The continuous dissemination of the novel emerging SARS-CoV-2 virus, with more than 78 million infected cases worldwide and higher than 1,700,000 deaths as of 23 December 2020, highlights the urgent need for the development of novel vaccines against COVID-19. With this aim, we have developed novel vaccine candidates based on the poxvirus modified vaccinia virus Ankara (MVA) strain expressing the full-length SARS-CoV-2 spike (S) protein, and we have evaluated their immunogenicity in mice using DNA/MVA or MVA/MVA prime/boost immunization protocols.
KEYWORDS: SARS-CoV-2, COVID-19, MVA, poxvirus, vaccines, S protein, T cell responses, neutralizing antibodies, K18-hACE2 mice, SARS-CoV-2 vaccine efficacy
ABSTRACT
Vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, are urgently needed. We developed two COVID-19 vaccines based on modified vaccinia virus Ankara (MVA) vectors expressing the entire SARS-CoV-2 spike (S) protein (MVA-CoV2-S); their immunogenicity was evaluated in mice using DNA/MVA or MVA/MVA prime/boost immunizations. Both vaccines induced robust, broad, and polyfunctional S-specific CD4+ (mainly Th1) and CD8+ T-cell responses, with a T effector memory phenotype. DNA/MVA immunizations elicited higher T-cell responses. All vaccine regimens triggered high titers of IgG antibodies specific for the S, as well as for the receptor-binding domain; the predominance of the IgG2c isotype was indicative of Th1 immunity. Notably, serum samples from vaccinated mice neutralized SARS-CoV-2 in cell cultures, and those from MVA/MVA immunizations showed a higher neutralizing capacity. Remarkably, one or two doses of MVA-CoV2-S protect humanized K18-hACE2 mice from a lethal dose of SARS-CoV-2. In addition, two doses of MVA-CoV2-S confer full inhibition of virus replication in the lungs. These results demonstrate the robust immunogenicity and full efficacy of MVA-based COVID-19 vaccines in animal models and support its translation to the clinic.
IMPORTANCE The continuous dissemination of the novel emerging SARS-CoV-2 virus, with more than 78 million infected cases worldwide and higher than 1,700,000 deaths as of 23 December 2020, highlights the urgent need for the development of novel vaccines against COVID-19. With this aim, we have developed novel vaccine candidates based on the poxvirus modified vaccinia virus Ankara (MVA) strain expressing the full-length SARS-CoV-2 spike (S) protein, and we have evaluated their immunogenicity in mice using DNA/MVA or MVA/MVA prime/boost immunization protocols. The results showed the induction of a potent S-specific T-cell response and high titers of neutralizing antibodies. Remarkably, humanized K18-hACE2 mice immunized with one or two doses of the MVA-based vaccine were 100% protected from SARS-CoV-2 lethality. Moreover, two doses of the vaccine prevented virus replication in lungs. Our findings prove the robust immunogenicity and efficacy of MVA-based COVID-19 vaccines in animal models and support its translation to the clinic.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped virus with a positive-sense, single-stranded RNA genome of approximately 30 kb belonging to the family Coronaviridae, genus Betacoronavirus (1, 2). SARS-CoV-2 jumped from animals to humans, as did related SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (3), causing the coronavirus disease 2019 (COVID-19) pandemic, which is making a major impact on human health (4). COVID-19 symptoms range from asymptomatic or slight flu-like manifestations to life-threatening pneumonia (5). The first cases appeared in the Chinese city of Wuhan in December 2019, and by 10 January 2020, the SARS-CoV-2 sequence was published (2, 6). SARS-CoV-2 infection quickly spread worldwide, and on 11 March 2020, the World Health Organization declared COVID-19 as a global pandemic. More than 78 million infections and 1,700,000 deaths have been reported (https://www.gisaid.org/epiflu-applications/global-cases-covid-19/).
This global pandemic has driven an unprecedented worldwide effort to find antivirals and a rapidly producible and scalable vaccine to prevent the disease. A large number of vaccine candidates are under development, based either on the whole virus (inactivated or attenuated viruses), nonreplicating viral vectors expressing SARS-CoV-2 antigens, nucleic acid (mRNA or DNA), and subunits (proteins or virus-like particles) (7, 8). COVID-19 vaccine development is predominantly based on SARS-CoV-2 spike (S) expression. The envelope S protein mediates virus cell entry through binding to its host receptor, angiotensin-converting enzyme 2 (ACE2) (9), and is the main target for neutralizing antibodies (nAbs) (10–12). Some vaccine candidates have shown immunogenicity and ability to reduce the severity of disease in preclinical animal models, like mice (13–18), and nonhuman primates (NHP) (15, 19–22). Furthermore, some of the most advanced candidate vaccines, such as those based on adenovirus vectors, mRNA, and inactivated SARS-CoV-2 have entered phase I/II clinical trials showing immunogenicity and apparent safety (23, 24). The urgent need of a safe and efficacious vaccine is accelerating the phase III clinical trials of some of the aforementioned vaccine candidates, and by the end of 2020, high-efficacy phase III clinical trials with adenovirus-based vaccine ChAdOx1 nCoV-19 (25) and for the landmark phase III mRNA vaccine trial from Pfizer/BioNTech (26) have been published; the mRNA vaccines from Pfizer and Moderna were approved on December 2020 by FDA for emergency human vaccination use against SARS-CoV-2.
One of the most promising and established candidate vector for a vaccine is vaccinia virus (VACV), which served to eradicate smallpox (27). Since then, highly attenuated VACV strains, like modified vaccinia virus Ankara (MVA), have been produced and shown in several preclinical and clinical trials to be an excellent vector to generate efficacious vaccines against a variety of pathogens (28–32). Our group has generated MVA-based vaccine candidates expressing antigens for several emerging viruses, such as Chikungunya (33), Zika (34), and Ebolavirus (35), which are highly immunogenic in animal models (mice or NHP), and induce pathogen-specific T-cell responses and nAbs. Of note, one single dose of those vaccines provided protection against infection in animal models of disease (33–38). Remarkably, an MVA vaccine against smallpox has been approved for human use by the FDA and European Medicines Agency (EMA) agencies (39).
Therefore, we took advantage of our MVA-based vaccine platform to generate novel COVID-19 vaccines. Here, we describe the preclinical testing of novel COVID-19 vaccine candidates based on MVA expressing the full-length SARS-CoV-2 S protein. We generated two MVA-based vaccines, one using the attenuated wild-type MVA (MVA-WT) as parental virus (termed MVA-CoV2-S, abbreviated as MVA-S) and other from an optimized MVA lacking VACV immunomodulatory genes C6L, K7R, and A46R (termed MVA-Δ-CoV2-S, abbreviated as MVA-Δ-S), whose deletion was able to increase immune responses against HIV-1 antigens (40). We characterized the biochemical, genetic, and stability properties of both MVA vectors and compared their ability to induce SARS-CoV-2 S-specific cell and humoral immune responses in a mouse model following homologous MVA/MVA or heterologous DNA/MVA prime/boost protocols. Both vaccine candidates were highly immunogenic triggering S-specific T- and B-cell responses. Importantly, K18-hACE2 mice immunized with one or two doses of MVA-CoV2-S were completely protected against SARS-CoV-2 lethality, and two doses of the vaccine prevented virus replication in the lungs. Our findings highlight the use of MVA expressing the full-length S antigen as a potential vaccine candidate against SARS-CoV-2, worth further safety and efficacy analysis in NHP and clinical trials.
RESULTS
Generation and in vitro characterization of MVA-S vaccine candidates expressing the SARS-CoV-2 S protein.
To generate novel vaccines against COVID-19 that stimulate SARS-CoV-2-specific T- and B-cell immune responses, we have developed two MVA-based vaccine candidates expressing the SARS-CoV-2 full-length S structural gene. The MVA-S candidate was generated by inserting the S gene into the VACV thymidine kinase (TK) locus of MVA-WT, whereas the MVA-Δ-S construct was generated using an optimized MVA (termed MVA-Δ-GFP) lacking VACV immunomodulatory genes C6L, K7R, and A46R (33–35) (Fig. 1A).
FIG 1.
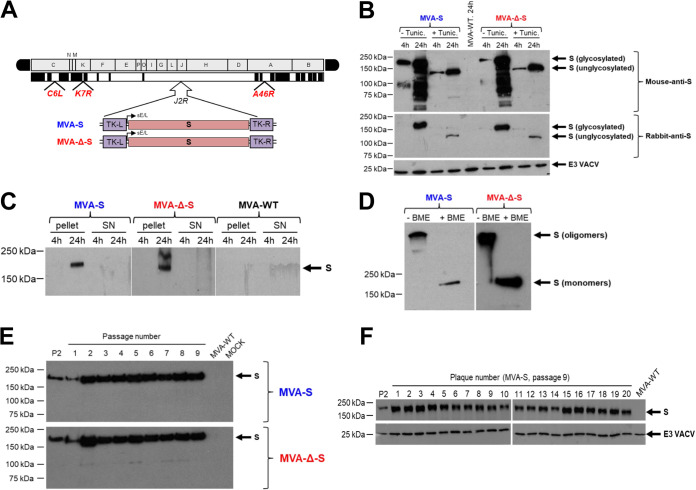
Design, generation, and in vitro characterization of MVA-S and MVA-Δ-S vaccine candidates. (A) Scheme of the MVA-S and MVA-Δ-S genomes with the SARS-CoV-2 S gene inserted within the MVA TK viral locus (J2R). The MVA genome regions are indicated by capital letters. Deleted or fragmented MVA genes are represented as black boxes. The locations of C6L, K7R, and A46R genes deleted in the MVA-Δ-S construct are indicated. (B) Expression of SARS-CoV-2 S protein by MVA-S vaccine candidates. Western blotting of the MVA-infected DF-1 cell samples (5 PFU/cell), treated or untreated with tunicamycin, at 4 and 24 h postinfection (hpi). Mouse monoclonal or rabbit polyclonal anti-S antibodies were used to identify the protein as described in Materials and Methods. A rabbit polyclonal antibody against the VACV E3 protein was used to control the amount of viral proteins loaded on the 7% SDS-PAGE under reducing conditions. The size (in kilodaltons [kDa]) and migration of the molecular weight markers are indicated. (C) Expression kinetics of S protein in cellular pellets and supernatants from cells infected with MVA-S and MVA-Δ-S vaccine candidates. Western blotting of MVA-infected cell samples and corresponding cell supernatants (SN) at 4 and 24 hpi, as in panel B. A rabbit polyclonal anti-S antibody was used for protein identification. (D) Expression of S protein under reducing and nonreducing conditions. Western blotting of MVA-infected DF-1 cell samples (24 hpi) under reducing (+ β-mercaptoethanol [BME]) and nonreducing (- BME) conditions. A rabbit polyclonal anti-S antibody was used for protein identification. The presumed state of the S protein based on its migration is indicated. (E and F) Genetic stability of MVA-S and MVA-Δ-S vaccine candidates. Western blotting of DF-1 cell samples (24 hpi) infected with initial P2 stocks and with 9 successive passages of MVA-S and MVA-Δ-S viruses (E) or from 20 individual virus plaques picked after 9 consecutive cell infection cycles of the MVA-S construct (F). Samples were analyzed under reducing conditions. Rabbit polyclonal anti-S and anti-VACV E3 antibodies were used for protein identification.
The correct generation and purity of the MVA-S constructs were analyzed by PCR using oligonucleotides annealing in the VACV TK-flanking regions, which demonstrated the proper insertion of the SARS-CoV-2 S gene and no parental MVA contamination, and DNA sequencing confirmed the S gene sequence fidelity (data not shown). Western blotting showed S protein expression in permissive chicken DF-1 cells infected with both MVA-S vaccine candidates and an increase in protein expression with time postinfection (Fig. 1B). Furthermore, the S electrophoretic mobility reduction in the presence of tunicamycin indicated that the S protein was glycosylated (Fig. 1B). Additionally, analysis of cell extracts and supernatants showed exclusively cell-associated protein (Fig. 1C), as expected for an S protein that contains the transmembrane domain. Interestingly, in the absence of reducing agents, the S protein exhibited electrophoretic migration consistent with the formation of oligomers, presumably trimers (Fig. 1D), whereas the reduced S protein migrated with a size of ∼180 kDa, indicating lack of furin S protein cleavage into S1 and S2 regions.
