Many severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serology tests have proven to be less accurate than expected and do not assess antibody function as neutralizing, correlating with protection from reinfection. A new assay technology measuring the interaction of the purified SARS-CoV-2 spike protein receptor binding domain (RBD) with the extracellular domain of the human angiotensin-converting enzyme 2 (hACE2) receptor detects these important antibodies.
KEYWORDS: ELISA, SARS-CoV-2, neutralizing antibodies, serology, vaccines
ABSTRACT
Many severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serology tests have proven to be less accurate than expected and do not assess antibody function as neutralizing, correlating with protection from reinfection. A new assay technology measuring the interaction of the purified SARS-CoV-2 spike protein receptor binding domain (RBD) with the extracellular domain of the human angiotensin-converting enzyme 2 (hACE2) receptor detects these important antibodies. The cPass surrogate virus neutralization test (sVNT), compared directly with eight SARS-CoV-2 IgG serology and two live-cell neutralization tests, gives similar or improved accuracy for qualitative delineation between positive and negative individuals in a fast, scalable, and high-throughput assay. The combined data support the cPass sVNT as a tool for highly accurate SARS-CoV-2 immunity surveillance of infected/recovered and/or vaccinated individuals as well as drug and convalescent-phase donor screening. The data also preview a novel application for the cPass sVNT in calibrating the stringency of live-cell neutralization tests and its use in longitudinal testing of recovered and/or vaccinated patients.
INTRODUCTION
Molecular and serological tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are a critical component of disease control strategies globally (1, 2). Consequently, the demand for test kits is high, and the market has responded with a growing number of commercially available tests but without clear global guidelines to ensure their efficacy and accuracy (3). Unfortunately, the data generated from these tests vary widely in terms of sensitivity, specificity, and accuracy, leading to concerns regarding the actual number of disease carriers who can unknowingly spread the virus throughout the population (4–6).
The majority of serology tests on the market primarily detect the natural IgM and IgG antibodies that are generated in response to SARS-CoV-2 infection (5). These assays are typically enzyme-linked immunosorbent assay (ELISA) based, with the plate surface coated with either full-length or truncated purified spike or nucleocapsid protein, with detection via anti-IgG or anti-IgM conjugated to horseradish peroxidase (HRP) or other fluorophores (7). In fact, many of these kits use plates coated with the receptor binding domain (RBD) of the spike protein due to its high immunogenicity (8, 9). Since the coating process involves the passive adsorption of proteins via hydrophobic interactions, conformational changes of the coated protein molecules may occur, resulting in newly exposed or altered epitopes that may not be present in the native state (10–14). This can lead to nonspecific binding of immunoglobulins to the coated surface and reduced specificity (11, 15).
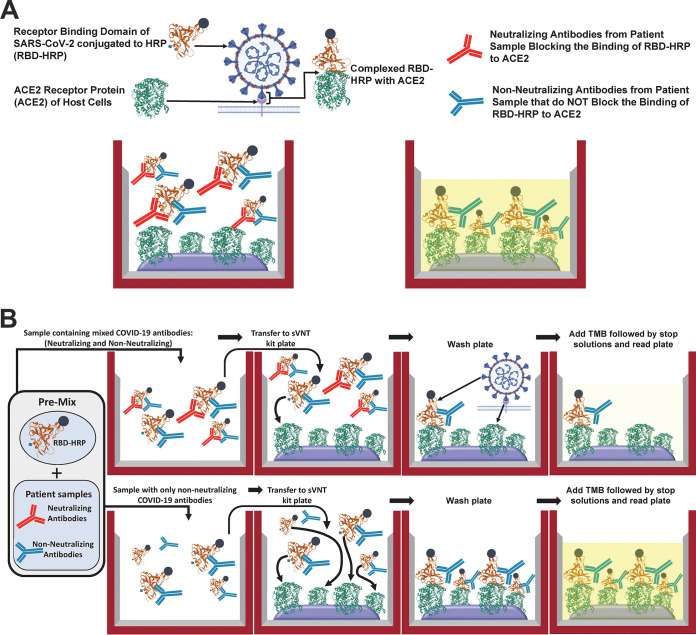
A novel serology assay termed the cPass surrogate virus neutralization test (sVNT) directly addresses the potential problems associated with the preexisting technologies while providing additional functional data (16). Utilizing 96-well plates coated with the purified extracellular domain of the human angiotensin-converting enzyme 2 (hACE2) receptor and a purified, solubilized, recombinant RBD conjugated to HRP (RBD-HRP), the assay capitalizes on the strong interaction between the hACE2 receptor and the RBD coupled with the high immunogenicity of the RBD (8, 17). This permits the direct assessment of the inhibitory capacity of immunoglobulins, antibody-based drugs, and compounds that block (or neutralize) this binding event (Fig. 1) (8, 9, 17, 18).
FIG 1.
cPass sVNT design and description. (A) sVNT design. The test consists of a purified RBD-HRP conjugate (brown) in solution and ELISA plates coated with the hACE2 receptor (green), which form a strong complex. When mixed with a sample containing proteins, small molecules, or antibodies that block the interaction between the RBD and the hACE2 receptor, a low OD450 will be measured after incubation with TMB and stop solution. (B) Performing the sVNT. Sample dilutions are initially mixed with the RBD-HRP solution, with incubation for 30 min at 37°C to permit the binding of components to the RBD. If the sample does not contain constituents that bind and block the RBD-hACE2 interaction (bottom four wells), the RBD-HRP will bind to the hACE2-coated wells, giving a yellow color after incubation with TMB for 15 min at 37°C followed by stop solution. If the sample contains blocking constituents, they will bind to the RBD during the initial 30 min and inhibit the interaction with hACE (top four wells), giving a light-yellow color after the addition of stop solution.
There are several advantages to this assay format over traditional serology tests. It measures the interaction between the hACE2 receptor and the RBD to elucidate the function of antibodies (and other molecules) as neutralizing (16). The test is amenable to the indirect detection of immunoglobulins that abrogate the interaction between the RBD and the hACE2 receptor and is therefore not specific to any isotype (e.g., IgG, IgM, or IgA, etc.) (i.e., isotype agnostic). Antibodies generated in all species infected with SARS-CoV-2 are detectable with this assay (16, 19). For vaccine and drug development organizations, this test offers potential application as a high-throughput, safe, and practical methodology for screening antibodies, proteins, peptides, or small molecules that block the interaction between the RBD and the hACE2 receptor. As opposed to the more traditional virus neutralization tests (20), this assay does not require a biosafety level 3 (BSL3) containment laboratory. Also, the cPass sVNT can be performed in about 1.5 h per 96-well plate, compared with 2 to 4 days for virus and pseudovirus tests.
