Non-albicans Candida species are emerging in the nosocomial environment, with the multidrug-resistant (MDR) species Candida auris being the most notorious example. Consequently, rapid and accurate species identification has become essential.
KEYWORDS: Candida, chromogenic media, selective media, rapid identification, Candida auris, coinfection
ABSTRACT
Non-albicans Candida species are emerging in the nosocomial environment, with the multidrug-resistant (MDR) species Candida auris being the most notorious example. Consequently, rapid and accurate species identification has become essential. The objective of this study was to evaluate five commercially available chromogenic media for the presumptive identification of C. auris. Two novel chromogenic formulations, CHROMagar Candida Plus (CHROMagar) and HiCrome C. auris MDR selective agar (HiMedia), and three reference media, CandiSelect (Bio-Rad), CHROMagar Candida (CHROMagar), and Chromatic Candida (Liofilchem), were inoculated with a collection of 9 genetically diverse C. auris strains and 35 strains from closely related comparator species. After 48 h of incubation, the media were evaluated for their ability to detect and identify C. auris. All media had the same limitations in the differentiation of the more common species Candida dubliniensis and Candida glabrata. Only on CHROMagar Candida Plus did C. auris colonies develop a species-specific coloration. Nevertheless, the closely related pathogenic species Candida pseudohaemulonii and Candida vulturna developed a similar appearance as C. auris on this medium. CHROMagar Candida Plus was shown to be superior in the detection and identification of C. auris, with 100% inclusivity for C. auris compared to 0% and 33% for the reference media and HiCrome C. auris MDR selective agar, respectively. Although C. vulturna and C. pseudohaemulonii can cause false positives, CHROMagar Candida Plus was shown to be a valuable addition to the plethora of mostly molecular methods for C. auris detection and identification.
INTRODUCTION
Candida species are the most frequently encountered fungi in hospital settings. Candida yeasts are among the top four of nosocomial bloodstream pathogens and cause more than 400,000 bloodstream infections annually (1). Therefore, Candida contributes significantly to morbidity and mortality (2, 3). Although Candida albicans is still the main cause of candidiasis, a shift is taking place. An increasing amount of non-C. albicans Candida species is causing life-threatening invasive infections (4, 5). One of the main contributors to this epidemiological shift is the unprecedented emergence of the multidrug-resistant (MDR) species Candida auris. This yeast was first described in 2009 as a novel member of the Candida haemulonii species complex (6, 7). C. auris has rapidly become widespread since its first description, causing difficult to control nosocomial outbreaks around the world (8, 9). Whole-genome sequencing of clinical C. auris isolates revealed four closely related but unique clades predominate on different continents (East Asian, South Asian, South African, and South American) (8, 10). In South Africa, C. auris already causes 14% of all reported candidemia cases (11). The shift from generally antifungal-susceptible C. albicans to more resistant species, like C. auris, calls for rapid and reliable identification in order to provide the right treatment on time (5, 12).
To detect pathogenic yeasts in polymicrobial clinical samples properly, selective media are crucial. However, conventional selective media, like Sabouraud dextrose agar and malt extract agar, do not differentiate between species. Chromogenic agars, conversely, allow for the identification of Candida species in polymicrobial samples due to generation of a species-specific color (13–15). As a result, they not only allow for rapid identification of single species but are also a great tool to detect the increasing number of yeast-yeast coinfections (16, 17). Currently available chromogenic media enable the identification of four of the common Candida pathogens, including C. albicans, Candida tropicalis, Nakaseomyces glabrata (syn. Candida glabrata), and Pichia kudriavzevii (syn. Candida krusei) (13, 14, 18). However, most chromogenic media do not differentiate emerging Candida species, including C. auris. Other diagnostic technologies like matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), PCR, and internal transcribed spacer (ITS) sequencing are available to reliably identify C. auris and its siblings (19–21) but require expertise and costly apparatus. In contrast, chromogenic media allow for rapid direct identification just by the color and morphology of the colony, usually within 24 to 48 h of incubation.
Here, we have evaluated and compared five commercially available chromogenic media, including two novel formulations specifically developed to differentiate C. auris. The performances of these five media were tested using a panel of genetically diverse C. auris strains and closely related comparator species. Only CHROMagar Candida Plus reliably differentiated C. auris from most of its sibling species and the common Candida pathogens. Exceptions with a similar phenotype on this novel chromogenic medium included Candida vulturna and Candida pseudohaemulonii.
