Whole-genome sequences of Candida auris isolates from nosocomial and nonnosocomial infections were compared. The average numbers of single nucleotide variations were different between the two groups. The small amount of genetic variability between intra- or interhost isolates suggests recovery of all colonizing or infecting genomes for comparison is required for outbreaks.
KEYWORDS: Candida auris, whole-genome sequencing, nosocomial transmission
ABSTRACT
Whole-genome sequences of Candida auris isolates from nosocomial and nonnosocomial infections were compared. The average numbers of single nucleotide variations were different between the two groups. The small amount of genetic variability between intra- or interhost isolates suggests recovery of all colonizing or infecting genomes for comparison is required for outbreaks.
INTRODUCTION
At our hospital, the fungal pathogen Candida auris was first recovered from a patient’s deep wound culture in June 2018 and identified in three more patients in late 2018. These cases were followed by 20 more cases in early 2019. It was unclear whether this surge of 20 cases of C. auris at our institution was a result of hospital transmission. Therefore, genomes of C. auris isolates that were nosocomially acquired and those that were present on hospital admission were analyzed, followed by comparison to local and international strains, to elucidate the molecular epidemiology and evaluate the utility of whole-genome sequencing (WGS) in local outbreak and transmission investigation.
MATERIALS AND METHODS
C. auris nosocomial acquisition, defined as detection of C. auris >48 h from the time of admission, was identified in 5 of 20 patients. Fifteen nonnosocomial cases included patients with C. auris present on admission (POA). Clinical information was collected for all patients with positive isolates at our hospital, Northwestern Memorial Hospital (NMH), for admissions in the 90 days preceding C. auris detection. Descriptive and comparative statistics were performed using IBM SPSS Statistics version 25.0.
WGS was performed on 20 isolates from 14 NMH patients (5 from five nosocomial patients, 15 from nine POA patients). Subsequently, the 20 NMH isolates were compared to five isolates from three non-NMH clinical sites (ACL Laboratories) separated by a minimum and maximum distance of 5.5 and 25.2 miles from each other, respectively, and two isolates from the Centers for Diseases Control and Prevention (CDC) and the Food and Drug Administration Antibiotic Resistance Isolate Bank (Table 1). Species identification was performed using Vitek 2 (software V8.0; bioMérieux, Durham, NC, USA) and verified with sequencing of an internal transcribed spacer (ITS) (1).
TABLE 1.
Comparison of clinical characteristics of nosocomial and POA patients
| Isolate | Location | Patient | Source | Fluconazole MIC (µg/ml) | SRA accession no. |
|---|---|---|---|---|---|
| CA01 | NMH | Nosocomial 1 | Pleural fluid | 2 | SRR12363130, SRR12363136 |
| CA02 | NMH | POA1 | Blood | 4 | SRR12363129 |
| CA03 | NMH | POA2 | Urine | 2 | SRR12363163 |
| CA04 | NMH | Nosocomial 2 | Respiratory | 4 | SRR12363162 |
| CA05 | NMH | POA3 | Blood | 2 | SRR12363161 |
| CA06 | NMH | Nosocomial 3 | Blood | 4 | SRR12363160 |
| CA07 | NMH | Nosocomial 4 | Wound | 2 | SRR12363159 |
| CA08 | NMH | POA4 | Patient room | Unknown | SRR12363158 |
| CA09 | NMH | POA4 | Patient room | Unknown | SRR12363157 |
| CA10 | NMH | POA4 | Patient room | Unknown | SRR12363156 |
| CA11 | NMH | POA4 | Urine | 4 | SRR12363155 |
