Abstract
The analysis of protein-protein interactions (PPIs) is essential for the understanding of cellular signaling. Besides probing PPIs with immunoprecipitation-based techniques, peptide pull-downs are an alternative tool specifically useful to study interactome changes induced by post-translational modifications. Peptides for pull-downs can be chemically synthesized and thus offer the possibility to include amino acid exchanges and post-translational modifications (PTMs) in the pull-down reaction. The combination of peptide pull-down and analysis of the binding partners with mass spectrometry offers the direct measurement of interactome changes induced by PTMs or by amino acid exchanges in the interaction site. The possibility of large-scale peptide synthesis on a membrane surface opened the possibility to systematically analyze interactome changes for mutations of many proteins at the same time. Short linear motifs (SLiMs) are amino acid patterns that can mediate protein binding. A significant number of SLiMs are located in regions of proteins, which are lacking a secondary structure, making the interaction motifs readily available for binding reactions. Peptides are particularly well suited to study protein interactions, which are based on SLiM-mediated binding. New technologies using arrayed peptides for interaction studies are able to identify SLIM-based interaction and identify the interaction motifs.
Graphical abstract
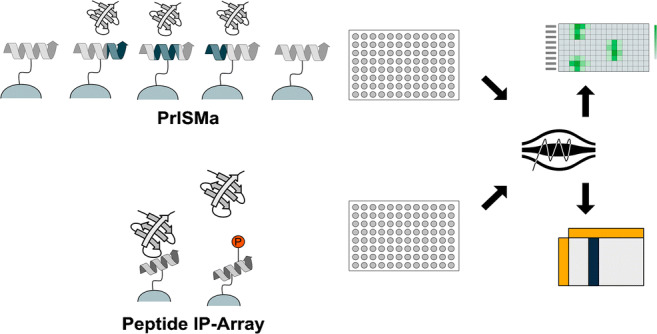
Keywords: PRISMA, Proteomics, Interactomics, PTM, IDR, SLiM
Introduction
Cellular signaling is in large parts based on a complex network of protein interactions with other proteins or other biological molecules. Studying protein-protein interaction (PPI) networks is pivotal for the understanding of cellular signaling [1–3]. Protein-protein interactions can be studied in different ways, including the genetic modification of the protein sequence, measurements by two-hybrid interactions, or chromatographic comigration [4–6]. The direct detection of interaction partners after isolating the protein of interest is the most common one [7]. The mass spectrometric measurement of interacting proteins has increased in popularity due to the increased sensitivity and possibilities of modern mass spectrometry–based proteomics [6, 8–10]. While many studies use whole proteins for interaction studies, the use of peptides in such studies has increased over time [11–16]. In this article, we will focus on the newly developing field of peptide array–based interaction studies.
Immunoprecipitations and peptide pull-downs
Since its development in the early 1980s, immunoprecipitations have increasingly been used to study PPIs [17]. Using specific antibodies against the protein of interest allowed the rapid isolation and detection of interaction partners by western blotting. Despite its advantage, the technique required additional knowledge about the interaction partners, so a proper antibody could be selected (Fig. 1a). This strategy to detect PPI turned out to be of particular interest when it was combined with the introduction of protein tags as fusion proteins, using molecular biology techniques. Using fusions for all proteins in an organism allows the systematic analysis of PPI in an arrayed manner by combining protein pull-down techniques with mass spectrometric identification of the interactors [18].
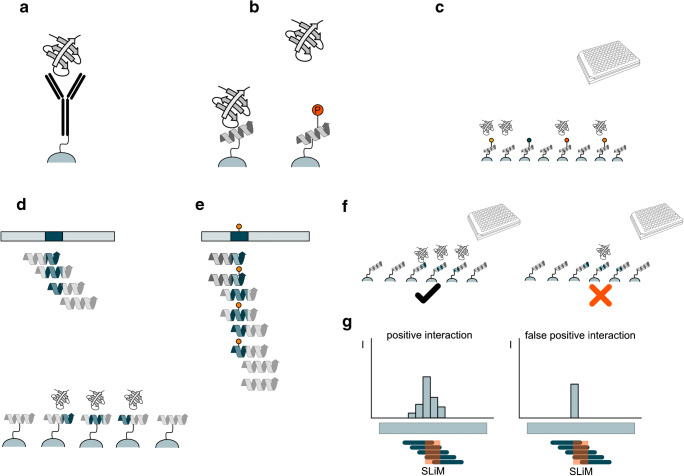
Fig. 1.