MATERIALS AND METHODS
Media.
Two novel chromogenic media specifically designed to detect C. auris, CHROMagar Candida Plus (CCP) (bioTRADING, Mijdrecht, The Netherlands) and HiCrome C. auris MDR selective agar (HAMA) (HiMedia, Mumbai, India), were evaluated for their ability to differentiate C. auris from its close relatives and the major clinical Candida species. Three other commercially available chromogenic media, CandiSelect (CS) (Bio-Rad, Veenendaal, The Netherlands), CHROMagar Candida (CAC) (bioTRADING), and Chromatic Candida (CC) (Liofilchem, Roseto degli Abruzzi, Italy), were used as a reference for specificity of the tested yeasts.
Fungal strains, identification, and culture.
Nine C. auris strains were selected, as follows: two strains per clade, representing the four clades that are specific to each geographic region and one strain representing the potential fifth clade. Strains came from reputable international culture collections, including our own Centraalbureau voor Schimmelcultures (CBS) yeast collection located at the Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands), and included C. auris CDC-AR0387 and CDC-AR0388 (clade I), CBS10913 and CBS12373 (clade II), CDC-AR0383 and CDC-AR0384 (clade III), CDC-AR0385 and CDC-AR0386 (clade IV), and CDC-AR1097 (potential clade V, Iran). To evaluate the specificity for C. auris of the tested (improved) chromogenic agars, the phenotype of these nine C. auris strains was compared to that of closely related yeast species (n = 35). The selected comparator species included a wide range of Candida yeasts belonging to the Metschnikowia clade and, in particular, members of the C. haemulonii species complex that contains C. auris's closest relatives as follows: Candida akabanensis (n = 2), Candida citri (n = 1), Candida duobushaemulonii (n = 3), Candida ecuadorensis (n = 1), Candida fructus (n = 1), Candida haemulonii (n = 3), Candida haemulonii var. vulnera (n = 3), Candida intermedia (n = 2), Candida kutaonensis (n = 1), Clavispora lusitaniae (n = 6), Clavispora opuntiae (n = 2), Candida phyllophila (n = 1), Candida pseudoflosculorum (n = 1), Candida pseudohaemulonii (n = 2), Candida pseudointermedia (n = 2), Candida sharkiensis (n = 1), Candida vitiphila (n = 1), and Candida vulturna (n = 2). Additionally, one strain of the major clinical Candida species, C. albicans, Candida dubliniensis, C. tropicalis, Nakaseomyces glabrata (syn. C. glabrata), and Pichia kudriavzevii (syn. C. krusei) were included (22). All strains were subjected to ITS sequencing to confirm species identity (21, 23).
Strains were cultured on 2% glucose, 0.5% yeast extract, 1% peptone, and 1.5% agar (GYPA) at 25°C before testing. The ability of the tested chromogenic media to differentiate C. auris from its close relatives and the common clinical Candida species was evaluated as described previously (24) with minor modifications. In brief, cells were suspended in phosphate-buffered saline (PBS; without Ca, Mg, pH 7.3 to 7.5) (Lonza, Basel, Switzerland) to make aqueous suspensions for the nine C. auris strains and 40 comparator strains listed above. Subsequently, 2-μl cell suspensions were spotted onto the chromogenic media plates and incubated for 48 h at 35°C (CCP, CS, CAC, CC) and 42°C (HAMA) as recommended by the manufacturers. To check for organism viability, the same suspensions were spotted in parallel onto GYPA plates, which were incubated for 48 h at 35°C and 30°C (as some of the species included did not grow well at 35°C).
To assess the ability of the tested chromogenic media to detect C. auris from coinfections with other yeasts, mixtures containing 2.0 × 103 cells/ml of C. auris CBS10913 plus major clinical Candida species or members of the C. haemulonii species complex were prepared 1:1 in PBS. One hundred microliters of the resulting mixtures, containing ∼200 cells, was plated onto the different chromogenic media. Pure cultures were used as a control. Plates were incubated at 35°C for 48 h unless stated otherwise.
