Diagnosis of COVID-19 by PCR offers high sensitivity, but the utility of detecting samples with high cycle threshold (CT) values remains controversial. Currently available rapid diagnostic tests (RDTs) for SARS-CoV-2 nucleocapsid antigens (Ag) have sensitivity well below PCR.
KEYWORDS: COVID-19, SARS-CoV-2, diagnostic, nucleoprotein, antigen
ABSTRACT
Diagnosis of COVID-19 by PCR offers high sensitivity, but the utility of detecting samples with high cycle threshold (CT) values remains controversial. Currently available rapid diagnostic tests (RDTs) for SARS-CoV-2 nucleocapsid antigens (Ag) have sensitivity well below PCR. The correlation of Ag and RNA quantities in clinical nasopharyngeal (NP) samples is unknown. An ultrasensitive, quantitative electrochemiluminescence immunoassay for SARS-CoV-2 nucleocapsid (the MSD S-PLEX SARS-CoV-2 N assay) was used to measure Ag in clinical NP samples from adults and children previously tested by PCR. The S-PLEX Ag assay had a limit of detection (LOD) of 0.16 pg/ml and a cutoff of 0.32 pg/ml. Ag concentrations measured in clinical NP samples (collected in 3.0 ml of media) ranged from less than 160 fg/ml to 2.7 μg/ml. Log-transformed Ag concentrations correlated tightly with CT values. In 35 adult and 101 pediatric PCR-positive samples, the sensitivities were 91% (95% confidence interval, 77 to 98%) and 79% (70 to 87%), respectively. In samples with a CT of ≤35, the sensitivities were 100% (88 to 100%) and 96% (88 to 99%), respectively. In 50 adult and 40 pediatric PCR-negative specimens, the specificities were 100% (93 to 100%) and 98% (87 to 100%), respectively. Nucleocapsid concentrations in clinical NP samples span 8 orders of magnitude and correlate closely with RNA concentrations (CT values). The S-PLEX Ag assay showed 96 to 100% sensitivity in samples from children and adults with CT values of ≤35, and a specificity of 98 to 100%. These results clarify Ag concentration distributions in clinical samples, providing insight into the performance of Ag RDTs and offering a new approach to diagnosis of COVID-19.
INTRODUCTION
Nucleic acid amplification tests (NAATs) for SARS-CoV-2, the etiologic agent of COVID-19, can be highly sensitive for diagnosis of COVID-19 and are being performed in centralized laboratories around the globe in staggering numbers (1, 2). However, NAAT capacity and utility have been impacted worldwide by recurring shortages of testing reagents and logistic barriers, causing long delays in results turnaround time. The need for more rapid and decentralized testing options has led to swift development of rapid diagnostic tests (RDTs) for point-of-care (POC) use that detect SARS-CoV-2 nucleocapsid antigen (Ag) in as little as 15 min. Reported clinical sensitivities for Ag RDTs vary widely, ranging from 74 to 97% versus PCR (3–7) when performed at the POC in patients suspected of COVID. Concerns about both false-negative and false-positive Ag RDT results (8) have raised caution regarding implementation in many settings in which their use is being considered, including K-12 schools, nursing homes, and community testing centers.
In symptomatic adults, viral loads in nasopharyngeal (NP) samples (as measured by NAAT) increase approximately 2 days prior to symptom onset, peak in 2 to 4 days, remain high over the first 7 days of symptoms, then decrease to undetectable levels over a variable time frame. RNA levels in asymptomatically infected adults appear to follow similar kinetics (9, 10). However, the time course for Ag concentration is not as well understood due to the lack of highly sensitive and quantitative assays for Ag measurement.
We have developed quantitative immunoassays for SARS-CoV-2 nucleocapsid Ag using conventional R-PLEX and enhanced ultrasensitive S-PLEX electrochemiluminescence (ECL) assay formats (Meso Scale Discovery [MSD]) and used these assays to measure the Ag concentration in clinical NP swab eluates previously tested by PCR for clinical diagnosis. Our goal was to assess Ag concentration distributions in clinical samples, correlate Ag concentrations with cycle threshold (CT) values as a measure of RNA concentration, and assess the sensitivity and specificity of the S-PLEX Ag assay for the detection of SARS-CoV-2 in clinical samples versus PCR as a reference method.
MATERIALS AND METHODS
Immunoassay protocols.
ECL immunoassays for SARS-CoV-2 nucleocapsid Ag, employing sandwich immunoassay formats, were carried out using assay kits, instrumentation, and multiwell plate consumables from MSD (11). The multiwell plate consumables have integrated screen-printed carbon ink electrodes on the bottom of each well that are used as solid-phase supports for binding reactions and as the source of electrical energy for inducing ECL from ECL labels in binding complexes on their surfaces. Two ECL assay formats were employed: a conventional ECL format (12) (the MSD R-PLEX SARS-CoV-2 N assay kit) and an ultrasensitive ECL format (13–15) that provides additional signal enhancement and sensitivity (the MSD S-PLEX SARS-CoV-2 N assay kit). The assays were run according to protocols that are provided in the kit package inserts. Details of the immunoassay protocol are provided in Methods S1 in the supplemental material.
