Reports of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection have raised important questions about the strength and durability of the immune response to primary infection, which are key factors in predicting the course of the pandemic. Identifying reinfection requires detecting the virus at two different time points and using viral genomic data to distinguish reinfection from persistent viral carriage.
KEYWORDS: reinfection, COVID-19, SARS-CoV-2, viral immunity
ABSTRACT
Reports of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection have raised important questions about the strength and durability of the immune response to primary infection, which are key factors in predicting the course of the pandemic. Identifying reinfection requires detecting the virus at two different time points and using viral genomic data to distinguish reinfection from persistent viral carriage. This process is hindered by challenges of logistics and capacity, such as banking samples from primary infection and performing viral genome sequencing. These challenges may help to explain why very few cases have been described to date. In addition, reinfection may be a rare phenomenon, but detailed prospective studies are needed to rigorously assess its frequency. To provide context for future investigations of SARS-CoV-2 reinfection, we review 16 cases that have been published to date or are available in preprint. Reinfection occurred across demographic spectra and in patients whose initial infections were both asymptomatic/mild and moderate/severe. For cases in which severity could be compared between episodes, half of reinfections were less severe, raising the possibility of partial immune protection. Although many patients had a positive total immunoglobulin or IgG result at the time of reinfection, very little examination of their immune response was performed. Further work is needed to elucidate the frequency, determinants, and consequences of SARS-CoV-2 reinfection. Establishing the necessary frameworks for surveillance and investigation will rely heavily on clinical laboratories and clinical investigators, and we propose several considerations to guide the medical community in identifying and characterizing SARS-CoV-2 reinfections.
INTRODUCTION
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), has had profound effects not only on human health but also on the collective mental well-being, social fabric, and economy of communities across the globe. There have been more than 64 million cases and 1.4 million deaths globally as of 2 December 2020 (1). While a highly susceptible population is assumed to be a key factor responsible for the explosiveness of the pandemic, one of the main questions in predicting its course is how well and for how long the immune response to an initial SARS-CoV-2 infection protects from reinfection.
On 25 August, the first case of reinfection by a phylogenetically distinct variant of SARS-CoV-2 was reported in the medical literature (2) and was rapidly followed by additional cases across the globe (3–8). These cases have garnered considerable media and academic attention (9) because they indicate that infection by SARS-CoV-2 does not uniformly confer protective immunity to all individuals. They raise several critical questions. Is SARS-CoV-2 reinfection a widespread phenomenon or is it limited to a small number of individuals who may have immune deficits? Does reinfection indicate that the natural immune response to SARS-CoV-2 is too weak, too short, or too narrow to protect against subsequent exposure? What are the clinical consequences for patients who experience reinfection, and to what extent might reinfection contribute to forward transmission? Understanding the frequency, determinants, and consequences of SARS-CoV-2 reinfection is essential to predicting the course of the COVID-19 pandemic, gaining important insight into the pathophysiology of this new disease, and guiding ongoing vaccine development efforts. However, there are considerable logistic challenges to identifying reinfection cases. Here, we review emerging data and concepts regarding SARS-CoV-2 reinfection, highlight important knowledge gaps, and offer suggestions for future surveillance and investigation.
CHALLENGES IN DETECTING SARS-CoV-2 REINFECTION
Identification of SARS-CoV-2 reinfection currently relies upon molecular detection of the virus at two different time points, often with intervening negative tests, as well as viral genetic sequencing data to support reinfection rather than persistent viral carriage. Because of the limited availability of routine sequencing capabilities at hospital and public health laboratories, clinical and laboratory criteria must be used to prioritize suspected reinfection cases for detailed investigation. Recently, the Centers for Disease Control and Prevention (CDC) released a guidance protocol designed to support public health laboratory investigation into suspected SARS-CoV-2 reinfections (10). This guidance defines epidemiological criteria for suspected reinfections, as well as cycle threshold (CT) value cutoffs and sequencing parameters (Fig. 1). Specifically, investigative criteria include a positive real-time reverse transcription-PCR (RT-PCR) test more than 90 days after the initial test (with CT of <33) or a positive RT-PCR test more than 45 days after the initial test (with CT of <33) that is accompanied by compatible symptoms or epidemiological exposure.
FIG 1.