Data availability.
Sequence data was deposited into GenBank under accession numbers MT974606 to MT974688.
RESULTS
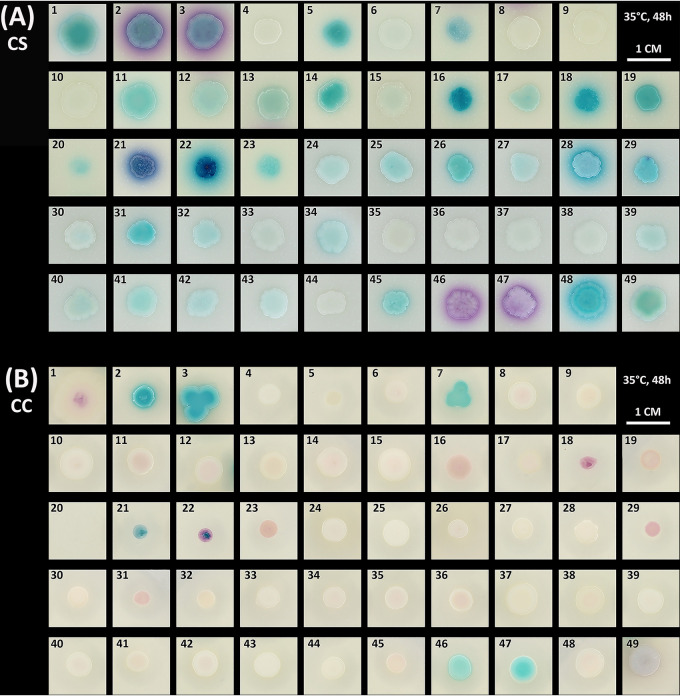
After 48 h of incubation at 35°C on the reference chromogenic media (CS, CC, CAC), C. auris colonies appeared pale cream to light blue (CS) or beige to pink with pale borders (CC, CAC) (Fig. 1A to C, spots 33 to 41). With the CS, CC, and CAC media, it was not possible to distinguish C. auris from other closely related pathogenic siblings (C. haemulonii [spots 27, 28, 29], C. duobushaemulonii [spots 24, 25, 26], C. pseudohaemulonii [spots 44, 45], C. haemulonii var. vulnera [spots 30, 31, 32], and C. vulturna [spots 42, 43]). In addition, when compared to most other closely related yeast species tested, C. auris colonies displayed a similar phenotype on CS, CC, and CAC media as well. Only Clavispora opuntiae (spots 2, 3) produced distinctive coloration on all three media, being violet on CS and light to dark blue on CC and CAC, respectively. Since most chromogenic media are designed to detect the major clinical species, C. albicans (spot 46), C. dubliniensis (spot 47), N. glabrata (spot 48), C. tropicalis (spot 49), and P. kudriavzevii (spot 1), these yeasts all had a distinctive phenotype. Nonetheless, C. albicans and C. dubliniensis behaved similarly on all tested media, both producing green colonies on CC and CAC and purple on CS. Interestingly, C. lusitaniae (spots 10 to 15) showed a high intrastrain variability of colony phenotypes, especially on CAC medium, ranging from pale pink to dark purple. Because of this variability, some strains had a phenotype similar to N. glabrata on all three media. Similar to C. lusitaniae, the C. haemulonii strains tested also produced different phenotypes, with strain MC153 (spot 28) being the most distinctive.
FIG 1.