The analysis of the growth kinetics in permissive DF-1 cells showed that MVA-S and MVA-Δ-S had a similar viral growth, comparable to their MVA-WT and MVA-Δ-GFP parental viruses, respectively (data not shown). Therefore, the constitutive expression of SARS-CoV-2 S protein does not impair MVA vector replication under permissive conditions. Next, MVA-S and MVA-Δ-S virus stability was determined by analysis of SARS-CoV-2 S protein expression during successive virus passages in cell cultures (Fig. 1E). The results showed that MVA-S and MVA-Δ-S efficiently expressed the S protein up to 9 successive passages, indicating virus genetic stability in cell cultures (Fig. 1E). Moreover, all the individual plaques isolated from MVA-S at passage 9 showed stable S protein expression (Fig. 1F). Furthermore, the S protein expressed from viruses obtained from the 9 serial passages or from the 20 individual plaques was not cleaved into S1 and S2 regions (Fig. 1E and F).
Subcellular distribution of the SARS-CoV-2 S protein in cells infected with MVA-S and MVA-Δ-S.
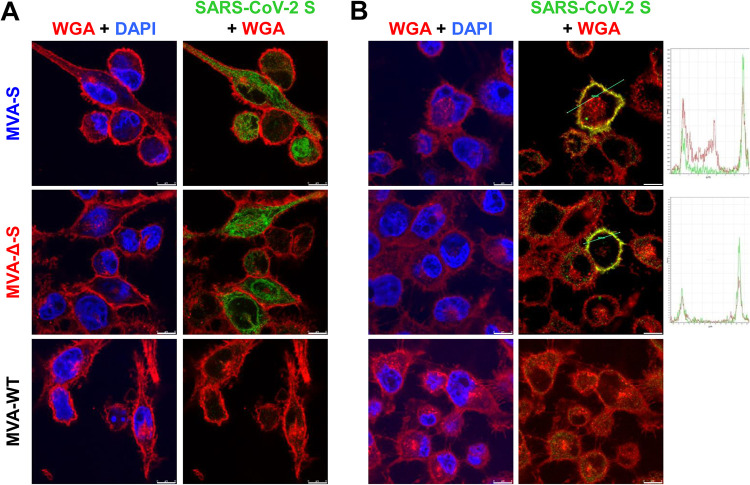
Expression and cellular distribution of the SARS-CoV-2 S protein produced by MVA-S and MVA-Δ-S constructs were studied by confocal immunofluorescence microscopy in nonpermissive human HeLa cells at 24 h postinfection (hpi) (Fig. 2). In permeabilized cells, large amounts of S protein were detected in the cytoplasm of infected cells (Fig. 2A). Moreover, in nonpermeabilized cells, we observed S protein cell surface expression, which colocalized with the wheat germ agglutinin (WGA) cell membrane probe (Fig. 2B). These results further confirm SARS-CoV-2 S protein expression in cells infected with both MVA-S vaccine candidates and showed that it was located at the cell surface and cytoplasm.
FIG 2.
Subcellular distribution of the SARS-CoV-2 S protein in MVA-S- and MVA-Δ-S-infected cells. Confocal immunofluorescence microscopy of infected and permeabilized (A) or nonpermeabilized (B) HeLa cells. Cells were infected with 0.5 PFU/cell with the indicated viruses, fixed at 24 hpi, permeabilized or nonpermeabilized, and stained with Alexa Fluor 594-conjugated WGA probe (red) and with a rabbit polyclonal anti-S antibody further detected with a rabbit Alexa Fluor 488-conjugated antibody (green). Cell nuclei were stained using DAPI (blue). A diagram with the S and WGA signal profiles through a cell slice is shown on the right and shows colocalization (yellow) on the cell surface. Bars, 8 μm.
Immunization of mice with MVA-S vaccine candidates induces strong, broad, and polyfunctional SARS-CoV-2 S-specific T cell immunity.
We next analyzed the SARS-CoV-2-specific immunogenicity elicited by MVA-S and MVA-Δ-S in groups of C57BL/6 mice (n = 6) immunized following heterologous DNA/MVA or homologous MVA/MVA prime/boost immunization regimens, as these protocols are more immunogenic and superior in efficacy to immunizations with one single dose, as previously reported for Zika and Chikungunya (33, 34). In the heterologous DNA/MVA protocols, mice received 100 μg of DNA-S prime by the intramuscular (i.m.) route, and 15 days later were boosted with 1 × 107 PFU of MVA-S or MVA-Δ-S by the i.m. route. In the homologous MVA/MVA protocols, mice were immunized similarly with two doses of MVA-S or MVA-Δ-S. Animals primed and boosted with nonrecombinant MVA-WT were used as a control group. To evaluate the adaptive SARS-CoV-2-specific T-cell immune responses elicited by the different immunization groups at 11 days postboost, we stimulated splenocytes with S1 and S2 peptide pools, spanning the entire S protein.
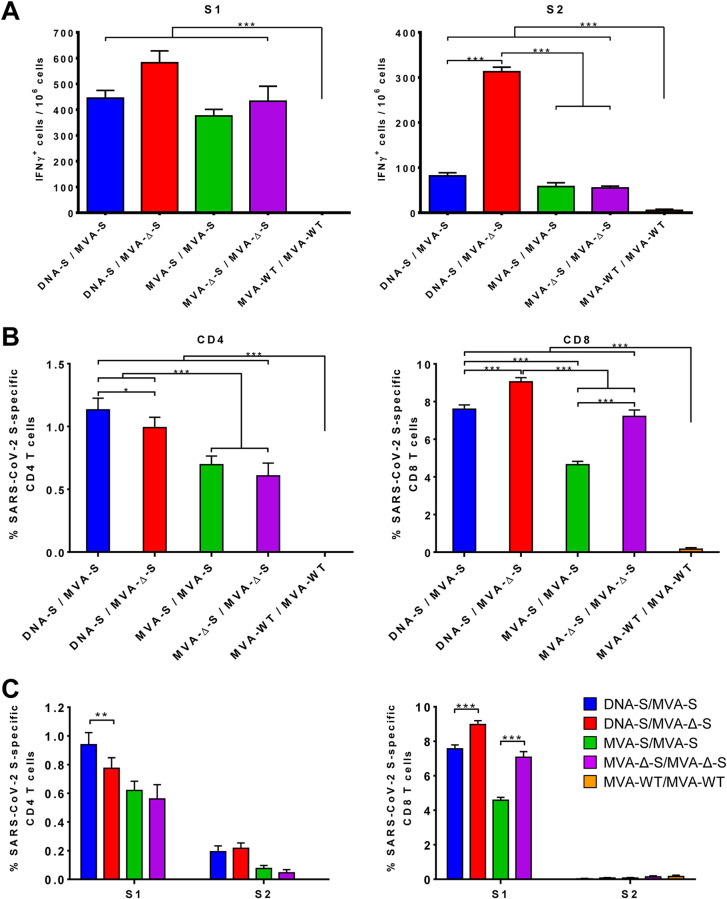
SARS-CoV-2-specific cell responses were measured by an enzyme-linked immunosorbent spot (ELISpot) assay. All MVA-S- or MVA-Δ-S-vaccinated groups induced significant high levels of gamma interferon (IFN-γ)-secreting cells against SARS-CoV-2 S1 and S2 peptide pools compared to control group MVA-WT/MVA-WT with the response mainly toward S1 and to a lesser extent against S2 (Fig. 3A). The DNA-S/MVA-Δ-S group elicited the highest levels of IFN-γ-secreting cells, being significantly greater compared to DNA-S/MVA-S or to the homologous MVA/MVA regimens in the case of S2 (Fig. 3A).
FIG 3.
MVA-S and MVA-Δ-S elicit SARS-CoV-2 S-specific T-cell immune responses in immunized mice. (A) Magnitude of SARS-CoV-2-specific cell responses directed against S1 and S2 regions. Cells secreting IFN-γ per million of splenocytes and directed against S1 (left panel) or S2 (right panel) peptide pools in mice immunized with the indicate prime/boost protocols. IFN-γ was evaluated by an ELISpot assay from a pool of splenocytes derived from six immunized mice per group at 11 days postboost as described in Materials and Methods. Samples were analyzed in triplicate; bars show the triplicate mean values and the standard deviation. P values from one-way ANOVA followed by post hoc pairwise comparisons by Student’s t tests with Holm correction for multiple comparisons (***, P < 0.001). (B) Magnitude of total SARS-CoV-2 S-specific T-cell immune responses. Percentages of CD4+ or CD8+ T cells expressing CD107a and/or producing IFN-γ and/or TNF-α and/or IL-2 against a mixture of S1 and S2 peptide pools in mice immunized with the indicative regimen. Cell percentages were determined by ICS from splenocyte pools as described in Materials and Methods. (C) SARS-CoV-2 S-specific T-cell immune responses against S1 and S2 regions. The percentages of S1- or S2-specific CD4+ and CD8+ T cells were determined as described above for panel B. P values were determined as described in Materials and Methods using an approach that corrects measurements for the medium response, calculating confidence intervals (*, P < 0.05; **, P < 0.005; ***, P < 0.001).
Considering the relevant role of T-cell responses during SARS-CoV-2 infection (41, 42), we next evaluated by an intracellular cytokine staining (ICS) assay the induction of SARS-CoV-2 S-specific CD4+ and CD8+ T cells expressing CD107a, IFN-γ, tumor necrosis factor alpha (TNF-α), and/or interleukin 2 (IL-2) upon stimulation with S1 and S2 peptide pools. All vaccinated groups elicited robust S-specific CD4+ and CD8+ T-cell responses, being greater in heterologous DNA/MVA than in homologous MVA/MVA regimens, with a higher overall response in the CD8+ T-cell repertoire and reflecting a CD4+ Th1 profile (Fig. 3B). The frequencies of S-specific CD4+ T cells induced by DNA-S/MVA-S were significantly higher than those induced by the DNA-S/MVA-Δ-S immunization group but were similar between MVA-S/MVA-S and MVA-Δ-S/MVA-Δ-S groups (Fig. 3B, left panel). However, boosting with MVA-Δ-S elicited significantly higher levels of S-specific CD8+ T cells than with MVA-S in both prime/boost regimens (Fig. 3B, right panel). Furthermore, S-specific CD4+ and CD8+ T-cell responses were broad and directed mainly against the S1 peptide pool in all vaccinated groups (Fig. 3C), as in the ELIspot results (Fig. 3A).
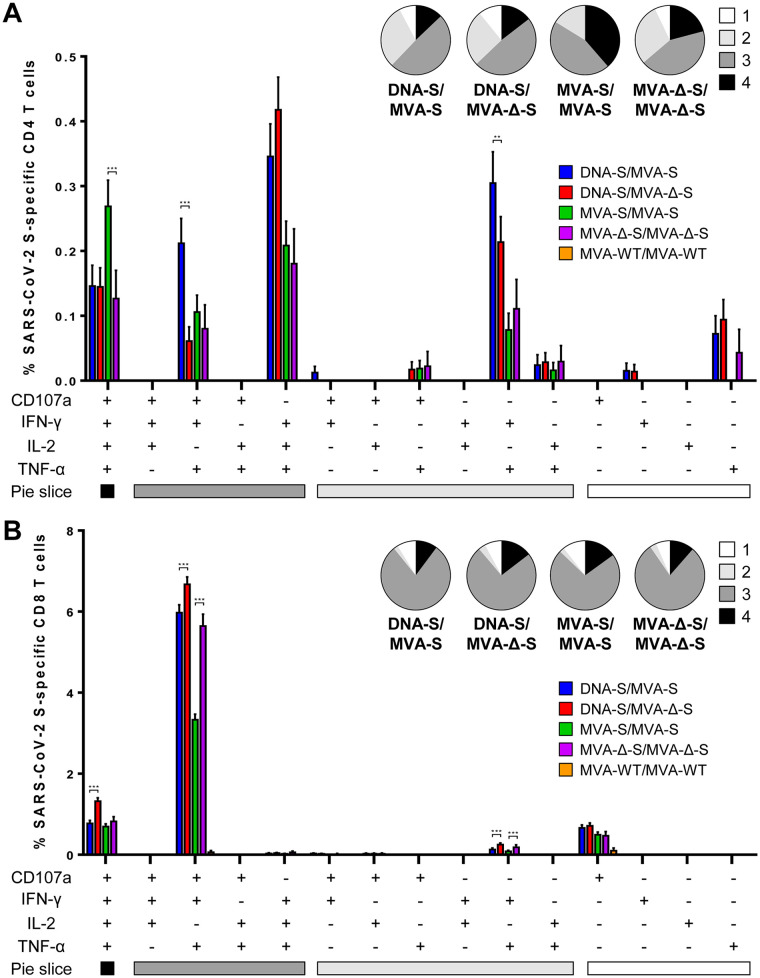
The quality of the S-specific T-cell response was next characterized by analyzing the pattern of cytokine production (IFN-γ, and/or TNF-α, and/or IL-2) and its cytotoxic potential (CD107a) (Fig. 4). The S-specific CD4+ T-cell responses were similar and highly polyfunctional in all vaccinated groups, with around 90% of the cells exhibiting two or more functions (Fig. 4A, pie charts). CD4+ T cells expressing IFN-γ, TNF-α, and IL-2 (IFN-γ-TNF-α-IL-2), CD107a-IFN-γ-TNF-α-IL-2, CD107a-IFN-γ-TNF-α, and IFN-γ-TNF-α were the most induced populations (Fig. 4A, bars), showing a Th1 profile and maintaining the differences between vaccinated groups previously shown in Fig. 3. On the other hand, all vaccinated groups had a similar and robust polyfunctional profile of S-specific CD8+ T-cell responses, with around 90% of the cells exhibiting three or four functions (Fig. 4B, pie charts). CD8+ T cells expressing CD107a-IFN-γ-TNF-α and CD107a-IFN-γ-TNF-α-IL-2 were the most elicited populations (Fig. 4B, bars), preserving the differences among vaccinated groups (Fig. 3).