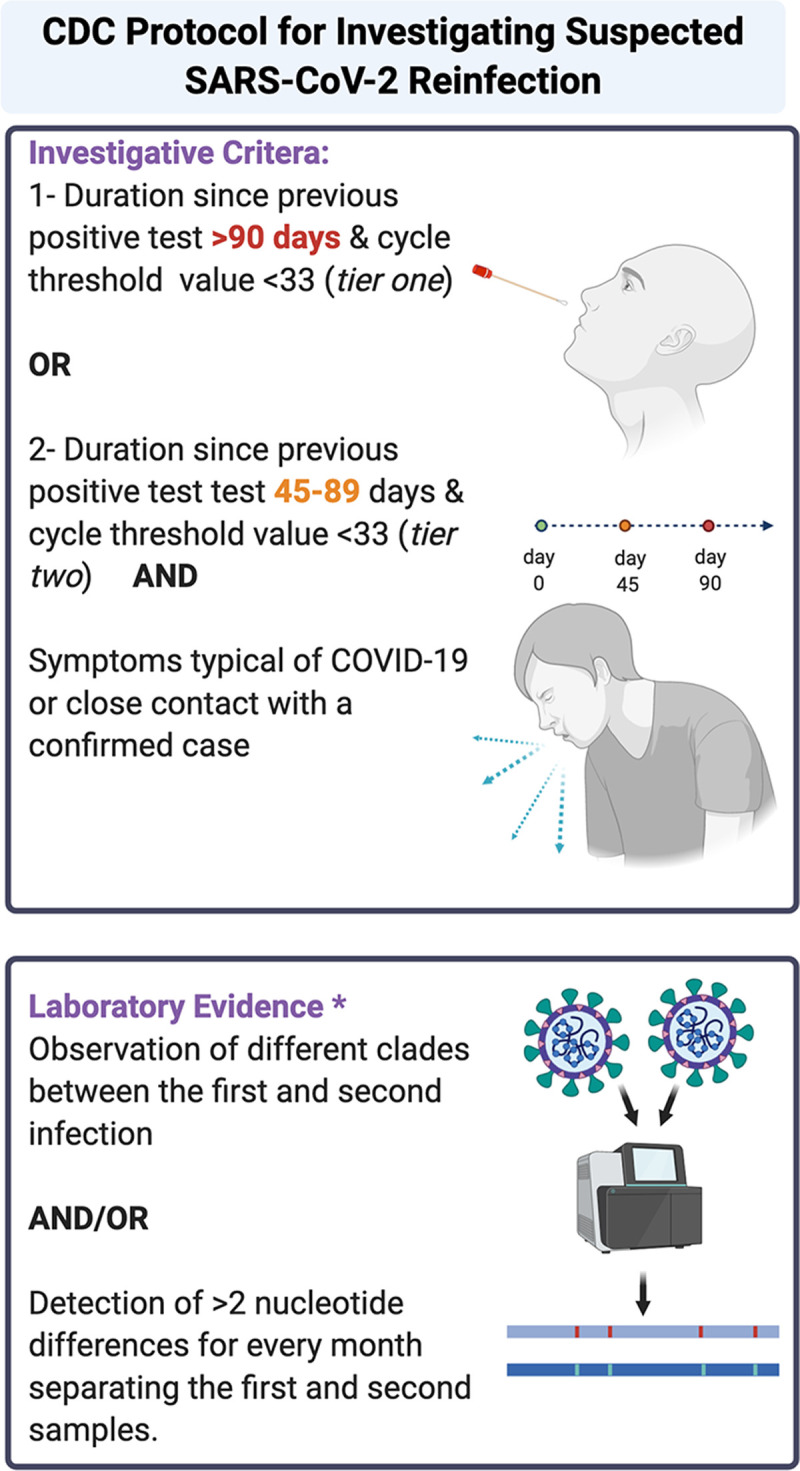
Centers for Diseases Control and Prevention investigation protocol for investigating suspected SARS-CoV-2 reinfection (10). Figure created using BioRender. * CDC also defined poor evidence but possible category as ≤2 nucleotide differences per month in consensus between sequences that meet quality metrics or >2 nucleotide differences per month in consensus between sequences that do not meet the quality metrics, ideally coupled with other evidence of actual infection (e.g., high viral titers in each sample or positive for subgenomic mRNA [sgmRNA] and culture). COVID-19, coronavirus disease 2019.
These guidelines help address one of the most important challenges in identifying reinfections, which is accounting for the fact that RT-PCR test positivity can persist for weeks following the resolution of clinical symptoms (11–13). A meta-analysis of 43 studies and 3,229 individuals (excluding case reports and case series with fewer than 5 patients) found the mean duration between first and last positive RT-PCR test to be 17 days, with a maximum duration of 83 days (14). Our experience in the Emory Healthcare system between 15 March 2020 and November 2020 is consistent with this. Of 22,443 unique patients who had at least two tests each (for a total of 51,134 tests), 456 patients had at least 2 positive tests. The median (interquartile range [IQR]) duration between first and last positive test was 19 days (12, 15), and durations of 45 and 90 days represented the 88th and 97th percentiles, respectively. Applying the CDC investigative criteria would thus identify 58 cases of potential reinfection in our system, a tractable number to study, assuming that all initial samples and clinical data are available for investigation. An important caveat to the investigative criteria is that they likely do not apply to immunocompromised individuals, who can have prolonged virus replication (16). In addition, imposing a cutoff CT of <33 may miss cases in which partial immune protection leads to lower viral loads during reinfection, though this cutoff is sensible in selecting cases for which viral genome sequencing is likely to be successful.
The second challenge addressed by CDC guidance is how to use viral genome sequencing to distinguish reinfection from within-patient virus evolution. Compared to many RNA viruses, SARS-CoV-2 has a relatively stable genome due to inherent proofreading activity by a 3′-to-5′ exoribonuclease (17). Because there is limited viral diversity, reinfection is considered confirmed when the viruses from the first and second infections are different enough to belong to different clades (18) or lineages (19) or when they differ by more than 2 substitutions per month, which is the general population-level viral substitution rate as assessed by multiple studies (10). This comparison is dependent on the availability of isolates from both the first and second infections, which can only be achieved through extensive biobanking during primary infection. Of note, these criteria may miss cases of reinfection by closely related viruses, which would have important implications for understanding natural immunity to SARS-CoV-2.
