Accurate diagnosis of acute severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is critical for appropriate management of patients with this disease. We examined the possible complementary role of laboratory-developed class-specific clinical serology in assessing SARS-CoV-2 infection in hospitalized patients.
KEYWORDS: antibodies, COVID-19, SARS-CoV-2, serology, spike, immunodiagnostics
ABSTRACT
Accurate diagnosis of acute severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is critical for appropriate management of patients with this disease. We examined the possible complementary role of laboratory-developed class-specific clinical serology in assessing SARS-CoV-2 infection in hospitalized patients. Serological tests for immunoglobulin G (IgG), IgA, and IgM antibodies against the receptor binding domain (RBD) of SARS-CoV-2 were evaluated using samples from real-time reverse transcription-quantitative PCR (qRT-PCR)-confirmed inpatient coronavirus disease 2019 (COVID-19) cases. We analyzed the influence of timing and clinical severity on the diagnostic value of class-specific COVID-19 serology testing. Cross-sectional analysis revealed higher sensitivity and specificity at lower optical density cutoffs for IgA in hospitalized patients than for IgG and IgM serology (IgG area under the curve [AUC] of 0.91 [95% confidence interval {CI}, 0.89 to 0.93] versus IgA AUC of 0.97 [95% CI, 0.96 to 0.98] versus IgM AUC of 0.95 [95% CI, 0.92 to 0.97]). The enhanced performance of IgA serology was apparent in the first 2 weeks after symptom onset and the first week after PCR testing. In patients requiring intubation, all three tests exhibit enhanced sensitivity. Among PCR-negative patients under investigation for SARS-CoV-2 infection, 2 out of 61 showed clear evidence of seroconversion IgG, IgA, and IgM. Suspected false-positive results in the latter population were most frequently observed in IgG and IgM serology tests. Our findings suggest the potential utility of IgA serology in the acute setting and explore the benefits and limitations of class-specific serology as a complementary diagnostic tool to PCR for COVID-19 in the acute setting.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19), first emerged in late 2019 in a cluster of atypical pneumonia cases linked to a seafood and poultry market in Wuhan, China (1). SARS-CoV-2 is a betacoronavirus, related to a lineage of bat coronaviruses as well as the zoonotic SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV). The virus targets cells through interactions between the receptor binding domain (RBD) of its spike (S) protein and human angiotensin-converting enzyme 2 (ACE2) (1, 2) in the respiratory tract and other target organs, where infection and immune-mediated damage lead to local and systemic disease (3). Sequencing of the viral RNA genome (4) in the early days of the pandemic enabled the rapid development of reverse transcription-PCR (RT-PCR)-based nucleic acid tests (NATs) (5), which have been widely implemented for the diagnosis of acute SARS-CoV-2 infection (6). However, SARS-CoV-2 PCR tests have shown limitations in sensitivity for a variety of reasons, including variability in nasopharyngeal swab acquisition and processing and kinetics of the viral infection itself (7–9). Thus, the focus has shifted toward the need to develop and validate serological assays, which detect antibody responses elicited by both current and past exposure to the virus and may therefore serve as complementary approaches in diagnosing COVID-19, even following acute presentation of the disease. To this end, as the early pandemic evolved, the Centers for Disease and Control and Prevention (CDC) tasked Emory University with the goal of developing a serological platform to assess anti-SARS-CoV-2 antibodies. In addition to playing a possible complementary role to molecular approaches in the diagnosis of COVID-19, the CDC and Emory (in addition to other institutions) thought that a serological platform may serve additional purposes, including (i) seroepidemiological surveys, (ii) screening of donors for convalescent-phase plasma therapy, and (iii) assessment of vaccine immunogenicity. In addition, since the development of RBD-specific antibodies correlates strongly with in vitro neutralizing activity in hospitalized patients and the development and implementation of high-throughput neutralizing assays have remained challenging in convalescent-phase plasma donor centers, RBD-specific serology may provide some insight into the virus neutralization capacity when seeking to optimally select convalescent-phase plasma donors (10, 11).
Most clinical serological platforms for the detection of pathogen exposure or infection examine the reactivity of patient immunoglobulin M (IgM), IgG, or both against antigenic determinants of the pathogen; some also include direct detection of pathogen antigens. Serological tests for SARS-CoV-2 have largely been no different, with platforms described that test for virus-specific IgG, IgM, or pan-Ig. The rationale for this approach is understandable, as the serological responses to novel infectious organisms often result in an early IgM response followed by subsequent class switching to IgG. However, given the respiratory nature of the pathogen and the specific immune response predicted to form within respiratory mucosal tissues, examination of IgA SARS-CoV-2 antibodies may hold promise in the serological assessment of this disease. Dimeric IgA anti-SARS-CoV-2 antibodies have also recently been reported to exhibit an enhanced neutralization capacity compared to IgG antibodies (12), suggesting that evaluation of IgA in general may provide additional insight when selecting convalescent-phase plasma donors. It is now well established that the kinetics of IgG, IgM, and IgA responses differ among COVID-19 patients, with some reporting the unusual, early onset of an IgG response and persistence of IgM (13). Several recent studies likewise suggest that IgA responses may be useful in the evaluation of COVID-19 (12, 14–16), providing further evidence in support of IgA incorporation into serological diagnostic assays. However, additional data using longitudinal sampling are needed to accurately assess the class-specific responses and their clinical correlates. This is especially important when considering that while patients can present with a variety of symptoms, symptom onset itself can provide physicians with useful information when considering different diagnostic approaches. Such studies will refine the ability of serology in general, in addition to the performance of individual antibody classes, to aid in the diagnosis of COVID-19.
To balance the throughput needs of a clinical diagnostics laboratory with the value of a semiquantitative platform, we developed single-dilution enzyme-linked immunosorbent assay (ELISA)-based screening assays to detect IgG, IgA, or IgM specific for the RBD of SARS-CoV-2 spike (S). We then validated and compared these tests using samples collected from PCR-confirmed COVID-19 patients and prepandemic samples from healthy blood donors and patients being screened for other viral infections or HLA antibodies. Early development and validation data were submitted to the Food and Drug Administration and resulted in emergency use authorization (EUA) approval. Using receiver operating characteristic (ROC) analysis, we found that the IgA serology assay exhibited favorable performance characteristics, especially within the first 2 weeks after symptom onset.
