CASE
A 55-year-old woman with an unexplored 1-month history of rectal bleeding presented to the emergency department with abdominal pain, chills, and fever. The patient had received rituximab, a monoclonal antibody targeting the CD20 antigen expressed on B cells, for rheumatoid arthritis. An abdominal computed tomography (CT) scan highlighted a right perirectal collection and focal sigmoiditis with few diverticula (Fig. 1). Owing to sepsis, the patient received piperacillin-tazobactam in association with gentamicin and underwent an early laparoscopy to drain the collection a few hours after the initiation of antimicrobial therapy. Gram staining of the perirectal collection revealed numerous polymorphonuclear leukocytes with no visible microorganisms. Surgical samples and blood cultures remained sterile even after 5 days of incubation. The patient presented no improvement in her clinical condition. Persistent fever and recurring chills along with high levels of inflammatory blood markers resulted in a treatment change to vancomycin, cefepime, and metronidazole. A CT scan on day 10 showed a stable rectal abscess. On day 14, the antibiotics were replaced by meropenem, amikacin, and fluconazole due to the persistent fever. Other sets of blood cultures remained negative. The fever persisted with no explanation other than the rectal abscess.
FIG 1.
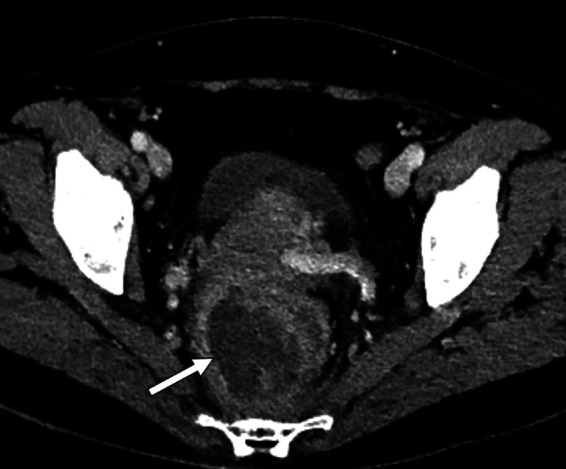
Contrast-enhanced abdominopelvic CT scan. White arrow indicates a perirectal abscess.
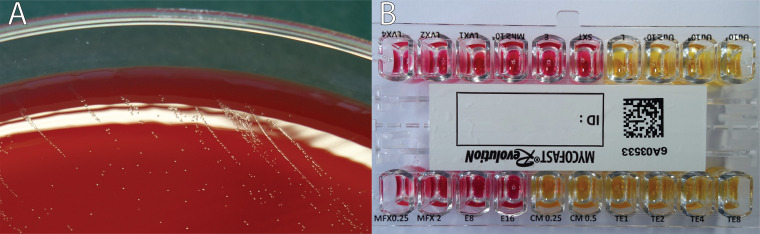
On day 20, the patient underwent an exploratory laparotomy and a low Hartmann’s resection of the rectum. Pathological examination of the resected specimen led to a diagnosis of perforated rectal endometriosis. Gram staining of the perirectal collection again showed numerous polymorphonuclear leukocytes and no visible microorganisms. However, 4 days of incubation on blood agar at 35°C under 5% CO2 resulted in the formation of pinpoint-sized colonies resembling water droplets (Fig. 2A). These colonies could not be identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (“no peaks found”). Gram staining performed on the colonies showed no bacteria. These results led us to suspect Mycoplasma hominis, and the colonies were transferred to differential agar medium (A7) for further analysis.
FIG 2.
Microbial analysis images. (A) Pinpoint-sized colonies, later identified as M. hominis, on blood agar following 4 days of incubation under 5% CO2 at 35°C. (B) Colorimetric antibiotic susceptibility results. MFX, moxifloxacin; E, erythromycin; CM, clindamycin; TE, tetracycline; LVX, levofloxacin.
M. hominis identification was confirmed after 48 h of incubation on A7 agar. In addition, after a total of 6 days of incubation on blood agar, MALDI-TOF MS (Bruker Daltonics, Wissembourg, France) identified the larger colonies as M. hominis with a score of 1.97 (database MBT IVD, Library 9.0).