PUBLISHED CASES OF SARS-CoV-2 REINFECTION
To synthesize lessons from the cases of SARS-CoV-2 reinfection that have been described to date, we searched MEDLINE, EMBASE, and preprint servers (MedRxiv, BioRxiv, and SSRN) on 15 November 2020 for reports of SARS-CoV-2 reinfection, using keywords “reinfection,” “re-infection,” “SARS-CoV-2,” and “secondary infection.” We restricted our search to publications in English and limited our review to those confirmed by viral genome sequencing and analysis (Table 1). At the time of the search, there were 16 reported cases of reinfection confirmed by sequencing, 10 of which were in preprint (3, 4, 6–8, 20–24).
TABLE 1.
Features of reported SARS-CoV-2 reinfectionsa
| Authors (reference) | Country | Duration between infections (days or mo) | Initial infection severity (CT value) | Negative intermittent RT-PCR testing (day[s]) | Reinfection severity (CT value) | Genomic feature(s) |
Serology resultsb | |
|---|---|---|---|---|---|---|---|---|
| Infection | Reinfectionc | |||||||
| Tillett et al. (3) | US (NV) | 48 | Mild (35.2) | Yes (38) | Severe (35.3) | Clade 20C | Clade 20C; 7 SNVs compared to reference not seen in initial infection strain | Roche Elecsys anti-SARS-CoV-2 IgM/IgG positive on day 8 of reinfection |
| To et al. (2) | China (Hong Kong) | 142 | Mild (NA) | Yes (20) | Asymptomatic (26.7) | Clade 19A | Clade 20A | Abbott SARS-CoV-2 negative on day 1 of reinfection then positive on day 5 |
| Goldman et al. (8)d | US (WA) | 140 | Severe (26.5) | Yes (39, 40) | Severe (39.6) | Clade 19B | Clade 20A | RBD, spike and NC IgG, spike IgM, spike and NC IgA positive on day 14 of reinfection; nAbs detected on days 14 and 42 of reinfection |
| Gupta et al. (4) | ||||||||
| Case 1 | India | 108 | Asymptomatic (36) | Yes (8) | Asymptomatic (16.6) | NA | 9 SNVs compared to initial infection | NA |
| Case 2 | India | 111 | Asymptomatic (28.2) | Yes (10) | Asymptomatic (16.9) | NA | 10 SNVs compared to initial infection | NA |
| Larson et al. (7) | US (VA) | 64 | Moderate (NA) | NA | Severe (NA) | Partial genome obtainede | Lineage B.1.26; several potential variations and one high-confidence variation compared to initial infection, including D614G | Spike IgG positive on day 8 of reinfection |
| Van Elslande et al. (24) | Belgium | 3 mo | Moderate (25.6) | NA | Mild (32.6) | Lineage B.1.1 | Lineage A | Roche nucleocapsid IgG positive on day 7 of reinfection |
| Prado-Vivar et al. (21)d | Ecuador | 63 | Mild (36.9) | Yes (21) | Moderate (NA) | Clade 20A | Clade 19 B | IgM positive, IgG negative on day 7 of initial infection; IgM and IgG positive on day 28 of reinfection |
| Shastri et al. (22)d | ||||||||
| Case 1 | India | 66 | Mild (32) | Yes (4) | Mild (25) | Lineage B.1 | Lineage B | Abbott anti-NC IgG negative on day 5 of reinfection |
| Case 2 | India | 65 | Asymptomatic (33) | Yes (3) | Mild (36) | Lineage B.1.1 | Lineage B.1.1; 7 SNVs in initial strain compared to reference not present in reinfection strain, including D614G | Abbott NC IgG negative on day 7 of reinfection |
| Case 3 | India | 19 | Asymptomatic (36) | Yes (2) | Mild (21) | Lineage B.1.1 | Lineage B.1.1; 5 SNVs compared to reference not present in initial infection strain, including D614G | NA |
| Case 4 | India | 55 | Mild (32) | NA | Mild (17) | Lineage B.1.1 | Lineage B.1.1; 8 SNVs compared to reference not present in initial infection strain, including D614G | Roche NC total antibody negative on day 20 of reinfection |
| Abu-Raddad et al. (23)d | ||||||||
| Case 1 | Qatar | 46 | Asymptomatic/mild (36) | NA | Asymptomatic/mild (28) | NA | 9 SNVs compared to initial infection strain, including D614G | NA |
| Case 2 | Qatar | 71 | Asymptomatic/mild (17) | NA | Asymptomatic/mild (29) | NA | 11 SNVs compared to initial infection strain, including D614G | NA |
| Case 3 | Qatar | 88 | Asymptomatic/mild (36) | NA | Asymptomatic/mild (25) | NA | Partial genome obtainede; 3 SNVs compared to initial strain, including D614Ge | Roche Elecsys anti-SARS-CoV-2 negative at time of reinfection |
| Case 4 | Qatar | 55 | Asymptomatic/mild (30) | NA | Asymptomatic/mild (32) | Partial genome obtainede | 1 SNV compared to initial infection strain, including D614G | NA |
CT, cycle threshold; NA, not available; nAbs, neutralizing antibodies; NC, nucleocapsid; RBD, receptor binding domain; RT-PCR, reverse transcription-PCR; SNV, single nucleotide variant.
Serology results reported relative to the day of symptom onset if reported; if not reported or patient asymptomatic, then serology results relative to day of RT-PCR testing.
Preprint study.