The cPass sVNT was compared to eight traditional SARS-CoV-2 IgG ELISAs in two separate studies that utilized protein-coated plates and to two cell-based, live-virus neutralization tests using human serum and plasma samples collected from several cohorts of SARS-CoV-2 PCR-confirmed positive, negative, and prepandemic deidentified samples. Finally, an approach to the use of the cPass sVNT for longitudinal studies to assess changes in neutralizing antibody titers in patients who recovered from coronavirus disease 2019 (COVID-19) or vaccinated subjects is described.
MATERIALS AND METHODS
Samples.
For study 1, plasma and serum samples from The Children’s Hospital Colorado’s COVID-19 convalescent-phase plasma (CCP) donor program registered with the FDA as eligible to collect CCP on 31 March 2020 were collected. Eligible individuals for the CCP donor program were confirmed to be PCR positive for SARS-CoV-2, were symptom free for at least 14 days prior to plasma donation, and met all standard blood donation criteria according to FDA requirements. For each donor, the number of days from the PCR-positive SARS-CoV-2 test to the day of plasma donation and the number of donations were tracked. Positive and prepandemic presumed negative samples were deidentified, tested, and tabulated (Fig. 2 and Table 1).
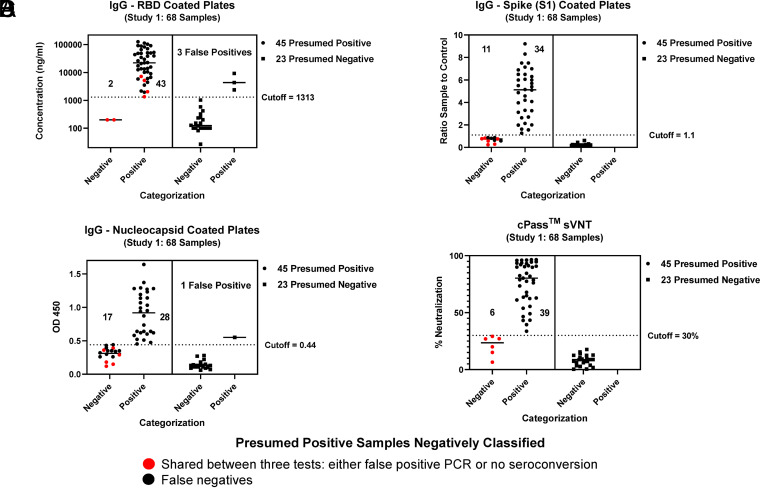
FIG 2.
Study 1: direct comparison between nucleocapsid-, RBD-, and spike (S1)-coated IgG ELISA plates with the cPass sVNT. (A) RBD-coated plate; (B) nucleocapsid-coated plate; (C) spike (S1)-coated plate; (D) cPass sVNT. Forty-five PCR-positive samples from patients with blood drawn more than 14 days after PCR testing were categorized by the four tests (round symbols at the left side of each chart). Of the negative delineated samples, six were shared between three tests (red dots). Twenty-three prepandemic presumed negative samples were categorized by the four tests (square symbols at the right side of each chart). Both the nucleocapsid- and RBD-coated IgG ELISA plates gave false positives.
TABLE 1.
Combined data from study 1 comparing assay performances of three commercial serology tests and the cPass sVNTa
| Parameter | Value for IgG detection in study 1 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Nucleocapsid-coated plates |
Spike (S1)-coated plates |
RBD-coated plates |
cPass sVNT |
|||||
| Positive (n = 45) | Negative (n = 23) | Positive (n = 45) | Negative (n = 23) | Positive (n = 45) | Negative (n = 23) | Positive (n = 45) | Negative(n = 23) | |
| No. of positive samples | 28 | 1 | 34 | 0 | 43 | 3 | 39 | 0 |
| No. of negative | 17 | 22 | 11 | 23 | 2 | 20 | 6 | 23 |
| Sensitivity (%) | 62.22 | 75.56 | 95.56 | 86.67 | ||||
| Specificity (%) | 95.65 | 100.00 | 86.96 | 100.00 | ||||
| Accuracy (%) | 92.31 | 97.36 | 87.82 | 98.67 | ||||
| PPV (%) | 61.39 | 100.00 | 44.87 | 100.00 | ||||
| NPV (%) | 95.80 | 97.36 | 99.44 | 98.54 | ||||
See Fig. 2. Sensitivity, specificity, accuracy, positive predictive values (PPV), and negative predictive values (NPV) are shown. A prevalence of 10% was used for the calculations.
For study 2, a subset of the identical serum samples collected for a previously published article comparing six commercial serology assays were tested and delineated with the cPass sVNT (21). The collection and description of the deidentified patient cohorts for both the positive and prepandemic samples are well described (21). The data for the positive samples from patients between 48 and 80 years of age were grouped, summarized, and tabulated by sampling days after symptom onset along with the prepandemic samples (Table 2).
TABLE 2.