Appearance of Candida auris and comparator species on CandiSelect (CS) (A), Chromatic Candida (CC) (B), CHROMagar Candida (CAC) (C), CHROMagar Candida Plus (CCP) (D), and HiCrome C. auris MDR selective agar (HAMA) (E). Two microliters of aqueous suspension of each organism was spotted onto each medium. Plates were incubated at 35°C (CS, CAC, CC, CCP) and 42°C (HAMA) for 48 h. Tested yeast species were as follows: (1) Pichia kudriavzevii CBS2050 (syn. Candia krusei), (2) Clavispora opuntiae CBS11046, (3) Clavispora opuntiae CBS11164, (4) Candida sharkiensis CBS11368, (5) Candida kutaonensis CBS11388, (6) Candida citri CBS11858, (7) Candida ecuadorensis CBS12653, (8) Candida phyllophila CBS12671, (9) Candida vitiphila CBS12672, (10) Clavispora lusitaniae CBS5094, (11) Clavispora lusitaniae CBS4870, (12) Clavispora lusitaniae CBS11356, (13) Clavispora lusitaniae CBS5901, (14) Clavispora lusitaniae CBS1944, (15) Clavispora lusitaniae CBS2866, (16) Candida intermedia CBS2291, (17) Candida intermedia CBS5311, (18) Candida pseudointermedia CBS6918, (19) Candida pseudointermedia CBS2879, (20) Clavispora fructus CBS6380, (21) Candida akabanensis CBS5039, (22) Candida akabanensis CBS7878, (23) Candida pseudoflosculorum CBS8584, (24) Candida duobushaemulonii CBS7800, (25) Candida duobushaemulonii CBS7799, (26) Candida duobushaemulonii CBS7098, (27) Candida haemulonii CBS6590, (28) Candida haemulonii MC153, (29) Candida haemulonii CBS12371, (30) Candida haemulonii var. vulnera CDC-AR0391, (31) Candida haemulonii var. vulnera CBS6332, (32) Candida haemulonii var. vulnera CBS12438, (33) Candida auris CDC-AR0383 (clade III), (34) Candida auris CDC-AR0384 (clade III), (35) Candida auris CDC-AR0385 (clade IV), (36) Candida auris CDC-AR0386 (clade IV), (37) Candida auris CDC-AR0387 (clade I), (38) Candida auris CDC-AR0388 (clade I), (39) Candida auris CBS10913 (clade II), (40) Candida auris CBS12373 (clade II), (41) Candida auris CDC-AR1097 (potential clade V), (42) Candida vulturna CBS14366, (43) Candida vulturna CBS15630, (44) Candida pseudohaemulonii CBS 10004, (45) Candida pseudohaemulonii CBS12370, (46) Candida albicans CBS562, (47) Candida dubliniensis CBS7987, (48) Nakaseomyces glabrata (syn. Candida glabrata) CBS138, and (49) Candida tropicalis CBS94.
The two novel chromogenic media evaluated in this study use different approaches for the isolation and differentiation of C. auris. CCP works like a traditional chromogenic medium, allowing for identification by color and colony morphology of the species. HAMA, conversely, inhibits growth of species other than multidrug-resistant C. auris and uses a chromogenic mixture to impart purple color to C. auris for identification. On CCP, all C. auris strains appeared pale cream to lavender with a distinctive blue halo surrounding the colony, independent of the clade they belong to (Fig. 1D, spots 33 to 41). C. auris could be easily differentiated from all species that were tested in parallel, except C. vulturna (spots 42, 43) and C. pseudohaemulonii (spots 44, 45). Both species are closely related to C. auris and produced similar colonies on the novel CCP medium.
Incubation of the yeast species at 42°C for 48 h on HAMA medium effectively inhibited the growth of all comparator species. Most species produced very faint colonies with a light to dark purple phenotype (Fig. 1E). Of the nine C. auris strains, only three showed good growth. Strains CDC-AR0385 and CDC-AR0386 (spot 35, 36), both belonging to the South American clade (clade IV, Venezuela), produced cream colonies with a pink center. Strain CDC-AR0388 (spot 38) of the South Asian clade (clade I, Pakistan) produced a colony with a different phenotype being much darker purple.