FIG 4.
MVA-S and MVA-Δ-S induce polyfunctional SARS-CoV-2 S-specific T-cell immune responses in immunized mice. Polyfunctional profiles (based on expression of selected markers CD107a, IFN-γ, TNF-α, and IL-2) of total SARS-CoV-2-specific CD4+ (A) or CD8+ (B) T-cell immune responses directed against a mixture of S1 and S2 peptide pools. T-cell responses were analyzed by ICS in splenocyte pools from six mice per group at 11 days postboost as described in Materials and Methods. The response profiles are shown on the x axis, and the percentages of T cells for each of the immunization regimens are shown on the y axis. The pie charts summarize the percentage of S-specific T cells exhibiting one, two, three, or four markers, which are shown color coded. P values were determined as described in Materials and Methods using an approach that corrects measurements for the medium response, calculating confidence intervals (**, P < 0.005; ***, P < 0.001).
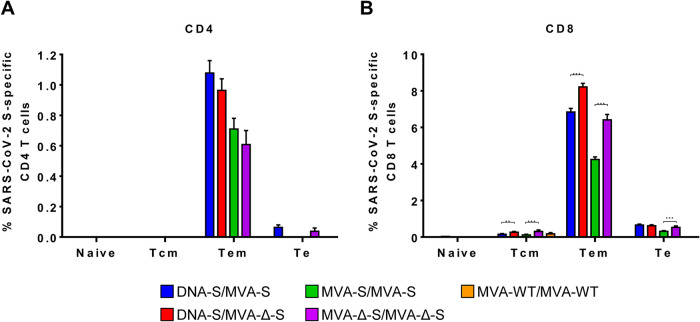
Next, we determined the memory phenotype of SARS-CoV-2 S-specific CD4+ and CD8+ T cells by measuring the expression of CD127 and CD62L surface markers (which defined different memory subpopulations, T central memory (Tcm) (CD127+/CD62L+), T effector memory (Tem) (CD127+/CD62L−), and T effector (Te) (CD127−/CD62L−) cells (43), and determined the percentage of cells expressing CD107a, IFN-γ, TNF-α, and/or IL-2 upon stimulation with S1 and S2 peptide pools (Fig. 5). The results showed that in all vaccinated groups, SARS-CoV-2 S-specific CD4+ and CD8+ T cells were mainly of the Tem phenotype (Fig. 5), and those differences between vaccinated groups were maintained (Fig. 3 and 4).
FIG 5.
Memory phenotypic profiles of the SARS-CoV-2 S-specific CD4+ and CD8+ T cells elicited in mice immunized with the MVA-S and MVA-Δ-S vaccine candidates. Percentages of naive (CD127−/CD62L−), T central memory (Tcm) (CD127+/CD62L+), T effector memory (Tem) (CD127+/CD62L−), and T effector (Te) (CD127−/CD62L−) CD4+ (A) or CD8+ (B) T cells specific for S1 and S2 peptide pools, and expressing any of the markers, CD107a, IFN-γ, TNF-α, and IL-2. T-cell responses were analyzed by ICS in splenocyte pools from six mice per group at 11 days postboost as described in Materials and Methods. P values were determined as described in Materials and Methods using an approach that corrects measurements for the medium response, calculating confidence intervals (**, P < 0.005; ***, P < 0.001).
Induction of SARS-CoV-2 S-specific CD4+ Tfh cells, CD4+ Treg cells, and CD8+ CD103+ T resident-like memory cells in mice immunized with the MVA-S vaccine candidates.
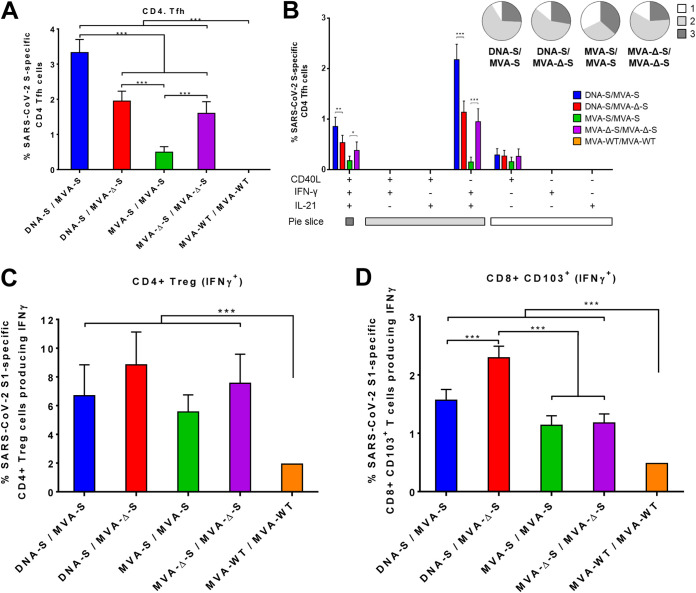
To further extend the analysis of T-cell immune responses, we next evaluated by ICS the SARS-CoV-2 S-specific CD4+ T follicular helper (Tfh) cells elicited by the different immunization groups in splenocytes stimulated ex vivo with S protein plus S1 and S2 peptide pools. All vaccinated groups induced S-specific CD4+ Tfh cells (CXCR5+, PD1+) expressing CD40L (Te marker) on their surface and/or secreting IFN-γ and/or IL-21 (Fig. 6A), with the absence of IL-4 secretion (data not shown), supporting a Th1-like response. Heterologous DNA-S/MVA-S immunization was the most effective regimen to induce CD4+ Tfh cells (Fig. 6A). S-specific CD4+ Tfh cells were highly polyfunctional, with around 70 to 90% of the cells exhibiting two or three functions (Fig. 6B, pie charts), and cells expressing IFN-γ-IL-21 and CD40L-IFN-γ-IL-21 being the most abundant populations elicited by all vaccinated groups (Fig. 6B, bars).
FIG 6.
Characterization of the SARS-CoV-2 S-specific CD4+ Tfh, CD4+ Treg, and CD8+, CD103+ T resident-like memory cell immune responses elicited by MVA-S and MVA-Δ-S vaccine candidates in immunized mice. (A) Magnitude of the S-specific CD4+ Tfh cell immune responses directed against a mixture of S protein plus S1 and S2 peptide pools. Percentages of CD4+ Tfh cells (CXCR5+, PD1+) expressing CD40L and/or producing IFN-γ and/or IL-21 are shown. (B) Polyfunctionality of the S-specific CD4+ Tfh cells. The response profiles based on the T cell expression of the CD40L, IFN-γ, and/or IL-21 markers are shown as in Fig. 4. The pie charts summarize the percentage of S-specific T cells expressing one, two, or three markers, which are shown color coded. Percentages of IFN-γ-producing CD4+ Treg (CD4+, FOXP3+) cells or CD8+ CD103+ T resident-like memory specific for S1 peptide pool are shown in panels C and D, respectively. T-cell responses were analyzed by ICS in splenocyte pools from six mice per group at 11 days postboost as described in Materials and Methods. P values were determined as described in Materials and Methods using an approach that corrects measurements for the medium response, calculating confidence intervals (*, P < 0.05; **, P < 0.005; ***, P < 0.001).
Furthermore, all vaccinated groups induced SARS-CoV-2 S-specific CD4+ T regulatory (Treg) cells (CD4+, FOXP3+) secreting IFN-γ (Fig. 6C), with no secretion of IL-10 and IL-17 (data not shown), which is also indicative of a Th1-like immunity. The response was directed against S1 peptide pool, with no S2-specific CD4+ Treg cells detected. Although not significant, immunizations with the MVA-Δ-S construct elicited slightly higher responses than those carried out with the MVA-S construct (Fig. 6C).
Moreover, all vaccination regimens generated SARS-CoV-2 S-specific CD8+ CD103+ T resident-like memory cells secreting IFN-γ (Fig. 6D). Heterologous DNA-S/MVA-Δ-S immunization induced a significantly higher magnitude of S-specific CD8+ CD103+ T resident-like memory cells than DNA-S/MVA-S or the homologous MVA/MVA immunizations (Fig. 6D).
Mouse immunization with MVA-S vaccine candidates induces IgG antibodies against SARS-CoV-2 S and receptor-binding domain (RBD) proteins.
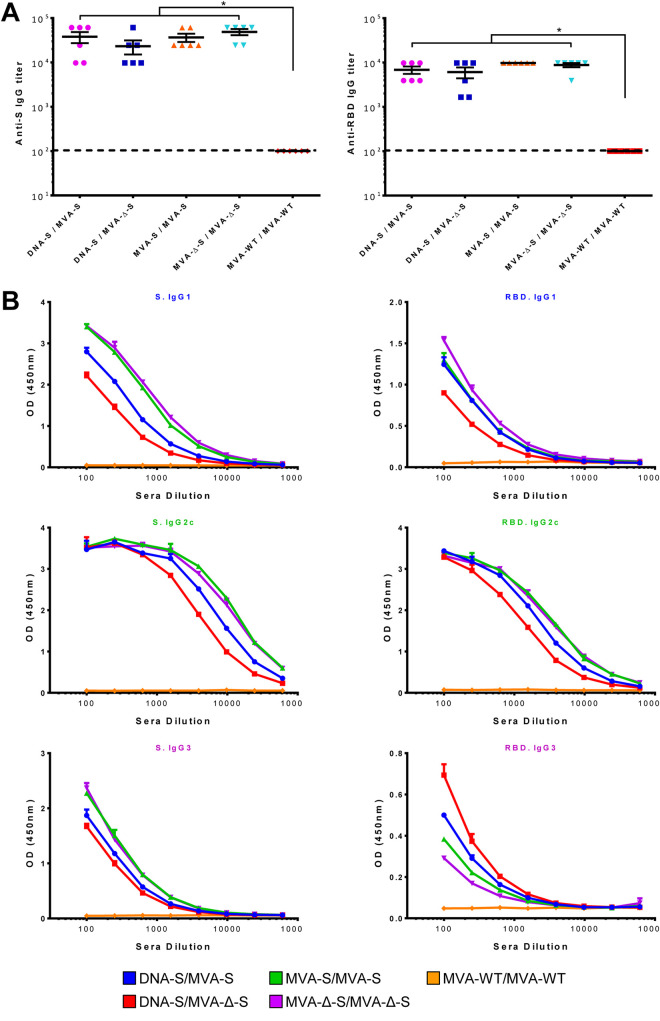
Antibodies against SARS-CoV-2 S protein are decisive to control the COVID-19 progression (10–12). Thus, to evaluate the ability of the MVA-S vaccine candidates to elicit humoral immune responses against SARS-CoV-2, we determined the presence of S- and RBD-specific IgGs in the sera of immunized mice by an enzyme-linked immunosorbent assay (ELISA) (Fig. 7). All vaccinated groups induced high IgG antibody titers against the S (Fig. 7A, left), and the RBD proteins (Fig. 7A, right). The IgG2c subtype prevailed in the isotype characterization, followed by IgG1 subtype, with homologous MVA/MVA prime/boost immunizations inducing higher levels than heterologous DNA/MVA regimens (Fig. 7B). The IgG2c/IgG1 ratio was above 1 in all vaccinated groups, likely suggestive of Th1-like immune responses.
FIG 7.