Combined data from study 2 comparing the assay performances of six commercial serology tests and the cPass sVNTa
| Assay | Analyte | Time after symptom onset |
Negative samples (serum collected before Nov 2019) |
PPV (%) | NPV (%) | ACC (%) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5–9 days |
10–19 days |
>19 days |
All time points |
No. of Neg samples | No. of Equ samples | No. of Pos samples | Spec (%) | CI (%) | ||||||||||||||||||
| No. of Neg samples | No. of Pos samples | Sens (%) | CI (%) | No. of Neg samples | No. of Pos samples | Sens (%) | CI (%) | No. of Neg samples | No. of Pos samples | Sens (%) | CI (%) | No. of Neg samples | No. of Pos samples | Sens (%) | CI (%) | Total no. of samples | ||||||||||
| Abbott | IgG | 0 | 6 | 100 | 54–100 | 3 | 9 | 75 | 43–95 | 0 | 9 | 100 | 66–100 | 3 | 24 | 89 | 71–98 | 27 | 49 | 0 | 1 | 98 | 89–100 | 83 | 99 | 97 |
| Affinity | IgG | 0 | 7 | 100 | 59–100 | 2 | 11 | 85 | 55–98 | 0 | 8 | 100 | 63–100 | 2 | 26 | 93 | 71–98 | 28 | 47 | 0 | 0 | 100 | 92–100 | 100 | 99 | 99 |
| Bio-Rad | IgG | 0 | 6 | 100 | 54–100 | 3 | 9 | 75 | 43–95 | 0 | 9 | 100 | 66–100 | 3 | 24 | 89 | 71–98 | 27 | 50 | 0 | 0 | 100 | 93–100 | 100 | 99 | 99 |
| DiaSorin | IgG | 4 | 2 | 33 | 4–78 | 4 | 8 | 67 | 35–90 | 0 | 9 | 100 | 66–100 | 8 | 19 | 70 | 71–98 | 27 | 48 | 1 | 1 | 96 | 86–100 | 66 | 97 | 93 |
| Euroimmun | IgG | 3 | 3 | 50 | 12–88 | 4 | 8 | 67 | 35–90 | 0 | 8 | 100 | 63–100 | 7 | 19 | 73 | 71–98 | 26 | 50 | 0 | 0 | 100 | 93–100 | 100 | 97 | 97 |
| Roche | Total Ab | 1 | 6 | 86 | 42–100 | 4 | 9 | 69 | 39–91 | 0 | 9 | 100 | 66–100 | 5 | 24 | 83 | 71–98 | 29 | 50 | 0 | 0 | 100 | 93–100 | 100 | 98 | 98 |
| GenScript | RBD-hACE2 | 1 | 6 | 86 | 42–100 | 1 | 12 | 92 | 64–100 | 0 | 9 | 100 | 66–100 | 2 | 27 | 93 | 71–98 | 29 | 50 | 0 | 0 | 100 | 93–100 | 100 | 99 | 99 |
Sensitivity (Sens), specificity (Spec), accuracy (ACC), positive predictive values (PPV), and negative predictive values (NPV) are shown. Pos, positive; Neg, negative; CI, 95% confidence interval; Equ, equivocal result; Ab antibody. A prevalence of 10% was used for the calculations.
The data in Table 3 were derived from PCR-positive and -negative deidentified samples collected and tested in Singapore (Health Sciences Authority, conducted by Diagnostic Development Hub [DxD Hub]), DukeNUS, commercial vendors, and Granger Genetics, with data collated and analyzed by Corgenix Clinical Laboratory.
TABLE 3.
Combined clinical data for the cPass sVNTa
| GenScript cPass SARS-CoV-2 sVNT parameter | RT-qPCR result |
|
|---|---|---|
| Positive (n = 186) | Negative (n = 480) | |
| No. of positive results | 181 | 3 |
| No. of negative results | 5 | 477 |
| Sensitivity (%) | 97.30 | |
| Specificity (%) | 99.40 | |
| Accuracy (%) | 99.20 | |
| PPV (%) | 94.50 | |
| NPV (%) | 99.70 | |
Based on overall clinical data collected by 1 June 2020. Totals of 186 positive and 480 negative samples were verified by PCR and then screened by the sVNT. Sensitivity, specificity, accuracy, and positive and negative predictive values are shown. A prevalence of 10% was used for the calculations.
SARS-CoV-2 IgG ELISAs.
(i) Study 1. The CE-marked Epitope Diagnostics Inc. (EDI) (San Diego, CA) ELISA (catalog number KT-1032) utilizes the SARS-CoV-2 recombinant nucleocapsid antigen, and samples were diluted, tested, and analyzed according to the kit instructions for IgG. The CE-marked and FDA emergency-use authorization (EUA)-approved Euroimmun (Lubeck, Germany) ELISA (catalog number 2606) utilizes the S1 domain, including the receptor binding domain (RBD), of the SARS-CoV-2 spike protein, and samples were diluted, tested, and analyzed according to the kit instructions for IgG. The FDA Policy D, in vitro diagnostic (IVD)-status Akston Biosciences (Beverly, MA) ELISA (catalog number 600016) utilizes the recombinant RBD antigen of the SARS-CoV-2 spike protein, with samples diluted, tested, and analyzed according to the kit instructions for IgG.
For the EDI assay, positive, negative, and borderline results were calculated based on the average optical density at 450 nm (OD450) value for the negative control assayed in triplicate for the specific assay. The positive cutoff value was calculated using the formula positive cutoff = 1.1 × (NC + 0.18), where NC is the average OD450 of triplicate negative-control OD values. For study 1, given the day-to-day fluctuation in OD450 values from both the positive cutoff and our own interplate positive-control calibrator, the median positive cutoff OD450 for several days of testing (0.44) was used to delineate positive and negative samples.
(ii) Study 2. The six commercial IgG ELISAs (Abbott Laboratories, Epitope Diagnostics Inc., Affinity Diagnostics Corp, DRG International Inc. [supplied by Bio-Rad], Euroimmun, and Roche Diagnostics) used for the detection of SARS-CoV-2 IgG antibodies and the associated protocols employed for screening the positive and negative study samples were previously described (21). The positive cutoff defined in the kit instructions for each assay was used to delineate positive and negative samples compared with the cPass sVNT using a 30% cutoff.
SARS-CoV-2 cPass surrogate virus-neutralizing test.
The GenScript (Piscataway, NJ) cPass sVNT (catalog number L00847) utilizes the recombinant RBD of the SARS-CoV-2 spike protein to detect antibodies that block the RBD from binding to the hACE2 receptor. Plasma or serum samples and the kit-supplied positive and negative controls were diluted 1:10 in kit-specific sample dilution buffer according to the kit insert. The diluted samples and controls were preincubated with RBD-HRP in a “neutralization reaction” mixture for 30 min at 37°C, permitting the interaction and binding of neutralizing antibodies with RBD-HRP (Fig. 1B). Each neutralization reaction mixture was then added to the capture plate precoated with the hACE2 protein whereby the free RBD-HRP as well as RBD-HRP bound to nonneutralizing antibodies strongly interact with hACE2 and were captured on the plate (Fig. 1B). RBD-HRP complexed with neutralizing antibodies (i.e., those blocking the interaction between the RBD and hACE2) remained in the supernatant and were removed in a subsequent wash step. After the wash steps, 3,3′,5,5′-tetramethylbenzidine (TMB) followed by stop solution was added to all wells, permitting the visualization of RBD-HRP bound to the plate based on the OD450 intensity. The color intensity is inversely proportional to the amount of neutralizing antibody in standards or samples (Fig. 1B).