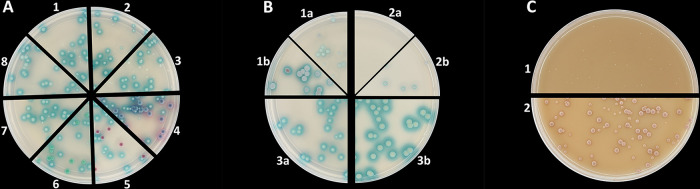
The performance data of the media tested in the spot assay (Fig. 1) are presented in Table 1. CCP showed the best capacity to discriminate between C. auris and most of the comparator yeast species (100% inclusivity), except C. vulturna and C. pseudohaemulonii (90% exclusivity). Therefore, we next evaluated the performance of this medium in the detection of C. auris from mixed microbial samples. To this end, suspensions of C. auris or C. auris mixed with other members of the C. haemulonii species complex (including C. vulturna and C. pseudohaemulonii) and the major clinical Candida species, C. albicans, C. tropicalis, and N. glabrata, were prepared in sterile saline. One hundred microliters of the resulting mixtures, containing approximately 200 cells, was inoculated on CCP and the other chromogenic media in parallel for comparison. After 48 h (the incubation time specified by the manufacturers), only CCP reliably discriminated C. auris from all comparator species (Fig. 2A). On CCP, distinctive colonies with blue haloes could be detected. This is in contrast to the reference media where C. auris appeared pale cream just like all other members of the C. haemulonii species complex (not shown). Although C. pseudohaemulonii produced a similar phenotype in the spot assay (Fig. 1), inoculated as a mixture, this species produced much smaller colonies compared to those of C. auris without the blue halo (Fig. 2A, 7). C. vulturna, in contrast, was harder to distinguish since colonies had the same size and morphology as C. auris (Fig. 2A, 8). Nonetheless, after 48 h, the blue haloes of C. auris colonies were much bigger and more intense. Only after further incubation for 72 h was C. vulturna able to produce similar haloes as C. auris (Fig. 2B, 1b). C. pseudohaemulonii colonies produced a blue halo as well after 72 h, but still these colonies were much smaller and therefore easy to distinguish (Fig. 2B, 2b). On the HAMA medium, none of the comparator species grew after inoculation with pure suspensions (not shown). Since most C. auris strains also did not grow on this medium, strain CDC-AR0385 was used to plate out as a mixture. This strain produced very small white colonies after 48 h of growth at 42°C. Further incubation for 96 h was needed before colonies started to produce a pink to purple color (Fig. 2C).
TABLE 1.
Performance data of Candida chromogenic media after 48 h of incubation in spot assay
| Strain and parameter | No. of samples using: |
||||
|---|---|---|---|---|---|
| CS | CC | CAC | CCP | HAMAd | |
| Candida auris (n = 9) | |||||
| True positive | 0 | 0 | 0 | 9 | 3 |
| False negative | 9 | 9 | 9 | 0 | 6 |
| Comparator strains (n = 40) | |||||
| Recovery | 40 | 39 | 40 | 40 | 3 |
| False positive (typical Candida auris morphology) | 20 | 24 | 13 | 4 | 0 |
| True negative | 20 | 16 | 27 | 36 | 40 |
| Accuracy (%)a | 40.8 | 32.7 | 55.1 | 91.8 | 87.8 |
| Exclusivity (%)b | 50 | 40 | 67.5 | 90 | 100 |
| Inclusivity (%)c | 0.0 | 0.0 | 0.0 | 100.0 | 33.3 |
Accuracy (%) = (true positive + true negative) × 100 / total.
Exclusivity (%) = true negative × 100 / (true negative + false positive).
Inclusivity (%) = true positive × 100 / (true positive + false negative).
Selective medium inhibiting growth of species other than C. auris causing low recovery.
FIG 2.
(A) Comparative detection of Candida auris from mixtures with other yeast on CHROMagar Candida Plus inoculated with mixtures of C. auris plus (1) Candida haemulonii var. vulnera CBS12438, (2) Candida haemulonii CBS6590, (3) Candida duobushaemulonii CBS7799, (4) Candida tropicalis CBS94, (5) Nakaseomyces glabrata (syn. Candida glabrata) CBS138, (6) Candida albicans CBS562, (7) Candida pseudohaemulonii CBS12370, and (8) Candida vulturna CBS14366 as described in Materials and Methods. (B) CHROMagar Candida Plus inoculated with pure suspensions of (1) C. vulturna CBS14366, (2) C. pseudohaemulonii CBS12370, and (3) C. auris CBS10913 (clade II) incubated at 35°C for (a) 48 h and (b) 72 h. (C) Recovery of C. auris CDC-AR0385 (clade I) on HiCrome C. auris MDR Selective Agar after (1) 48 h and (2) 96 h of incubation at 42°C.
DISCUSSION
Non-albicans Candida species are emerging in the nosocomial environment. Often, these species have a multidrug-resistant phenotype, with C. auris being the most notorious example. Consequently, rapid and accurate species identification has become essential in order to provide the optimal treatment. Chromogenic media are able to detect and identify Candida yeasts at species level in an easy and cost-effective way. In this study, we evaluated five commercially available chromogenic media for the detection and differentiation of C. auris from closely related comparator species and common clinical Candida.