| CA12 | NMH | Nosocomial 5 | Respiratory | 4 | SRR12363154 |
| CA13 | NMH | POA5 | Wound | 2 | SRR12363152 |
| CA14 | NMH | POA6 | Urine | 4 | SRR12363151 |
| CA15 | NMH | POA7 | Skin | 2 | SRR12363150 |
| CA16 | NMH | POA7 | Nose | Unknown | SRR12363149 |
| CA17 | NMH | POA7 | Axilla | Unknown | SRR12363148 |
| CA18 | NMH | POA7 | Groin | Unknown | SRR12363147 |
| CA19 | NMH | POA8 | Skin | 2 | SRR12363146 |
| CA20 | NMH | POA9 | Skin | Unknown | SRR12363145 |
| CA21 | ACL/5.5 milesa | ACL | Respiratory | 4 | SRR12363144 |
| CA22 | ACL/16.4 milesa | ACL | Blood | 2 | SRR12363143 |
| CA23 | ACL/16.4 milesa | ACL | Urine | 4 | SRR12363141 |
| CA24 | ACL/15.2 milesa | ACL | Urine | 256 | SRR12363140 |
| CA25 | ACL/15.2 milesa | ACL | Synovial | 128 | SRR12363139 |
| CA26 | CDC/Venezuela | CDC | AR-0385 | 256 | SRR12363138 |
| CA27 | CDC/Venezuela | CDC | AR-0386 | 256 | SRR12363137 |
Miles indicate distance from NMH.
MIC values (in micrograms per milliliter) were obtained using Sensititre YeastOne YO3IVD AST plates (Thermo Scientific, Waltham, MA, USA) for NMH isolates. Fluconazole MIC values were obtained from ACL Laboratories by written report. Fluconazole MIC values for the CDC isolates were obtained from the CDC & FDA Antibiotic Resistance Isolate Bank (2).
Genomic DNA was isolated with the Qiagen genomic DNA extraction kit (Qiagen, Hilden, German). DNA libraries were prepared, pooled, and sequenced on the Illumina MiSeq platform (Nextera XT; Illumina, CA, USA) using V3 chemistry to generate paired-end 300-bp reads. Raw reads were trimmed with Trimmomatic v0.32. As there were no publicly available complete reference sequences for C. auris isolates in the same global clade as the isolates in this study, complete genome sequences were generated for isolate CA01 using a combination of long-read and short-read sequencing technologies. Nanopore libraries were generated (SQK-LSK109 kit; Oxford Nanopore Technologies, Oxford, UK) and sequenced on the GridION platform using FLO-MIN106D flow cells. Our hybrid long-read/short-read genome assembly approach was adapted from a previously published method (3).
Single nucleotide variants (SNVs) were determined by alignment (Burrows-Wheeler Aligner bwa v0.7.15) (4) to CA01. Duplicate reads were removed (SAMtools v1.9), and variants relative to the reference were determined using mpileup function of bcftools v1.9 with the following options: -E (recalculate extended base alignment quality [BAQ]), -Q 25 (skip bases with BAQ less than 25), -q 30 (skip alignments with mapQ less than 30), and -m 2 (minimum gapped reads for indel candidates of 2) (5). Variants were called using the call function of bcftools v1.9 with the following options: -m (multiallelic-caller model) and –ploidy 1 (haploid). Variants were masked if they failed to meet one or more of the following criteria: minimum SNV quality score of 200; minimum read consensus of 75%; minimum of 5 reads covering the SNV position; maximum of 3× the median read depth of the total alignment; minimum of 1 read in either direction covering the SNV position; homozygous under the diploid model; and not within a repetitive region as determined by BLAST alignment of the fragmented reference sequence CA01 against itself. Variant positions in the genome were manually examined and validated using Tablet v 1.19.09.03 (6). Maximum likelihood phylogenetic trees were produced using FastTree v2.1.9 using the generalized time-reversible (GTR) model and gamma likelihood and annotated using the Interactive Tree of Life (iTOL) v4 (7, 8).