Data are interpreted by the percent inhibition of RBD-HRP binding, calculated as follows: percent inhibition = (1 − OD value of sample/OD value of background) × 100%. A 30% cutoff is used to delineate positive and negative samples where this cutoff has been calibrated against the gold-standard plaque reduction neutralization test (PRNT) using high-stringency PRNT90 (90% plaque reduction) data analysis. Percent inhibition of ≥30% indicates the presence of SARS-CoV-2 RBD-interacting antibodies blocking the RBD-hACE2 interaction.
Plaque reduction neutralization test.
The PRNT is considered the gold standard for characterizing neutralizing antibodies to most viruses, including SARS-CoV-2. Serum samples were heat inactivated for 30 min at 56°C. Serial 2-fold dilutions of the inactivated samples were prepared in a 96-well plate (Greiner Bio-One, Monroe, NC). A viral stock (strain hCoV-19/USA/WA1/2020; BEI Resources, Manassas, VA) containing approximately 200 PFU per 0.1 ml was added to each well containing serum dilutions. Following a 1-h incubation period at 37°C in a CO2 incubator, 6-well plates (Greiner Bio-One, Monroe, NC) containing recently confluent Vero cells (ATCC, Manassas, VA) were inoculated with the virus-serum mixtures. After a second incubation period of 45 min at 37°C in a CO2 incubator, 2 ml of an overlay (2× agarose [melt 1% agarose in water using a microwave oven and cool to 45°C prior to mixing with 2× minimal essential medium {MEM} with 4% fetal bovine {FBS} {Peak Serum, Wellington, CO} and 3 ml of 7.5% sodium bicarbonate per 100 ml of solution]) was added to each well. Finally, the plates were incubated for 24 h at 37°C in a CO2 incubator, upon which a second overlay containing neutral red (Millipore Sigma, St. Louis, MO) was dispensed into each well, followed by a 24-h incubation at 37°C in a CO2 incubator. The number of plaques was counted 48 to 72 h after the initial inoculation. The highest dilution of serum that inhibits (reduces) plaque formation by 50%, 75%, or 90% (PRNT50, PRNT75, or PRNT90, respectively) was calculated based on the titer of the viral stock and the number of plaques present at each dilution.
Focus reduction neutralization assay.
For the focus reduction neutralization test (FRNT), Vero E6 cells (ATCC, Manassas, VA) were seeded into 96-well plates. Serum samples were heat inactivated and serially diluted (2-fold, starting at 1:10) in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher, Pittsburgh, PA, USA) plus 1% FBS in 96-well plates. Approximately 100 focus-forming units (FFU) of SARS-CoV-2 USA-WA1/2020 (deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH) were added to each well, and the serum-virus mixture was incubated for 1 h at 37°C. After incubation, medium was removed from cells, and the serum-virus mixture was added for 1 h at 37°C. After 1 h, samples were removed, and cells were overlaid with 1% methylcellulose (Millipore Sigma, St. Louis, MO) in MEM (Thermo Fisher, Pittsburgh, PA, USA)–2% FBS and incubated for 30 h at 37°C. Cells were fixed with 4% paraformaldehyde (Acros Organics, Pittsburgh, PA, USA) and probed with 1 μg/ml of an anti-SARS-CoV spike monoclonal antibody (CR3022; Absolute Antibody, Boston, MA, USA) in Perm wash (1× phosphate-buffered saline [PBS]–0.1% saponin–0.1% bovine serum albumin [BSA]) for 2 h at room temperature (RT). After washing, cells were incubated with horseradish peroxidase (HRP)-conjugated goat anti-human IgG (Southern Biotech, Birmingham, AL, USA) (1:1,000) for 1.5 h at RT. After washing, SARS-CoV-2-positive foci were visualized with TrueBlue substrate (Thermo Fisher, Pittsburgh, PA, USA) and counted using a CTL Biospot analyzer and Biospot software (Cellular Technology Ltd., Shaker Heights, OH, USA). The FRNT50, FRNT75, and FRNT90 titers were calculated relative to a virus-only control (no serum) set at 100%, using GraphPad Prism 8 (GraphPad, La Jolla, CA, USA) default nonlinear curve fit constrained between 0 and 100%.
RESULTS
RBD soluble cPass sVNT versus eight IgG-specific serology tests, including RBD-, nucleocapsid-, or spike (S1)-coated plates, for COVID-19 diagnosis and classification.
For study 1, 68 (45 SARS-CoV-2 PCR presumed positive and 23 prepandemic presumed negative) human serum samples were directly compared across four tests (see Materials and Methods) using either RBD (Fig. 2A)-, nucleocapsid (Fig. 2B)-, or spike (S1-RBD) (Fig. 2C)-coated ELISA plates and the RBD soluble cPass sVNT (Fig. 2D). Of the PCR-positive samples (Fig 2, circles at the left side of each graph), six were categorized as false negatives and shared between three tests (nucleocapsid [Fig. 2B], spike S1 [Fig. 2C], and cPass sVNT [Fig. 2D] [red circles]). The RBD-coated ELISA (Fig. 2A) plates gave only two false-negative samples (shared between the four tests [red circles]) but also exhibited three false-positive samples (Fig. 2A, squares at the right side of graph above the cutoff line), suggesting a lower specificity for this assay. The remaining negatively classified samples (i.e., false negatives) among the 45 PCR presumed positive samples (Fig. 2, black circles at the left side of each graph below the cutoff line) for the nucleocapsid (Fig. 2B) and spike S1 (Fig. 2C) protein-coated ELISA plates were likely undetectable by these assays. For the 23 prepandemic presumed negative samples (Fig. 2, squares at the right side of each graph), the RBD (Fig. 2A)- and nucleocapsid (Fig. 2B)-coated ELISA plates misclassified 3 and 1 samples, respectively, as positive (squares above the cutoff line), whereas spike S1 (Fig. 2C) and the cPass sVNT (Fig. 2D) classified all samples correctly as negative (squares below the cutoff line). In summary, for study 1, the GenScript cPass sVNT delivered comparable or improved accuracy and negative and positive predictive values versus the other serology tests (Table 1).
For study 2, the data from previously tested human serum samples (21) (see Materials and Methods) were directly compared with the data for the cPass sVNT for SARS-CoV-2-positive (blood samples drawn at different intervals after symptom onset) and prepandemic deidentified individuals (Table 2). Consistent with study 1, the cPass sVNT gave results that were similar or superior to those of the other serological tests and also demonstrated the presence of neutralizing antibodies within 5 days after symptom onset.
