We compared the performance of the Abbott BinaxNOW COVID-19 antigen card to that of a standard reverse transcription-PCR (RT-PCR) assay (Thermo Fisher TaqPath COVID-19 Combo kit) for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2,645 asymptomatic students presenting for screening at the University of Utah. SARS-CoV-2 RNA was detected in 1.7% of the study participants by RT-PCR.
KEYWORDS: asymptomatic screening, BinaxNOW COVID-19 antigen card, rapid antigen tests, SARS-CoV-2
ABSTRACT
We compared the performance of the Abbott BinaxNOW COVID-19 antigen card to that of a standard reverse transcription-PCR (RT-PCR) assay (Thermo Fisher TaqPath COVID-19 Combo kit) for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2,645 asymptomatic students presenting for screening at the University of Utah. SARS-CoV-2 RNA was detected in 1.7% of the study participants by RT-PCR. BinaxNOW identified 24 infections but missed 21 infections that were detected by RT-PCR. The analytical sensitivity (positive agreement) and analytical specificity (negative agreement) for the BinaxNOW were 53.3% and 100%, respectively, compared to the RT-PCR assay. The median cycle threshold (CT) value in the specimens that had concordant positive BinaxNOW antigen results was significantly lower than that of specimens that were discordant (CT of 17.6 versus 29.6; P < 0.001). In individuals with presumably high viral loads (CT of <23.0), a 95.8% positive agreement was observed between the RT-PCR assay and BinaxNOW. Due to the possibility of false-negative results, caution must be taken when utilizing rapid antigen testing for screening asymptomatic individuals.
INTRODUCTION
With its high degree of transmissibility, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen for the novel 2019 coronavirus disease (COVID-19), has undoubtedly led to one of the most remarkable global public health epidemics in recent history. Timely identification and isolation of infected individuals are crucial in mitigating the rampant community spread of SARS-CoV-2. The gold-standard method for COVID-19 diagnosis remains the detection of SARS-CoV-2 RNA in respiratory tract specimens using nucleic acid amplification techniques such as reverse transcription-PCR (RT-PCR). However, SARS-CoV-2 nucleic acid amplification tests (NAATs) are generally more expensive than alternative methodologies and may have prolonged turnaround times due to limited test supplies, reagent allocation, and fixed laboratory capacity, which have been exacerbated by extremely high demand.
Efforts to expand testing capacity have led to the development of several rapid antigen (Ag) tests designed to detect SARS-CoV-2 nucleocapsid antigen, primarily in symptomatic individuals (1). At the time of this writing, the U.S. Food and Drug Administration (FDA) has granted emergency-use authorization (EUA) to 11 SARS-CoV-2 antigen tests (2). Although these antigen tests are intended to be utilized in symptomatic individuals (within the first 5 to 7 days of symptom onset), the U.S. Department of Health and Human Services (HHS), through the Public Readiness and Emergency Preparedness Act (PREP Act), permits their use for screening asymptomatic individuals in congregate facilities, including schools (3). However, there are limited data on the performance characteristics of rapid antigen tests in asymptomatic or presymptomatic individuals. A recent meta-analysis of published literature on rapid, point-of-care antigen tests reported an average sensitivity and specificity of 56.2% and 99.5%, respectively, compared to NAAT (1). However, these studies were not limited exclusively to asymptomatic individuals, the specimen type was primarily nasopharyngeal and/or oropharyngeal, and none of the antigen tests included have received EUA approval from the FDA.
In this study, we evaluated the diagnostic performance characteristics of the Abbott BinaxNOW COVID-19 antigen card (referred to here as BinaxNOW) in a population of college-age students who were asymptomatic at the time of testing. BinaxNOW is a rapid lateral flow immunoassay that qualitatively detects SARS-CoV-2 nucleocapsid antigen in direct nasal swab specimens. The package insert cites a positive agreement of 97.1% and a negative agreement of 98.5% compared to an EUA RT-PCR assay (4). These data were based on a clinical study involving a total of 102 patients, of whom 95 had symptoms consistent with COVID-19 and only 7 were asymptomatic. This was recently updated to a positive agreement of 84.6%, based on a larger study involving 460 symptomatic individuals. Of note, the U.S. federal government has distributed 150 million BinaxNOW antigen cards to states across the country (5). BinaxNOW also received EUA for at-home use under the supervision of a telehealth proctor (6). Therefore, characterizing the performance characteristics of BinaxNOW for off-label use in an asymptomatic population is essential given its potential widespread application for asymptomatic screening in a variety of settings.