As a quality control step and to further elucidate the genomic dynamics of clonal isolate evolution over time, several isolates from different passages and multiple colonies from a single colony passage were simultaneously sequenced. Two strains, CA01 from NMH and CA21 from ACL, were used. Frozen stocks were subcultured three times on inhibitory mold agar plates using a single colony pick. Three colonies from the third passage along with the corresponding colony from the second passage were sequenced simultaneously and also compared to the original strain.
Data availability.
Sequence data are deposited at NCBI under BioProject accession number PRJNA649943. The complete genome sequence of CA01 is available under accession number SAMN15689540. Sequencing read file accession numbers are given in Table 1.
RESULTS
Table S1 in the supplemental material summarizes patient demographic and clinical characteristics. Two patients with nosocomial isolates (CA06 and CA07) shared 39 days in the same intensive care unit; otherwise, no clinical overlap was identified. Risk factors for nosocomial acquisition were female gender, higher Charlson comorbidity index (9), use of hemodialysis, and any occurrence of diarrhea in the 90 days preceding diagnosis. Mortality rates at 1 year were higher in nosocomial than in POA patients (Table S1).
Twenty isolates from 14 NMH patients and 7 isolates from outside sources underwent WGS. The NMH isolates included 5 from five nosocomial patients (CA01, CA04, CA06, CA07, and CA12) and 15 from nine POA patients, including 4 isolates from a patient and their room (CA08 to CA11) and 4 isolates from different body sites of one patient (CA15 to CA18) (Table 1).
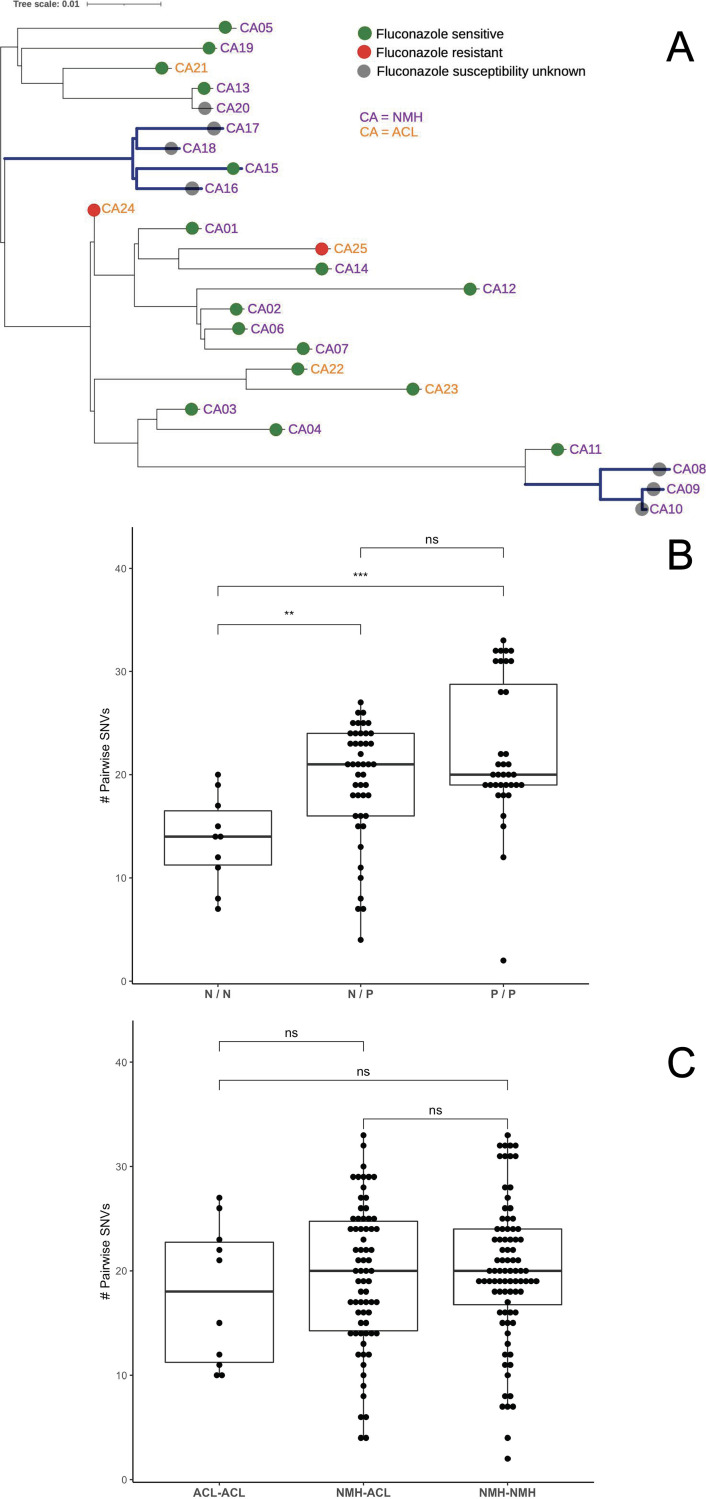
Phylogenetic analysis of nosocomial and POA isolates failed to separate the two groups (Fig. 1A). The variant positions were largely distributed across the seven chromosomes. The average number of SNV differences between nosocomial isolates was significantly less than either the average of POA compared with POA isolates or nosocomial compared with POA isolates (Fig. 1B; Table 1); however, the range of pairwise SNV values overlapped between the three comparisons, precluding the designation of a SNV difference cutoff that would allow differentiation between nosocomial or nonnosocomial isolate pairs.
FIG 1.