The commercial Mycofast Revolution colorimetric assay (ELITechGroup, Puteaux, France) was used to determine antibiotic susceptibility by using the Clinical and Laboratory Standards Institute (CLSI) breakpoints (1). The isolate was susceptible to both tetracycline (MIC, ≤1 mg/liter) and clindamycin (MIC, ≤0.25 mg/liter) but resistant to erythromycin and azithromycin (MIC, >16 mg/liter), levofloxacin (MIC, >4 mg/liter), and moxifloxacin (MIC, >2 mg/liter) (Fig. 2B). The patient was treated with doxycycline for 3 weeks with a favorable outcome.
DISCUSSION
Mycoplasma belongs to the class Mollicutes, which is characterized by the absence of a cell wall. M. hominis is a commensal bacterium colonizing the urogenital tract and is a causative agent for urogenital infections, such as pelvic inflammatory disease or chorioamnionitis. Although extragenital infections are less frequent, M. hominis has been reported to cause mediastinitis (2), abscesses, and bacteremia, particularly in postoperative patients and immunocompromised patients (3, 4).
Immunosuppression is the main risk factor for developing extragenital M. hominis infections (3). Approximately 50% of patients with extragenital M. hominis infections had an impaired cell-mediated immune system or hypogammaglobulinemia (3). In our case, the patient had received rituximab, which decreases B cell numbers for more than 6 months after administration and may also decrease immunoglobulin levels. Solid-organ transplant recipients are particularly at risk of developing extragenital M. hominis infections (4). M. hominis has also been identified in postoperative infections (1). Hyperammonemia syndrome, an encephalopathy due to high plasma ammonium levels, has also been linked to Mycoplasma and Ureaplasma species following lung, kidney, or hematopoietic stem cell transplantation (5). Notably, ammonium is produced from arginine by one of the major energy-producing pathways of M. hominis. Overall, M. hominis infection should be suspected in immunocompromised patients with extragenital abscesses, particularly when numerous leukocytes are present with no visible microorganisms.
The diagnosis of invasive M. hominis infections is challenging. Because M. hominis lacks a cell wall, it cannot be detected by Gram staining. DNA fluorochrome staining (acridine orange or Hoechst 33258) may be used to detect Mycoplasma in body fluids, but these stains are not specific. Without clinical suspicion of M. hominis infection, specific tests for Mycoplasma detection are not routinely performed. Our case shows the serendipitous diagnosis of M. hominis through the observation of pinpoint-sized colonies, which can grow on blood and chocolate agar after 2 to 7 days of incubation (2). The small size of these colonies (diameter, 0.2 mm) renders them difficult to detect without careful inspection under reflected light. This knowledge could prove useful to clinical microbiologists. In our case, no bacterial growth was detected from the first surgical samples. Even if M. hominis can grow on blood or chocolate agar, this is not always reliable. Translucent M. hominis colonies may be overlooked before the agar medium is discarded because they can easily be mistaken for water droplets. Prolonged incubation is necessary to allow M. hominis colonies to develop.
M. hominis should be suspected when Gram staining fails to detect microorganisms from pinpoint-sized colonies, warranting subculture onto mycoplasma medium. If mycoplasma medium is unavailable locally, another alternative would be to perform an acridine orange stain on the colonies in order to prove the presence of microorganisms and to send the isolate to a reference laboratory for identification by MALDI-TOF MS or 16S rRNA sequencing. There are several types of mycoplasma media, including SP4 agar supplemented with arginine, Hayflick agar, and A7 agar, with penicillin G generally added for selectivity. Agar plates should be incubated under 5 to 10% CO2 or under anaerobic conditions at 35°C for a minimum of 5 days. A stereomicroscope can aid in the visualization of the colonies, identifiable by their typical “fried egg” appearance. The biochemical profile helps to differentiate M. hominis from Ureaplasma spp.: M. hominis utilizes arginine (alkaline shift from orange to deep-red [Fig. 2B]), in contrast to Ureaplasma spp., which utilize urea. Other Mycoplasma species are also able to utilize arginine, but M. hominis is the only human-pathogenic species of Mycoplasma able to grow on blood or chocolate agar.