MATERIALS AND METHODS
Study population and specimen collection.
The participants of this study were primarily college-age (undergraduate and graduate) students at the University of Utah in Salt Lake City, UT. At the time of specimen collection, the students were first queried to ensure that they were not experiencing any signs and/or symptoms of COVID-19. Specimen collection occurred at a temporary indoor testing site from 13 to 20 November 2020. Two nasal swabs were collected from each participant, according to the technique recommended by the U.S. Centers for Disease Control and Prevention (CDC) (7). The study participants were instructed to swab both nares at the level of the midturbinate for each collection. Trained nonmedical personnel observed the specimen collection process. The first swab collected from the participants was randomly assigned to be tested with either BinaxNOW or the RT-PCR assay in an effort to minimize sampling bias.
Detection of SARS-CoV-2 viral antigen.
The BinaxNOW antigen cards utilized in this study were received from the Utah Department of Health as part of a U.S. federal government initiative to expand COVID-19 testing capacity. Testing was performed by trained nonmedical personnel (University of Utah Hope Corps interns) according to the manufacturer’s instructions (4). Each testing personnel was trained on the test procedure (including the appropriate use of personal protective equipment) and result interpretation using detailed step-by-step videos provided by the manufacturer. To evaluate for competence, each testing personnel was required to pass an assessment quiz and successfully perform external quality control using a positive-control swab and a sterile swab (negative control). External quality control was also performed for each new kit of BinaxNOW antigen cards.
Results were interpreted visually after 15 min. A specimen was deemed positive for SARS-CoV-2 viral antigen if two pink/purple lines (control line on the top and sample line on the bottom) were observed on the test card, as illustrated in the assay product insert (4). A faint pink/purple line in the sample region of the test card (in addition to a pink/purple control line) was also interpreted as a positive result. A single pink/purple line in the control region of the test card was interpreted as a negative result. If no line was observed in the control region or if the line remained blue, the result was interpreted as invalid.
Participants were notified of their BinaxNOW result using the Navica mobile app, which is a free mobile app provided by Abbott (8). Any participant who tested positive was contacted to return to the testing site within 24 h and submit a saliva specimen for SARS-CoV-2 NAAT at ARUP Laboratories. These individuals were instructed to self-isolate while awaiting NAAT confirmation. Individuals who received an invalid BinaxNOW result were also contacted for repeat antigen testing. Participants receiving a negative antigen test were counseled that these results were “presumptive” and did not negate the need for mitigation behaviors designed to reduce the spread of SARS-CoV-2.
Detection of SARS-CoV-2 nucleic acid.
The other nasal swab was placed into ARUP COVID-19 Transport Media (9) and tested at ARUP Laboratories using the Thermo Fisher TaqPath COVID-19 Combo kit, referred to here as the TaqPath COVID-19 kit (10). These specimens were stored frozen (−20°C) and tested within 10 days of receipt in the clinical laboratory. The TaqPath COVID-19 kit targets regions of three coronavirus genes: open reading frame 1ab (ORF1ab), the gene for the S protein, and the gene for the N protein. Forty amplification cycles are performed by the assay. At least two genes have to be detected for the result to be reported as positive for SARS-CoV-2. The cycle threshold (CT) value for each specimen was reported as the average of the CT values of the detected coronavirus genes. An inconclusive result is reported when only one gene is detected after consecutive repeat testing. Detection of SARS-CoV-2 RNA in the confirmatory saliva specimens was performed in real time using one of three FDA EUA assays (either the Hologic Panther Fusion SARS-CoV-2 assay, the Roche Cobas SARS-CoV-2 assay, or the Thermo Fisher TaqPath COVID-19 Combo kit). All participants were notified of their NAAT results.
Statistical analysis.
The TaqPath COVID-19 kit was used as the benchmark for assessing the diagnostic accuracy of BinaxNOW. The analytical performance characteristics (sensitivity, specificity, and predictive values) were calculated from a 2-by-2 contingency table using GraphPad Prism 8 software. Agreement between methods was assessed at various CT cutoffs reported in the package insert for BinaxNOW (4) and the published literature. The 95% confidence intervals (CIs) are based on the Wilson-Brown method. A nonparametric t test (Mann-Whitney test) was performed using GraphPad Prism 8 software to evaluate for statistical significance (P values) between median CT values. The kappa coefficient (κ) was calculated using the Microsoft Excel Analyse-it software package (version 5.20).