(A) Phylogenetic analysis of NMH and ACL isolates. Midpoint-rooted maximum likelihood phylogenetic analysis of all NMH and ACL isolates with corresponding fluconazole sensitivity profiles, when known. The first highlighted nodes represent four isolates from a single patient. The second highlighted nodes represent environmental strains from the room of patient CA11. Nosocomial strains are CA01, CA04, CA06, CA07, and CA12. (B) Pairwise SNV differences between nosocomial and POA isolates. N, nosocomial isolates; P, isolates from POA patients, ns, no significant difference. Statistical analysis was performed with unpaired Wilcoxon tests. **, P < 0.01; ***, P < 0.001. (C) Pairwise SNP differences between NMH and ACL strains. ACL, isolates from ACL Laboratories; NMH, isolates from NMH nosocomial and POA patients; ns, no significant difference. Statistical analysis was performed with unpaired Wilcoxon tests.
Isolates from the two patients who shared at least one clinical space during their hospitalizations (CA06 and CA07) differed by 7 SNVs, a value that exceeds the number of SNVs separating many other unrelated isolate pairs. The isolates from a patient and the patient’s hospital room (CA08 to CA11) differed by 1 to 8 SNVs despite a presumably similar origin (see Table S2). Genomes of the four isolates from the same patient (CA15 to CA18) were not identical, differing by 4 to 9 SNVs (Table S2). These data suggest that there can be considerable genetic variability between presumably related isolates, even when collected from the same environments.
Of the two isolates that underwent in vitro subculturing, one (CA21) was identical in all subcultures compared to the originally sequenced isolate. For the other isolate (CA01), there were two variants noted: the newly sequenced isolates all differed from the originally sequenced isolate (A→T) at position 755634 on chromosome 5 and one of the 3 second-passage isolates differed from the original and the other three new subcultures at position 3870964 on chromosome 1.
The 20 NMH isolates were compared to five isolates from ACL (CA21 to CA25) and two CDC isolates (CA26 and CA27) from the South American clade originally collected from Venezuela in 2012 and obtained from the CDC & FDA Antibiotic Resistance Isolate Bank (AR bank number 0385 and AR bank number 0386). The range of total pairwise SNV differences between all NMH and ACL isolates was 4 to 33 SNVs. Overall pairwise SNV differences were not significantly different when comparing NMH to ACL (Fig. 1C). All Chicagoland isolates belonged to the South American clade; however, the CDC isolates were phylogenetically distinct from the NMH and ACL isolates, differing by 185 to 204 SNVs. This is in contrast to the other recognized geographic clades, where isolates differed from the South Asian, East Asian, and South African clades by 149,813 to 167,488, 162,434 to 165,672, and 161,710 to 167,390 SNVs, respectively.