One of the genomes reported was of low quality.
Demographic and clinical features of reinfection cases.
Reinfection occurred across demographic spectra; half of the patients (50% [8/16]) were between 20 and 30 years old. Gender was reported in 15 cases, among which 11 patients (73%) were male and four (27%) were female. Eight cases (50%) occurred among high-risk groups, including 7 health care workers (HCWs) (4, 7, 22) and 1 nursing home resident (8). While a publication and detection bias may exist for high-risk groups due to increased scrutiny and access to testing, these groups also have a higher burden of exposure for potential reinfection.
Notably, reinfection occurred among patients whose initial infections were both asymptomatic/mild (75% [9/12]) and moderate/severe (25% [3/12]) (25). The demonstration that moderate/severe initial infections do not necessarily provide enhanced protection against reinfection is important because patients with more severe infection have been found to have higher neutralizing antibody titers (26), which may be expected to confer protection.
Also of note, the severity of the reinfection episode itself was asymptomatic/mild in 12 cases (75%) and moderate/severe in 4 cases (25%). Among cases in which severity could be compared across episodes (n = 12), half of the patients had less-severe disease during the second infection. The observation that many reinfection cases were less severe than initial cases is interesting because it may suggest partial protection from disease and argues against antibody-dependent immune enhancement, which can be seen with other viral pathogens (27). In the absence of routine surveillance, we would have expected a bias toward detection of symptomatic reinfection, underscoring the importance of prospective screening. Ultimately, increased efforts toward detection and clinical characterization of reinfection will allow a better understanding of its clinical consequences, including the potential impact of repeat infection on long-term outcomes such as “long COVID” (28).
SARS-CoV-2 viral loads in reinfection cases.
The SARS-CoV-2 RT-PCR CT value is a metric that may not only help identify reinfection cases, but also provide information about their clinical and public health implications. CT value is dependent on sample type (29), severity of infection (30), date of collection relative to symptom onset (15), and assay and platform used (31) and hence may not always be comparable across episodes (32). However, a low or lower CT value, obtained in the same laboratory with the same method, may provide supporting evidence for reinfection versus persistent viral carriage. Among the 16 published reinfection cases, 14 reported SARS-CoV-2 RT-PCR CT values at the time of second infection. The median (range) CT value was 27.3 (16.0 to 39.6), which was similar to the median (range) CT value at initial infection, 32.5 (17.0 to 38.0).
Beyond a single CT measurement, serial testing during the initial phase of a suspected reinfection to assess the CT value trajectory may be informative. This approach was evaluated in a recent study of patients with primary infection, among whom a decreasing CT over 2 days was found to provide strong evidence of acute infection (33); a similar evaluation may distinguish reinfection from prolonged viral carriage. Another potentially useful test is the detection of subgenomic RNAs, which are transcripts generated during the viral life cycle as the templates for protein synthesis but which are not carried in the viral particle along with genomic RNA. In several studies, detection of subgenomic RNA has been adopted as a surrogate for active replication (34, 35); however, subgenomic RNA has also been detected late in the clinical course and correlated poorly with viral culture, perhaps due to persistence in cellular vesicles (36). If serial CT testing and/or subgenomic RNA detection prove to be useful markers of reinfection, they may allow detection of reinfections even when isolates from the primary infection are not available for comparative genome sequencing.
Assessing the CT value during reinfection may also provide information regarding the public health implications of infection. The ability to culture virus (which is itself an imperfect marker of infectiousness) has been linked to CT value, and most culture-positive samples have CT values in the mid-20s (37, 38). Among the 16 described reinfection cases, 8 had CT values of less than 28 and 6 had CT values of less than or equal to 25, suggesting they may have been infectious and a potential source of transmission (37). While viral culture was only attempted in one of the cases (7) to assess potential infectiousness, some information may be derived from a population level assessment of previously infected residents of Wuhan, China, in May 2020. Among 34,424 patients with a prior documented positive RT-PCR test, 107 tested positive again (after an unclear time interval). Although most of these samples likely reflected persistent test positivity, some may have been reinfections, and notably, virus culture was negative in all cases (39).
In the future, enhanced screening for reinfection will be facilitated by ongoing efforts to increase testing and diagnostic capacity and the availability of different platforms (40). A multitude of rapid antigen and real-time loop-mediated isothermal amplification (LAMP) tests are becoming increasingly available and should be integrated into reinfection surveillance algorithms given their anticipated widespread availability and their ability to capture those with the highest viral loads.
Genomic features of reinfection cases.
The current gold standard for identifying reinfection is detection of a distinct virus by genome sequencing. Detection of reinfection is most straightforward when viruses belong to a different clade (18) or lineage (19), as this provides clear evidence of infection by a different virus. Among 16 published reinfection cases, 5 (31%) had a different clade or lineage detected between initial infection and reinfection. Eight (50%) were infected with the same clade but had differences of >2 substitutions/month between them, compatible with CDC criteria. Three cases (19%) had low-quality genome sequences but were found to harbor different D614G alleles between the initial and reinfection strains and, therefore, were considered to represent reinfection.
Given the challenge of detecting reinfection by closely related viruses, it is important to conduct further studies characterizing the within-host evolution of SARS-CoV-2 to better understand the diversity expected over time (41, 42). In addition, although reinfection is most apparent when viruses are different enough to distinguish by genome sequencing, it remains unclear whether these viral genomic differences play a causative role in reinfection. That is, does reinfection occur when viral genomic differences permit escape from an existing, but narrow, immune response to the initial infection? Answering this question will require detailed mapping of the relationship between virus substitutions and immune escape (43).