The novel CHROMagar Candida Plus medium showed the best performance in the rapid identification of C. auris compared to that of the reference media, having the highest accuracy and inclusivity (Table 1). This result is in line with the study performed by Borman and colleagues who used 52 comparator species recovered from routine clinical samples to test the specificity of CCP for C. auris (24). In that study, only the rare species Candida diddensiae had a similar appearance. By using only comparator strains of closely related yeasts, we here identified two new clinically relevant species that have a phenotype similar to C. auris on CCP. Both species, C. pseudohaemulonii and C. vulturna, are pathogenic siblings of C. auris and part of the C. haemulonii species complex. They have been isolated from bloodstream infections and show resistance against amphotericin B (6, 25). Although C. pseudohaemulonii looked similar to C. auris in the spot assay, in mixed culture colonies, it was much smaller and produced the distinctive blue halo only after 72 h. This would make it less likely to misidentify C. pseudohaemulonii for C. auris when encountered in clinical samples. In contrast, C. vulturna colonies could only be differentiated from C. auris based on the size of the blue halo. However, as depicted by Fig. 1D, this halo size is strain dependent and probably not a reliable characteristic for differentiation. Therefore, C. vulturna is likely to cause false positives for C. auris when detected on the CCP medium. Complementary to this, the CCP medium showed similar problems as the reference media, with the identification of the more common clinical species C. albicans, C. dubliniensis, and N. glabrata. On all media, C. albicans and C. dubliniensis produced almost identical colony phenotypes. Likewise, C. lusitaniae could be mistaken for N. glabrata. The low specificity of chromogenic media for these species has been reported before (14), and the new CCP medium could not solve this problem.
The HiCrome C. auris MDR selective agar showed very low sensitivity for C. auris compared to that of the CCP medium. Only three out of nine C. auris strains grew on this medium. This is probably because HAMA contains an inhibitory mixture, making it selective for multidrug-resistant yeasts. The composition and concentration of antifungals of the inhibitory mixture are not specified on the product data sheet though. Looking at the MIC metadata of the tested C. auris strains, all strains except CBS10913 have high MICs for one or more antifungals (26). However, only strains with fluconazole MIC values of >256 μg/ml were able to grow, suggesting HAMA only selects C. auris strains with high fluconazole resistance. HAMA successfully inhibited growth of all comparator species, indicating it could be used for the detection of (fluconazole-resistant) C. auris. Nevertheless, HAMA does not allow for reliable identification due to its poor chromogenic performance. The lack of a specific, and even variable, color for C. auris makes additional methods, such as MALDI-TOF, to confirm identification necessary. This implies that the use of HAMA does not have any advantages over selective media for C. auris that have been described previously (27, 28). Notably, all selective media need to be incubated at 42°C in order to inhibit the growth of most other clinical Candida species, except C. auris (7, 27, 28). Therefore, incubating the CCP medium at 42°C likely increases its selectivity for C. auris. Conversely, this would limit the detection of possible other clinically relevant yeasts in polymicrobial samples.
A possible limitation of this study is that we did not test the media with clinical samples from patients infected by C. auris or the other closely related clinically relevant species like C. pseudohaemulonii and C. vulturna. However, Bayona and coworkers recently confirmed the suitability of the CCP medium for the detection and identification of C. auris and other yeasts in the context of a hospital outbreak (29). Despite the lack of clinical samples, this study used a very broad panel of closely related yeasts with which C. auris can be misidentified by biochemical identification systems. This allowed for a thorough evaluation of the specificity of the tested chromogenic media.
In conclusion, the novel chromogenic medium CHROMagar Candida Plus was shown to be superior in the detection and identification of C. auris compared to the other chromogenic media tested. Nevertheless, C. vulturna and C. pseudohaemulonii produced a phenotype similar to C. auris on CCP. Both species have been reported to cause nosocomial infections and therefore could cause false positives. Moreover, the use of CCP medium could not overcome the limitations in the identification of N. glabrata and C. dubliniensis, previously reported for other chromogenic agars. However, despite some limitations, CCP showed to be a valuable addition to the plethora of mostly molecular methods for C. auris detection and identification.