MVA-S and MVA-Δ-S elicit SARS-CoV-2 S-specific humoral immune responses in immunized mice. (A) Titers of IgG antibodies specific for the S (left) and RBD (right) proteins. Titers were determined by ELISA in individual mouse serum samples collected 11 days postboost and were calculated as the serum dilution (y axis) at which the absorbance was three times higher than the naive serum value in each of the indicated immunization regimens, shown in the x axis. The dashed line represents the detection limit. P values from Kruskal-Wallis test followed by post hoc pairwise comparisons by Wilcoxon rank sum tests with Holm correction for multiple comparisons: (*, P < 0.05). (B) Levels of IgG1, IgG2c, and IgG3 isotypes specific for the S (left) and RBD (right) proteins. Mean optical density (OD at 450 nm) and standard deviations determined by ELISA with duplicate serum dilutions from pooled serum samples (n = 6) from immunized mice at 11 days postboost are represented.
MVA-S vaccine candidates elicits high levels of SARS-CoV-2 nAbs.
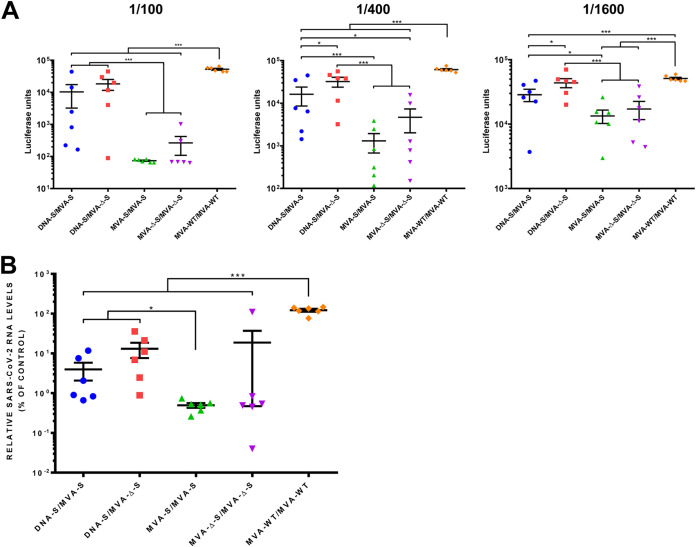
SARS-CoV-2 nAbs correlate with protection in humans (10, 11), and the presence of RBD-specific antibodies in the sera of mice immunized with the MVA-S vectors (Fig. 7) suggested the existence of SARS-CoV-2 nAbs. Thus, we next analyzed the ability of mouse serum samples to interfere with the entry of SARS-CoV-2 S-pseudotyped reporter retroviral vectors in Vero-E6 cells by a luciferase assay (Fig. 8A). The individual serum samples obtained from all mice vaccinated with MVA-S or MVA-Δ-S significantly neutralized S-pseudotyped retrovirus particles at a 1/100 dilution compared to sera from the MVA-WT/MVA-WT control group (Fig. 8A, left panel). Sera from both homologous MVA/MVA prime/boost immunizations were the most active in pseudovirus neutralization, with MVA-S/MVA-S inducing a significant 700-fold reduction in the luciferase units compared to that of the control group (Fig. 8A, left panel). The inhibition decreased with serum sample dilution (Fig. 8A, middle and right panels), but remarkably, serum samples from both MVA-S/MVA-S prime/boost immunizations showed a 50% inhibitory dilution (ID50) higher than 1,600 (Fig. 8A, right panel).
FIG 8.
MVA-S and MVA-Δ-S induce SARS-CoV-2 neutralizing antibodies in immunized mice. (A) Neutralization of retrovirus-based pseudoparticles expressing the SARS-CoV-2 S protein. Inhibition of S-pseudotype retrovirus entry in Vero-E6 cells was measured by a luciferase assay with serum samples collected 11 days postboost. Each dot represents the mean luciferase units determined from triplicates of individual mouse serum samples (diluted 1/100 [left panel], 1/400 [middle panel], and 1/1,600 [right panel]). Mean values and standard errors of the means (SEM) for each immunization group are represented. (B) Neutralization of SARS-CoV-2 cell infection. Virus RNA was quantified 6 h after infection of Vero-E6 cells with SARS-CoV-2 strain NL/2020 (MOI of 1 PFU/cell) mixed with a 1/50 dilution of mouse serum samples. Mean RNA levels and SEM from triplicates of individual serum samples are represented for each immunization group. The average RNA level from infections performed in the presence of mouse serum samples from the control group MVA-WT/MVA-WT was set at 100%. P values from Kruskal-Wallis test followed by post hoc pairwise comparisons by Wilcoxon rank sum tests with Holm correction for multiple comparisons: (*, P < 0.05; ***, P < 0.001).
To further analyze serum neutralization of live SARS-CoV-2 virus, we carried out experiments with bona fide infectious virus in a cell culture infection model. SARS-CoV-2 strain NL/2020 infections were carried out in Vero-E6 cells in the presence of serum samples (diluted 1/50), and the intracellular viral RNA was analyzed 6 hpi. The mouse serum samples from homologous MVA/MVA and heterologous DNA/MVA prime/boost immunizations reduced SARS-CoV-2 infection significantly compared to the MVA-WT/MVA-WT control group (Fig. 8B). Consistently, mouse serum samples from homologous MVA-S/MVA-S prime/boost immunization were the most effective in SARS-CoV-2 neutralization, with 245-fold reduction in viral RNA.
Morbidity, mortality, and virus replication induced by SARS-CoV-2 in humanized K18-hACE2 mice are completely prevented by vaccination with MVA-S.
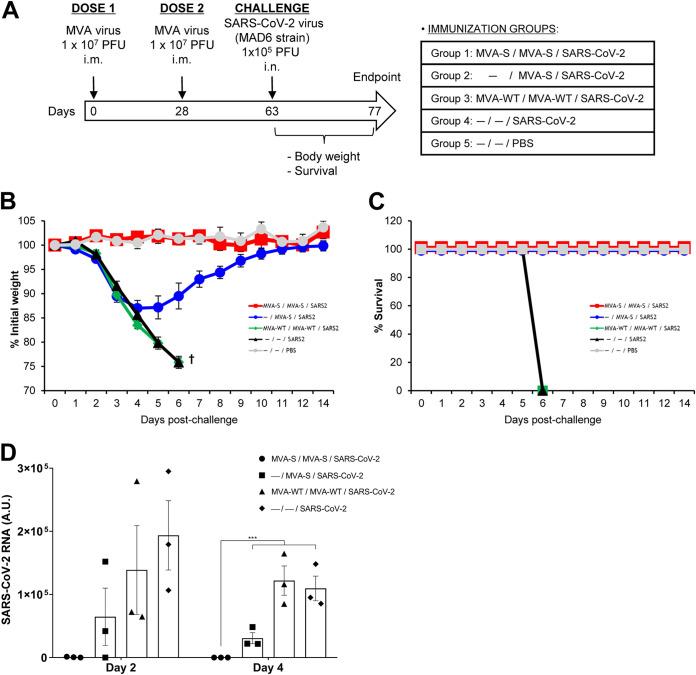
Next, we analyzed the efficacy of the MVA-S vaccine candidate in K18-hACE2 mice, a mouse model for SARS-CoV-2 infection and lethality (44, 45). For morbidity and mortality evaluation, groups of K18-hACE2 mice (n = 5) were immunized with one or two doses (1 × 107 PFU by the i.m. route) of MVA-S at weeks 0 and 4 and then challenged at week 9 with 1 × 105 PFU of SARS-CoV-2 (MAD6 isolate) by the intranasal route (Fig. 9A). Mice primed and boosted with MVA-WT or unvaccinated, and unchallenged mice were used as control groups. The challenged K18-hACE2 mice were monitored for change in body weight (Fig. 9B) and mortality (Fig. 9C) for 14 days postchallenge. All the K18-hACE2 mice immunized with two doses of MVA-S and challenged with SARS-CoV-2 did not show loss of body weight (Fig. 9B) and survived (Fig. 9C), whereas all the MVA-WT- immunized or nonimmunized and challenged K18-hACE2 mice showed loss of body weight (more than 25%) (Fig. 9B) and died (Fig. 9C) within 6 days postchallenge. On the other hand, all mice immunized with one single dose of MVA-S and challenged with SARS-CoV-2 lost body weight during the first days postchallenge (Fig. 9B), but by day 4, they recovered and all survived (Fig. 9C). To analyze the effect of the vaccine in virus replication, groups of humanized K18-hACE2 mice (n = 6) were vaccinated and infected as described above in Fig. 9A, and at 2 and 4 days after SARS-CoV-2 virus challenge, three mice per group were sacrificed, lung tissue samples were collected, RNA was extracted and analyzed by reverse transcription-quantitative PCR (RT-qPCR) for SARS-CoV-2 replication. As shown in Fig. 9D, SARS-CoV-2 replication was completely prevented at days 2 and 4 postchallenge in animals that received two doses of MVA-S in comparison with the high levels of SARS-CoV-2 viral RNA observed in the controls. Furthermore, a single dose of MVA-S vaccine candidate has also a major effect on virus replication. Thus, the MVA-S vaccine was highly efficacious to control SARS-CoV-2 morbidity, lethality, and virus replication in a mouse model of infection.
FIG 9.
MVA-S protects humanized K18-hACE2 mice from SARS-CoV-2 infection. (A) Efficacy schedule. K18-hACE2 mice (n = 5 per group) were immunized with one or two doses of 1 × 107 PFU of MVA-S or MVA-WT by the i.m. route at weeks 0 and 4. At week 9, mice were i.n. challenged with 1 × 105 PFU of SARS-CoV-2 (MAD6 isolate). Unvaccinated mice (n = 5) were also i.n. challenged with 1 × 105 PFU of SARS-CoV-2, and unvaccinated and unchallenged mice (n = 5) were used as a control. The challenged mice were monitored for change of body weights (B) and mortality (C) for 14 days. (D) SARS-CoV-2 RNA detected by RT-qPCR targeting viral E gene with normalization to 18S in lungs obtained at 2 (n = 3) and 4 (n = 3) days postchallenge from mice vaccinated and infected as in panel A. Mean RNA levels (in arbitrary units [A.U.]) and SEM from duplicates of each lung sample is represented for each immunization group; relative values are referred to uninfected mice. P values from two-way ANOVA test are shown (***, P < 0.001).
DISCUSSION
Development of a variety of safe and effective vaccines to counteract the SARS-CoV-2 pandemic is a major goal. Several clinical trials are ongoing with various candidate vaccines, and more will be added to the pipeline (23, 24). The critical issues are to fulfil the safety requirements of the corresponding drug agencies and to achieve highly protective and durable responses, which will be conditioned by the nature of the vaccine. As of late December 2020, two candidate mRNA vaccines have been approved by the FDA for emergency human use. One of the most promising vaccine candidates against pathogens is based on the poxvirus vector MVA, due to its safety record, stability, long-term immune responses, and ability to incorporate large fragments of foreign genetic material (28–31). With this in mind, we generated two MVA vectors expressing the full-length SARS-CoV-2 S protein. In cultured cells, both MVA-based vaccine candidates mediated expression of intracellular and cell surface S protein, which it is glycosylated and likely forming trimers. In mice, we compared their immunogenicity and observed that MVA-S/MVA-S or DNA-S/MVA-S prime/boost regimens induced potent levels of SARS-CoV-2 S-specific T-cell and humoral immune responses. Importantly, these levels correlated with complete protection of MVA-S-vaccinated humanized K18-hACE2 mice after challenge with a lethal dose of SARS-CoV-2.
SARS-CoV-2-specific T-cell responses are observed in individuals that recovered from COVID-19 (41), as well as in samples obtained years before the appearance of the SARS-CoV-2 pandemic (42), reflecting the important role played by T cells in the fight against SARS-CoV-2 infection. Along this line, we observed that the two MVA-based vaccine candidates induced a robust, broad, and polyfunctional SARS-CoV-2 S-specific T-cell response, mainly mediated by CD8+ T cells, which suggested higher cytotoxicity to eliminate virus-infected cells, but also with the induction of CD4+ T cells of a Th1 profile. Overall, MVA-Δ-S was superior to MVA-S in terms of activating S-specific CD8+ T-cell responses, a phenomenon mainly due to the deletion of VACV immunomodulatory genes C6L, K7R, and A46R, as we previously described with other MVA-based vaccines (33–35). Moreover, the S-specific CD4+ and CD8+ T-cell responses were broad, directed mainly against the S1 region of the S protein, which is the most distinct S portion among coronaviruses. Furthermore, those S-specific T-cell responses induced were of a T effector memory phenotype. Additionally, the T-cell responses were highly polyfunctional, with cells secreting simultaneously IFN-γ, TNF-α, and/or IL-2, and producing CD107, which is a good quality immune parameter to fight viral infections (46). Moreover, the T-cell activation profile induced by all vaccinated groups was broad and distinct, and we observed the induction of S-specific CD4+ Tfh cells expressing IFN-γ and IL-21 (with no secretion of IL-4), CD4+ Treg cells secreting IFN-γ (with no secretion of IL-10 and IL-17), and the activation of IFN-γ-producing S-specific CD8+ CD103+ T resident-like memory cells. Remarkably, the detection of CD4+ T cells secreting CD107, IFN-γ, TNF-α, and IL-2, but not IL-4, IL-10, and IL-17 indicate a bias toward Th1 immunity, a desirable response to control the SARS-CoV-2 infection, together with the absence of a potentially deleterious Th2-biased immune response that might induce vaccine-associated enhanced respiratory disease (47).