a Protein immunoprecipitation. The protein of interest is captured by an immobilized antibody for the pull-down. b Peptide pull-down. A peptide and a PTM-modified version of the same peptide are immobilized on a bead. The PTM prevents in this case the binding of the protein. c Highly parallelized peptide pull-down. The peptides are carrying different PTMs or mutations (indicated by the colored pins), which are enabling (orange and red) or preventing (black) interaction. d Peptide array designed for a PrISMa screen. A SLiM-containing area in a protein of interest (dark blue) is covered by different tiling peptides. Each of the peptides covers a different part and only the second peptide contains the entire SLiM (dark blue shade). e Inclusion of PTMs in the peptide array designed for PrISMa analysis. For each of the tiling peptides, a peptide with and one without the PTM is included. f + g Identification of false-positive binders in a PrISMa setup. Neighboring peptides cover different parts of the SLiM allowing only partial binding. A protein peptide not showing this binding behavior is excluded as it has a high probability to be nonspecific binding
In parallel, advances in the chemical synthesis of peptides allowed the production of larger quantities and longer peptides. This enabled the use of peptides in pull-down assays as an alternative to the genetic generation of tagged protein fragments [19]. The chemical synthesis of peptides carrying amino acid exchanges provided a fast alternative to cloning techniques [20–22]. Several studies used arrays of synthetic peptides containing alanine exchanges for the corresponding amino acids as a binding matrix. Incubation of the matrix and detection of the protein interactors allowed the identification of essential amino acids for the interaction [23–26]. An alternative technique for the screening for the best interacting peptides is the phage display technique, where peptides are presented on a cell using phage expressing protein fragments [21, 27–29].
Peptide pull-down meets mass spectrometry
The full potential of a peptide pull-down is only utilized in combination with a powerful detection methodology, which allows the identification of new interaction partners. With the rise of mass spectrometry–based proteomics, interacting proteins can be identified in a peptide pull-down (reviewed in [13]). Differently from protein-based interaction studies, peptide-protein interactions are usually of low affinity, thus preferring high-affinity interactions with more abundant proteins [30]. This preference introduces a bias against the detection of low abundant interaction partners or transient interactions and thus limits the use of peptide pull-downs. At the same time, a peptide bait permits narrowing the interaction site down to a fraction of a protein, allowing the precise mapping of the interaction site without the need to generate protein truncations [31, 32].
A significant advantage of peptides over protein pull-downs is the possibility of including post-translational modifications (PTMs) in the peptide. As peptides are chemically synthesized, any PTM can be included as long as it is compatible with the synthesis technique. The binding surface is thus fully modified, which is usually not the case with complete proteins [33–36].
Fast regulation of biological networks relies on the rapid addition and removal of PTMs during signaling, leading in many cases to the formation or loss of protein interactions. Capturing these transient interactions is challenging [36, 37]. Dissecting the recruitment of proteins using PTM-containing peptides allows identifying different complexes involved in the interaction (Fig. 1b). The interaction of SH2 domains with the phosphorylated C-terminal tail of the epidermal growth receptor is such an example. Synthetic phosphotyrosine-containing peptides were used to show the specificity of Grp2’s SH2 domains [38, 39]. To discriminate the different interactomes of the ErbB receptors for the phosphorylated versus the unphosphorylated state, phosphorylated peptides and their unmodified counterparts derived from the C-terminal region of ErbB receptors were used in a pull-down study, revealing the specific recruitment of Stat5 to the double phosphorylated C-terminus [40]. A systematic study of all 99 human SH2 domains probed their specificity in a system biological study. The binding patterns were confirmed by peptide pull-down experiments showing the regulation in a time-resolved manner [41]. Besides phosphorylations, other PTMs have been shown to change interactions. An example is the methylation at position three of the transcription factor C/EBPß, which leads to the loss of interaction with the SWI/SNF complex [42–44].