For definite identification at the species level, MALDI-TOF MS can be used. M. hominis is present in both the Bruker MALDI Biotyper and Vitek MS databases. In total, the Bruker MBT 8326 IVD database contains 12 species of Mycoplasma, and the Vitek MS database (v3.2) contains 14 species of Mycoplasma. However, the main issue is the low biomass of the Mycoplasma colonies, especially after incubation on blood or chocolate agar instead of a specific mycoplasma medium. In our case, MALDI-TOF MS identification failed after 4 days of incubation on blood agar but was successful after 6 days of incubation on blood agar or 2 days on A7 agar, with a score of 1.97. Pereyre et al. suggested that accurate species-level identification was achievable for Mycoplasma with a score of ≥1.70 for Bruker MALDI-TOF MS, instead of the classical threshold of ≥2.00 (6).
Molecular methods provide an alternative for diagnosing extragenital M. hominis infections, not only for identifying suspect colonies but also for detecting the organism directly from clinical samples. Several in-house real-time PCR assays have been developed for M. hominis detection, targeting either the 16S rRNA, gap, or yidC gene. However, minor sequence variations have been reported in 16S rRNA or gap genes, which may lower clinical sensitivity (7). Real-time PCR targeting the yidC gene was estimated to have a limit of detection of 7 copies/μl, making it more sensitive than culture of urogenital samples (7). Direct 16S rRNA sequencing from clinical samples has been used successfully to diagnose extragenital M. hominis infections (2). Extending this to next-generation sequencing or shotgun metagenomic sequencing is promising for diagnosing pathogens in abscesses.
M. hominis is naturally resistant to all antibiotics targeting cell wall synthesis (β-lactams, glycopeptides, fosfomycin), erythromycin and azithromycin, sulfamides, and rifampin. Without a definitive diagnosis, it is unlikely that patients suffering from extragenital M. hominis infections will receive effective antimicrobial therapy. For example, patients with intra-abdominal abscesses or mediastinitis typically receive a broad-spectrum β-lactam, associated with an antibiotic covering Gram-positive bacteria (a glycopeptide or an oxazolidinone). These antibiotics are not active against M. hominis. Inadequate antimicrobial therapy can lead to poor outcomes, including complications and iterative readmissions with prolonged hospital stays (4). M. hominis is potentially susceptible to tetracyclines, clindamycin, and fluoroquinolones (levofloxacin or moxifloxacin). However, acquired resistance has been reported. High-level resistance to tetracyclines is carried by the tet(M) gene, and isolates resistant to fluoroquinolones harbor mutations in the gyrA, parC, or parE gene (8). In France, the resistance rate was 15% for tetracyclines, 3% for levofloxacin, and 2% for moxifloxacin (8). Therefore, antimicrobial susceptibility testing must be performed in order to select adequate antimicrobial therapy.
The disk diffusion method is not recommended for testing antimicrobial susceptibility, since there is no correlation between the inhibition diameter and the MIC. The Clinical and Laboratory Standards Institute (CLSI) recognizes agar dilution and broth microdilution as the reference methods for antimicrobial susceptibility (9). Agar gradient diffusion (Etest) represents a potentially comparable method (10) but is not endorsed by the CLSI (9). Commercial antimicrobial susceptibility assays using M. hominis have also been reported to perform similarly to the reference methods (8). These commercial assays provide a simple method for screening several concentrations of antimicrobials in a microwell plate format.
Extragenital infections due to M. hominis are rare but not exceptional, particularly in immunocompromised patients, and are likely underdiagnosed. Clinical microbiologists should be aware of the appearance of M. hominis colonies on blood agar after prolonged incubation so as to ensure appropriate testing for M. hominis in the case of culture-negative abscesses, particularly in immunocompromised individuals.
SELF-ASSESSMENT QUESTIONS
-
1.Which of the following conditions is associated with extragenital Mycoplasma hominis infections?