Our work also revealed an immunogenic role for a DNA vector expressing the S protein when delivered in prime/boost protocols. In fact, DNA vaccines expressing the S protein are important candidates against SARS-CoV-2, and clinical trials are ongoing. Results in NHP have shown protective immune responses with vectors expressing the full-length S, which were more effective than those producing truncated versions of the antigen (22). Our results showed that priming with a DNA-S vector is an effective approach to trigger an enhanced T-cell response against the S antigen after a boost with MVA-S. This effect is in line with our previous observations in the HIV-1 field, where DNA priming induced a preferential HIV-1-specific T-cell activation when used in different DNA/poxvirus vector combinations in NHP (48). These observations suggested the combination of DNA and poxvirus vectors as the preferred vaccination strategy to implement T-cell responses.
In terms of antibodies, heterologous DNA-S/MVA-S and homologous MVA-S/MVA-S immunizations induced high titers of S- and RBD-specific IgG antibodies, with IgG2c isotype antibodies being the most induced, reflecting a Th1-like immune response. Remarkably, antibodies that efficiently neutralized retrovirus-based pseudoparticles expressing the SARS-CoV-2 S protein, as well as a SARS-CoV-2 natural isolate (strain NL/2020), were induced. The homologous MVA-S/MVA-S vector combinations triggered the highest nAb ID50 titers, which were similar to those induced in immunized mice by other COVID-19 vaccine candidates (16) and to those mediated by SARS-CoV-2 immune human convalescent plasma samples (49, 50), reflecting a potent humoral immune response.
All these findings established a strong capacity of both MVA-based vaccine candidates to activate SARS-CoV-2-specific key immune T-cell populations and antibodies that will be necessary for protection against SARS-CoV-2 (10, 51). Therefore, of the combination of vectors used and based on the immunogenic T- and B-cell properties elicited, particularly on the levels of neutralizing antibodies, we selected MVA-S as a lead vaccine candidate for challenge studies in humanized K18-hACE2 mice. Remarkably, we found that humanized mice vaccinated with one or two doses of MVA-S were fully protected from a lethal dose of live SARS-CoV-2. Two controls (phosphate-buffered saline [PBS] and MVA-WT) develop more than 25% body weight loss and animals have to be sacrificed by day 6 postchallenge, while those mice that received a single dose lost weight but recovered completely; those mice vaccinated twice did not lose weight and were fully protected. The animals that receive one or two doses of the MVA-S vaccine were in good health for the whole study (14 days). In addition, the MVA-S vaccine fully protected against SARS-CoV-2 as shown by inhibition of SARS-CoV-2 virus replication in the lungs of vaccinated and infected humanized K18-hACE2 mice, correlating with the morbidity and mortality observed.
Others have recently described the generation of synthetic MVA vectors coexpressing SARS-CoV-2 S and nucleocapsid proteins, showing similar to us the induction of SARS-CoV-2-specific T-cell and humoral responses but not as detail as induced by our MVA-based vaccine candidates; however, they did not define the vaccine efficacy of those synthetic MVA vectors (13).
The data presented here demonstrated that MVA-based vaccine candidates expressing the SARS-CoV-2 full-length S protein are highly immunogenic in mice, with a robust activation of both arms of the adaptive immune system, T and B cells. They triggered Th1-skewed immune responses that can facilitate virus clearance without respiratory complications. Furthermore, we identified two prime/boost regimens to elicit preferentially either T-cell (DNA/MVA) or antibodies (MVA/MVA), which could be used differentially in vaccination. Importantly, two doses of MVA-S were able to completely protect humanized K18-hACE2 mice from SARS-CoV-2 infection (morbidity, mortality, and virus replication), reinforcing the translation of MVA-S to clinical trials.
MATERIALS AND METHODS
Ethics statement.
Female C57BL/6OlaHsd mice (6 to 8 weeks old) used for immunogenicity assays were purchased from Envigo Laboratories and stored in the animal facility of the Centro Nacional de Biotecnología (CNB) (Madrid, Spain). Female transgenic humanized K18-hACE2 mice, expressing human ACE2, were obtained from the Jackson Laboratory [034860-B6.Cg-Tg(K18-ACE2)2Prlmn/J, genetic background C57BL/6J × SJL/J)F2], and efficacy experiments were performed in the biosafety level 3 (BSL-3) facilities at the Centro de Investigación en Sanidad Animal (CISA)-Instituto Nacional De Investigaciones Agrarias (INIA) (Madrid, Spain). The immunogenicity and efficacy animal studies were approved by the Ethical Committee of Animal Experimentation (CEEA) of the CNB (Madrid, Spain) and by the Division of Animal Protection of the Comunidad de Madrid (PROEX 49/20, 169.4/20, and 161.5/20). Animal procedures conformed with international guidelines and with Spanish law under the Royal Decree (RD 53/2013).
Cells.
DF-1 cells (a spontaneously immortalized chicken embryo fibroblast [CEF] cell line, ATCC catalog no. CRL-12203) and HeLa cells (a human epithelial cervix adenocarcinoma; ATCC catalog no. CCL-2) were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-Life Technologies) supplemented with penicillin (100 U/ml; Sigma-Aldrich), streptomycin (100 μg/ml; Sigma-Aldrich), l-glutamine (2 mM; Sigma-Aldrich), nonessential amino acids (0.1 mM; Sigma-Aldrich), gentamicin (50 μg/ml; Sigma-Aldrich), amphotericin B (Fungizone, 0.5 μg/ml; Gibco-Life Technologies), and 10% heat-inactivated fetal calf serum (FCS) (Gibco-Life Technologies). HEK-293T cells (ATCC catalog no. CRL-3216; for the production of pseudotyped retroviral particles) and Vero-E6 cells (ATCC catalog no. CRL-1586) were maintained in subconfluent cultures in complete medium (DMEM supplemented with 10 mM HEPES [Gibco-Life Technologies], 1× nonessential amino acids [Gibco-Life Technologies], penicillin [100 U/ml; Sigma-Aldrich], streptomycin [100 μg/ml; Sigma-Aldrich], and 10% fetal bovine serum [FBS] [heat inactivated at 56°C for 30 min, Gibco-Life Technologies]). Cell cultures were maintained at 37°C in a humidified incubator containing 5% CO2.
Viruses.
The poxviruses used in this study included the attenuated MVA-WT strain (kindly provided by G. Sutter) obtained from the Chorioallantois vaccinia virus Ankara (CVA) strain after 586 serial passages in CEF cells (52), and the recombinant MVA-Δ-GFP virus expressing green fluorescent protein (GFP), which is inserted into the TK locus of the MVA-WT genome under the transcriptional control of a VACV synthetic early/late (sE/L) promoter, and containing deletions in VACV immunomodulatory genes C6L, K7R, and A46R (33). MVA-WT and MVA-Δ-GFP were used as the parental viruses for the generation of MVA-S and MVA-Δ-S vaccine candidates, respectively. All viruses were grown in DF-1 cells to obtain a master seed stock (passage 2 [P2] stock) and titrated in DF-1 cells by a plaque immunostaining assay, using a rabbit polyclonal antibody against VACV strain WR (CNB; diluted 1:1,000), followed by an anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Sigma-Aldrich; diluted 1:1,000), as previously described (53). Determinations of the titers of the different viruses were performed at least two times. Furthermore, viruses grown in DF-1 cells were purified by centrifugation through two 36% (wt/vol) sucrose cushions in 10 mM Tris-HCl (pH 9). All viral stocks were free of contamination with mycoplasma (checked by PCR specific for mycoplasma), bacteria (checked by growth in LB plates without ampicillin), or fungi (checked by growth in Columbia blood agar plates; Oxoid).
SARS-CoV-2 strain NL/2020 (kindly provided by R. Molenkamp, Erasmus University Medical Center, Rotterdam, the Netherlands) and SARS-CoV-2 strain MAD6 (kindly provided by José M. Honrubia and Luis Enjuanes, CNB-CSIC, Madrid, Spain) were propagated in Vero-E6 cells by inoculation at a multiplicity of infection (MOI) of 0.001 PFU/cell (passage 2). Cell supernatants were harvested at 72 hpi, cleared by centrifugation, aliquoted, and stored at −80°C. Virus infectivity titers were determined by standard plaque assay in Vero-E6 cells.
Construction of plasmid transfer vector pCyA-S.
The plasmid transfer vector pCyA-S was constructed and used for the generation of MVA-S and MVA-Δ-S vaccine candidates, in which the SARS-CoV-2 full-length S gene was inserted into the TK locus of parental viruses MVA-WT and MVA-Δ-GFP, respectively, under the transcriptional control of the viral sE/L promoter. In detail, a 3,822-kbp DNA fragment encoding the SARS-CoV-2 full-length S gene (Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, GenBank accession number MN908947.3; the most contemporary isolate available at the time we initiated this work) was synthesized and human codon optimized by GeneArt and inserted into plasmid vector pCyA (previously described in reference 54) to generate the plasmid transfer vector pCyA-S (11,322 bp). This plasmid also contains a β-galactosidase (β-Gal) reporter gene sequence between two repetitions of the left TK-flanking arm, which allows the reporter gene to be deleted from the final recombinant virus by homologous recombination after successive passages.
Generation of MVA-S and MVA-Δ-S vaccine candidates.
Cultured DF-1 cells (3 × 106 cells) were infected with parental MVA-WT or MVA-Δ-GFP viruses at an MOI of 0.05 PFU/cell and transfected 1 h later with 10 μg of DNA plasmid pCyA-S, using Lipofectamine reagent according to the manufacturer’s recommendations (Invitrogen). At 48 hpi, the cells were harvested, lysed by freeze-thaw cycling, sonicated, and used for recombinant virus screening. Recombinant MVA-S and MVA-Δ-S vaccine candidates containing the 3,822-kb SARS-CoV-2 full-length S gene, inserted in the TK locus, and transiently coexpressing the β-Gal marker gene were selected by consecutive rounds of plaque purification in DF-1 cells stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 400 μg/ml, for three passages in total). In the following plaque purification steps, recombinant MVA-S and MVA-Δ-S vaccine candidates containing the 3,822-kb SARS-CoV-2 full-length S gene and with the β-Gal gene deleted by homologous recombination between the left TK arm and the short left TK arm repeat flanking the marker were isolated by three additional consecutive rounds of plaque purification screening for nonstaining viral foci in DF-1 cells in the presence of X-Gal (400 μg/ml). In each round of purification, the isolated plaques were expanded in DF-1 cells, and the crude viruses obtained were used for the next plaque purification round. The resulting recombinant viruses, MVA-S and MVA-Δ-S (passage 2 [P2] stock after the last plaque purification), were grown in DF-1 cells, purified by centrifugation through two 36% (wt/vol) sucrose cushions, and titrated by plaque immunostaining assay (53).
PCR analysis of recombinant vectors.
To verify that the SARS-CoV-2 S gene had been correctly inserted in MVA-S and MVA-Δ-S vaccine candidates, viral DNA was extracted from DF-1 cells mock infected or infected at 5 PFU/cell for 24 h with the different viruses, as previously described (55), and the correct insertion of the S gene was confirmed by PCR analysis using primers TK-L and TK-R, annealing in the MVA TK gene-flanking regions (previously described in reference 54). The amplification protocol was performed using PuReTaq Ready-To-Go PCR beads (GE Healthcare), in accordance with the manufacturer’s protocol. PCR products were run in 1% agarose gel and visualized by SYBR Safe staining (Invitrogen). Moreover, the SARS-CoV-2 S gene insertion was also confirmed by DNA sequence analysis (Macrogen).
Expression of SARS-CoV-2 S protein by Western blotting.