A large cohort of well-characterized, PCR-verified positive and negative samples was screened with the cPass sVNT (Table 3). The comparable or superior specificity and sensitivity compared to other commercial serology tests (7, 22) translate to comparable or higher positive (94.5%) and negative (99.7%) predictive values and overall accuracy (99.2%), which is critical in population monitoring and contact tracing.
cPass sVNT ELISA versus live-cell viral neutralization tests (PRNT and FRNT).
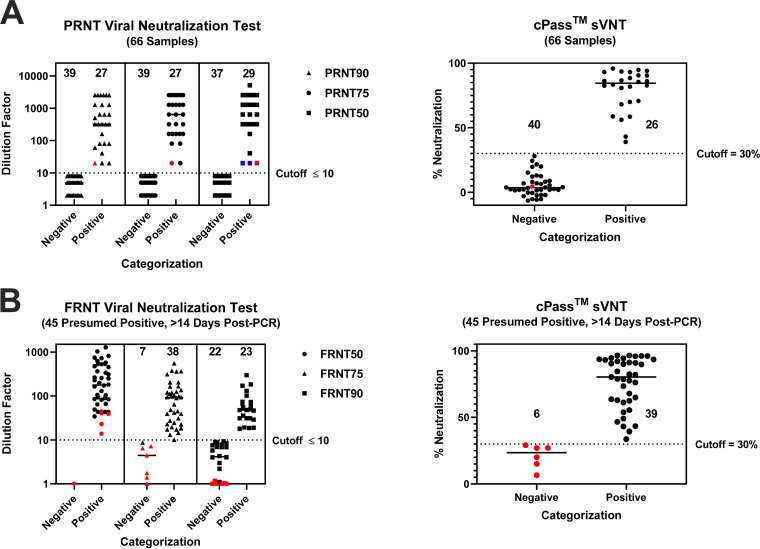
The CDC’s interim guidelines for COVID-19 antibody testing specific for SARS-CoV-2 neutralizing antibody detection include two assays for neutralizing antibody screening: (i) a virus-neutralizing test (VNT) such as the plaque reduction neutralization test (PRNT) and the focus reduction neutralization test (FRNT) and (ii) the pseudovirus neutralization test (pVNT) (23). These tests require live cells and virus with a multiday procedure that necessitates a BSL2 or BSL3 containment laboratory. Since the RBD for both SARS-CoV-1 and SARS-CoV-2 is immunodominant (8, 17), it has been postulated and shown that the cPass sVNT gives comparable results (19) and can potentially be used in lieu of the VNT or pVNT. Comparison of the PRNT50, PRNT75, and PRNT90 values with the sVNT values on 66 well-characterized samples gave a high correlation in delineating positive and negative samples for PRNT75 and PRNT90, where one sample (red dot) of 66 did not corroborate (Fig. 3A). However, when using a lower-stringency analysis for the PRNT (i.e., the reciprocal dilution that inhibited 50% of infection), two of the samples found to be negative by the cPass sVNT had detectable PRNT50 titers (Fig. 3A, blue dots).
FIG 3.
Direct comparison between the PRNT, FRNT (at different analysis stringencies), and sVNT. (A) PRNT. Sixty-six samples were assayed between the PRNT50, PRNT75, PRNT90, and sVNT. One sample was discordant between the sVNT and the PRNT75 and PRNT90 (red dot). Two samples were discordant between the PRNT50 and PRNT75 (blue dots). For the PRNT, negative samples with values below 10 were randomly assigned values of 2, 5, or 8 to more easily visualize the number of negative samples. (B) FRNT. Forty-five presumed positive samples were tested between the FRNT50, FRNT75, and FRNT90 and the sVNT. The same six samples were categorized as negative by the sVNT, FRNT75, and FRNT90 (red dots). One sample was discordant between the FRNT75 and sVNT. For the FRNT, all samples with a value of zero were assigned a value of 1 to more easily visualize the negative samples.
The same 45 PCR-confirmed, presumed positive samples from study 1 (Fig. 2 and Table 1) were also tested for live-virus-neutralizing activity using a SARS-CoV-2-specific FRNT. FRNT50, FRNT75, and FRNT90 titers were determined, giving excellent correlation between the cPass sVNT and FRNT75 (Fig. 3B). However, when comparing FRNT75 to FRNT50, five of the six samples found to be negative by the cPass sVNT had detectable FRNT50 titers. Reciprocally, 16 samples found to be positive by the cPass sVNT did not have detectable FRNT90 titers.
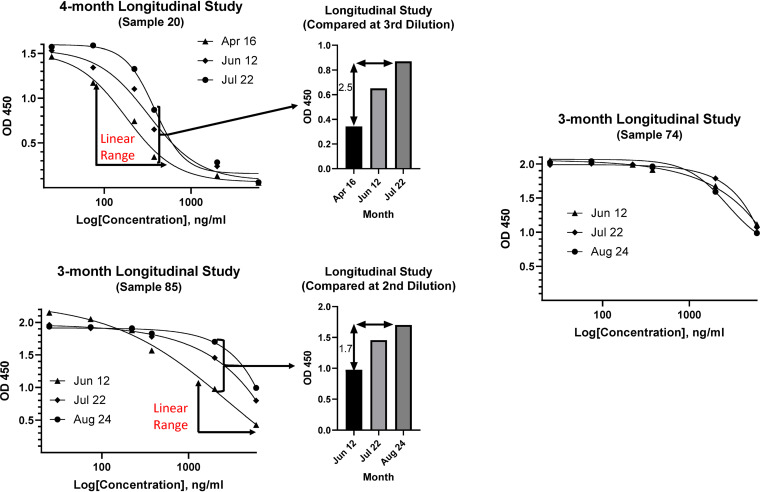
Temporal persistence of circulating neutralization antibodies in longitudinal studies (experimental design is critical).
Serum samples from three individuals who recovered from COVID-19 were collected over 3 months to assess the persistence of inhibitory antibodies using the cPass sVNT (Fig. 4). In order to determine the true quantitative difference between the time points, a serial dilution series of each sample was performed on the same plate to uncover a dilution whereby the signals were within the linear, quantitative range. For sample 20, the third dilution (1:90) was within the linear range of each dilution series, and a decrease in inhibitory antibodies of approximately 2.5-fold was measured over a 4-month period. For sample 85, the second dilution (1:30) was within the linear range and gave a 1.7-fold decrease over 3 months. However, if the samples had been diluted by only 10-fold, where the signal was within the lower plateau of the dilution curves for all three time points, very little difference in inhibitory antibodies would be quantified for sample 20 (compare the first point at the highest concentration in the dilution series at each month for sample 20). Sample 74 exhibited almost overlapping dilution curves over 3 months, indicating no change and, thus, persistence of immunity over that time period.