Immune features of reinfection cases.
One of the most important questions about SARS-CoV-2 reinfections is whether they occur in the face of existing immune responses. Among the 16 described cases, the median (range) duration between the first and second infection was 66 (19 to 142) days, suggesting ample time for the development of neutralizing antibodies (44) and cellular immune responses (45). Ten cases reported results of serology testing at the time of the second infection, 6 of which had a positive total immunoglobulin (Ig) or IgG result. None of the patients had a known immunodeficient state. Beyond assessing IgG levels, very little examination of these patients’ immune responses has been performed. In one case, neutralizing antibody levels were measured at the time of the second infection and were comparable to those observed after boosted vaccination (8). Further investigation of immune parameters in patients who experience reinfection is critical to understanding its implications for the future of the pandemic (46).
SARS-CoV-2 IMMUNITY AND ITS ROLE IN REINFECTION
The relatively small amount of data currently available from reinfection cases must be considered in the context of what is known about SARS-CoV-2 immunity more broadly. Protection against reinfection by viral pathogens is largely mediated by adaptive immune memory, which has the long-term potential to maintain and reinforce pathogen-specific antibodies and effector cells (47). Adaptive immune responses to secondary antigen or pathogen exposures are more rapid and potent than primary responses and may substantially mitigate disease or prevent reinfection altogether, particularly via neutralizing or opsonizing antibodies (47, 48). Why this phenomenon is so highly effective and endures for decades for some pathogens (e.g., smallpox and measles) and is shorter lived for others (respiratory syncytial virus [RSV] and rotavirus) remains a fundamental question for immunologists and vaccinologists.
A growing body of literature describes features of the human immune response during asymptomatic, acute, and early convalescent SARS-CoV-2 infection. The vast majority of humans infected by SARS-CoV-2 generate virus-specific antibody responses, including neutralizing antibodies targeting the spike protein (in addition to other viral antigens). There is less population-level information on T cell responses, but several studies indicate SARS-CoV-2 infection consistently elicits CD8+ and CD4+ T cell responses (49, 50). Interestingly, up to 50% of people harbor preexisting SARS-CoV-2-reactive memory T cells (mostly CD4+ T cells) that have been primed via exposure to endemic CoVs (51). T cell immunity rarely if ever provides sterilizing immunity against infection or reinfection per se, but it can have beneficial effects, including more rapid viral clearance resulting in decreased disease severity or duration of infectiousness. Importantly, robust CD4+ T cells help may favor generation and maintenance of affinity-matured antibodies and memory B cell responses that mediate long-term protection. Finally, recent data indicate that SARS-CoV-2 infection may stimulate some innate immune signaling pathways differently or less strongly than other viral infections (52). It remains unclear what effect these early innate immune events will have on the quality and longevity of ensuing memory responses.
The immunologic determinants of protection against SARS-CoV-2 infection remain under investigation, but neutralizing antibodies are clearly the leading contender. Strong data from animal models indicate that the presence of neutralizing antibodies prevents infection and disease (such as lung pathology) and attenuates virus replication in airway epithelia (53, 54). Anecdotal evidence for protection from neutralizing antibodies was derived from an interesting natural experiment on a fishing vessel that suffered an outbreak with a very high attack rate (55). Three passengers known to have neutralizing antibodies to SARS-CoV-2 due to prior infection were spared, suggesting that neutralizing antibodies are very likely a key mediator of protective immunity to SARS-CoV-2. Samples for study of cellular immune responses were not available. Phase III vaccine studies will give a clearer picture of how neutralizing antibody levels correlate with protection in humans.
Despite evidence for protection from neutralizing antibodies, a major concern during the COVID-19 pandemic has been that protective immunity may be transient. This concern is largely driven by inconsistent findings regarding the duration of seropositivity. Some studies have emphasized “rapid (antibody) decay” (56), with large portions of a study population seroreverting within a few months. Others have found that antibody levels plateau (57) or are maintained at steady-state levels that are lower than initial peak responses (45). It is not clear to what extent these antibody trajectories will affect susceptibility to reinfection. Drawing inferences outside SARS-CoV-2 itself, the duration of protective immunity against seasonal CoVs ranges from a few months to a few years, with reinfections known to occur in that time frame. Detection of antibody responses to SARS and Middle East respiratory syndrome (MERS) also dissipates over approximately 3 to 5 years (58). Of note, animal CoVs are also known to cause reinfection, including in hosts with measurable antibodies (59). Collectively, this information suggests that it would not be surprising to find waning immunity and reversion to a SARS-CoV-2-susceptible state over months to years. To address this, it is critical to establish prospective studies that allow real-time capture of reinfection cases and intensive study of immunologic parameters before, during, and after the reinfection event. In addition, new tools measuring both humoral and cell-mediated immune responses are needed to support the detailed widespread testing necessary for defining the future susceptibility of individuals to SARS-CoV-2 reinfection (60).