To check the correct expression of SARS-CoV-2 S protein by MVA-S and MVA-Δ-S vaccine candidates, monolayers of DF-1 cells were mock infected or infected at 5 PFU/cell with the different viruses in the presence or absence of tunicamycin (10 μg/ml), an inhibitor of N-glycosylation. At 4 and 24 hpi, cell extracts or supernatants were lysed in reducing (in the presence of 1× Laemmli plus β-mercaptoethanol) or nonreducing conditions (in the presence of 1× Laemmli without β-mercaptoethanol), fractionated by 7% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by Western blotting with a rabbit polyclonal anti-SARS-CoV-2 S antibody (Genetex; diluted 1:2,000; recognizing SARS-CoV-2 S1 region) or a mouse polyclonal anti-SARS-CoV-1 antibody (Genetex; diluted 1:2,000; recognizing SARS-CoV-1 and SARS-CoV-2 S2 regions) to analyze the expression of the SARS-CoV-2 S protein. For a viral loading control, we used a rabbit anti-VACV E3 (CNB; diluted 1:1,000) antibody. HRP-conjugated anti-rabbit or anti-mouse antibodies (Sigma-Aldrich; diluted 1:5,000 or 1:2,000, respectively) were used as the secondary antibodies. The immunocomplexes were detected with an HRP-luminol enhanced-chemiluminescence system (ECL Plus) (GE Healthcare).
Analysis of virus growth.
To study the virus growth profile of MVA-S and MVA-Δ-S vaccine candidates, monolayers of DF-1 cells grown in 12-well plates were infected in duplicate at 0.01 PFU/cell with parental viruses MVA-WT and MVA-Δ-GFP and recombinant MVA-S and MVA-Δ-S vaccine candidates. Following virus adsorption for 1 h at 37°C, the inoculum was removed, and the infected cells were washed with DMEM and incubated with fresh DMEM-2% FCS at 37°C in a 5% CO2 atmosphere. At different times (0, 24, 48, and 72 hpi), cells were collected by scraping, freeze-thawed three times, and briefly sonicated. Virus titers in cell lysates were determined by a plaque immunostaining assay in DF-1 cells as previously described (53).
Genetic stability.
The genetic stability of MVA-S and MVA-Δ-S vaccine candidates was analyzed as previously described (33). Monolayers of DF-1 cells were infected at 0.05 PFU/cell with recombinant MVA-S and MVA-Δ-S vaccine candidates (P2 stocks). At 72 hpi, cells were collected by being scraped. After three freeze-thaw cycles and a brief sonication, the cellular extract was centrifuged at 1,500 rpm for 5 min, and the supernatant was used for a new round of infection at a low MOI. The same procedure was repeated 9 times. Moreover, 20 individual plaques from MVA-S virus derived from passage 9 were picked. Then, expression of SARS-CoV-2 S protein at all 9 passages and in the 20 individual plaques from passage 9 was detected by Western blotting after infection of DF-1 cells with virus stocks from each passage and from the 20 plaques, using a rabbit polyclonal anti-SARS-CoV-2 S antibody (Genetex; diluted 1:2,000). An anti-rabbit HRP-conjugated antibody (Sigma; diluted 1:5,000) was used as the secondary antibody. The immunocomplexes were detected using an enhanced HRP-luminol chemiluminescence system (ECL Plus; GE Healthcare).
Expression of SARS-CoV-2 S protein by confocal immunofluorescence microscopy.
Immunofluorescence experiments were performed in HeLa cells infected at an MOI of 0.5 PFU/cell for 24 h as previously described (33–35). Then, permeabilized and nonpermeabilized infected HeLa cells were stained with WGA probe conjugated to the red fluorescent dye Alexa Fluor 594 (Invitrogen; diluted 1:200) and the rabbit polyclonal antibody against SARS-CoV-2 S protein (Genetex; diluted 1:200) to label the cell membrane and the SARS-CoV-2 S protein, respectively. Anti-SARS-CoV-2 S was detected with a rabbit secondary antibody conjugated with the fluorochrome Alexa Fluor 488 (green) (Invitrogen; diluted 1:500). We used DAPI (4′,6′-diamidino-2-phenylindole; Sigma) to stain the cell nuclei. Images of sections of the cells were taken with a Leica TCS SP5 microscope.
DNA recombinant vector.
A DNA plasmid expressing SARS-CoV-2 S protein (termed DNA-S) was generated by inserting the human codon-optimized full-length SARS-CoV-2 S gene into the pcDNA-3.1 plasmid. DNA-S was purified with the EndoFree Plasmid Mega kit (Qiagen) in accordance with the manufacturer’s protocol and was used in the heterologous prime/boost immunization protocols as a prime.
Peptides and proteins.
SARS-CoV-2 S peptide pools were used in the immunogenicity analysis and purchased from JPT Peptide Technologies (Berlin, Germany). The S peptide pools were distributed in two groups spanning the S1 and S2 regions of the S protein, with each peptide pool containing 158 (S1) or 157 peptides (S2) as consecutive 15-mers overlapping by 11 amino acids. Furthermore, a purified recombinant SARS-CoV-2 S(ΔTM) protein expressed in HEK293 cells was also used in the immunogenicity analysis and purchased from Creative Biomart.
SARS-CoV-2 S and RBD proteins produced in mammalian cells were used in the ELISA analysis of antibodies from serum samples. To prepare the soluble S, a recombinant cDNA coding for residues 1 to 1208 of the S (soluble portion) protein was cloned in the pcDNA3.1 vector for expression in HEK-293F cells. It comprised the endogenous signal and S1 human codon-optimized sequences, followed for the S2 coding region from the Wuhan-Hu-1 strain (GenBank accession number MN908947.3), a T4 fibritin trimerization sequence, a Flag epitope, and a 8×His tag. To improve soluble S protein stability and expression, the furin recognition motif (RRAR) was replaced by the GSAS sequence, and the K986P and V987P substitutions were introduced in the S2 portion. Protein was purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography from transfected cell supernatants. RBD expression and purification will be described in detail elsewhere (under review). Briefly, the RBD region that comprises S residues 334 to 528 was expressed fused to the human IgG1 Fc portion and purified by affinity chromatography from either HEK-293F or CHO cell supernatants. The Fc portion and additional tags were released from the RBD by thrombin treatment and removed using a protein A column and size exclusion chromatography. Protein concentrations were determined by absorbance at 280 nm.
Mouse immunization schedule.
DNA prime/MVA boost or MVA prime/MVA boost immunization protocols were performed in female C57BL/6 mice (6 to 8 weeks old) as previously described (34, 56) to evaluate the immunogenicity of MVA-S and MVA-Δ-S vaccine candidates. On the one hand, groups of animals (n = 6) received 100 μg of DNA-S in 100 μl of PBS (50 μg/leg) by the i.m. route, and 2 weeks later, they received an i.m. inoculation of 1 × 107 PFU of the sucrose-purified MVA virus (MVA-S or MVA-Δ-S) in 100 μl of PBS (50 μl/leg). On the other hand, groups of animals (n = 6) received two doses of 1 × 107 PFU of MVA-S or MVA-Δ-S by the i.m. route in 100 μl of PBS (50 μl/leg) at 0 and 2 weeks. Mice primed and boosted with nonrecombinant MVA-WT were used as a control group. At 11 days after the last immunization, mice were sacrificed using carbon dioxide (CO2). Blood from each individual mouse was collected and processed to obtain serum samples to analyze the titers of IgG antibodies, IgG isotypes, and neutralizing antibodies against SARS-CoV-2; the spleens were extracted and processed to measure the adaptive T-cell immune responses to the SARS-CoV-2 S antigen by ELISpot and ICS assays. No adverse effects were detected in immunized mice.
Efficacy studies in K18-hACE2 transgenic mice.
Female K18-hACE2 mice (10 weeks old at the beginning of the study) immunized with one or two doses of MVA-S were used to evaluate the efficacy of MVA-S vaccine candidates. For morbidity and mortality evaluation, groups of animals (n = 5) received one or two doses of 1 × 107 PFU of MVA-S by the i.m. route in 100 μl of PBS (50 μl/leg) at 0 and 4 weeks. Mice primed and boosted with nonrecombinant MVA-WT or not vaccinated were used as a control group. At week 9, mice were challenged with 1 × 105 PFU of SARS-CoV-2 MAD6 by the intranasal route in 50 μl of PBS, and one group of unvaccinated mice were left uninfected. Mice were monitored for body weight change and mortality for 14 days postchallenge. Animals approaching 25% of weight loss were euthanized. For the analysis of SARS-CoV-2 virus replication in lungs, groups of animals (n = 6) were similarly vaccinated and infected as described above, and at days 2 and 4 postchallenge, three mice per group were euthanized and necropsied. Lungs were collected, placed in RNALater stabilization reagent (Sigma-Aldrich), and stored at −80°C until RNA extraction.
ELISpot assay.
The ELISpot assay was used to detect SARS-CoV-2 S-specific IFN-γ-secreting cells. First, 96-well nitrocellulose-bottom plates (Millipore) were covered with 75 μl/well of a solution of the rat anti-mouse IFN-γ monoclonal antibody (Pharmingen) at a concentration of 6 μg/ml in PBS. After incubating overnight at room temperature, the wells were washed three times with RPMI medium and blocked with RPMI-10% FCS for at least 1 h at 37°C in a 5% CO2 atmosphere. After spleen processing, 106 splenocytes per condition were restimulated with 1 μg/ml of the SARS-CoV-2 S1 or S2 peptide pools or with RPMI-10% FCS. The plates were incubated with the peptides for 48 h at 37°C in a 5% CO2 atmosphere, washed five times with PBS-Tween 20, and incubated with 2 μg/ml of biotinylated rat anti-mouse IFN-γ monoclonal antibody XMG1.2 (Pharmingen) diluted in PBS-Tween 20 for 2 h at room temperature. The plates were then washed five times with PBS-Tween 20, and a 1:800 dilution of HRP-conjugated streptavidin (0.5 mg/ml; Sigma-Aldrich) was added. After 1 h at room temperature, it was washed three times with PBS-Tween 20 and two times with PBS, and finally 1 μg/ml of the diaminobenzidine (DAB) substrate (Sigma-Aldrich) resuspended in 50 mM Tris-Cl (pH 7.5) and 0.015% H2O2 was added to develop the plates. The reaction was stopped by washing the plate with abundant water, and once it was dry, the spots were counted using the ELISpot Reader System ELR02 plate reader (AID Autoimmun Diagnostika GmbH) with the aid of AID ELISpot reader system software (Vitro).
ICS assay.
The magnitude, breadth, polyfunctionality, and phenotypes of the SARS-CoV-2 S-specific CD4+ and CD8+ T-cell adaptive immune responses were analyzed by ICS as previously described (33, 40, 57–59) with some modifications. After spleen processing, 4 × 106 fresh splenocytes (depleted of red blood cells) were seeded on M96 plates and stimulated for 6 h in complete RPMI 1640 medium supplemented with 10% FCS containing 1 μl/ml Golgiplug (BD Biosciences) to inhibit cytokine secretion, 1× monensin (eBioscience, Thermo Fisher Scientific), anti-CD107a–fluorescein isothiocyanate (FITC) (BD Biosciences), and the SARS-CoV-2 S1 and S2 peptide pools (1 μg/ml). Cells were then washed, stained for surface markers, fixed, permeabilized (Cytofix/Cytoperm kit; BD Biosciences), and stained intracellularly with the appropriate fluorochromes. Dead cells were excluded using the violet LIVE/DEAD stain kit (Invitrogen). The fluorochrome-conjugated antibodies used for functional analyses were CD3-phycoerythrin (PE)-CF594, CD4-allophycocyanin (APC)-Cy7, CD8-V500, IFN-γ–PE-Cy7, TNF-α–PE, and IL-2–APC. In addition, the antibodies used for phenotypic analyses were CD62L-Alexa Fluor 700 and CD127-peridinin chlorophyll protein (PerCP)-Cy5.5. All antibodies were from BD Biosciences.