Linear motifs as interaction mediators
Many proteins adapt a defined fold, which is determined by their amino acid sequence. These folded domains are contrasted by regions lacking a specific fold, which are called intrinsically disordered regions (IDRs) [45]. Despite their lack of structure, IDRs are important docking sites for many proteins [46, 47]. They are often decorated with many PTMs indicating their importance for regulated interactions [45, 48–50]. IDRs can harbor many interaction motifs, which fall into three groups: short linear motifs (SLiMs), molecular recognition features (MoRFs), and intrinsically disordered domains (IDDs). The groups differ in length and how they support interactions. SLiMs are usually between three and 10 amino acids in length while MoRFs are slightly longer with 10 to 70 amino acids. MoRFs undergo a transition from the unordered to an ordered state while the ligand-binding takes place [51]. IDDs fold upon interaction with the binding partner [52].
SLiMs promote interactions via specific amino acid patterns, which are recognized by their interaction partners (Fig. 2). Because of their short dimensions, SLiMs represent a compact module that is structurally and functionally autonomous [53–59]. Several SLiMs within a protein can mediate the interaction with a large variety of proteins. The compact structure and defined amino acid patterns make SLiMs preferred targets for PTM modification. A single PTM can mask a SLiM and thus prevent or mediate binding [11, 60–62].
Fig. 2.
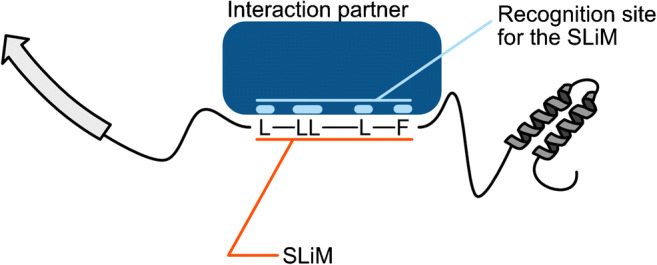
Interaction of a SLiM with a specific binding site of an interacting protein. The SLiM consists of a specific amino acid pattern, which, in this case, is defined for a set of amino acids and interspaced with amino acids (X) with no contribution to the binding (here: LXXLLXXXLXXF)
Synthesized peptides covering short parts of a protein can contain complete SLiMs, making them the perfect carrier for SLiM-based interaction studies. This has been used in studies, where a known sequence with and without the PTM was used to identify the interaction partners, as demonstrated in a study using two different peptides derived from EGFR and Sos1 and 2. The peptides were synthesized in phosphorylated and unmodified form and used in a pull-down experiment utilizing SILAC-labeled (stable isotope labeling in cell culture) cell extracts. The interaction partners were identified using mass spectrometry [63].
Peptide array–based interaction screens
The advancements of peptide synthesis by SPOT synthesis allow the creation of arrays holding many different peptides on a single membrane surface [23, 64, 65]. Cellulose membranes are an attractive alternative support to bead-based technologies, which can directly be used in biological assays, including immunological assays or parallelized peptide pull-downs [66]. The synthesis allows the inclusion of different PTMs in the membrane, permitting the systematic comparison of the interactome of a peptide sequence and its modified counterpart.
While peptide matrices have been used to find the optimal binding sequence [67–69], the true power of the approach emerges when it is combined with a proteomics readout. Meyer and coworkers use this principle to analyze mutations that cause neurological diseases [12]. One hundred twenty known disease-causing mutations were selected in extensive bioinformatics analysis. Peptides for the wild-type and mutated sequence were used to construct a peptide array and probed for differential binding with a proteomics readout (Fig. 1c). This created a PPI network of gained and lost interactions. A subnetwork of five interactors was related to clathrin-mediated transport. Three of the interaction nodes created a dileucine motif which is necessary for clathrin-dependent transport. In case of the glucose transporter GLUT1, the mutated version was wrongly localized to the endocytic compartment [12].