RESULTS
Positivity rates of the rapid antigen test and nucleic acid amplification test.
Two nasal swab specimens were collected from 2,645 individuals. Among the study participants, 1,369 (51.8%) identified as female, 1,274 (48.2%) identified as male, and 2 (0.1%) iidentified as nonbinary. The average age of the study participants was 24 years (range, 15 to 86 years). Table 1 summarizes the results from BinaxNOW and the TaqPath COVID-19 kit. A negative result with BinaxNOW was observed in 2,618 (99.0%) individuals, while a positive result was observed in 24 (0.9%) individuals. An invalid BinaxNOW result was initially observed in 3 (0.1%) individuals; however, repeat testing using a new nasal swab specimen from these individuals yielded a negative result. For the TaqPath COVID-19 kit, SARS-CoV-2 RNA was not detected in 2,595 (98.1%) individuals, and 46 (1.7%) individuals had detectable SARS-CoV-2 RNA, while 4 (0.2%) individuals had an inconclusive result.
TABLE 1.
Summary of results from the BinaxNOW antigen card and the TaqPath COVID-19 kit
| Result | No. of samples with result |
|
|---|---|---|
| BinaxNOW antigen card | TaqPath COVID-19 kit | |
| Positive | 24 | 46 |
| Negative | 2,618 | 2,595 |
| Inconclusive/invalid | 3a | 4b |
| Total | 2,645 | 2,645 |
Repeat testing yielded a negative result.
Only the N protein gene was detected in these specimens (the CT value was >30).
Concordance between the rapid antigen test and the nucleic acid amplification test.
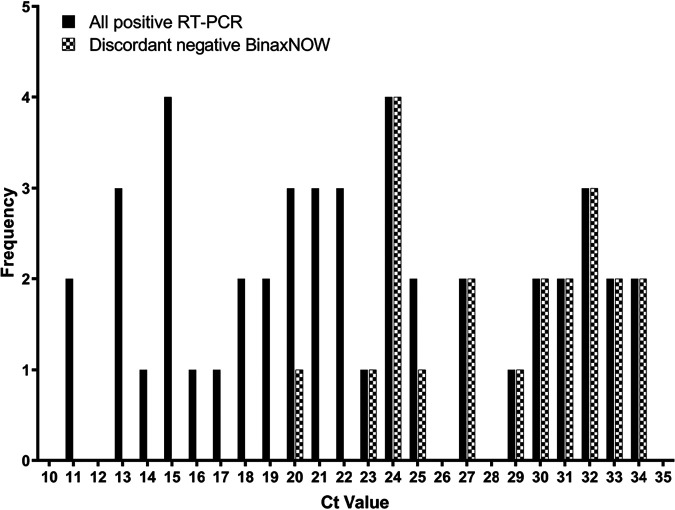
The analytical sensitivity and specificity of BinaxNOW are summarized in Table 2. Of the 46 individuals who had detectable SARS-CoV-2 RNA, 24 had a concordant positive antigen result, indicating a positive agreement of 53.3% between the two tests. The kappa coefficient (κ = 0.69; 95% CI, 0.57 to 0.82) indicates substantial agreement between the methods. The median cycle threshold (CT) value in the specimens that had concordant positive results was significantly lower (CT of 17.6) than that in the specimens that were discordant (CT of 29.6) (P < 0.001), as illustrated in Fig. 1. In specimens with presumably high viral loads (CT of <23.0), a 95.8% positive agreement was observed (Table 3). A 0% positive agreement was observed in samples with both a CT of ≥33 and a CT of ≥30, as shown in Table 3.
TABLE 2.
Diagnostic performance of the BinaxNOW antigen card compared to the TaqPath COVID-19 kit for detection of SARS-CoV-2a
| BinaxNOW antigen card result | No. of samples tested with TaqPath COVID-19 kit |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 24 | 0 | 24 |
| Negative | 21 | 2,593 | 2,614 |
| Total | 45 | 2,593 | 2,638 |
The analytical sensitivity (positive agreement) was 53.3% (95% CI, 39.1% to 67.1%), the analytical specificity (negative agreement) was 100% (95% CI, 99.9% to 100%), the positive predictive value was 100% (95% CI, 86.2% to 100%), the negative predictive value was 99.2% (95% CI, 98.7% to 99.4%), and the kappa coefficient was 0.69 (95% CI, 0.57 to 0.82) (predictive values are assuming a disease prevalence of 1.7%). Note that 4 inconclusive RT-PCR results and 3 invalid BinaxNOW results were excluded from the calculations in the table.