Fluconazole-susceptible isolates were not phylogenetically distinct from fluconazole-resistant isolates (Fig. 1A). Among fluconazole-susceptible isolates, a maximum of 39 SNVs differentiated any two isolates. The two fluconazole-resistant isolates, CA24 and CA25, differed by 10 SNVs. Interestingly, fluconazole-susceptible CA01 differed from fluconazole-resistant CA24 by only 4 SNVs. These two isolates were cultured from institutions 15 miles apart.
DISCUSSION
Previous studies of C. auris outbreaks were focused on establishing epidemiological links and identifying transmission sources. Our study provides a comparison of the genetic diversity of C. auris isolates among patients with and without nosocomial acquisition. We additionally compared institutional isolates with local and international strains to better understand regional C. auris transmission dynamics.
Examination of the genomic diversity of C. auris from our hospital, across a region, and outside the United States showed low levels of variation among the isolates from the Chicago area and no significant difference in the overall number of pairwise SNVs across hospital systems. These results suggest C. auris in Chicago may have originated from a single introduction event followed by local transmission, as was previously hypothesized and was further supported by the finding that there were minimal single nucleotide variations between isolates from different institutions in the Chicagoland area (10). Prior to this study, only one C. auris strain from a patient in Illinois had undergone WGS and belonged to the South American clade, differing by fewer than 150 SNVs (11). We found that all our Chicagoland area isolates were related to the South American clade. Within the four known clades, there has been described a wide genetic variability between clades but relatively little genetic distance within clades; previously described South American isolates differed by fewer than 16 SNVs between isolates yet differed from other clade isolates by tens of thousands of SNVs (12). Additional studies have confirmed clade clonality on a continental level, even between isolates from hospitals separated by thousands of miles (12, 13). WGS of all our Chicago area isolates supports this observation; however, further study is needed to elucidate the biology behind this unusual intra- and interclade variability.
Interestingly, despite the clustering of Chicago isolates, we found the average number of SNVs between nosocomial isolates was significantly less than either the average of POA compared with POA isolates or nosocomial compared with POA isolates, supporting possible nosocomial acquisition of C. auris by some patients. However, no distinct cluster was associated with nosocomial isolates and no genome sequence was shared among nosocomial isolates, as is commonly seen in WGS of nosocomial transmission events with other pathogens. These results are not likely explained by intrahost heterogeneous populations, as an investigation of a single center outbreak showed multiple unique sequences were shared by two or more patients (14). It was likely that nosocomially infected patients acquired C. auris at the hospital where more than one strain was present. Considering that ranges of pairwise SNV values of nosocomial isolates overlapped with the POA isolates in our study, it is possible that colonization with C. auris was not detected in nosocomial cases upon admission or an epidemiological link was not identified in patients with genome variation of <10 SNVs.
Intrahost diversity of C. auris has been reported by several studies (12–17). Patients may host a mixture of C. auris genomes at a given time and subsequently acquire a different group of genomes. In these studies, genome variation of the heterogeneous populations ranged from between 6 and 9 SNPs to >60 SNPs. We found similar results. As the rate of C. auris evolution was estimated to be 5.75 mutations per genome per year (14), the intrahost diversity of C. auris was considered to be the consequence of mixed colonization or infections rather than mutation accumulation. As we noted limited in vitro variability on repeated subculturing of two isolates, the results support the possibility of mixed colonization or accelerated mutagenesis within hosts, and accumulation of mutations during lab culture or sequencing or analysis error is less likely to account for the findings. Within-host mutation is further favored by our finding that isolates from the same patient or from a patient’s room were more closely related to each other than to other isolates in this study, but this will require further study to determine. This intrahost diversity indicates that the traditional strategy for outbreak investigation, i.e., comparing a single genome from each patient, is insufficient to establish strain transmission for C. auris. In order to fully evaluate the genome diversity of C. auris populations in a host, the sampling of multiple body sites and the recovery of a sampling of the population of the colonizing/infecting genomes for comparison will likely be required for outbreak investigation.