FIG 4.
Longitudinal assessment of viral titers by the cPass sVNT for serum samples taken at different time points postinfection. Samples were initially diluted 1:10 according to the kit instructions and then serially diluted 1:3 for an additional five dilutions to generate a competition curve for each sample at each time point. Samples 20 and 85 were compared for titers within the linear range of each curve or by the OD450 ratio.
DISCUSSION
The quality of serology test data has been widely variable and resulted in lower levels of sensitivity and specificity for some commercial tests, which has led to reduced confidence in serological testing. This can directly contribute to the increased spread of disease (5, 7, 20, 22, 24). The root cause of reduced accuracy is likely a consequence of the choice of antigen, associated posttranslational modifications (25, 26), and/or the protein-coated surface of the ELISA plates. Since the process for coating ELISA plates relies primarily on hydrophobic interactions, coating plates with proteins such as the spike S1 and S2 domains, the RBD, or nucleocapsid proteins of SARS-CoV-2 can lead to various subpopulations of structurally altered antigens in each well (14). This in turn can lead to the exposure of antigenic sites that would not otherwise be present in the native state, giving increased false positives from nonspecific immunoglobulin binding (11, 15). This was observed for nucleocapsid- and RBD-coated plates with prepandemic samples in study 1 (Fig. 2A and Table 1) and for the Abbott and DiaSorin tests in study 2 (Table 2). This issue has also been described for other serology assays (4, 7, 22, 24). Ideally, the “bait” protein used to capture circulating immunoglobulins should be in a native or near-native conformation to ensure that the antigenic sites of the protein are correctly and consistently exposed to the disease-related antibodies. This is likely the case for the cPass sVNT because the purified RBD-HRP is supplied and applied in solution (Fig. 1) (16) and evidenced by the high specificity of the assay in this work (Tables 1 to 3). Although the cPass sVNT utilizes hACE2 protein-coated plates, which can lead to structural perturbations of this protein, there is evidence that the immobilized hACE2 receptor maintains a strong interaction with the RBD, suggesting minimal loss of structural integrity (16, 26–28). Furthermore, since immunoglobulins from any isotypes that recognize RBD antigenic sites can bind and will be measured as a total antibody response, the sensitivity and negative predictive value of this versus immunoglobulin-specific tests (i.e., IgG/IgM) should be similar or improved for the cPass sVNT, as was shown here (Fig. 2 and Tables 1 and 2) and by others (29).
Taken together, these points help explain the similar or improved specificity, sensitivity, positive and negative predictive values, and accuracy obtained for the cPass sVNT versus other popular commercial SARS-CoV-2 IgG tests (Tables 1 and 2) (7, 29). Furthermore, these data support the notion that a binding antibody response as measured by the presence of circulating immunoglobulins coincides with neutralizing antibodies.
Although for study 1, the cPass sVNT categorized a total of six PCR-positive samples as negative (Fig. 2D, red circles), the spike (S1) (Fig. 2C) and nucleocapsid (Fig. 2B) assays also coincided with their negative classification, suggesting that these “false negatives” were, in fact, true negatives or possibly did not seroconvert. Since these samples were categorized as positive by quantitative PCR (qPCR) testing, this raises the question about the accuracy of qPCR. Some recent SARS-CoV-2 and Middle East respiratory syndrome (MERS) studies suggest that rates of PCR false positives can range from about 2% to 30%, with an average of 8% (30, 31). This may be attributed to using a cycle threshold cutoff that is too high and beyond the limit of detection for qPCR (6), accounting in part for the six false-negative samples delineated by the cPass sVNT and the other two serology tests (Fig. 2B to D, red dots). In fact, at quantification cycle (Cq) values above 35, many of the technical replicates for a given sample are negative, and single copies of contaminating DNA can result in a false-positive call (32).
Application of the cPass sVNT as a high-throughput screening tool for COVID-19 drug or vaccine development.
In order to abrogate viral entry, replication, and spread of infection, a vaccine should induce the production of antibodies that block (or neutralize) the interaction between the RBD and the hACE2 receptor (33–35). Some antibody-based drug candidates are similarly targeting this interaction (36, 37).
To date, the gold and silver standards in assessing the neutralization activity from drugs or antibodies are viral neutralization tests (VNTs) and pseudovirus neutralization tests (pVNTs) (38, 39). The VNT requires live SARS-CoV-2 and cells that express the hACE2 receptor and therefore requires a BSL3 containment laboratory, personal protective equipment, and highly trained personnel to conduct the experiments, whereas the pVNT can be performed in a BSL2 laboratory. Both tests involve sample incubations and manipulations that give results in 2 and 4 days and are therefore of relatively low throughput, expensive, and time-consuming, requiring aseptic techniques and personal protective equipment. The early phases of vaccine or drug development typically require the screening of large numbers of compounds and/or serum samples from candidate vaccine clinical trials to uncover those that neutralize the virus-host cell interaction with the greatest efficiency and efficacy (40). Furthermore, once a good potential vaccine or drug candidate has been selected, clinical trials involving thousands of individuals are required to assess protection against infection and the longevity of the neutralizing antibody response postvaccination (41). Thus, thousands of samples must be collected at regular time points and screened for neutralizing antibody titers, which would be challenging, expensive, and time-consuming using the VNT or pVNT.
Since the cPass sVNT utilizes the purified protein components of the RBD-hACE2 interaction in a high-throughput ELISA requiring about 1.5 h for each 96-well plate assay in a BSL2 laboratory (Fig. 1), it can potentially be used to screen for the best neutralizing drug and/or antibodies generated by vaccination (16). The cPass sVNT was compared directly with the FRNT and PRNT using serum from patients who recovered from COVID-19. An excellent correlation with FRNT75, PRNT75, and PRNT90 in detecting the presence of neutralizing antibodies postinfection (Fig. 3) (19) was revealed, supporting its application as a reliable tool for vaccine development and longitudinal studies tracking immune responses postvaccination. These data are likely owing to the immunodominance of the RBD versus other antigenic sites of the spike protein (8, 9).