CONCLUSION
Identifying and studying SARS-CoV-2 reinfections will provide critical clinical and public health information for addressing the COVID-19 pandemic. Current data from published reinfection cases and studies of the immune response after initial SARS-CoV-2 infection raise the possibility that reinfection may be common. Prospective studies, including extensive biobanking of samples from primary infection, are necessary to elucidate the full determinants and consequences of reinfection. Establishing these frameworks will rely heavily on clinical laboratories and clinical investigators. We propose several actionable steps for the medical community to consider in the effort to identify, characterize, and contain the impact of SARS-CoV-2 reinfections (Table 2).
TABLE 2.
Actionable suggestions for SARS-CoV-2 reinfection response
| Category | Recommendation |
|---|---|
| Case definition implementation | Implementation of screening for reinfection based on readily available data points (i.e., laboratory and epidemiological variables) according to CDC guidance. Case definition should be periodically reviewed and updated based on emerging data. |
| Establishment of surveillance frameworks | Based on case definitions, surveillance systems should be constructed for the identification of reinfections and for prospective follow-up. Extensive biobanking from primary infections is necessary to confirm reinfections (through viral genome sequencing) and fully characterize immune parameters. |
| Prospective follow-up reinfection cases | To determine the individual clinical burden and public health implications, reinfection cases should be followed prospectively. Predefined clinical endpoints should be measured as well as contact tracing of reinfection patients. |
| Evaluation of the immunologic and virologic determinants of reinfection | Understanding immune kinetics immediately prior to and following natural reexposure will expand our understanding of correlates of protection and provide guidance for vaccine development and administration. |
With positive results recently released from interim analyses of multiple phase III trials, continued study of reinfection cases as they relate to vaccine efficacy is of critical importance. For example, monitoring of patients for reinfection or postvaccination infection is necessary to assess whether viral escape mutations arise, requiring vaccine modification. This may be relatively simple to achieve given current vaccine constructs, such as mRNA vaccines, and proceed in a manner similar to the annual review and update of influenza vaccines. Ideally, studies of SARS-CoV-2 reinfection should be integrated into efforts to characterize vaccine-elicited immunity compared to that of natural infection, with the goal of developing safe vaccines and efficacious administration schedules that elicit robust and durable immune responses to curb the COVID-19 pandemic.
ACKNOWLEDGMENT
Ahmed Babiker received consulting fees from Arc Bio.
REFERENCES
- 1.Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. doi: 10.1016/s1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.To KK-W, Hung IF-N, Ip JD, Chu AW-H, Chan W-M, Tam AR, Fong CH-Y, Yuan S, Tsoi H-W, Ng AC-K, Lee LL-Y, Wan P, Tso EY-K, To W-K, Tsang DN-C, Chan K-H, Huang J-D, Kok K-H, Cheng VC-C, Yuen K-Y. 25 August 2020. Coronavirus disease 2019 (COVID-19) re-infection by a phylogenetically distinct severe acute respiratory syndrome coronavirus 2 strain confirmed by whole genome sequencing. Clin Infect Dis doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, Laverdure C, Verma SC, Rossetto CC, Jackson D, Farrell MJ, Van Hooser S, Pandori M. 2020. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis 21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta V, Bhoyar RC, Jain A, Srivastava S, Upadhayay R, Imran M, Jolly B, Divakar MK, Sharma D, Sehgal P, Ranjan G, Gupta R, Scaria V, Sivasubbu S. 23 September 2020. Asymptomatic reinfection in two healthcare workers from India with genetically distinct severe acute respiratory syndrome coronavirus 2. Clin Infect Dis doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suthar MS, Zimmerman M, Kauffman R, Mantus G, Linderman S, Vanderheiden A, Nyhoff L, Davis C, Adekunle S, Affer M, Sherman M, Reynolds S, Verkerke H, Alter DN, Guarner J, Bryksin J, Horwath M, Arthur C, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson E, Menachery V, Rouphael N, Metha A, Stephens DS, Ahmed R, Roback J, Wrammert J. 8 May 2020. Rapid generation of neutralizing antibody responses in COVID-19 patients. medRxiv doi: 10.1101/2020.05.03.20084442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, Laenen L, André E, Van Ranst M, Lagrou K, Maes P. 5 September 2020. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, Long KA, Potocki R, Smith DR, Lanteri C, Burgess T, Bishop-Lilly KA. 19 September 2020. a case of early re-infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis doi: 10.1093/cid/ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman JD, Wang K, Roltgen K, Nielsen SCA, Roach JC, Naccache SN, Yang F, Wirz OF, Yost KE, Lee J-Y, Chun K, Wrin T, Petropoulos CJ, Lee I, Fallen S, Manner PM, Wallick JA, Algren HA, Murray KM, Su Y, Hadlock J, Jeharajah J, Berrington WR, Pappas GP, Nyatsatsang ST, Greninger AL, Satpathy AT, Pauk JS, Boyd SD, Heath JR. 25 September 2020. Reinfection with SARS-CoV-2 and failure of humoral immunity: a case report. medRxiv doi: 10.1101/2020.09.22.20192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandavilli A. 13 October 2020. Coronavirus reinfections are real but very, very rare. (The New York Times.) https://www.nytimes.com/2020/10/13/health/coronavirus-reinfection.html.
- 10.Centers for Disease Control and Prevention. 2020. Common investigation protocol for investigating suspected SARS-CoV-2 reinfection. https://www.cdc.gov/coronavirus/2019-ncov/php/reinfection.html. Accessed 3 November 2020.