MATERIALS AND METHODS
Sample collection and processing.
Hospitalized patients diagnosed with or under investigation for COVID-19 who were seen in the emergency department and/or admitted at Emory University Hospital and Emory University Hospital Midtown from 9 March 2020 to 15 May 2020 were identified by SARS-CoV-2 PCR testing records. In-house-validated RT-quantitative PCR (qRT-PCR) PCR results were obtained from the medical records of each admitting institution. Nasopharyngeal swabs were collected by the admitting medical team according to standard hospital procedures for each hospital. Residual serum and heparinized plasma samples from fully resulted clinical laboratory tests were identified and set aside as “discarded tissue” samples in accordance with clinical laboratory director approval. Residual samples were aliquoted by research staff and stored at −80°C prior to research use. The sample cohort utilized in this study partially overlapped the smaller cohort utilized for the separate, previously reported clinical IgG ELISA performed in the Emory clinical laboratory (10).
Chart review.
A retrospective chart review of patients in the study cohort was initially performed by Emory medical students and clinical staff who were in at least year 3 of medical doctorate training or currently hold a medical doctorate or an equivalent degree, followed by symptom onset adjudication by at least 2 independent physicians. Reviewers were blind to the ELISA results at the time of chart review. Patient information and clinical course details were entered into a REDCap database.
For analysis of disease severity at presentation, four categories were utilized based on COVID-19-specific severity categories developed by the National Health Commission of China and reported in multiple previous studies (17). These categories were as follows:
-
(1)
mild (mild clinical symptoms and no pulmonary changes on imaging),
-
(2)
moderate (fever and signs of respiratory infection/pneumonia changes upon imaging),
-
(3)
severe (at least one of the following: respiratory rate of ≥30 breaths/min, oxygen saturation of ≤93% under resting conditions, and arterial partial pressure of oxygen [PaO2]/oxygen concentration [FiO2] of ≤300 mm Hg [1 mm Hg = 0.133 kPa]), and
-
(4)
critical (respiratory failure requiring mechanical ventilation, shock, multiple-organ dysfunction/failure, and requiring ICU [intensive care unit] admission).
A retrospective chart review of symptom onset dates was performed using defined criteria. At least one of the following symptoms must have been reported as a new symptom or a significant change from the patient’s baseline to be considered for symptom onset: cough, shortness of breath or difficulty breathing, fever (including subjective fever), chills, muscle pain, headache, sore throat, loss of taste or smell, rash, or diarrhea. Symptom onset dates were considered valid for this study if a patient-reported exact date or a date within approximately ±2 days could be determined with reasonable clinical confidence. For consistency, all symptom onset data entered into the REDCap database were rechecked by one of two reviewers holding medical doctorates (M. Horwath and H. Nakahara), with the determination of some equivocal dates resolved by consensus.
Coronavirus spike and RBD enzyme-linked immunosorbent assays.
The purified recombinant 6× RBD from SARS-CoV-2, Wuhan-Hu-1 (GenBank GenPept accession number QHD43416), was kindly provided to the Emory Medical Lab (EML) by Jens Wrammert of the Emory Department of Pediatrics and Vaccine Center (purified as described in reference 10). A research protocol from the Wrammert group was used as a starting point in the development of the EML assays. HKU1 and OC43 recombinant S1 domains were obtained from the Centers for Disease Control and Prevention. RBDs from alphacoronaviruses 229E and NL63 were obtained from Sino Biological. Briefly, high-binding ELISA plates were coated with coronavirus RBD or S1 proteins at 1 μg/ml in phosphate-buffered saline (PBS) at 4°C overnight or at 37°C for 1 h. Plates were then washed three times with 0.5% PBS-Tween (PBST) and blocked for 30 min at room temperature (RT) in ELISA buffer (1% bovine serum albumin [BSA] and 0.2% Tween 20 in PBS). Plates were then tapped out after blocking, and serum or plasma samples were prediluted at 1:20 in ELISA buffer before addition to the test plate at final dilutions of 1:200 for the IgG assays and 1:100 for the IgA and IgM assays. Samples were incubated at room temperature for 30 min and washed three times in 0.5% PBST. Horseradish peroxidase (HRP)-conjugated anti-human IgG (catalog number 62-8420; Invitrogen), IgA (catalog number 2050-05; SouthernBiotech), and IgM (catalog number 31415; Invitrogen) were used for detection. The specificity of each conjugate was tested using IgG, IgM, and IgA purified from human serum (catalog numbers I2511, I8260, and I4036; Sigma) immobilized on high-binding plates (see Fig. S1A to C in the supplemental material). Conjugate and sample dilutions were selected to minimize signal loss while avoiding high overall background signals in prepandemic negative samples. SigmaFast OPD was used for development according to the manufacturer’s instructions, and reactions were stopped using 1 N HCl before reading on a BioTek Synergy plate reader at a wavelength of 492 nm.
Statistics.
ROC analysis was performed using Prism 8 (GraphPad). Areas under the curve (AUCs) were compared by generating Z-scores using the following formula:
To calculate a two-tailed P value, we used the above-described Z-scores for each comparison in the normal distribution function [NORMSDIST(Z)] of Microsoft Excel. Statistical comparisons of the means for multiple groups were done using one-way analysis of variance (ANOVA) with correction by the Tukey test of P values for multiple comparisons.
Study approval and ethical statement.
Serum and plasma samples from patients diagnosed with SARS-CoV-2 infection by PCR or under suspicion for COVID-19 (PCR tested with a negative result) were collected in Atlanta, GA, at Emory University Hospital and Emory University Hospital Midtown. Collection, processing, and storage of these samples were approved under a waiver for the use of discarded samples by the University Institutional Review Board (IRB) (approval number 00022371).
RESULTS
Characteristics of PCR-confirmed and PCR-negative cohorts.
Between March and May 2020, we evaluated longitudinal samples from patients under investigation for COVID-19 at two Emory Healthcare-affiliated hospitals: Emory University Hospital and Emory University Hospital Midtown. A total of 139 individuals were tested for SARS-CoV-2 by PCR; 78 patients who tested positive and 61 who tested negative were included in this study. Compiled data for each group from a retrospective chart review are summarized in Table 1. On average, PCR-confirmed cases were older (64.3 versus 59.8 years) and more likely to be African American (79.5% versus 63.9%). Clinically, PCR-confirmed COVID-19 cases in this cohort were more likely to present with moderate (57.7% versus 41.0%) or severe (11.5% versus 0%) signs and symptoms. Cases were also more likely to require intubation (46.2% versus 18.0%) and exhibited higher mortality rates (19.2% versus 3.3%). Overviews of each patient and summarized testing results from PCR-positive and -negative cohorts are shown in Fig. S4A and B, respectively, in the supplemental material.
