PLoS Pathog 12:e1005900. doi: 10.1371/journal.ppat.1005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Hu M, Wang C, Lu H, Chen F, Xu J, Shang Y, Wang F, Qin J, Yan Q, Krueger BJ, Renne R, Gao SJ, Lu C. 2017. A viral microRNA downregulates metastasis suppressor CD82 and induces cell invasion and angiogenesis by activating the c-Met signaling. Oncogene 36:5407–5420. doi: 10.1038/onc.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramalingam D, Happel C, Ziegelbauer JM. 2015. Kaposi's sarcoma-associated herpesvirus microRNAs repress breakpoint cluster region protein expression, enhance Rac1 activity, and increase in vitro angiogenesis. J Virol 89:4249–4261. doi: 10.1128/JVI.03687-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warburg O. 1956. On respiratory impairment in cancer cells. Science 124:269–270. [PubMed] [Google Scholar]
- 33.Chen D, Gao Y, Nicholas J. 2014. Human herpesvirus 8 interleukin-6 contributes to primary effusion lymphoma cell viability via suppression of proapoptotic cathepsin D, a cointeraction partner of vitamin K epoxide reductase complex subunit 1 variant 2. J Virol 88:1025–1038. doi: 10.1128/JVI.02830-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samols MA, Hu J, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog 3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahlke C, Maul K, Christalla T, Walz N, Schult P, Stocking C, Grundhoff A. 2012. A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS One 7:e49435. doi: 10.1371/journal.pone.0049435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abekhoukh S, Sahin HB, Grossi M, Zongaro S, Maurin T, Madrigal I, Kazue-Sugioka D, Raas-Rothschild A, Doulazmi M, Carrera P, Stachon A, Scherer S, Drula Do Nascimento MR, Trembleau A, Arroyo I, Szatmari P, Smith IM, Mila M, Smith AC, Giangrande A, Caille I, Bardoni B. 2017. New insights into the regulatory function of CYFIP1 in the context of WAVE- and FMRP-containing complexes. Dis Model Mech 10:463–474. doi: 10.1242/dmm.025809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellare P, Ganem D. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall V, Parks T, Bagni R, Wang CD, Samols MA, Hu J, Wyvil KM, Aleman K, Little RF, Yarchoan R, Renne R, Whitby D. 2007. Conservation of virally encoded microRNAs in Kaposi sarcoma–associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis 195:645–659. doi: 10.1086/511434. [DOI] [PubMed] [Google Scholar]
- 39.Marshall V, Martro E, Labo N, Ray A, Wang D, Mbisa G, Bagni RK, Volfovsky N, Casabona J, Whitby D, EURO-SHAKS Study Group . 2010. Kaposi sarcoma (KS)-associated herpesvirus microRNA sequence analysis and KS risk in a European AIDS-KS case control study. J Infect Dis 202:1126–1135. doi: 10.1086/656045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray A, Marshall V, Uldrick T, Leighty R, Labo N, Wyvill K, Aleman K, Polizzotto MN, Little RF, Yarchoan R, Whitby D. 2012. Sequence analysis of Kaposi sarcoma-associated herpesvirus (KSHV) microRNAs in patients with multicentric Castleman disease and KSHV-associated inflammatory cytokine syndrome. J Infect Dis 205:1665–1676. doi: 10.1093/infdis/jis249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han S-J, Marshall V, Barsov E, Quiñones O, Ray A, Labo N, Trivett M, Ott D, Renne R, Whitby D. 2013. Kaposi's sarcoma-associated herpesvirus microRNA single-nucleotide polymorphisms identified in clinical samples can affect microRNA processing, level of expression, and silencing activity. J Virol 87:12237–12248. doi: 10.1128/JVI.01202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minarowska A, Minarowski L, Karwowska A, Gacko M. 2007. Regulatory role of cathepsin D in apoptosis. Folia Histochem Cytobiol 45:159–163. [PubMed] [Google Scholar]
- 43.Sturzl M, Gaus D, Dirks WG, Ganem D, Jochmann R. 2013. Kaposi's sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int J Cancer 132:1954–1958. doi: 10.1002/ijc.27849. [DOI] [PubMed] [Google Scholar]