A screening technique targeting transient SLiM-based interactions along the primary structure was recently developed. The technique, PrISMa (Protein Interaction Screen on a Peptide Matrix), is based on a membrane-bound array of overlapping peptides, spanning the entire sequence of the protein of interest, creating a sliding window for the detection of SLiM-mediated interactions [11] (Fig. 1d, f). The interaction partners for each peptide were identified using mass spectrometry–based proteomics. The PrISMa technique provides a number of advantages over peptide pull-downs. The high concentration of immobilized peptides on the matrix allows the stabilization of the transient interactions, thus increasing the sensitivity for detecting these interactions. The overlapping peptide structure of the matrix offers the implementation of powerful filtering methods, based on the partial presence of the SLiM in the peptide adjacent to the main binding peptide. This allows separating unspecific background binding from SLiM-mediated interactions (Fig. 1g). The technique was used to map the interactome of the transcription factor C/EBPβ, identifying a large number of new interaction partners [11].
Additionally, the setup of the peptide array allows the inclusion of PTMs in the matrix to probe simultaneously for PTM-mediated interactions (Fig. 1e). For the C/EBPß study, several PTMs including methylation, citrullination, acetylation, and phosphorylation were included. This revealed the binding of the TLE3 complex specifically to the methylated form of C/EBPß.
A limitation of the method is the restriction of the screen: it can be only applied to intrinsically disordered regions, in which the structure depends only on the amino acid sequence. For interactions that depend on three-dimensional structures, this cannot be applied. To address this limitation, Ramberger and coworkers combined the PrISMa method with a BioID interaction screen to explore the C/EBPα interactome. They observed a significant overlap of the interaction partners between both technical approaches, and interestingly, common protein binders with C/EBPβ interactome [60].
Outlook
Over time, peptide-based interaction studies have significantly helped to reveal interaction sites or confirm specific PTM-regulated interactions. The new matrix-based interaction screens open a new era of interaction studies allowing to test many SLiM-based interactions at the same time. Besides the costs for the peptide matrix, using a mass spectrometric readout, the time of the measurement for all the interaction screens is a major restriction. Although the measurement of certain interactions can be parallelized by the use of isotopic labeling techniques, like SILAC and maybe TMT/iTRAQ in the future, the measurement of a peptide matrix can consume weeks on data acquisition in the mass spectrometer. Here, the technical developments of fast liquid chromatography systems might reduce the time constraints and open the use of peptide matrices for more laboratories. This will broaden the use of these techniques to identify more, potential druggable, PPI-driven diseases and will promote the deeper understanding of PPI networks, which depend on low affinity interactions.
Acknowledgements
We thank Marta L. Mendes for critically reading this manuscript.
Funding
This work was supported by the CovSerum (COVID-19/2020-1/14707743) grant of the Luxembourg Fonds National de la Recherche (FNR) to GD.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47:D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes H-W. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 5.Dong M, Yang LL, Williams K, Fisher SJ, Hall SC, Biggin MD, Jin J, Witkowska HE. A “tagless” strategy for identification of stable protein complexes genome-wide by multidimensional orthogonal chromatographic separation and iTRAQ reagent tracking. J Proteome Res. 2008;7:1836–1849. doi: 10.1021/pr700624e. [DOI] [PubMed] [Google Scholar]