TABLE 1.
Patient characteristics
| Parameter | Value for group |
|
|---|---|---|
| SARS-CoV-2 PCR positive (n = 78) | SARS-CoV-2 PCR negative (n = 61) | |
| Demographics | ||
| Mean age (yrs) (range) | 64.3 (22–100) | 59.8 (20–97) |
| No. (%) of female patients | 29 (47.5) | 33 (42.3) |
| No. (%) of male patients | 32 (52.5) | 45 (57.7) |
| No. (%) of patients of race | ||
| African-American or black | 61 (78.2) | 40 (65.6) |
| Asian | 0 (0.0) | 1 (1.6) |
| Caucasian or white | 15 (19.2) | 20 (32.8) |
| Unknown, unavailable, or unreported | 2 (2.6) | 0 (0.0) |
| No. (%) of patients of ethnicity | ||
| Hispanic or Latino | 0 (0.0) | 2 (3.3) |
| Non-Hispanic or non-Latino | 73 (93.6) | 56 (91.8) |
| Unknown or unavailable | 5 (6.4) | 3 (4.9) |
| No. (%) of patients with severity score at presentationa | ||
| 1 (mild) | 15 (19.2) | 19 (31.1) |
| 2 (moderate) | 45 (57.7) | 25 (41.0) |
| 3 (severe) | 9 (11.5) | 0 (0.0) |
| 4 (critical) | 9 (11.5) | 17 (27.9) |
| Clinical course | ||
| No. (%) of patients with intensive care unit admission | 43 (55.1) | 31 (50.8) |
| No. (%) of patients with intubation | 36 (46.2) | 11 (18.0) |
| Mean length of hospital stay (days) (range)b | 10.2 (0–39) | 17.4 (1–48) |
| No. (%) of patients with discharge status | ||
| Discharge to home | 46 (59.0) | 48 (78.7) |
| Transfer to another facility | 14 (17.9) | 6 (9.8) |
| Transfer to hospice | 1 (1.3) | 2 (3.3) |
| Deceased | 15 (19.2) | 2 (3.3) |
| Other or still in hospital | 2 (2.6) | 3 (4.9) |
| Sample set characteristics | ||
| No. (%) of patients with symptom start date available in chart | 54 (69.2) | 39 (63.9) |
| Mean no. of study samples per patient (range) | 6.5 (1–33) | 5.1 (1–16) |
| Mean no. of days from symptoms to 1st study sample (range)c | 9.4 (1–25) | 5.3 (−2–18) |
| Mean no. of days from PCR test to 1st study sample (range)d | 3.6 (−1–19) | 1.9 (−3–14) |
See Materials and Methods for severity criteria.
Calculated for patients discharged at the time of chart review.
Calculated for patients with an available symptom start date.
Calculated by the earliest positive SARS-CoV-2 PCR test or the earliest negative test for PCR-negative patients.
Cross-sectional comparison of IgG, IgA, and IgM serology assays in hospitalized COVID-19 patients.
We evaluated three enzyme-linked immunosorbent screening assays, developed and characterized in-house (Fig. S1), for the detection of SARS-CoV-2 receptor binding domain (RBD)-specific immunoglobulin G (IgG), IgA, and IgM in 508 samples from 78 PCR-confirmed COVID-19 cases. A total of 131 prepandemic controls, including blood donors and patients being screened for HLA antibodies and antibodies against other viruses, were used as true-negative cases in this analysis. In addition to optimizing overall assay conditions (Fig. S1) and given concerns regarding features of the assay that may limit specificity (a common challenge with serological assays), we also assessed several commercially available secondary antibodies for possible differences in background reactivity. This approach failed to reveal significant differences in background reactivity between distinct secondaries (Fig. S2), allowing us to select conjugated secondary anti-human immunoglobulin reagents based on availability and cost-effectiveness.
To test for potential cross-reactivity in the IgG and IgA assays with common coronavirus strains, we measured reactivity with purified S1 domains from two human betacoronaviruses that cause human cold (OC43 and HKU1) in a subset of samples from our overall analysis. The latter analysis revealed little correlation (Fig. S3K) in reactivity by optical density (OD) values of the SARS-CoV-2 RBD ELISAs for IgG and IgA with the common cold coronavirus protein (Fig. S3C to E), consistent with the polymorphic nature of the SARS-CoV-2 RBD (Fig. S3A and B). We repeated the latter analyses using recombinant RBDs from two endemic alphacoronaviruses, 229E and NL63, finding a similar lack of a strong correlation (Fig. S3G to K).