- 11.Carmo A, Pereira-Vaz J, Mota V, Mendes A, Morais C, da Silva AC, Camilo E, Pinto CS, Cunha E, Pereira J, Coucelo M, Martinho P, Correia L, Marques G, Araújo L, Rodrigues F. 2020. Clearance and persistence of SARS-CoV-2 RNA in patients with COVID-19. J Med Virol 92:2227–2231. doi: 10.1002/jmv.26103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou B, She J, Wang Y, Ma X. 2020. Duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis 71:2240–2242. doi: 10.1093/cid/ciaa451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao AT, Tong YX, Zhang S. 2020. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis 71:2249–2251. doi: 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. 2021. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babiker A, Bradley HL, Stittleburg VD, Ingersoll JM, Key A, Kraft CS, Waggoner JJ, Piantadosi A. 2020. Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. J Clin Microbiol 59:e02142-20. doi: 10.1128/JCM.02142-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TYT, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodcroft EH, Neher RA, Bedford T. 2020. Year-letter genetic clade naming for SARS-CoV-2 on Nextstain.org. https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming. Accessed 30 November 2020.
- 19.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To KK, Hung IF, Ip JD, Chu AW, Chan WM, Tam AR, Fong CH, Yuan S, Tsoi HW, Ng AC, Lee LL, Wan P, Tso E, To WK, Tsang D, Chan KH, Huang JD, Kok KH, Cheng VC, Yuen KY. 25 August 2020. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prado-Vivar BA, Monica B-W, Guadalupe JJ, Marquez S, Gutierrez B, Rojas-Silva P, Grunauer M, Trueba G, Barragan V, Cardenas P. 3 September 2020. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. SSRN doi: 10.2139/ssrn.3686174. [DOI]
- 22.Shastri JA, Swapneil P, Agarwal S, Chatterjee N, Pathak M, Sharma C, Kanakan A, Vivekanand A, Vasudevan JS, Maurya R, Fatihi S, Thukral L, Agrawal A, Pinto L, Pandey R, Sunil S. 21 September 2020. Whole genome sequencing confirmed SARS-CoV-2 reinfections among healthcare workers in India with increased severity in the second episode. SSRN doi: 10.2139/ssrn.3688220. [DOI]
- 23.Abu-Raddad LJ, Chemaitelly H, Malek JA, Ahmed AA, Mohamoud YA, Younuskunju S, Ayoub HH, Al Kanaani Z, Al Khal A, Al Kuwari E, Butt AA, Coyle P, Jeremijenko A, Kaleeckal AH, Latif AN, Shaik RM, Rahim HFA, Yassine HM, Al Kuwari MG, Al Romaihi HE, Al Thani SM, Bertollini R. 28 September 2020. Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. medRxiv doi: 10.1101/2020.08.24.20179457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, Laenen L, André E, Van Ranst M, Lagrou K, Maes P. 5 September 2020. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandhi RT, Lynch JB, Del Rio C. 2020. Mild or moderate COVID-19. N Engl J Med 383:1757–1766. doi: 10.1056/NEJMcp2009249. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Pan Z, Yue S, Yu F, Zhang J, Yang Y, Li R, Liu B, Yang X, Gao L, Li Z, Lin Y, Huang Q, Xu L, Tang J, Hu L, Zhao J, Liu P, Zhang G, Chen Y, Deng K, Ye L. 2020. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther 5:180. doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirado SM, Yoon KJ. 2003. Antibody-dependent enhancement of virus infection and disease. Viral Immunol 16:69–86. doi: 10.1089/088282403763635465. [DOI] [PubMed] [Google Scholar]
- 28.del Rio C, Collins LF, Malani P. 2020. Long-term health consequences of COVID-19. JAMA 324:1723–1724. doi: 10.1001/jama.2020.19719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. 2020. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Yan L-M, Wan L, Xiang T-X, Le A, Liu J-M, Peiris M, Poon LLM, Zhang W. 2020. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broder K, Babiker A, Myers C, White T, Jones H, Cardella J, Burd EM, Hill CE, Kraft CS. 2020. Test agreement between Roche Cobas 6800 and Cepheid GeneXpert Xpress SARS-CoV-2 assays at high cycle threshold ranges. J Clin Microbiol 58:e01187-20. doi: 10.1128/JCM.01187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhoads D, Peaper DR, She RC, Nolte FS, Wojewoda CM, Anderson NW, Pritt BS. 12 August 2020. College of American Pathologists (CAP) Microbiology Committee Perspective: caution must be used in interpreting the cycle threshold (Ct) value. Clin Infect Dis doi: 10.1093/cid/ciaa1199. [DOI] [PubMed] [Google Scholar]
- 33.Kissler SM, Fauver JR, Mack C, Tai C, Shiue KY, Kalinich CC, Jednak S, Ott IM, Vogels CBF, Wohlgemuth J, Weisberger J, DiFiori J, Anderson DJ, Mancell J, Ho DD, Grubaugh ND, Grad YH. 1 December 2020. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. medRxiv doi: 10.1101/2020.10.21.20217042. [DOI] [Google Scholar]