FIG 1.

Distribution of the RT-PCR cycle threshold (CT) values in specimens with positive and negative BinaxNOW results. The P value is based on the Mann-Whitney test. The lines signify medians and interquartile ranges.
TABLE 3.
BinaxNOW antigen card diagnostic performance against the comparator RT-PCR method by cycle threshold counts
| Parameter | No. of positive results with TaqPath COVID-19 kit by CT category |
|||||
|---|---|---|---|---|---|---|
| CT < 33.0 | CT ≥ 33.0 | CT < 30.0 | CT ≥ 30.0 | CT < 23.0 | CT ≥ 23.0 | |
| BinaxNOW antigen card result | ||||||
| Positive | 24 | 0 | 24 | 0 | 23 | 1 |
| Negative | 18 | 3 | 12 | 9 | 1 | 20 |
| Total | 42 | 3 | 36 | 9 | 24 | 21 |
| % positive agreement (95% CI) | 57.1 (42.2–70.9) | 0 | 66.7 (50.3–79.8) | 0 | 95.8 (79.8–99.3) | 4.8 (0.8–22.7) |
The collection of two consecutive bilateral nasal swab specimens did not significantly affect the detection of SARS-CoV-2 using either NAAT or the rapid antigen test (P = 0.5683 by Fisher’s exact test). The rapid antigen test was performed using the first nasal swab specimen in 12 (50%) out of the 24 individuals with concordant positive results. No statistically significant difference in median CT values was observed in concordant positive samples regardless of whether the rapid antigen test was performed using the first nasal swab or the second nasal swab (Fig. 2) (P = 0.5800). A discordant result between the rapid antigen test and NAAT (i.e., antigen negative/NAAT positive) was observed in 21 individuals. Discordant results between BinaxNOW and the RT-PCR assay were more likely at CT values of >23.0, as shown in Fig. 3. The antigen test was performed using the first nasal swab specimen in 9 (40.9%) out of the 21 individuals with discordant results. While a slightly higher median CT value was observed when the antigen test was performed using the second nasal swab than using the first nasal swab, the difference was not statistically significant (P = 0.1752), as shown in Fig. 2. In one individual with a discordant result, an invalid BinaxNOW antigen result was initially obtained, with a negative result observed upon repeat testing using a new nasal swab specimen. It is worth mentioning that for this individual, the initial invalid BinaxNOW result was obtained using the second nasal swab specimen, while the negative result from the repeat test was obtained from a third nasal swab. Hence, the validity of the negative BinaxNOW result in this individual could be questionable due to sampling bias. Invalid results were excluded in the diagnostic performance characteristic calculations.
FIG 2.
Distribution of the RT-PCR cycle threshold (CT) values in specimens with concordant positive BinaxNOW results (A) and discordant negative BinaxNOW results (B) sorted by order of nasal swab collection. The P value is based on the Mann-Whitney test. The lines signify medians and interquartile ranges.
FIG 3.
Frequency distribution of RT-PCR cycle threshold (CT) values in all specimens with detectable SARS-CoV-2 and specimens with discordant BinaxNOW results.
Twenty-two out of the 24 individuals (91.7%) with a positive antigen result returned to the testing site and submitted a follow-up saliva specimen. There was 100% agreement between these positive BinaxNOW specimens and saliva NAAT.
DISCUSSION
Compared to NAAT, the BinaxNOW antigen card showed low analytical sensitivity (53.3%) for detecting SARS-CoV-2 infection in an asymptomatic or presymptomatic population. This observation is consistent with the findings of other recent studies conducted using different SARS-CoV-2 antigen assays in unselected populations (11–13). The collection of two consecutive bilateral nasal swab specimens did not statistically affect the detection of SARS-CoV-2 using either the RT-PCR assay or the rapid antigen test. However, there was a trend toward higher CT values for the second swab, indicating a smaller amount of virus present, which may have disproportionally affected the antigen positivity rate. One study found a difference of 6 to 7 CTs between the limits of detection of the BinaxNOW antigen test and RT-PCR tests, indicating an ∼100-fold difference in sensitivity (14).