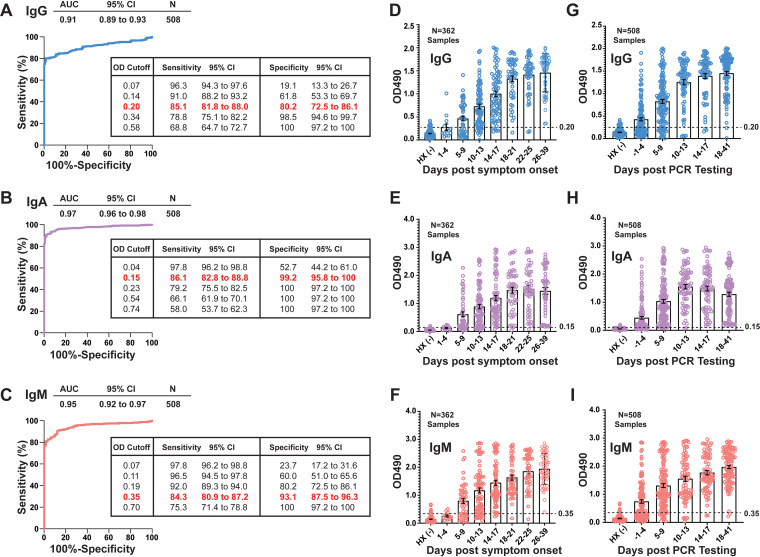
Assay performance was determined in the overall COVID-19 PCR-positive cohort using receiver operating characteristic (ROC) analysis. In a ROC analysis, the area under the curve (AUC) for each assay inversely correlates with rates of false positivity and negativity at increasing OD value cutoffs. In the overall sample set, the IgA assay exhibited significantly improved characteristics compared to the IgM and IgG assays (Fig. 1A to C) (AUC of 0.97 versus 0.91 for IgG and 0.95 for IgM [P < 0.0001 for IgA versus IgG, and P = 0.01 for IgA versus IgM]). This finding was partly due to higher levels of false positivity in the IgG and IgM assays than in the IgA assay over a range of cutoff values (Fig. S4). For continuous variables like OD values, the output of a ROC analysis aids in the selection of optimal cutoffs, from which clinical laboratories can determine and report an alpha response (positive or negative) with corresponding sensitivity and specificity at the chosen cutoff. The inset tables in Fig. 1A to C illustrate the trade-offs involved in selecting such a cutoff. For this analysis, we selected OD cutoffs with the goal of maintaining a sensitivity above 80% at an optimal specificity for each assay (0.2 for IgG, 0.15 for IgA, and 0.35 for IgM). At an OD cutoff of 0.2, our IgG serology assay achieved a sensitivity of 85.10% (95% confidence interval [CI], 81.75% to 87.93%) and a specificity of 80.15% (95% CI, 72.51% to 86.08%). At an OD cutoff of >0.15, the IgA assay achieved a sensitivity of 87.25% (95% CI, 84.08% to 89.87%) and a specificity of 99.24% (95% CI, 95.80% to 99.96%). At an OD cutoff of >0.35, the IgM assay reached a sensitivity of 84.31% (95% CI, 80.90% to 87.21%) and a specificity of 93.89% (95% CI, 88.41% to 96.87%).
FIG 1.
Evaluation of class-specific SARS-CoV-2 serology assay performance. (A to C) Receiver operating characteristic (ROC) analyses of RBD-specific IgG, IgA, and IgM serology in serum and plasma samples from a cohort of hospitalized patients with PCR-confirmed SARS-CoV-2 infection (n = 508 samples from 78 individuals). Areas under the curve (AUCs), correlative with overall assay performance, are shown with 95% confidence intervals (CIs). Inset tables indicate sensitivities and specificities at various OD cutoffs, with the selected cutoff for each assay highlighted in red. (D to F) IgG, IgA, and IgM OD values binned and plotted by timing after symptom onset (n = 362 samples from 54 individuals). OD values from 131 prepandemic serum and plasma samples, which served as negative historical controls (HX −) in these analyses, are plotted to the left of each time series. (G to I) OD values plotted as described above for panels D to F but instead binning samples using time after PCR testing (n = 508 samples from 78 individuals). OD cutoffs are indicated by a dashed line for each assay.
Because class-specific antibody responses depend on the onset, magnitude, and duration of the antiviral immune response, we hypothesized that the diagnostic performances of our IgG, IgA, and IgM serology assays would change with time after symptom onset. Similar to recent reports (10, 18, 19), improved performance was observed when serology was performed more than 7 days after PCR testing (10). To address the question of timing in our cohort, we conducted a systematic chart review and estimated the date of symptom onset and time to serology for a subset of samples for which this information was available (n = 362 samples from 54 patients). By binning these samples into 4-day increments after symptom onset (Fig. 1D to F) or PCR testing (Fig. 1G to I), it was clear that the average OD value in each assay increased over time for each class.
Serology performance improves with time after symptom onset and PCR testing.
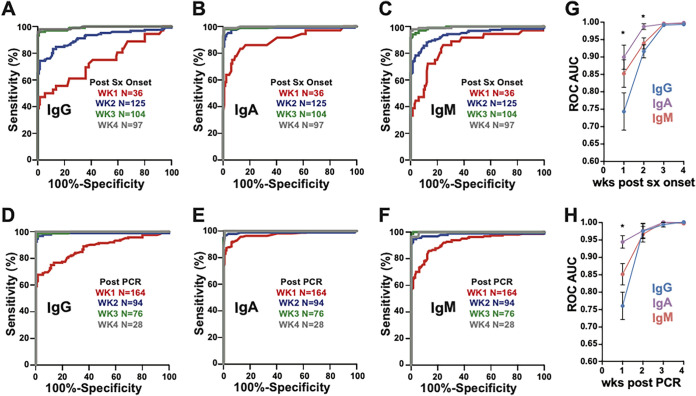
Based on the observation in the overall cohort that the OD increased with time after symptom onset and PCR testing, we repeated ROC analyses by binning samples by week after symptom onset (Fig. 2A to C) or by time after PCR testing (Fig. 2D to F). For all three assays, performance improved significantly over time, achieving AUCs of >0.99 at 3 or more weeks after onset or 2 or more weeks after PCR testing. However, IgA exhibited superior performance in the first and second weeks after symptom onset compared to IgG serology (week 1 AUC of 0.90 versus 0.74 [P = 0.01] and week 2 AUC of 0.99 versus 0.92 [P = 0.0005]). Together, these data corroborate observations for this and other viral infections that time after symptom onset correlates strongly with serology assay performance. This furthermore demonstrates that the overall performance advantage of IgA testing observed in Fig. 1 is likely due to superior detection of true-positive samples early in the clinical course.
FIG 2.
Performance of class-specific SARS-CoV-2 serology testing increases over time. (A to F) ROC analysis of antibody class-specific serology with samples binned into weeks after symptom (sx) onset (A to C) or time after PCR (D to F). (G and H) Areas under the curve (AUCs) with standard errors (SE) plotted for these analyses for weeks after symptom onset (G) and weeks after PCR testing (H). Statistical significance was determined by using Student’s t tests on Z-scores of AUC and SE values for each ROC curve (* indicates a P value of <0.05). The IgA serology test performed significantly better than IgG serology in samples collected within 1 week of symptom onset (AUC of 0.90 versus 0.74 [P = 0.01]). In addition, IgA (AUC of 0.99) performed significantly better than IgG (AUC of 0.92) in the second week following symptom onset (P = 0.0005). All of the tests exhibited superior performance (AUC of >0.99) in samples collected 3 or >4 weeks after symptom onset or >2 weeks after PCR testing.