-
a.Neutropenia
-
b.Hypogammaglobulinemia
-
c.Obesity
-
d.Diabetes
-
a.
-
2.Which of the following statements concerning Mycoplasma hominis identification is correct?
-
a.M. hominis is unable to grow on blood or chocolate agar under 5% CO2.
-
b.M. hominis is undetectable by Gram staining.
-
c.M. hominis is absent from MALDI-TOF MS databases.
-
d.M. hominis produces large colonies (>1 mm) on mycoplasma medium.
-
a.
-
3.Mycoplasma hominis is naturally resistant to which class of antibiotics?
-
a.Tetracyclines
-
b.β-Lactams
-
c.Lincosamides
-
d.Fluoroquinolones
-
a.
ACKNOWLEDGMENTS
We declare that we have no conflicts of interest.
Funding for this work was received from CHU Lille.
Footnotes
For answers to the self-assessment questions and take-home points, see https://doi.org/10.1128/JCM.02344-20 in this issue.
REFERENCES
- 1.Waites KB, Duffy LB, Bebear CM, Matlow A, Talkington DF, Kenny GE, Totten PA, Bade DJ, Zheng X, Davidson MK, Shortridge VD, Watts JL, Brown SD. 2012. Standardized methods and quality control limits for agar and broth microdilution susceptibility testing of Mycoplasma pneumoniae, Mycoplasma hominis, and Ureaplasma urealyticum. J Clin Microbiol 50:3542–3547. doi: 10.1128/JCM.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Guern R, Loiez C, Loobuyck V, Rousse N, Courcol R, Wallet F. 2015. A new case of Mycoplasma hominis mediastinitis and sternal osteitis after cardiac surgery. Int J Infect Dis 31:53–55. doi: 10.1016/j.ijid.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 3.Meyer RD, Clough W. 1993. Extragenital Mycoplasma hominis infections in adults: emphasis on immunosuppression. Clin Infect Dis 17(Suppl 1):S243–S249. doi: 10.1093/clinids/17.supplement_1.s243. [DOI] [PubMed] [Google Scholar]
- 4.Adams M, Bouzigard R, Al-Obaidi M, Zangeneh TT. 2020. Perinephric abscess in a renal transplant recipient due to Mycoplasma hominis: case report and review of the literature. Transpl Infect Dis 22:e13308. doi: 10.1111/tid.13308. [DOI] [PubMed] [Google Scholar]
- 5.Wylam ME, Kennedy CC, Hernandez NM, Peters SG, Maleszewski JJ, Cassivi SD, Scott JP. 2013. Fatal hyperammonaemia caused by Mycoplasma hominis. Lancet 382:1956. doi: 10.1016/S0140-6736(13)62115-7. [DOI] [PubMed] [Google Scholar]
- 6.Pereyre S, Tardy F, Renaudin H, Cauvin E, Del Pra Netto Machado L, Tricot A, Benoit F, Treilles M, Bebear C. 2013. Identification and subtyping of clinically relevant human and ruminant mycoplasmas by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:3314–3323. doi: 10.1128/JCM.01573-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferandon C, Peuchant O, Janis C, Benard A, Renaudin H, Pereyre S, Bebear C. 2011. Development of a real-time PCR targeting the yidC gene for the detection of Mycoplasma hominis and comparison with quantitative culture. Clin Microbiol Infect 17:155–159. doi: 10.1111/j.1469-0691.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- 8.Meygret A, Le Roy C, Renaudin H, Bebear C, Pereyre S. 2018. Tetracycline and fluoroquinolone resistance in clinical Ureaplasma spp. and Mycoplasma hominis isolates in France between 2010 and 2015. J Antimicrob Chemother 73:2696–2703. doi: 10.1093/jac/dky238. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2011. Methods for antimicrobial susceptibility testing for human mycoplasmas; approved guideline M43-A. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 10.Waites KB, Canupp KC, Kenny GE. 1999. In vitro susceptibilities of Mycoplasma hominis to six fluoroquinolones as determined by E test. Antimicrob Agents Chemother 43:2571–2573. doi: 10.1128/AAC.43.10.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]