Our results indicate that a relatively high viral load (and a corresponding low CT value of <23) must be present to generate a positive BinaxNOW result. At the onset of our study, the BinaxNOW product insert reported a positive agreement of 83.3% in specimens with a CT of ≥33 (4). The manufacturer has recently updated this information to a positive agreement of 37.8%. CT values are a relative approximation of virus loads. Differences in assay design and other important preanalytic variables (e.g., specimen source, collection method, and volume of transport medium, etc.) impact reported CT values such that these measurements are not directly comparable across real-time NAAT platforms (15).
In contrast to analytical sensitivity, the specificity of BinaxNOW testing was excellent (100%). The test was able to be performed successfully at the point of care by nonmedical personnel, with a relatively low rate of invalid results (0.1%), supporting the findings of another recent study (16). These observations raise the question of whether confirmation of positive BinaxNOW results is necessary, as cautioned in a recent warning by the FDA regarding the potential for false-positive results from rapid SARS-CoV-2 antigen tests (17). It is important to note, however, that operators underwent comprehensive training and that quality control testing was performed regularly on-site. This is especially important in the context of at-home testing. Additional studies are needed to determine whether BinaxNOW test performance will be comparable in a telehealth-observed home setting.
Despite its relatively low analytical sensitivity, BinaxNOW may still be beneficial for surveillance testing in selected settings where testing resources are limited, especially when weighed against the alternative of no screening testing. Rapid antigen testing identified 24 infections in asymptomatic individuals, with qualitatively high viral loads, who may be more likely to be infectious to others (18, 19). These infections were all confirmed by saliva NAAT, and individuals were instructed to self-isolate. Given the relatively low prevalence (1.7%) in our student population, the negative predictive value of BinaxNOW was excellent (99.2%).
A total of 21 asymptomatic students had false-negative antigen test results. We do not know if these individuals developed symptoms in the days following the negative antigen result. We also cannot speculate as to how infectious these individuals were; presumably, the risk of viral transmission to others is not zero (18, 19), although the higher CT values associated with these samples may indicate a low risk of transmission. However, it is well established that asymptomatic carriers of SARS-CoV-2 can efficiently transmit the infection (20, 21). Thus, all participants were counseled to continue with physical distancing, face masking, and proper hand hygiene despite a negative iBinaxNOW result. The public health implications of a false-negative screening result in an asymptomatic population will depend on the population to which the test is applied. For example, tolerance for false-negative results may be greater in a congregate setting consisting of young, otherwise healthy individuals (e.g., a college campus) with few risk factors for severe clinical outcomes from COVID-19 versus a long-term-care facility setting or other demographics with one or multiple risk factors for poor COVID-19-associated outcomes.
The limitations of this study include the relatively small number of positive results and the lack of serial repeat testing data for the asymptomatic student cohort to determine if the 21 false-negative results would eventually test positive after subsequent assessments. This would be useful for validating the effectiveness of the proposed strategy of repeat serial testing using less sensitive antigen tests as an infection prevention and control measure (22, 23).
To the best of our knowledge, this is the first study evaluating the performance of a rapid SARS-CoV-2 antigen test in an exclusively asymptomatic population. The analytical sensitivity of BinaxNOW for off-label use in an asymptomatic population is lower than the performance claims for symptomatic patients reported by the manufacturer. As recommended by the manufacturer, negative results should be interpreted as presumptive negative. Careful assessment of the impact of false-negative results is warranted before a testing strategy utilizing rapid SARS-CoV-2 antigen tests is implemented. The specificity of BinaxNOW, however, was excellent.
ACKNOWLEDGMENTS
We thank the ARUP Institute for Clinical and Experimental Pathology for their support with performing the RT-PCR tests. We also acknowledge the University of Utah Health outpatient point-of-care testing team, the University of Utah Hope Corps interns, and the general student body for participating in this study.