Longitudinal analysis of combinatorial and individual class-specific serology results.
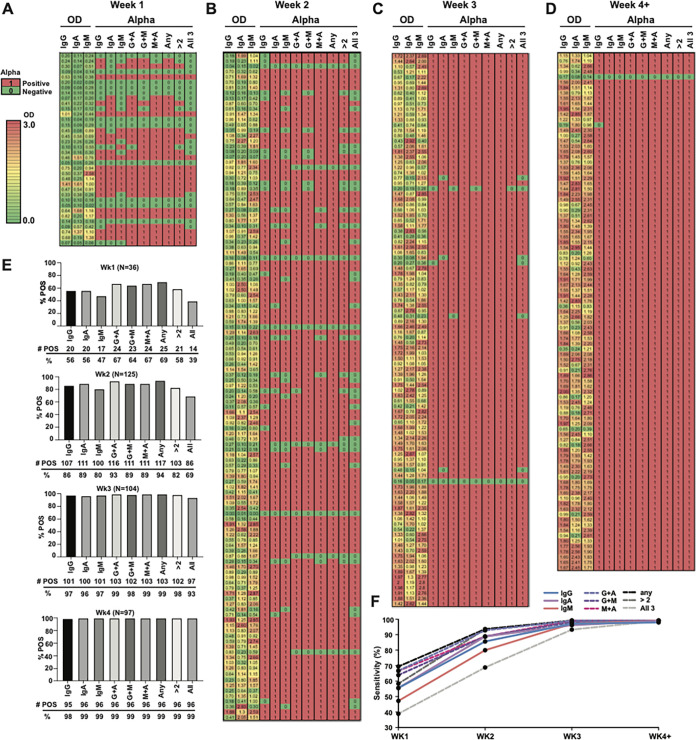
To visualize the trends observed in Fig. 1 and 2 and assess the effect of combining class-specific serology results, we generated heat maps of OD values and alpha responses for samples collected 1, 2, 3, or ≥4 weeks after symptom onset from patients who tested positive for SARS-CoV-2 by PCR and had reliable, adjudicated symptom onset data after chart review (n = 54) (Fig. 3A to D). Figure 3E shows the corresponding percentages of samples in each week after symptom onset that tested positive by individual or different combinations of assays (i.e., IgG plus IgA [G+A], G+M, or M+A [any combination of 2 or all 3]). Consistent with our findings in Fig. 2, sensitivities were high and comparable using any combination of testing more than 2 weeks after symptom onset (Fig. 3E and F). Some advantage in sensitivity was observed for combinations of G+A testing or the use of any positive result in the first 2 weeks after symptom onset. However, the improved sensitivity must be weighed against the combined loss of specificity resulting from combining IgA serology with less specific IgG or IgM (see insets in Fig. 1A to C for specificity comparisons at the chosen cutoff).
FIG 3.
Sensitivity of antibody class-specific SARS-CoV-2 serology increases over time after symptom onset with an associated rise in the OD reading and alpha response. (A to D) OD values of 362 samples from 54 individuals for whom reliable symptom onset information could be obtained by chart review divided by week after symptom onset into heat maps. The far left of each heat map shows the OD value result for each serology test performed. To the right is the alpha response for each test using the OD cutoffs for each assay determined in Fig. 1 (0.2 for IgG, 0.15 for IgA, and 0.35 for IgM), followed by different combinations of testing (G+A, G+M, M+A, any, >2 tests, or all tests). (E) Percent positivity of the samples plotted for each of the weekly heat maps and for each individual test or testing combination. (F) Sensitivity of each individual test or testing combination plotted over weeks after symptom onset. The specificity for each assay was determined by testing 131 historical negative samples and is listed for each cutoff in Fig. 1A to C.
Analysis of IgG, IgA, and IgM OD values in individuals with longitudinal sampling over time.
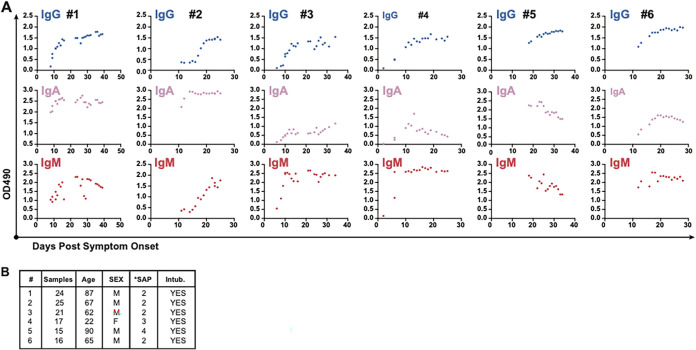
While determining titers with known standards is the preferred method for quantifying serological responses in ELISA-based assays, ELISA OD values correlate with the level of the antibody being detected within the linear range of the test. We therefore plotted individual OD responses over time after symptom onset for 6 PCR-positive patients for whom nearly daily sampling was available in our data set (Fig. 4). Overall, the OD values increased with time within individuals, suggesting that the trends observed in Fig. 1 are at least partly due to the evolution of individual immune responses over time. In addition, patterns of IgG, IgA, and IgM were not always correlated in individuals. In patient 1, for instance, a low level of increasing IgG is observed alongside stable high IgA levels and a parabolic IgM response. Patient 2 exhibits what appears to be a late IgM response with a simultaneous sigmoidal increase in the IgG signal, again with a stable and high IgA signal. Patients 3 and 4 both exhibit a lower-level IgA signal with robust and early IgM occurring prior to a sigmoidal rise in the IgG signal. Patients 5 and 6 did not have samples available earlier than day 10 after symptom onset, but both patients showed declining IgM and IgA with a stable rise in the IgG signal between 20 and 30 days after onset. While limitations due to assay variability and other patient factors may influence the level of antibody detection at different time points in distinct patients, overall, these data suggest that unique, class-specific patterns likely contribute to the overall performance of serology tests over time.
FIG 4.
Individual antibody responses by OD value over time after symptom onset. (A) Individual OD values for SARS-CoV-2 RBD-specific IgG, IgA, and IgM plotted over time after symptom onset for 6 individuals for whom >10 simultaneously tested longitudinal samples were available. (B) Basic characteristics of each individual displayed in panel A. SAP, symptom severity at presentation.
Examination of assay performance in samples from patients requiring intubation.