- 34.Perera R, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, Chin AWH, Chu DKW, Cheng SMS, Poon LLM, Chuang VWM, Peiris M. 2020. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 36.Alexandersen S, Chamings A, Bhatta TR. 2020. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat Commun 11:6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 2020. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveillance 25:2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao S, Gan Y, Wang C, Bachmann M, Wei S, Gong J, Huang Y, Wang T, Li L, Lu K, Jiang H, Gong Y, Xu H, Shen X, Tian Q, Lv C, Song F, Yin X, Lu Z. 2020. Post-lockdown SARS-CoV-2 nucleic acid screening in nearly ten million residents of Wuhan, China. Nat Commun 11:5917. doi: 10.1038/s41467-020-19802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babiker A, Myers CW, Hill CE, Guarner J. 2020. SARS-CoV-2 testing: trials and tribulations. Am J Clin Pathol 153:706–708. doi: 10.1093/ajcp/aqaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, Bachman MA, Wobus CE, Adams M, Washer L, Martin ET, Lauring AS. 2020. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 223:23–27. doi: 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ. 2020. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 183:1901.e9–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. 1 December 2020. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. bioRxiv doi: 10.1101/2020.11.30.405472. [DOI] [PMC free article] [PubMed]
- 44.Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, Vanderheiden A, Nyhoff L, Davis CW, Adekunle O, Affer M, Sherman M, Reynolds S, Verkerke HP, Alter DN, Guarner J, Bryksin J, Horwath MC, Arthur CM, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson EJ, Menachery VD, Rouphael N, Mehta AK, Stephens DS, Ahmed R, Roback JD, Wrammert J. 2020. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med 1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dan JM, Mateus J, Kato Y, Hastie KM, Faliti CE, Ramirez SI, Frazier A, Yu ED, Grifoni A, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S. 18 December 2020. Immunological memory to SARS-CoV-2 assessed for up to eight months after infection. bioRxiv doi: 10.1101/2020.11.15.383323. [DOI] [PMC free article] [PubMed]
- 46.Overbaugh J. 2020. Understanding protection from SARS-CoV-2 by studying reinfection. Nat Med 26:1680–1681. doi: 10.1038/s41591-020-1121-z. [DOI] [PubMed] [Google Scholar]
- 47.Ahmed R, Gray D. 1996. Immunological memory and protective immunity: understanding their relation. Science 272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 48.Amanna IJ, Slifka MK, Crotty S. 2006. Immunity and immunological memory following smallpox vaccination. Immunol Rev 211:320–337. doi: 10.1111/j.0105-2896.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 49.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, Dong C. 2020. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52:971.e3–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, Crotty S. 2020. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183:996.e19–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181:1489.e15–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mick E, Kamm J, Pisco AO, Ratnasiri K, Babik JM, Castañeda G, DeRisi JL, Detweiler AM, Hao SL, Kangelaris KN, Kumar GR, Li LM, Mann SA, Neff N, Prasad PA, Serpa PH, Shah SJ, Spottiswoode N, Tan M, Calfee CS, Christenson SA, Kistler A, Langelier C. 2020. Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses. Nat Commun 11:5854. doi: 10.1038/s41467-020-19587-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O'Connell S, Bock KW, Minai M, Nagata BM, Andersen H, Martinez DR, Noe AT, Douek N, Donaldson MM, Nji NN, Alvarado GS, Edwards DK, Flebbe DR, Lamb E, Doria-Rose NA, Lin BC, Louder MK, O'Dell S, Schmidt SD, Phung E, Chang LA, Yap C, Todd J-PM, Pessaint L, Van Ry A, Browne S, Greenhouse J, Putman-Taylor T, Strasbaugh A, Campbell T-A, Cook A, Dodson A, Steingrebe K, Shi W, Zhang Y, Abiona OM, Wang L, Pegu A, Yang ES, Leung K, Zhou T, Teng I-T, Widge A, et al. 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med 383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, Yang L, Abbott RK, Callaghan S, Garcia E, Hurtado J, Parren M, Peng L, Ramirez S, Ricketts J, Ricciardi MJ, Rawlings SA, Wu NC, Yuan M, Smith DM, Nemazee D, Teijaro JR, Voss JE, Wilson IA, Andrabi R, Briney B, Landais E, Sok D, Jardine JG, Burton DR. 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang M-L, Jerome KR, Bloom JD, Greninger AL. 2020. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol 58:e02107-20. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, Elliott J, Hofmann C, Hausner MA, Ferbas KG, Tobin NH, Aldrovandi GM, Yang OO. 2020. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild COVID-19. N Engl J Med 383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, Arnthorsson AO, Helgason D, Bjarnadottir K, Ingvarsson RF, Thorsteinsdottir B, Kristjansdottir S, Birgisdottir K, Kristinsdottir AM, Sigurdsson MI, Arnadottir GA, Ivarsdottir EV, Andresdottir M, Jonsson F, Agustsdottir AB, Berglund J, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Gottfredsson M, Gretarsdottir OS, Gudmundsdottir S, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Helgason A, Jensson BO, Jonasdottir A, Jonsson H, Kristjansson T, Kristinsson KG, Magnusdottir DN, Magnusson OT, Olafsdottir LB, Rognvaldsson S, Le Roux L, Sigmundsdottir G, Sigurdsson A, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Thordardottir M, Saemundsdottir J, et al. 2020. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poland GA, Ovsyannikova IG, Kennedy RB. 2020. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet 396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sariol A, Perlman S. 2020. Lessons for COVID-19 immunity from other coronavirus infections. Immunity 53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K. 2020. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol 58:e00797-20. doi: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]