REFERENCES
- 1.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, Spijker R, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group . 2020. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev 8:CD013705. 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.US Food and Drug Administration. 22 December 2020, accession date. Individual EUAs for antigen diagnostic tests for SARS-CoV-2. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas#individual-antigen. [Google Scholar]
- 3.US Department of Health and Human Services. 2020. Guidance for PREP Act coverage for COVID-19 screening tests at nursing homes, assisted-living facilities, long-term-care facilities, and other congregate facilities. US Department of Health and Human Services, Washington, DC. https://www.hhs.gov/guidance/sites/default/files/hhs-guidance-documents//prep-act-coverage-for-screening-in-congregate-settings.pdf. Accessed 19 December 2020. [Google Scholar]
- 4.Abbott Diagnostics Scarborough, Inc. 2020. BinaxNOW COVID-19 Ag card product insert. Abbott Diagnostics Scarborough, Inc, Scarborough, ME. [Google Scholar]
- 5.US Department of Health and Human Services. 2020. Trump administration will deploy 150 million rapid tests in 2020. US Department of Health and Human Services, Washington, DC. https://www.hhs.gov/about/news/2020/08/27/trump-administration-will-deploy-150-million-rapid-tests-in-2020.html. Accessed 19 December 2020. [Google Scholar]
- 6.US Food and Drug Administration. 2020. BinaxNOW COVID-19 Ag card home test. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/media/144576/download. Accessed 21 December 2020. [Google Scholar]
- 7.US Centers for Disease Control and Prevention. 19 December 2020, accession date. Interim guidelines for collecting, handling, and testing clinical specimens for COVID-19. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. [Google Scholar]
- 8.Abbott. 2020. Your NAVICA app questions answered. Abbott, Chicago, IL. https://www.abbott.com/BinaxNOW-Test-NAVICA-App/NAVICA-FAQ.html. Accessed 23 December 2020. [Google Scholar]
- 9.Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, Shakir SM. 2020. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol 58:e01824-20. 10.1128/JCM.01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Food and Drug Administration. 2020. TaqPath COVID-19 Combo kit. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/media/136113/download. Accessed 19 December 2020. [Google Scholar]
- 11.Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso A-M, Le Goff J, Delaugerre C. 2020. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol 58:e00977-20. 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. 2020. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol 129:104455. 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mak GCK, Lau SSY, Wong KKY, Chow NLS, Lau CS, Lam ETK, Chan RCW, Tsang DNC. 2020. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J Clin Virol 133:104684. 10.1016/j.jcv.2020.104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perchetti GA, Huang M-L, Mills MG, Jerome KR, Greninger AL. 11 December 2020. Analytical sensitivity of the Abbott BinaxNOW COVID-19 Ag CARD. J Clin Microbiol 10.1128/JCM.02880-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhoads D, Peaper DR, She RC, Nolte FS, Wojewoda CM, Anderson NW, Pritt BS. 12 August 2020. College of American Pathologists (CAP) Microbiology Committee perspective: caution must be used in interpreting the cycle threshold (Ct) value. Clin Infect Dis 10.1093/cid/ciaa1199. [DOI] [PubMed] [Google Scholar]
- 16.Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, Rubio L, Chamie G, Martinez J, Peng J, Black D, Wu W, Pak J, Laurie MT, Jones D, Miller S, Jacobo J, Rojas S, Rojas S, Nakamura R, Tulier-Laiwa V, Petersen M, Havlir DV, DeRisi J. 2020. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. medRxiv 10.1101/2020.11.02.20223891. [DOI] [PMC free article] [PubMed]
- 17.US Food and Drug Administration. 2020. Potential for false positive results with antigen tests for rapid detection of SARS-CoV-2—letter to clinical laboratory staff and health care providers. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/medical-devices/letters-health-care-providers/potential-false-positive-results-antigen-tests-rapid-detection-sars-cov-2-letter-clinical-laboratory. Accessed 19 December 2020. [Google Scholar]
- 18.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 2020. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 71:2663–2666. 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basile K, McPhie K, Carter I, Alderson S, Rahman H, Donovan L, Kumar S, Tran T, Ko D, Sivaruban T, Ngo C, Toi C, O’Sullivan MV, Sintchenko V, Chen SC-A, Maddocks S, Dwyer DE, Kok J. 24 October 2020. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis 10.1093/cid/ciaa1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugano N, Ando W, Fukushima W. 2020. Cluster of severe acute respiratory syndrome coronavirus 2 infections linked to music clubs in Osaka, Japan. J Infect Dis 222:1635–1640. 10.1093/infdis/jiaa542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA, Public Health-Seattle and King County and CDC COVID-19 Investigation Team . 2020. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 382:2081–2090. 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mina MJ, Parker R, Larremore DB. 2020. Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med 383:e120. 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 23.US Centers for Disease Control and Prevention. 22 December 2020, accession date. Considerations for use of SARS-CoV-2 antigen testing in nursing homes. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/hcp/nursing-homes-antigen-testing.html. [Google Scholar]