There is no consistency in the literature concerning the analysis stringency that should be applied to cell-based neutralization assays (i.e., PRNT50 versus PRNT90 or FRNT50 versus FRNT90) to ensure the accurate delineation of positive and negative samples. Recent concerns have emerged concerning the ensuing variability and confidence in the results when different stringencies are applied to the data analysis of these live-cell assays (42, 43). The correlation of the cPass sVNT with the FRNT and PRNT was examined between 50% and 90% foci and plaque reduction. Significant changes in the analysis were observed between the PRNT50 and PRNT75 (Fig. 3A shows two samples shifting from negative to positive [blue dots]) but with no change between the PRNT75 and PRNT90. For the FRNT, there were large differences between the FRNT50, FRNT75, and FRNT90 (Fig. 3B), making it difficult to accurately determine the true delineation of positive and negative samples. The corroboration of the cPass sVNT with the higher-stringency PRNT75 and PRNT90 (Fig. 3A) is supported by recent work (19) and underlines the benefit of this test in accurately delineating neutralization antibody-positive and -negative individuals.
Experimental design for comparative drug or vaccine testing.
The cPass sVNT was used to assess dynamic changes in neutralizing antibodies from samples from recovered SARS-CoV-2 patients. Within the linear range, there was a significant decrease in inhibitory antibodies over time for samples 20 and 85, with no decrease observed for sample 74 (Fig. 4). However, outside the linear range, near the upper or lower plateau of the dilution curves, the data points for each time point were almost overlapping. This underlines the importance of producing a dilution series from each sample when dissecting the quantitative difference in neutralization titers over time. The cPass sVNT offers a much-higher-throughput, lower-cost, and safer option to achieve high-quality longitudinal data versus the more traditional VNT and pVNT, especially considering the close correlation with the high-stringency PRNT90 (19).
Conclusion.
The cPass sVNT provides a newly structured, high-throughput assay (Fig. 1) that permits augmented specificity, sensitivity, and accuracy for serological assessment of disease versus preexisting IgG tests (Fig. 2 and Tables 1 to 3) (7). The test also permits the functional delineation of virus neutralization for patient recovery that correlates strongly with live-cell neutralization (PRNT90) (Fig. 3) (19) and for high-throughput screening of drug and vaccine immune response antibodies that neutralize the interaction between the RBD and the hACE2 receptor (Fig. 4).
Regulatory status.
The cPass SARS-CoV-2 neutralization antibody test is CE marked for diagnostic use in the European Union and authorized for emergency use by Health Sciences Authority in Singapore and the U.S. Food and Drug Administration for qualitative delineation between positive and negative patient samples. The quantitation and automation protocols have not yet been authorized by the FDA, the European Union, or Singapore and are for research use only.
REFERENCES
- 1.Brotherhood L, Kircher P, Santos C, Tertilt M. 2020. An economic model of the Covid-19 epidemic: the importance of testing and age-specific policies. CRC TR 224 Discussion Paper Series crctr224_2020_175v1, University of Bonn and University of Mannheim, Germany. https://ideas.repec.org/p/bon/boncrc/crctr224_2020_175v1.html. [Google Scholar]
- 2.Salathé M, Althaus CL, Neher R, Stringhini S, Hodcroft E, Fellay J, Zwahlen M, Senti G, Battegay M, Wilder-Smith A, Eckerle I, Egger M, Low N. 2020. COVID-19 epidemic in Switzerland: on the importance of testing, contact tracing and isolation. Swiss Med Wkly 150:w20225. 10.4414/smw.2020.20225. [DOI] [PubMed] [Google Scholar]
- 3.Kyhlstedt M, Andersson SW. 2020. Diagnostic and digital solutions to address the COVID-19 pandemic: the need for international collaboration to close the gap. Health Policy Technol 9:126–128. 10.1016/j.hlpt.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West CP, Montori VM, Sampathkumar P. 2020. COVID-19 testing: the threat of false-negative results. Mayo Clin Proc 95:1127–1129. 10.1016/j.mayocp.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathur G, Mathur S. 2020. Antibody testing for COVID-19: can it be used as a screening tool in areas with low prevalence? Am J Clin Pathol 154:1–3. 10.1093/ajcp/aqaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tom MR, Mina MJ. 2020. To interpret the SARS-CoV-2 test, consider the cycle threshold value. Clin Infect Dis 71:2252–2254. 10.1093/cid/ciaa619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lisoba Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui L-P, Johnston JC, Lan Z, Law S, MacLean E, Trajman A, Menzies D, Benedetti A, Ahmad Khan F. 2020. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ 370:m2516. 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A, Cornaby C, Bartelt L, Weiss S, Park Y, Edwards CE, Weimer E, Scherer EM, Rouphael N, Edupuganti S, Weiskopf D, Tse LV, Hou YJ, Margolis D, Sette A, Collins MH, Schmitz J, Baric RS, de Silva AM. 2020. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 5:eabc8413. 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, Chai X, He R, Li X, Lv Q, Zhu H, Deng W, Xu Y, Wang Y, Qiao L, Tan Y, Song L, Wang G, Du X, Gao N, Liu J, Xiao J, Su X-D, Du Z, Feng Y, Qin C, Qin C, Jin R, Xie XS. 2020. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182:73–84.e16. 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CS, Belfort G. 1989. Changing activity of ribonuclease A during adsorption: a molecular explanation. Proc Natl Acad Sci U S A 86:8392–8396. 10.1073/pnas.86.21.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Güven E, Duus K, Lydolph MC, Jørgensen CS, Laursen I, Houen G. 2014. Non-specific binding in solid phase immunoassays for autoantibodies correlates with inflammation markers. J Immunol Methods 403:26–36. 10.1016/j.jim.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Raffaini G, Ganazzoli F. 2010. Protein adsorption on a hydrophobic surface: a molecular dynamics study of lysozyme on graphite. Langmuir 26:5679–5689. 10.1021/la903769c. [DOI] [PubMed] [Google Scholar]
- 13.Sethuraman A, Belfort G. 2005. Protein structural perturbation and aggregation on homogeneous surfaces. Biophys J 88:1322–1333. 10.1529/biophysj.104.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Thier P, Bacharouche J, Duval JFL, Skali-Lami S, Francius G. 2015. Atomic force microscopy analysis of IgG films at hydrophobic surfaces: a promising method to probe IgG orientations and optimize ELISA tests performance. Biochim Biophys Acta 1854:138–145. 10.1016/j.bbapap.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Mannik M, Kapil S, Merrill CE. 1997. In patients with rheumatoid arthritis IgG binding to denatured collagen type II is in part mediated by IgG-fibronectin complexes. J Immunol 158:1446–1452. [PubMed] [Google Scholar]