ACKNOWLEDGMENTS
Candida auris reference strains were provided by the Centers for Disease Control and Prevention (Atlanta, GA, USA) Antimicrobial Resistance Isolate Bank. HiCrome C. auris MDR selective agar (HiMedia, Mumbai, India) was provided for free by EWC Diagnostics (Steenwijk, The Netherlands). CHROMagar Candida Plus (CHROMagar, Paris, France) was provided at a discounted price by bioTRADING (Mijdrecht, The Netherlands). No other external funding was received.
We declare no conflict of interest.
Conceptualization, A.W.D.J. and F.H.; Methodology, A.W.D.J. and F.H.; Investigation, A.W.D.J., C.D., and F.H.; Resources, M.C., R.M.T., and F.H.; Data Curation, A.W.D.J. and F.H.; Writing – Original Draft, A.W.D.J. and F.H.; Writing – Review & Editing, A.W.D.J., C.D., M.C., R.M.T., and F.H.; Visualization, A.W.D.J.; Supervision, F.H.; Funding Acquisition, F.H.
REFERENCES
- 1.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel) 3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassetti M, Giacobbe DR, Vena A, Trucchi C, Ansaldi F, Antonelli M, Adamkova V, Alicino C, Almyroudi M-P, Atchade E, Azzini AM, Carannante N, Carnelutti A, Corcione S, Cortegiani A, Dimopoulos G, Dubler S, García-Garmendia JL, Girardis M, Cornely OA, Ianniruberto S, Kullberg BJ, Lagrou K, Le Bihan C, Luzzati R, Malbrain MLNG, Merelli M, Marques AJ, Martin-Loeches I, Mesini A, Paiva J-A, Peghin M, Raineri SM, Rautemaa-Richardson R, Schouten J, Brugnaro P, Spapen H, Tasioudis P, Timsit J-F, Tisa V, Tumbarello M, van den Berg CHSB, Veber B, Venditti M, Voiriot G, Wauters J, Montravers P. 2019. Incidence and outcome of invasive candidiasis in intensive care units (ICUs) in Europe: results of the EUCANDICU project. Crit Care 23:219. doi: 10.1186/s13054-019-2497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 19:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open Forum Infect Dis 6:S79–S94. doi: 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stavrou AA, Lackner M, Lass-Flörl C, Boekhout T. 2019. The changing spectrum of Saccharomycotina yeasts causing candidemia: phylogeny mirrors antifungal susceptibility patterns for azole drugs and amphothericin B. FEMS Yeast Res 19:foz037. doi: 10.1093/femsyr/foz037. [DOI] [PubMed] [Google Scholar]
- 6.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gómez-López A, Boekhout T. 2012. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol 50:3641–3651. doi: 10.1128/JCM.02248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 8.Rhodes J, Fisher MC. 2019. Global epidemiology of emerging Candida auris. Curr Opin Microbiol 52:84–89. doi: 10.1016/j.mib.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 9.de Jong AW, Hagen F. 2019. Attack, defend and persist: how the fungal pathogen Candida auris was able to emerge globally in healthcare environments. Mycopathologia 184:353–365. doi: 10.1007/s11046-019-00351-w. [DOI] [PubMed] [Google Scholar]
- 10.Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Araúz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandón P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. doi: 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Schalkwyk E, Mpembe RS, Thomas J, Shuping L, Ismail H, Lowman W, Karstaedt AS, Chibabhai V, Wadula J, Avenant T, Messina A, Govind CN, Moodley K, Dawood H, Ramjathan P, Govender NP, GERMS-SA . 2019. Epidemiologic shift in candidemia driven by Candida auris, South Africa, 2016–2017. Emerg Infect Dis 25:1698–1707. doi: 10.3201/eid2509.190040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry JD. 2017. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin Microbiol Rev 30:449–479. doi: 10.1128/CMR.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scharmann U, Kirchhoff L, Chapot VLS, Dziobaka J, Verhasselt HL, Stauf R, Buer J, Steinmann J, Rath PM. 2020. Comparison of four commercially available chromogenic media to identify Candida albicans and other medically relevant Candida species. Mycoses 63:823–831. doi: 10.1111/myc.13119. [DOI] [PubMed] [Google Scholar]