- 6.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nooren IMA, Thornton JM. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunham WH, Mullin M, Gingras A-C. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 9.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmar G, Hernandez DP, Kowenz-Leutz E, Kirchner M, Kahlert G, Wesolowski R, Baum K, Knoblich M, Hofstätter M, Muller A, Wolf J, Reimer U, Leutz A. PRISMA: protein interaction screen on peptide matrix reveals interaction footprints and modifications- dependent interactome of intrinsically disordered C/EBPβ. iScience. 2019;13:351–370. doi: 10.1016/j.isci.2019.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer K, Kirchner M, Uyar B, Cheng J-Y, Russo G, Hernandez-Miranda LR, Szymborska A, Zauber H, Rudolph I-M, Willnow TE, Akalin A, Haucke V, Gerhardt H, Birchmeier C, Kühn R, Krauss M, Diecke S, Pascual JM, Selbach M. Mutations in disordered regions can cause disease by creating dileucine motifs. Cell. 2018;175:239–253.e17. doi: 10.1016/j.cell.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Schulze WX, Mann M. A novel proteomic screen for peptide-protein interactions. J Biol Chem. 2004;279:10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 14.Vermeulen M. Identifying chromatin readers using a SILAC-based histone peptide pull-down approach. Methods Enzymol. 2012;512:137–160. doi: 10.1016/B978-0-12-391940-3.00007-X. [DOI] [PubMed] [Google Scholar]
- 15.Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Otto JJ, Lee S-W. Chapter 7 Immunoprecipitation methods. In: Methods in cell biology. Elsevier; 1993. p. 119–27. [PubMed]
- 18.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 19.Geysen HM, Mason TJ. Screening chemically synthesized peptide libraries for biologically-relevant molecules. Bioorg Med Chem Lett. 1993;3:397–404. doi: 10.1016/S0960-894X(01)80221-3. [DOI] [Google Scholar]
- 20.Groll M, Bajorek M, Köhler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 21.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 22.Krupnick JG, Gurevich VV, Schepers T, Hamm HE, Benovic JL. Arrestin-rhodopsin interaction. Multi-site binding delineated by peptide inhibition. J Biol Chem. 1994;269:3226–3232. doi: 10.1016/S0021-9258(17)41852-7. [DOI] [PubMed] [Google Scholar]
- 23.Frank R. The SPOT-synthesis technique: synthetic peptide arrays on membrane supports—principles and applications. J Immunol Methods. 2002;267:13–26. doi: 10.1016/S0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 24.Geysen HM, Meloen RH, Barteling SJ. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci U S A. 1984;81:3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Titz B, Rajagopala SV, Ester C, Häuser R, Uetz P. Novel conserved assembly factor of the bacterial flagellum. J Bacteriol. 2006;188:7700–7706. doi: 10.1128/JB.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salinas VH, Ranganathan R. Coevolution-based inference of amino acid interactions underlying protein function. Elife. 2018;7. 10.7554/eLife.34300. [DOI] [PMC free article] [PubMed]
- 27.Kay BK, Kasanov J, Yamabhai M. Screening phage-displayed combinatorial peptide libraries. Methods. 2001;24:240–246. doi: 10.1006/meth.2001.1185. [DOI] [PubMed] [Google Scholar]
- 28.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 29.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 30.Yang M, Wu Z, Fields S. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 1995;23:1152–1156. doi: 10.1093/nar/23.7.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 33.Rivas JDL, De Las RJ, Fontanillo C. Protein–protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6:e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 35.Seo J, Lee K-J. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol. 2004;37:35–44. doi: 10.5483/bmbrep.2004.37.1.035. [DOI] [PubMed] [Google Scholar]
- 36.Yakubu RR, Nieves E, Weiss LM. The methods employed in mass spectrometric analysis of posttranslational modifications (PTMs) and protein-protein interactions (PPIs) Adv Exp Med Biol. 2019;1140:169–198. doi: 10.1007/978-3-030-15950-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deshaies RJ, Ferrell JE., Jr Multisite phosphorylation and the countdown to S phase. Cell. 2001;107:819–822. doi: 10.1016/S0092-8674(01)00620-1. [DOI] [PubMed] [Google Scholar]
- 38.Lemmon MA, Ladbury JE, Mandiyan V, Zhou M, Schlessinger J. Independent binding of peptide ligands to the SH2 and SH3 domains of Grb2. J Biol Chem. 1994;269:31653–31658. doi: 10.1016/S0021-9258(18)31745-9. [DOI] [PubMed] [Google Scholar]
- 39.Ward CW, Gough KH, Rashke M, Wan SS, Tribbick G, Wang J-X. Systematic mapping of potential binding sites for Shc and Grb2 SH2 domains on insulin receptor substrate-1 and the receptors for insulin, epidermal growth factor, platelet-derived growth factor, and fibroblast growth factor. J Biol Chem. 1996;271:5603–5609. doi: 10.1074/jbc.271.10.5603. [DOI] [PubMed] [Google Scholar]