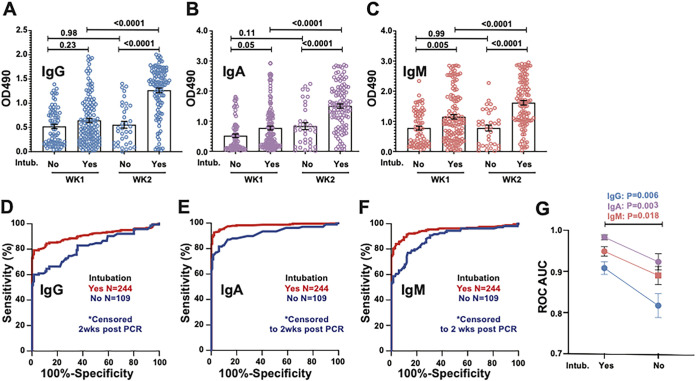
Given the variable serological responses observed among patients, we next compared OD values in samples collected from patients requiring intubation during their hospital course and investigated serology test performance in these samples relative to samples from patients who never required intubation. Because patients with more severe disease were more likely to have longer hospital stays and therefore more sampling from later in the clinical course (correlating with higher antibody levels), we censored the data in both groups to include only samples from the first 2 weeks after symptom onset. In patients requiring intubation, OD values for IgG, IgA, and IgM were higher in the second week than in the first week (P < 0.0001), whereas no significant increase was observed in samples from week 2 compared to week 1 for samples from patients who did not require intubation. Interestingly, IgA and IgM but not IgG OD values were higher in week 1 for samples from individuals who required intubation than for samples from those who did not (P = 0.05 and P = 0.005, respectively) (Fig. 5A to C). For all three assays, for samples collected during the first 2 weeks following symptom onset, ROC analysis revealed a higher overall performance for samples collected from those who required intubation than for samples from those who did not (statistical comparison summarized in Fig. 5G).
FIG 5.
Analysis of assay performance and OD values in samples from COVID-19 patients requiring intubation. (A to C) Plotted OD values binned by week after PCR testing from 244 samples from COVID-19 patients requiring intubation and 109 samples from patients who did not. To avoid sampling bias due to patients with more severe disease having longer hospital stays, only samples from within the first 2 weeks after PCR testing were included in this analysis. P values with correction for multiple comparisons from one-way analysis of variance (ANOVA) are displayed. (D to F) ROC analysis in the above-described samples binned by intubation status. (G) Statistical comparison of ROC areas under the curve (AUCs) by Student’s t test using Z-scores derived from AUC values and standard errors (SE).
Analysis of seroconversion among PCR-negative patients under high suspicion of SARS-CoV-2 infection.
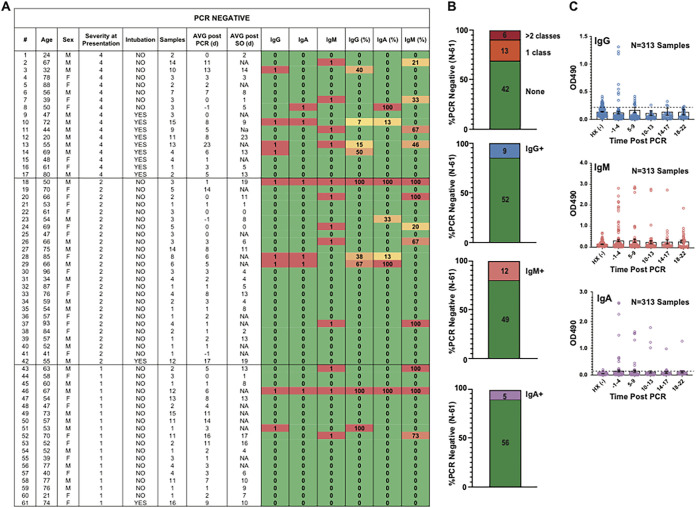
Due to concerns about the sensitivity of PCR testing for SARS-CoV-2 infection, particularly with respect to preanalytical variables that may significantly influence the likelihood of SARS-CoV-2 nucleic acid detection, we evaluated a cohort of 61 patients admitted to the hospital under high suspicion for COVID-19 for whom some degree of longitudinal sampling was available. Two PCR-negative individuals with longitudinal sampling showed clear evidence of seroconversion (sustained, high levels of SARS-CoV-2 RBD-specific IgG, IgA, and IgM). Rates of suspected false-positive results (low-OD-value positive results) were lowest for the IgA (n = 5/61) serology assay compared to the IgG (n = 9/61) and IgM (n = 12/61) assays (Fig. 6A to C). For suspected false-positive serology results, OD values were low and did not show the clear pattern of seroconversion seen in specimens collected from PCR-positive individuals.
FIG 6.
Analysis of seroconversion among PCR-negative patients under high suspicion of SARS-CoV-2 infection. A total of 313 samples from 61 patients under high suspicion for COVID-19 were tested for SARS-CoV-2 by PCR during the same time period as when the PCR-positive cohort was tested by all three in-house RBD serology assays. (A) PCR-negative cohort and description of sampling organized by severity score at presentation. Alpha responses (1, positive [red]; 0, negative [green]) for IgG, IgA, and IgM serology are shown along with the percentages of samples from a given individual that tested positive by a given assay (also heat mapped to red [high], orange/yellow [intermediate/low], or green [no positive results]). NA, not applicable. (B) Proportion of individuals who showed evidence of serological positivity by each assay. (C) OD values plotted over time, binned by time after PCR since very few of these patients provided reliable symptom onset (SO) data.
DISCUSSION
We report a comparative analysis of antibody-class-specific SARS-CoV-2 serology testing for the diagnosis of COVID-19 in the inpatient setting. Overall, the performance of serology testing improved with time after symptom onset and PCR testing in this population. Our results also suggest that IgA serology may provide added value when using serological tools to aid in the diagnosis of COVID-19 in hospitalized patients. Most COVID-19 patients show evidence of virus-neutralizing antibodies at 7 to 11 days postexposure or within the first 2 weeks of symptom onset (10). While the timing of seroconversion may seem to preclude the use of serology testing in the acute setting, severe COVID-19 typically presents in the second week after symptom onset (20), coinciding closely with the window in which diagnostic serology testing would be clinically useful, particularly in patients with a high pretest probability (PTP). It is reasonable, therefore, to consider and compare the utilities of antibody testing for the diagnosis of SARS-CoV-2 infection in the hospital setting. Serology testing in patients being admitted to the hospital for suspected SARS-CoV-2 infection may therefore complement PCR testing and improve the diagnostic capacity and confidence of health care systems and treating physicians.