- 16.Tan CW, Chia WN, Qin X, Liu P, Chen MI-C, Tiu C, Hu Z, Chen VC-W, Young BE, Sia WR, Tan Y-J, Foo R, Yi Y, Lye DC, Anderson DE, Wang L-F. 2020. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol 38:1073–1078. 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 17.Cao Z, Liu L, Du L, Zhang C, Jiang S, Li T, He Y. 2010. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virol J 7:299. 10.1186/1743-422X-7-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera RAPM, Ko R, Tsang OTY, Hui DSC, Kwan MYM, Brackman CJ, To EMW, Yen H-L, Leung K, Cheng SMS, Chan KH, Chan KCK, Li K-C, Saif L, Barrs VR, Wu JT, Sit THC, Poon LLM, Peiris M. 2021. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat, and hamster sera. J Clin Microbiol 59:e02504-20. 10.1128/JCM.02504-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Younes N, Al-Sadeq DW, Al-Jighefee H, Younes S, Al-Jamal O, Daas HI, Yassine HM, Nasrallah GK. 2020. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses 12:582. 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlton CL, Kanji JN, Johal K, Bailey A, Plitt SS, MacDonald C, Kunst A, Buss E, Burnes LE, Fonseca K, Berenger BM, Schnabl K, Hu J, Stokes W, Zelyas N, Tipples G. 2020. Evaluation of six commercial mid- to high-volume antibody and six point-of-care lateral flow assays for detection of SARS-CoV-2 antibodies. J Clin Microbiol 58:e01361-20. 10.1128/JCM.01361-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krüttgen A, Cornelissen CG, Dreher M, Hornef M, Imöhl M, Kleines M. 2020. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J Clin Virol 128:104394. 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention. 2020. Interim guidelines for COVID-19 antibody testing. Centers for Disease Control and Prevention, Atlanta, GA. Accessed 18 June 2020. [Google Scholar]
- 24.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma SA, Lyons AM, Li S, Wong AW-Y, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, II, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington LA, Loudermilk RP, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, et al. 2020. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol 38:1174–1183. 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V, Kopp M, Haynes BF, Acharya P. 2020. Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. bioRxiv 10.1101/2020.06.26.173765. [DOI]
- 26.Cao W, Dong C, Kim S, Hou D, Tai W, Du L, Im W, Zhang XF. 2020. Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. bioRxiv 10.1101/2020.07.31.230730. [DOI] [PMC free article] [PubMed]
- 27.Wu K, Chen L, Peng G, Zhou W, Pennell CA, Mansky LM, Geraghty RJ, Li F. 2011. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J Virol 85:5331–5337. 10.1128/JVI.02274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Petitjean SJL, Koehler M, Zhang Q, Dumitru AC, Chen W, Derclaye S, Vincent SP, Soumillion P, Alsteens D. 2020. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun 11:4541. 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan SS, Saw S, Chew KL, Huak CY, Khoo C, Pajarillaga A, Wang W, Tambyah P, Ong L, Jureen R, Sethi SK. 2020. Head-to-head evaluation on diagnostic accuracies of six SARS-CoV-2 serological assays. Pathology 52:770–777. 10.1016/j.pathol.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SH. 2020. Testing for SARS-CoV-2 in cellular components by routine nested RT-PCR followed by DNA sequencing. Int J Geriatr Rehabil 2:69–96. [Google Scholar]
- 31.Pas SD, Patel P, Reusken C, Domingo C, Corman VM, Drosten C, Dijkman R, Thiel V, Nowotny N, Koopmans MP, Niedrig M. 2015. First international external quality assessment of molecular diagnostics for Mers-CoV. J Clin Virol 69:81–85. 10.1016/j.jcv.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Casanovas-Massana A, Muenker MC, Moore AJ, Klein J, Lu P, Lu-Culligan A, Jiang X, Kim DJ, Kudo E, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Tokuyama M, Venkataraman A, Weizman O-E, Wong P, Yang Y, Cheemarla NR, White EB, Lapidus S, Earnest R, Geng B, Vijayakumar P, Odio C, Fournier J, Bermejo S, Farhadian S, Dela Cruz CS, Iwasaki A, Ko AI, Landry ML, Foxman EF, Grubaugh ND. 2020. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol 5:1299–1305. 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amanat F, Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity 52:583–589. 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA, II, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, mRNA-1273 Study Group. 2020. An mRNA vaccine against SARS-CoV-2—preliminary report. New Engl J Med 383:1920–1931. 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hotez PJ, Corry DB, Strych U, Bottazzi ME. 2020. COVID-19 vaccines: neutralizing antibodies and the alum advantage. Nat Rev Immunol 20:399–400. 10.1038/s41577-020-0358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 37.Tai W, Zhang X, He Y, Jiang S, Du L. 2020. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res 179:104820. 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y, Chang L, Wang L. 2020. Laboratory testing of SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2 (2019‐nCoV): current status, challenges, and countermeasures. Rev Med Virol 30:e2106. 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12:513. 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharpe HR, Gilbride C, Allen E, Belij‐Rammerstorfer S, Bissett C, Ewer K, Lambe T. 2020. The early landscape of coronavirus disease 2019 vaccine development in the UK and rest of the world. Immunology 160:223–232. 10.1111/imm.13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullard A. 2020. COVID-19 vaccine development pipeline gears up. Lancet 395:1751–1752. 10.1016/S0140-6736(20)31252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salje H, Rodríguez-Barraquer I, Rainwater-Lovett K, Nisalak A, Thaisomboonsuk B, Thomas SJ, Fernandez S, Jarman RG, Yoon I-K, Cummings DAT. 2014. Variability in dengue titer estimates from plaque reduction neutralization tests poses a challenge to epidemiological studies and vaccine development. PLoS Negl Trop Dis 8:e2952. 10.1371/journal.pntd.0002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee WT, Girardin RC, Dupuis AP, Kulas KE, Payne AF, Wong SJ, Arinsburg S, Nguyen FT, Mendu DR, Firpo-Betancourt A, Jhang J, Wajnberg A, Krammer F, Cordon-Cardo C, Amler S, Montecalvo M, Hutton B, Taylor J, McDonough KA. 2021. Neutralizing antibody responses in COVID-19 convalescent sera. J Infect Dis 223:47–57. 10.1093/infdis/jiaa673. [DOI] [PMC free article] [PubMed] [Google Scholar]