- 40.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005.0008. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tinti M, Kiemer L, Costa S, Miller ML, Sacco F, Olsen JV, Carducci M, Paoluzi S, Langone F, Workman CT, Blom N, Machida K, Thompson CM, Schutkowski M, Brunak S, Mann M, Mayer BJ, Castagnoli L, Cesareni G. The SH2 domain interaction landscape. Cell Rep. 2013;3:1293–1305. doi: 10.1016/j.celrep.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leutz A, Pless O, Lappe M, Dittmar G, Kowenz-Leutz E. Crosstalk between phosphorylation and multi-site arginine/lysine methylation in C/EBPs. Transcription. 2011;2:3–8. doi: 10.4161/trns.2.1.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pless O, Kowenz-Leutz E, Dittmar G, Leutz A. A differential proteome screening system for post-translational modification-dependent transcription factor interactions. Nat Protoc. 2011;6:359–364. doi: 10.1038/nprot.2011.303. [DOI] [PubMed] [Google Scholar]
- 44.Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010;29:1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 46.Forman-Kay JD, Mittag T. From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure. 2013;21:1492–1499. doi: 10.1016/j.str.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunker AK, Babu MM, Barbar E, Blackledge M, Bondos SE, Dosztányi Z, Dyson HJ, Forman-Kay J, Fuxreiter M, Gsponer J, Others Intrinsically disord. Proteins. 2013;1:e24157. doi: 10.4161/idp.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 49.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Oldfield CJ, Yan W, Shen B, Dunker AK. Intrinsically disordered domains: sequence ➔ disorder ➔ function relationships. Protein Sci. 2019;28:1652–1663. doi: 10.1002/pro.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davey NE, Cowan JL, Shields DC, Gibson TJ, Coldwell MJ, Edwards RJ. SLiMPrints: conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Res. 2012;40:10628–10641. doi: 10.1093/nar/gks854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 56.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins: Struct Funct Bioinf. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 57.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 58.Dunker AK, Keith Dunker A, Oldfield CJ, Meng J, Romero P, Yang JY, Chen J, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uversky VN. Intrinsically disordered proteins and their “mysterious” (meta)physics. Front Phys. 2019;7:10. doi: 10.3389/fphy.2019.00010. [DOI] [Google Scholar]
- 60.Ramberger E, Sapozhnikova V Kowenz-Leutz E, Zimmermann K, Nicot N, Nazarov Pet al. A comprehensive motifs-based interactome of the C/EBPα transcription factor. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 61.Owen I, Shewmaker F. The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int J Mol Sci. 2019;20. 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed]
- 62.Bah A, Forman-Kay JD. Modulation of intrinsically disordered protein function by post-translational modifications. J Biol Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 64.Kramer A, Schneider-Mergener J. Synthesis and screening of peptide libraries on continuous cellulose membrane supports. Methods Mol Biol. 1998;87:25–39. doi: 10.1385/0-89603-392-9:25. [DOI] [PubMed] [Google Scholar]
- 65.Frank R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217–9232. doi: 10.1016/S0040-4020(01)85612-X. [DOI] [Google Scholar]
- 66.Wenschuh H, Volkmer-Engert R, Schmidt M, Schulz M, Schneider-Mergener J, Reineke U. Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides. Biopolymers. 2000;55:188–206. doi: 10.1002/1097-0282(2000)55:3<188::AID-BIP20>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 67.Starr TN, Picton LK, Thornton JW. Alternative evolutionary histories in the sequence space of an ancient protein. Nature. 2017;549:409–413. doi: 10.1038/nature23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ha J-H, Presti MF, Loh SN. A single protein disruption site results in efficient reassembly by multiple engineering methods. Biophys J. 2019;117:56–65. doi: 10.1016/j.bpj.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dwyer MA, Hellinga HW. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr Opin Struct Biol. 2004;14:495–504. doi: 10.1016/j.sbi.2004.07.004. [DOI] [PubMed] [Google Scholar]