No isolate at our hospital was fluconazole resistant, yet NMH isolates were not clearly phylogenetically distinct from ACL isolates with higher fluconazole MICs. As fluconazole resistance in C. auris can occur through variations in the ERG11 gene, such as Y132F and K143R, it is possible that a single SNV in this gene could have led to resistance the ACL isolates (10, 18). Further study is necessary to reveal the resistance mechanism associated with Chicago area isolates. Limitations of our study include the small sample size and limited clinical information for patients outside NMH. Additionally, as admission surveillance cultures were not universally obtained, nosocomial acquisitions may have been designated POA.
In conclusion, we found that the average number of pairwise single nucleotide variants was significantly different between nosocomial and POA groups in spite of overall low genomic diversity of C. auris in the Chicago area. Genomic variations within and between hosts suggest that the recovery of a representation of the population of colonizing/infecting genomes for comparison may be required for C. auris outbreak investigations.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UIC Genome Research Core for the nanopore sequencing support. We also thank ACL Laboratories for providing C. auris isolates.
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (T32AI095207-07 to S.C.R.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2020. CDC & FDA Antibiotic Resistance Isolate Bank. https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=2. Accessed 14 May 2020.
- 3.Shin SC, Kim H, Lee JH, Kim HW, Park J, Choi BS, Lee SC, Kim JH, Lee H, Kim S. 2019. Nanopore sequencing reads improve assembly and gene annotation of the Parochlus steinenii genome. Sci Rep 9:5095. doi: 10.1038/s41598-019-41549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milne I, Bayer M, Stephen G, Cardle L, Marshall D. 2016. Tablet: visualizing next-generation sequence assemblies and mappings. Methods Mol Biol 1374:253–268. doi: 10.1007/978-1-4939-3167-5_14. [DOI] [PubMed] [Google Scholar]
- 7.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 10.Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, Forsberg K, Lockhart SR, Adam R, Alanio A, Alastruey-Izquierdo A, Althawadi S, Araúz AB, Ben-Ami R, Bharat A, Calvo B, Desnos-Ollivier M, Escandón P, Gardam D, Gunturu R, Heath CH, Kurzai O, Martin R, Litvintseva AP, Cuomo CA. 2020. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11:e03364-19. doi: 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM, MSD. 2016. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus - United States, May 2013-August 2016. MMWR Morb Mortal Wkly Rep 65:1234–1237. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 12.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM. 2018. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 379:1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 15.Biswas C, Wang Q, van Hal SJ, Eyre DW, Hudson B, Halliday CL, Mazsewska K, Kizny Gordon A, Lee A, Irinyi L, Heath CH, Chakrabarti A, Govender NP, Meyer W, Sintchenko V, Chen SC. 2020. Genetic heterogeneity of Australian Candida auris isolates: insights from a nonoutbreak setting using whole-genome sequencing. Open Forum Infect Dis 7:ofaa158. doi: 10.1093/ofid/ofaa158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, Litvintseva AP, US Candida auris Investigation Team. 2018. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18:1377–1384. doi: 10.1016/S1473-3099(18)30597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, Fisher MC, Schelenz S. 2018. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 7:43. doi: 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Healey KR, Kordalewska M, Jiménez Ortigosa C, Singh A, Berrío I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are deposited at NCBI under BioProject accession number PRJNA649943. The complete genome sequence of CA01 is available under accession number SAMN15689540. Sequencing read file accession numbers are given in Table 1.