The response of CD4+ Tfh cells against SARS-CoV-2 S antigens was analyzed by ICS as previously described (60) and similar to the analysis of SARS-CoV-2 S-specific CD4+ and CD8+ T cells. Splenocytes (4 × 106 cells) were stimulated for 6 h in complete RPMI 1640 medium supplemented with 10% FCS containing 1 μl/ml Golgiplug (BD Biosciences, Franklin Lakes, NJ, USA) to inhibit cytokine secretion, 1× monensin (eBioscience, Thermo Fisher Scientific), anti-CD154 (CD40L)–PE (BD Biosciences, Franklin Lakes, NJ, USA), and the SARS-CoV-2 S protein (5 μg/ml) plus S1 and S2 peptide pools (1 μg/ml). Cells were then processed similar to the analysis of SARS-CoV-2 S-specific CD4+ and CD8+ T cells. The following fluorochrome-conjugated antibodies were used: CD154-PE, IL-4-Alexa Fluor 488, IL-21-APC, and IFN-γ-PE-Cy7 for functional analyses and CD4-Alexa Fluor 700, CD8-V500, PD1 (CD279)-APC-efluor780, CXCR5-PE-CF594, and CD44-PE-Cy5 (SPRD) for phenotypic analyses. All antibodies were from BD Biosciences. The presence of CD4+ Foxp3+ Treg cells against SARS-CoV-2 S antigens was also analyzed by ICS, similar to the analysis of SARS-CoV-2 S-specific CD4+ and CD8+ T cells. The fluorochrome-conjugated antibodies used for functional analyses were CD44-V450, CD4-PE, CD62L-BV570, CD90.2-BV786, Foxp3-FITC, IFN-γ-APC-Cy7, IL-17-BV605, and IL-10-APC from either BD Biosciences or Biolegend. The response of CD8+ CD103+ T resident-like memory cells against SARS-CoV-2 S antigens was also analyzed by ICS, similarly to the analysis of SARS-CoV-2 S-specific CD4+ and CD8+ T cells. The fluorochrome-conjugated antibodies used for functional analyses were CD8-FITC, CD44-V450, CD62L-BV570, CD103-PE, CD90.2-BV786, and IFN-γ-APC-Cy7.
Cells were acquired with a Gallios flow cytometer (Beckman Coulter) or with a LSR Fortessa (BD Biosciences). Analyses of the data were performed with the FlowJo software version 10.4.2 (Tree Star). After gating, boolean combinations of single functional gates were created using FlowJo software to determine the frequency of each response based on all possible combinations of cytokine expression or all possible combinations of differentiation marker expression. Background responses detected in negative-control samples (RPMI) were subtracted from those detected in stimulated samples for every specific functional combination.
Antibody measurements by ELISA.
The total levels of IgG, IgG1, IgG2c, and IgG3 anti-SARS-CoV-2 S and RBD antibodies in individual or pooled sera from immunized mice were measured by ELISA as previously described (33). Ninety-six-well Nunc MaxiSorp plates were coated with 50 μl of purified recombinant SARS-CoV-2 S or RBD proteins at a concentration of 2 μg/ml in PBS at 4°C overnight. Plates were washed with PBS (Gibco-Life Technologies) supplemented with 0.05% Tween 20 (PBS-Tween) and blocked with 5% milk in PBS for 2 h at room temperature (RT). Individual or pooled serum samples from each immunization group were diluted in PBS-Tween-1% milk, added to plates, and incubated for 1.5 h at RT. Plates were then washed, and secondary HRP-conjugated goat anti-mouse IgG, IgG1, IgG2c, or IgG3 antibodies (Southern Biotech; all diluted 1:1,000 in PBS-Tween-1% milk) were added and incubated for 1 h at RT. Plates were washed, the TMB substrate (Sigma-Aldrich) was added, and the reaction was stopped by adding 1 M H2SO4. Absorbance was read at 450 nm. Total IgG titers were measured as the last dilution that gives an absorbance at least three times higher the absorbance of a naive serum.
Neutralization.
The capacity of the sera obtained from mice immunized with vaccine candidates MVA-S and MVA-Δ-S to neutralize SARS-CoV-2 virus was initially determined using retrovirus-based pseudoparticles expressing SARS-CoV-2 S protein. Pseudotyped viruses were produced by transfection of the above-mentioned DNA-S plasmid together with packaging plasmids (kindly provided by F. L. Cosset [INSERM, Lyon, France]) in HEK-293T cells as previously described (61). Vero-E6 cells were seeded in 96-well plates at 10,000 cells/well. Twenty-four hours postseeding, a mix of retrovirus-based pseudoparticles expressing SARS-CoV-2 S protein and mouse serum samples (at a final dilution of 1:100, 1:400, or 1:1,600) were preincubated for 1 h at 37°C and then added to the cells in triplicates. The DMEM-2% FCS medium was replaced at 24 hpi, and then 24 h later, cells were lysed in passive lysis buffer (Promega) and the luciferase activity was measured in a luminometer (Thermo Appliskan multimode microplate reader; Thermo Fisher Scientific). The ID50 was determined as the highest dilution of serum which resulted in a 50% reduction of luciferase units compared with mouse serum samples from control group MVA-WT/MVA-WT.
Neutralization of SARS-CoV-2 virus (strain NL/2020) was carried out in Vero-E6 cells at the BSL3 facilities of the CNB-CSIC. In brief, Vero-E6 cells were seeded onto 96-well plates at 20,000 cells/well. The next day, a mixture of 20,000 PFU of SARS-CoV-2 (MOI of 1) with individual mouse serum samples (at a final dilution of 1:50) were preincubated for 1 h at 37°C and then added to the cells in triplicates. After 1 h of incubation at 37°C, the inoculum was removed; the cells were washed twice with PBS, fresh medium was added, and the cells were further incubated for 5 additional hours at 37°C. At that point, the supernatant was discarded, the cells were washed once with PBS, and cell lysates were prepared in TRIzol reagent (Thermo Scientific). RNA extraction was carried out following the manufacturer’s instructions, and the viral RNA content was determined by RT-qPCR using a previously validated set of primers and probes specific for SARS-CoV-2 E gene (62) and the cellular β-actin gene (63) for normalization. The ΔCt method was used for relative quantitation of viral RNA. The average RNA level from infections performed in the presence of control mouse serum samples (MVA-WT/MVA-WT) was set at 100%.
Analysis of SARS-CoV-2 RNA by quantitative RT-PCR (RT-qPCR).
Lungs from mice used in the efficacy study were harvested and stored in RNALater (Sigma-Aldrich) at −80°C until homogenized with a gentleMACS dissociator (Miltenyi Biotec) in 2 ml RLT buffer (Qiagen) plus β-mercaptoethanol. Then, 600 μl of homogenized lung tissue was used to isolate total RNA using the RNeasy minikit (Qiagen), according to the manufacturer's specifications. First-strand cDNA synthesis and subsequent real-time PCR were performed in one step using NZYSpeedy One-step RT-qPCR Master Mix (NZYTech) according to the manufacturer’s specifications using ROX as reference dye. SARS-CoV-2 viral RNA content was determined using previously validated set of primers and probes specific for the subgenomic RNA for the SARS-CoV-2 E protein (62) and the cellular 18S rRNA for normalization. Data were acquired with a 7500 real-time PCR system (Applied Biosystems) and analyzed with 7500 software v2.0.6. Relative RNA arbitrary units (A.U.) were quantified relative to the negative group (uninfected mice) and were performed using the 2−ΔΔCt method. All samples were tested in duplicate.
Statistical procedures.
One-way analysis of variance (ANOVA) followed by post hoc Student’s t test comparisons with Holm correction for multiple testing was used for ELISpot analysis to establish the differences between two groups. In ELISA and neutralization assays, we used a Kruskal-Wallis test to determine whether there were differences between groups, followed by post hoc pairwise comparisons with Wilcoxon rank sum tests with Holm correction for multiple testing. Statistical analysis of the ICS assay results was performed as previously described (58), using an approach that corrects measurements for the medium response (RPMI), calculating confidence intervals and P values. Only antigen response values significantly larger than the value for the corresponding RPMI are represented. Background values were subtracted from all of the values used to allow analysis of proportionate representation of responses. For statistical analysis of SARS-CoV-2 RNA by quantitative RT-qPCR, two-way ANOVA with Tukey’s honestly significant difference (HSD) post hoc tests was applied. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.005; ***, P < 0.001.
ACKNOWLEDGMENTS
We thank the Spanish Research Council (CSIC) and the Spanish Ministry of Science and Innovation for continuous support. SARS-CoV-2 NL/2020 virus isolate was kindly provided by R. Molenkamp (Erasmus University Medical Center, Rotterdam). SARS-CoV-2 MAD6 virus isolate was kindly provided by José M. Honrubia and Luis Enjuanes (CNB-CSIC, Madrid, Spain). A patent has been issued, EP20382558.3. We thank Iván Jareño from the CNB Animal Facilities and Ana Oña and Silvia Gutiérrez from the CNB confocal immunofluorescence service for their help and assistance.
This research was supported by the Spanish Health Ministry, Instituto de Salud Carlos III (ISCIII), Fondo COVID-19 grant COV20/00151, and Fondo Supera COVID-19 (Crue Universidades-Banco Santander) (to J.G.-A.), CSIC grant 2020E84 and Ferrovial and Mapfre donations (to M.E.), and Fondo Solidario Juntos and Banco de Santander (to D.S.). This publication was supported by the European Virus Archive GLOBAL (EVA-GLOBAL) project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement 871029.
Conceptualization: J.G.-A. and M.E.; formal analysis: J.G.-A. and C.O.S.S.; funding acquisition: J.G.-A., D.S., and M.E.; investigation: J.G.-A., U.G., P.G., C.D.F., P.P., A.L.-F., and C.Z.; methodology: J.G.-A., U.G., P.G., C.D.F., P.P., A.L.-F., and C.Z.; resources: J.M.C.; software: C.O.S.S.; supervision: J.G.-A. and M.E.; validation: J.G.-A. and M.E.; visualization: J.G.-A.; writing—original draft: J.G.-A. and M.E.; writing—review and editing: all authors. All authors have read and agreed to the published version of the manuscript.
We declare that we have no competing interests.