- 15.Willinger B, Hillowoth C, Selitsc B, Manafi M. 2001. Performance of Candida ID, a new chromogenic medium for presumptive identification of Candida species, in comparison to CHROMagar Candida. J Clin Microbiol 39:3793–3795. doi: 10.1128/JCM.39.10.3793-3795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen J, Muñoz P, Guinea J, Rodríguez-Créixems M, Peláez T, Bouza E. 2007. Mixed fungemia: incidence, risk factors, and mortality in a general hospital. Clin Infect Dis 44:e109–e114. doi: 10.1086/518175. [DOI] [PubMed] [Google Scholar]
- 17.Tap RM, Lim TC, Kamarudin NA, Ginsapu SJ, Abd Razak MF, Ahmad N, Amran F. 2018. A fatal case of Candida auris and Candida tropicalis candidemia in neutropenic patient. Mycopathologia 183:559–564. doi: 10.1007/s11046-018-0244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath LL, Hospenthal DR, Murray CK, Dooley DP. 2003. Direct isolation of Candida spp. from blood cultures on the chromogenic medium CHROMagar Candida. J Clin Microbiol 41:2629–2632. doi: 10.1128/jcm.41.6.2629-2632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e1–277.e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Vatanshenassan M, Boekhout T, Mauder N, Robert V, Maier T, Meis JF, Berman J, Then E, Kostrzewa M, Hagen F. 2020. Evaluation of microsatellite typing, ITS sequencing, AFLP fingerprinting, MALDI-TOF MS, and Fourier-transform infrared spectroscopy analysis of Candida auris. J Fungi (Basel) 6:146. doi: 10.3390/jof6030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borman AM, Johnson EM. 2021. Name changes for fungi of medical importance, 2018 to 2019. J Clin Microbiol 59:e01811-20. doi: 10.1128/JCM.01811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand AC, Giraldo A, da Cunha KC, Sandoval-Denis M, Hendrickx M, Nishikaku AS, de Azevedo Melo AS, Merseguel KB, Khan A, Parente Rocha JA, Sampaio P, da Silva Briones MR, e Ferreira RC, de Medeiros Muniz M, Castañón-Olivares LR, Estrada-Barcenas D, Cassagne C, Mary C, Duan SY, Kong F, Sun AY, Zeng X, Zhao Z, Gantois N, Botterel F, Robbertse B, Schoch C, Gams W, Ellis D, Halliday C, Chen S, Sorrell TC, Piarroux R, Colombo AL, Pais C, de Hoog S, Zancopé-Oliveira RM, Taylor ML, Toriello C, de Almeida Soares CM, Delhaes L, Stubbe D, et al. 2015. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol 53:313–337. doi: 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- 24.Borman AM, Fraser M, Johnson EM. CHROMagarTM Candida Plus: A novel chromogenic agar that permits the rapid identification of Candida auris. Med Mycol, in press. [DOI] [PubMed] [Google Scholar]
- 25.Sipiczki M, Tap RM. 2016. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. Int J Syst Evol Microbiol 66:4009–4015. doi: 10.1099/ijsem.0.001302. [DOI] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. 2021. CDC & FDA Antibiotic Resistance Isolate Bank. Candida auris panel. https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=2. Accessed 22 January 2021.
- 27.Das S, Singh S, Tawde Y, Chakrabarti A, Shankarnarayan SA, Rudramurthy SM, Kaur H, Ghosh A. 2021. A selective medium for isolation and detection of Candida auris, an emerging pathogen. J Clin Microbiol 59:e00326-20. doi: 10.1128/JCM.00326-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar A, Sachu A, Mohan K, Vinod V, Dinesh K, Karim S. 2017. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal's medium. Rev Iberoam Micol 34:109–111. doi: 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Mulet Bayona JV, Salvador García C, Tormo Palop N, Gimeno Cardona C. 2020. Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagar™ Candida for the detection of Candida auris in surveillance samples. Diagn Microbiol Infect Dis 98:115168. doi: 10.1016/j.diagmicrobio.2020.115168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data was deposited into GenBank under accession numbers MT974606 to MT974688.