The majority of tests that have been designed to examine seroreactivity with SARS-CoV-2 to date rely on IgM, IgG, or total Ig antibody levels (21). As IgA is the primary class of antibody produced during active mucosal infections (22), we elected to likewise examine IgA anti-SARS-CoV-2 antibodies when designing this clinical assay. Circulating IgA can come from multiple sources, including tissue-resident plasmablasts, bone marrow plasmablasts, and damaged mucosal tissues. Two recent studies of the humoral immune response during acute SARS-CoV-2 infection highlight the importance of the IgA response. Sterlin et al. show that the IgA responses occur shortly after symptom onset, peaking in the third week of infection and driven by the clonal expansion of IgA plasmablasts (16). Wang et al. further demonstrate the potential importance of IgA for protection in a study of convalescent patients, showing that dimeric virus-specific IgA more potently neutralizes SARS-CoV-2 than equivalent amounts of IgG (12). These findings suggest that the magnitude and nature of the IgA response are likely to contribute significantly to the long-term protection and potential efficacy of convalescent-phase plasma therapy.
The majority of commercial and laboratory-developed tests with EUA report sensitivities and specificities exceeding 95% (21) in samples collected from a relatively small number of patients or patients assessed >14 or sometimes >30 days after the onset of symptoms (23–29). As the timing of serological evaluation likely influences assay performance, evaluating samples acquired early in the disease may be important when considering the use of serological assays closer to disease presentation. To achieve this goal, we employed symptom onset data when analyzing assay performance. Given the changes in antibody levels that can occur over time, the relative timing of sample acquisition may be important when considering apparent differences in test characteristics. In addition, samples used to determine assay specificity often do not focus on prepandemic samples from patients under investigation for other infections but instead use samples obtained from healthy controls (30). As background serological reactivity can differ in distinct patient populations and this assay was intended in part for hospitalized patients, we focused our study on clinically relevant populations in our specificity analysis in an attempt to rigorously challenge assay performance. Using these measures, we sought to provide an accurate picture of the utility of class-specific serology in the acute setting.
As with any study, there are several important limitations that should be considered. Any ELISA-based test relies on the performance of the detection reagents employed, in this case HRP-conjugated anti-human Ig antibodies. In choosing reagents, as this test was rapidly implemented for clinical use, many practical and analytical variables were considered. First, the relative background signals of different secondary conjugates were evaluated and found to be similar. As a result, important considerations regarding the supply chain, cost-effectiveness, and storage requirements were employed to select appropriate secondary antibodies. This was especially important as supply chain interruptions had compromised PCR-based testing early in the pandemic. While little difference in background reactivity was observed among the secondaries evaluated, the secondary antibodies employed, the target antigen utilized, and other variables can certainly impact assay performance. Due to limitations in the volumes of samples collected that were coupled with symptom onset data, we were not able to assess different secondary agents, antigen targets, or other variables throughout the validation process. This was partly due to the need to rapidly implement a clinical test and submit the corresponding results to the FDA for EUA approval. Furthermore, as this assay was developed as the serological assay for Emory University Healthcare, implementation of a similar commercial platform was not undertaken, precluding a direct comparison of these results with those of other platforms using samples acquired in parallel. With respect to the antigen target, the RBD was chosen based on several key features, including the possibility of correlations with neutralizing activity, unique sequence features compared to those of other coronavirus RBDs that were predicted to reduce cross-reactivity, and practical considerations surrounding recombinant RBD production yields. Limitations in sample volume also unfortunately precluded the evaluation of other antigen targets, including the SARS-CoV-2 full spike protein or nucleocapsid. These antigens are common targets in other assays, and the use of these or other antigen targets could certainly influence assay performance. It should also be noted that sampling was less frequent over the clinical course for the PCR-negative population. This was partly due to the nature of our sample collection, which relied on collecting and aliquoting residual sample material from clinical laboratory tests. This lack of available samples, particularly later after symptom onset, may have resulted in an underreporting of serological positivity at later time points for PCR-negative individuals in the cohort. With any assay development, decisions regarding target antigens, detection agents, and overall assay configurations must be made in order to move forward toward assay validation and, ultimately, EUA approval. In different settings, a variety of factors likely influence decisions regarding these variables, as is evidenced by the wide variety of serological test configurations now available for COVID-19. In addition to these variables, the types of samples used to assess assay performance can also influence reported test characteristics. Importantly, as recent results appear to corroborate key findings of the present study (16), examination of IgA in the acute setting may indeed have clinical utility. However, as with the implementation of any new assay, additional studies will certainly be needed to determine whether similar findings are observed in distinct patient populations likewise under evaluation for COVID-19.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge with gratitude the patients who participated in our study. We also acknowledge the tireless efforts of medical technicians and clinical laboratory directors at Emory Hospital Midtown and Emory University Hospital, without whom these studies and the response to the pandemic in general would not be possible.
This work would not have been possible without resources provided by the CDC for the development and implementation of this assay.
REFERENCES
- 1.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv 2020.01.22.914952. https://www.biorxiv.org/content/10.1101/2020.01.22.914952v2.