REFERENCES
- 1.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty I, Maity P. 2020. COVID-19 outbreak: migration, effects on society, global environment and prevention. Sci Total Environ 728:138882. doi: 10.1016/j.scitotenv.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amanat F, Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity 52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funk CD, Laferriere C, Ardakani A. 2020. A snapshot of the global race for vaccines targeting SARS-CoV-2 and the COVID-19 pandemic. Front Pharmacol 11:937. doi: 10.3389/fphar.2020.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang ML, Jerome KR, Bloom JD, Greninger AL. 2020. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J Clin Microbiol 58:e02107-20. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, To KK, Chan KH, Wong YC, Zhou R, Kwan KY, Fong CH, Chen LL, Choi CY, Lu L, Tsang OT, Leung WS, To WK, Hung IF, Yuen KY, Chen Z. 2020. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg Microbes Infect 9:1664–1670. doi: 10.1080/22221751.2020.1791738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salazar E, Kuchipudi SV, Christensen PA, Eagar TN, Yi X, Zhao P, Jin Z, Long SW, Olsen RJ, Chen J, Castillo B, Leveque C, Towers DM, Lavinder J, Gollihar JD, Cardona J, Ippolito GC, Nissly RH, Bird IM, Greenawalt D, Rossi RM, Gontu A, Srinivasan S, Poojary IB, Cattadori IM, Hudson PJ, Joselyn N, Prugar L, Huie K, Herbert A, Bernard DW, Dye J, Kapur V, Musser JM. 2020. Relationship between anti-spike protein antibody titers and SARS-CoV-2 in vitro virus neutralization in convalescent plasma. bioRxiv 10.1101/2020.06.08.138990. [DOI] [PMC free article] [PubMed]
- 13.Chiuppesi F, Salazar MD, Contreras H, Nguyen VH, Martinez J, Park S, Nguyen J, Kha M, Iniguez A, Zhou Q, Kaltcheva T, Levytskyy R, Ebelt ND, Kang TH, Wu X, Rogers T, Manuel ER, Shostak Y, Diamond DJ, Wussow F. 2020. Development of a synthetic poxvirus-based SARS-CoV-2 vaccine. bioRxiv 10.1101/2020.07.01.183236. [DOI] [PMC free article] [PubMed]
- 14.Corbett KS, Edwards D, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schafer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong WP, Schmidt SD, Wang L, Zhang Y, Stevens LJ, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Nason MC, Ruckwardt TJ, et al. 2020. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. bioRxiv 10.1101/2020.06.11.145920. [DOI] [PMC free article] [PubMed]
- 15.Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, Li J, Gong X, Lou X, Shi W, Wu D, Zhang H, Zhu L, Deng W, Li Y, Lu J, Li C, Wang X, Yin W, Zhang Y, Qin C. 2020. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laczko D, Hogan MJ, Toulmin SA, Hicks P, Lederer K, Gaudette BT, Castano D, Amanat F, Muramatsu H, Oguin TH, III, Ojha A, Zhang L, Mu Z, Parks R, Manzoni TB, Roper B, Strohmeier S, Tombacz I, Arwood L, Nachbagauer R, Kariko K, Greenhouse J, Pessaint L, Porto M, Putman-Taylor T, Strasbaugh A, Campbell TA, Lin PJC, Tam YK, Sempowski GD, Farzan M, Choe H, Saunders KO, Haynes BF, Andersen H, Eisenlohr LC, Weissman D, Krammer F, Bates P, Allman D, Locci M, Pardi N. 2020. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity 53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKay PF, Hu K, Blakney AK, Samnuan K, Brown JC, Penn R, Zhou J, Bouton CR, Rogers P, Polra K, Lin PJC, Barbosa C, Tam YK, Barclay WS, Shattock RJ. 2020. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun 11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith TRF, Patel A, Ramos S, Elwood D, Zhu X, Yan J, Gary EN, Walker SN, Schultheis K, Purwar M, Xu Z, Walters J, Bhojnagarwala P, Yang M, Chokkalingam N, Pezzoli P, Parzych E, Reuschel EL, Doan A, Tursi N, Vasquez M, Choi J, Tello-Ruiz E, Maricic I, Bah MA, Wu Y, Amante D, Park DH, Dia Y, Ali AR, Zaidi FI, Generotti A, Kim KY, Herring TA, Reeder S, Andrade VM, Buttigieg K, Zhao G, Wu JM, Li D, Bao L, Liu J, Deng W, Qin C, Brown AS, Khoshnejad M, Wang N, Chu J, Wrapp D, McLellan JS, et al. 2020. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun 11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, He X, Martinez DR, Rutten L, Bos R, van Manen D, Vellinga J, Custers J, Langedijk JP, Kwaks T, Bakkers MJG, Zuijdgeest D, Huber SKR, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna TM, Bondzie EA, Dagotto G, Gebre MS, Hoffman E, Jacob-Dolan C, Kirilova M, Li Z, Lin Z, Mahrokhian SH, Maxfield LF, Nampanya F, Nityanandam R, Nkolola JP, Patel S, Ventura JD, Verrington K, Wan H, Pessaint L, Ry AV, Blade K, Strasbaugh A, Cabus M, et al. 2020. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren W, Sun H, Gao GF, Chen J, Sun S, Zhao R, Gao G, Hu Y, Zhao G, Chen Y, Jin X, Fang F, Chen J, Wang Q, Gong S, Gao W, Sun Y, Su J, He A, Cheng X, Li M, Xia C, Li M, Sun L. 2020. Recombinant SARS-CoV-2 spike S1-Fc fusion protein induced high levels of neutralizing responses in nonhuman primates. Vaccine 38:5653–5658. doi: 10.1016/j.vaccine.2020.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato V, Bushmaker T, Flaxman A, Ulaszewska M, Feldmann F, Allen ER, Sharpe H, Schulz J, Holbrook M, Okumura A, Meade-White K, Perez-Perez L, Bissett C, Gilbride C, Williamson BN, Rosenke R, Long D, Ishwarbhai A, Kailath R, Rose L, Morris S, Powers C, Lovaglio J, Hanley PW, Scott D, Saturday G, de Wit E, Gilbert SC, Munster VJ. 2020. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv 10.1101/2020.05.13.093195. [DOI]
- 22.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, et al. 2020. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, mRNA Study Group . 2020. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med 383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, Wu SP, Wang BS, Wang Z, Wang L, Jia SY, Jiang HD, Wang L, Jiang T, Hu Y, Gou JB, Xu SB, Xu JJ, Wang XW, Wang W, Chen W. 2020. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, et al. 2020. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397:99−111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group . 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr A. 2003. Smallpox vaccination and bioterrorism with pox viruses. Comp Immunol Microbiol Infect Dis 26:423–430. doi: 10.1016/S0147-9571(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 28.Gomez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. 2011. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr Gene Ther 11:189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 29.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2013. Clinical applications of attenuated MVA poxvirus strain. Expert Rev Vaccines 12:1395–1416. doi: 10.1586/14760584.2013.845531. [DOI] [PubMed] [Google Scholar]
- 30.Volz A, Sutter G. 2013. Protective efficacy of modified vaccinia virus Ankara in preclinical studies. Vaccine 31:4235–4240. doi: 10.1016/j.vaccine.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Volz A, Sutter G. 2017. Modified vaccinia virus Ankara: history, value in basic research, and current perspectives for vaccine development. Adv Virus Res 97:187–243. doi: 10.1016/bs.aivir.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Arriaza J, Esteban M. 2014. Enhancing poxvirus vectors vaccine immunogenicity. Hum Vaccin Immunother 10:2235–2244. doi: 10.4161/hv.28974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Arriaza J, Cepeda V, Hallengard D, Sorzano CO, Kummerer BM, Liljestrom P, Esteban M. 2014. A novel poxvirus-based vaccine, MVA-CHIKV, is highly immunogenic and protects mice against chikungunya infection. J Virol 88:3527–3547. doi: 10.1128/JVI.03418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez P, Marín MQ, Lázaro-Frías A, Jiménez de Oya N, Blázquez A-B, Escribano-Romero E, S Sorzano CÓ, Ortego J, Saiz J-C, Esteban M, Martín-Acebes MA, García-Arriaza J. 2018. A vaccine based on a modified vaccinia virus Ankara vector expressing Zika virus structural proteins controls Zika virus replication in mice. Sci Rep 8:17385. doi: 10.1038/s41598-018-35724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazaro-Frias A, Gomez-Medina S, Sanchez-Sampedro L, Ljungberg K, Ustav M, Liljestrom P, Munoz-Fontela C, Esteban M, Garcia-Arriaza J. 2018. Distinct immunogenicity and efficacy of poxvirus-based vaccine candidates against Ebola virus expressing GP and VP40 proteins. J Virol 92:e00363-18. doi: 10.1128/JVI.00363-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallengard D, Lum FM, Kummerer BM, Lulla A, Lulla V, Garcia-Arriaza J, Fazakerley JK, Roques P, Le Grand R, Merits A, Ng LF, Esteban M, Liljestrom P. 2014. Prime-boost immunization strategies against Chikungunya virus. J Virol 88:13333–13343. doi: 10.1128/JVI.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohlund P, Garcia-Arriaza J, Zusinaite E, Szurgot I, Mannik A, Kraus A, Ustav M, Merits A, Esteban M, Liljestrom P, Ljungberg K. 2018. DNA-launched RNA replicon vaccines induce potent anti-Ebolavirus immune responses that can be further improved by a recombinant MVA boost. Sci Rep 8:12459. doi: 10.1038/s41598-018-31003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roques P, Ljungberg K, Kummerer BM, Gosse L, Dereuddre-Bosquet N, Tchitchek N, Hallengard D, Garcia-Arriaza J, Meinke A, Esteban M, Merits A, Le Grand R, Liljestrom P. 2017. Attenuated and vectored vaccines protect nonhuman primates against Chikungunya virus. JCI Insight 2:e83527. doi: 10.1172/jci.insight.83527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittman PR, Hahn M, Lee HS, Koca C, Samy N, Schmidt D, Hornung J, Weidenthaler H, Heery CR, Meyer TPH, Silbernagl G, Maclennan J, Chaplin P. 2019. Phase 3 efficacy trial of modified vaccinia Ankara as a vaccine against smallpox. N Engl J Med 381:1897–1908. doi: 10.1056/NEJMoa1817307. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Arriaza J, Arnaez P, Gomez CE, Sorzano CO, Esteban M. 2013. Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate MVA-B by deletion of vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS One 8:e66894. doi: 10.1371/journal.pone.0066894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181:1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI, Wang LF, Ooi EE, Kalimuddin S, Tambyah PA, Low JG, Tan YJ, Bertoletti A. 2020. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 43.Bachmann MF, Wolint P, Schwarz K, Jager P, Oxenius A. 2005. Functional properties and lineage relationship of CD8+ T cell subsets identified by expression of IL-7 receptor alpha and CD62L. J Immunol 175:4686–4696. doi: 10.4049/jimmunol.175.7.4686. [DOI] [PubMed] [Google Scholar]
- 44.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng J, Wong LR, Li K, Verma AK, Ortiz M, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Jr, Perlman S. 2020. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panagioti E, Klenerman P, Lee LN, van der Burg SH, Arens R. 2018. Features of effective T cell-inducing vaccines against chronic viral infections. Front Immunol 9:276. doi: 10.3389/fimmu.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graham BS. 2020. Rapid COVID-19 vaccine development. Science 368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 48.Asbach B, Kibler KV, Kostler J, Perdiguero B, Yates NL, Stanfield-Oakley S, Tomaras GD, Kao SF, Foulds KE, Roederer M, Seaman MS, Montefiori DC, Parks R, Ferrari G, Forthal DN, Phogat S, Tartaglia J, Barnett SW, Self SG, Gottardo R, Cristillo AD, Weiss DE, Galmin L, Ding S, Heeney JL, Esteban M, Jacobs BL, Pantaleo G, Wagner R. 2018. Priming with a potent HIV-1 DNA vaccine frames the quality of immune responses prior to a poxvirus and protein boost. J Virol 93:e01529-18. doi: 10.1128/JVI.01529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, Garcia-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma WL. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manners C, Larios Bautista E, Sidoti H, Lopez OJ. 2020. Protective adaptive immunity against severe acute respiratory syndrome coronaviruses 2 (SARS-CoV-2) and implications for vaccines. Cureus 12:e8399. doi: 10.7759/cureus.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayr A, Stickl H, Muller HK, Danner K, Singer H. 1978. The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author's transl). Zentralbl Bakteriol B 167:375–390. [PubMed] [Google Scholar]
- 53.Ramirez JC, Gherardi MM, Esteban M. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J Virol 74:923–933. doi: 10.1128/jvi.74.2.923-933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomez CE, Perdiguero B, Cepeda MV, Mingorance L, Garcia-Arriaza J, Vandermeeren A, Sorzano CO, Esteban M. 2013. High, broad, polyfunctional, and durable T cell immune responses induced in mice by a novel hepatitis C virus (HCV) vaccine candidate (MVA-HCV) based on modified vaccinia virus Ankara expressing the nearly full-length HCV genome. J Virol 87:7282–7300. doi: 10.1128/JVI.03246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez CE, Najera JL, Jimenez EP, Jimenez V, Wagner R, Graf M, Frachette MJ, Liljestrom P, Pantaleo G, Esteban M. 2007. Head-to-head comparison on the immunogenicity of two HIV/AIDS vaccine candidates based on the attenuated poxvirus strains MVA and NYVAC co-expressing in a single locus the HIV-1BX08 gp120 and HIV-1(IIIB) Gag-Pol-Nef proteins of clade B. Vaccine 25:2863–2885. doi: 10.1016/j.vaccine.2006.09.090. [DOI] [PubMed] [Google Scholar]
- 56.Marin MQ, Perez P, Ljungberg K, Sorzano COS, Gomez CE, Liljestrom P, Esteban M, Garcia-Arriaza J. 2019. Potent anti-hepatitis C virus (HCV) T cell immune responses induced in mice vaccinated with DNA-launched RNA replicons and modified vaccinia virus Ankara-HCV. J Virol 93:e00055-19. doi: 10.1128/JVI.00055-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Arriaza J, Gomez CE, Sorzano CO, Esteban M. 2014. Deletion of the vaccinia virus N2L gene encoding an inhibitor of IRF3 improves the immunogenicity of modified vaccinia virus Ankara expressing HIV-1 antigens. J Virol 88:3392–3410. doi: 10.1128/JVI.02723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Arriaza J, Najera JL, Gomez CE, Sorzano CO, Esteban M. 2010. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS One 5:e12395. doi: 10.1371/journal.pone.0012395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia-Arriaza J, Najera JL, Gomez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M. 2011. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 6:e24244. doi: 10.1371/journal.pone.0024244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perdiguero B, Gomez CE, Garcia-Arriaza J, Sanchez-Corzo C, Sorzano COS, Wilmschen S, von Laer D, Asbach B, Schmalzl C, Peterhoff D, Ding S, Wagner R, Kimpel J, Levy Y, Pantaleo G, Esteban M. 2019. Heterologous combination of VSV-GP and NYVAC vectors expressing HIV-1 trimeric gp145 Env as vaccination strategy to induce balanced B and T cell immune responses. Front Immunol 10:2941. doi: 10.3389/fimmu.2019.02941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3):2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gong EY, Smets A, Verheyen N, Clynhens M, Gustin E, Lory P, Kraus G. 2013. A duplex real-time RT-PCR assay for profiling inhibitors of four dengue serotypes. Methods Mol Biol 1030:195–203. doi: 10.1007/978-1-62703-484-5_16. [DOI] [PubMed] [Google Scholar]