- 5.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. 2020. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. 2020. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol 58:e00512-20. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winichakoon P, Chaiwarith R, Liwsrisakun C, Salee P, Goonna A, Limsukon A, Kaewpoowat Q. 2020. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J Clin Microbiol 58:e00297-20. doi: 10.1128/JCM.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel R, Babady E, Theel ES, Storch GA, Pinsky BA, George KS, Smith TC, Bertuzzi S. 2020. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: value of diagnostic testing for SARS-CoV-2/COVID-19. mBio 11:e00722-20. doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen H-L, Peiris M, Wu J. 2020. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, Vanderheiden A, Nyhoff L, Davis CW, Adekunle O, Affer M, Sherman M, Reynolds S, Verkerke HP, Alter DN, Guarner J, Bryksin J, Horwath MC, Arthur CM, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson EJ, Menachery VD, Rouphael N, Mehta AK, Stephens DS, Ahmed R, Roback JD, Wrammert J. 2020. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med 1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W, Limbo O, Smith C, Song G, Woehl J, Yang L, Abbott RK, Callaghan S, Garcia E, Hurtado J, Parren M, Peng L, Ramirez S, Ricketts J, Ricciardi MJ, Rawlings SA, Wu NC, Yuan M, Smith DM, Nemazee D, Teijaro JR, Voss JE, Wilson IA, Andrabi R, Briney B, Landais E, Sok D, Jardine JG, Burton DR. 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Viant C, Gaebler C, Cipolla M, Hoffmann H-H, Oliveira TY, Oren DA, Ramos V, Nogueira L, Michailidis E, Robbiani DF, Gazumyan A, Rice CM, Hatziioannou T, Bieniasz PD, Caskey M, Nussenzweig MC. 7 December 2020. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci Transl Med doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, Liao P, Qiu J-F, Lin Y, Cai X-F, Wang D-Q, Hu Y, Ren J-H, Tang N, Xu Y-Y, Yu L-H, Mo Z, Gong F, Zhang X-L, Tian W-G, Hu L, Zhang X-X, Xiang J-L, Du H-X, Liu H-W, Lang C-H, Luo X-H, Wu S-B, Cui X-P, Zhou Z, Zhu M-M, Wang J, Xue C-J, Li X-F, Wang L, Li Z-J, Wang K, Niu C-C, Yang Q-J, Tang X-J, Zhang Y, Liu X-M, Li J-J, Zhang D-C, Zhang F, Liu P, Yuan J, Li Q, Hu J-L, Chen J, Huang A-L. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 14.Ma H, Zeng W, He H, Zhao D, Jiang D, Zhou P, Cheng L, Li Y, Ma X, Jin T. 2020. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol 17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlke C, Heidepriem J, Kobbe R, Santer R, Koch T, Fathi A, Ly ML, Schmiedel S, Seeberger PH, ID-UKE COVID-19 Study Group, Addo MM, Loeffler FF. 2020. Distinct early IgA profile may determine severity of COVID-19 symptoms: an immunological case series. medRxiv 2020.04.14.20059733. https://www.medrxiv.org/content/10.1101/2020.04.14.20059733v1.
- 16.Sterlin D, Mathian A, Miyara M, Mohr A, Anna F, Claër L, Quentric P, Fadlallah J, Devilliers H, Ghillani P, Gunn C, Hockett R, Mudumba S, Guihot A, Luyt C-E, Mayaux J, Beurton A, Fourati S, Bruel T, Schwartz O, Lacorte J-M, Yssel H, Parizot C, Dorgham K, Charneau P, Amoura Z, Gorochov G. 7 December 2020. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Chen X, Deng X, Chen Z, Gong H, Yan H, Wu Q, Shi H, Lai S, Ajelli M, Viboud C, Yu H. 2020. Disease burden and clinical severity of the first pandemic wave of COVID-19 in Wuhan, China. Nat Commun 11:5411. doi: 10.1038/s41467-020-19238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou H, Wang T, Zhang B, Luo Y, Mao L, Wang F, Wu S, Sun Z. 2020. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunol 9:e01136. doi: 10.1002/cti2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tré-Hardy M, Blairon L, Wilmet A, Beukinga I, Malonne H, Dogné J-M, Douxfils J. 2020. The role of serology for COVID-19 control: population, kinetics and test performance do matter. J Infect 81:e91–e92. doi: 10.1016/j.jinf.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berlin DA, Gulick RM, Martinez FJ. 2020. Severe Covid-19. N Engl J Med 383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 21.Center for Devices and Radiological Health. 2020. In vitro diagnostics EUAs. FDA, Silver Spring, MD. [Google Scholar]
- 22.Corthésy B. 2013. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol 4:185. doi: 10.3389/fimmu.2013.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brochot E, Demey B, Handala L, François C, Duverlie G, Castelain S. 2020. Comparison of different serological assays for SARS-CoV-2 in real life. J Clin Virol 130:104569. doi: 10.1016/j.jcv.2020.104569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, Adriano A, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Dittrich S, Emperador D, Hooft L, Leeflang MM, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group. 2020. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev 6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui L-P, Johnston JC, Lan Z, Law S, MacLean E, Trajman A, Menzies D, Benedetti A, Ahmad Khan F. 2020. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ 370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zainol Rashid Z, Othman SN, Abdul Samat MN, Ali UK, Wong KK. 2020. Diagnostic performance of COVID-19 serology assays. Malays J Pathol 42:13–21. [PubMed] [Google Scholar]
- 27.Prazuck T, Colin M, Giachè S, Gubavu C, Seve A, Rzepecki V, Chevereau-Choquet M, Kiani C, Rodot V, Lionnet E, Courtellemont L, Guinard J, Pialoux G, Hocqueloux L. 2020. Evaluation of performance of two SARS-CoV-2 rapid IgM-IgG combined antibody tests on capillary whole blood samples from the fingertip. PLoS One 15:e0237694. doi: 10.1371/journal.pone.0237694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R, Liu X, Yuan L, Han H, Shereen MA, Zhen J, Niu Z, Li D, Liu F, Wu K, Luo Z, Zhu C. 2020. Analysis of adjunctive serological detection to nucleic acid test for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection diagnosis. Int Immunopharmacol 86:106746. doi: 10.1016/j.intimp.2020.106746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paradiso AV, De Summa S, Loconsole D, Procacci V, Sallustio A, Centrone F, Silvestris N, Cafagna V, De Palma G, Tufaro A, Garrisi VM, Chironna M. 2020. Rapid serological assays and SARS-CoV-2 real-time polymerase chain reaction assays for the detection of SARS-CoV-2: comparative study. J Med Internet Res 22:e19152. doi: 10.2196/19152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rongqing Z, Li M, Song H, Chen J, Ren W, Feng Y, Gao GF, Song J, Peng Y, Su B, Guo X, Wang Y, Chen J, Li J, Sun H, Bai Z, Cao W, Zhu J, Zhang Q, Sun Y, Sun S, Mao X, Su J, Chen X, He A, Gao W, Jin R, Jiang Y, Sun L. 2020. Early detection of severe acute respiratory syndrome coronavirus 2 antibodies as a serologic marker of infection in patients with coronavirus disease 2019. Clin Infect Dis 71:2066–2072. doi: 10.1093/cid/ciaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.