Immunoassay quantitation and analysis.
To calculate the concentration of nucleocapsid in the samples, an eight-point calibration curve with recombinant SARS-CoV-2 nucleocapsid protein (full length with a C-terminal His6 epitope tag expressed in mammalian cells [provided with the assay kits]) in the kit assay diluent was run in duplicate on each plate. The calibration assay signals as a function of protein concentration were fit to a four-parameter logistic (4PL) curve using 1/Y2 weighting. Nucleocapsid concentrations in test samples were determined by backfitting the measured assay signals to the 4PL curve. In addition to the calibration samples, high and low concentration control samples were run in duplicate on each plate to ensure consistent quantitation.
Limits of detection (LODs) were determined as the concentration of recombinant nucleocapsid that provides a signal at least three standard deviations above the assay background generated using the kit assay diluent as the sample (as determined by backfitting to the 4PL calibration curve). For assessing sample positivity, we set a relatively conservative threshold of 2× LOD.
Testing of nasopharyngeal swab samples.
NP swab samples in Copan universal transport media (UTM) or saline were tested in duplicate. Concentrations were calculated for each replicate and then averaged. Samples with calculated concentrations below the LOD or above the top calibrator were assigned the LOD or top calibrator values, respectively, for statistical analyses and graphing. Samples were retested if they provided a signal above the assay threshold and the coefficient of variation (CV) for the replicate measurements was greater than 25%, or if there was a reported operator error during processing. Samples that provided a calculated concentration greater than 50% of the top calibrator in the R-PLEX assay were rerun in the R-PLEX assay at a 1:100 dilution. To characterize the distribution of nucleocapsid concentrations and their correlation to PCR CT values, we used the following approach to assign a concentration value to each sample, given that the range of concentrations was greater than the dynamic range of either assay: (i) we used the concentration measured using the S-PLEX assay if it was less than half the concentration of the top S-PLEX calibrator; (ii) otherwise, we used the concentration measured using the R-PLEX assay if it was less than half the concentration of the top R-PLEX calibrator; (iii) otherwise, we used the concentration measured using the R-PLEX assay for a 1:100-diluted sample.
Analysis of a viral culture preparation.
The concentration of nucleocapsid was measured in a reference cell culture preparation of inactivated (gamma irradiation) SARS-CoV-2 virus (isolate USA-WA1/2020, catalog number NR-52287; BEI Resources) with assigned values for the concentration of infectious virus [2.8 × 105 50% tissue culture infective dose(s) (TCID50)/ml prior to inactivation] and RNA (4.1 × 109 copies/ml). Serial dilutions that provided concentrations in the quantitation range of the assay were corrected for dilution and then averaged to determine the concentration of nucleocapsid in the sample. Assay cross-reactivity for circulating coronaviruses OC43 and 229E was measured in viral culture supernatants (diluted 1:100; titers not determined) obtained from the American Type Culture Collection (catalog numbers VR-740 and VR-1558, respectively).
Clinical samples.
All samples were clinical NP swab samples eluted in either 3.0 ml of UTM (Copan, Murietta, CA) or 3.0 ml of normal saline (Remel, Lenexa, KS). Samples were obtained from adult hospital employees or pediatric hospital patients (a small number of whom were young adults) undergoing clinical testing for suspected SARS-CoV-2. Clinical data were obtained by chart review.
The majority of the samples were tested by the Hologic Panther fusion assay (Hologic, San Diego, CA); two adult samples were tested by a laboratory-developed test utilizing Altona Diagnostics reagents (16). The majority of PCR results were reported approximately 4 h after sample collection and residual volumes were divided into aliquots and frozen (−80°C) shortly afterward. Deidentified aliquots were shipped frozen to MSD for testing.
This study was performed under approval from the Boston Children’s Hospital institutional review board with waiver of informed consent; only fully deidentified data were analyzed.
RESULTS
Assay analytical performance.
The calculated LODs for the R-PLEX and S-PLEX Ag assays were 2 pg/ml and 0.16 pg/ml, respectively, and the assay signal was linear with concentration across the full calibration range of the assay. Based on these LOD values, we set the assay thresholds for classifying positive samples as 2× LOD or 4 pg/ml and 0.32 pg/ml, respectively. Nucleocapsid concentration in an irradiated SARS-CoV-2 culture preparation (BEI; see Materials and Methods), as measured using the S-PLEX Ag assay, was compared to the concentrations provided by BEI in TCID50 and RNA copies/ml. We calculated the ratio of Ag to virion as 0.89 pg per TCID50 (or 1.1 × 107 protein molecules per TCID50) and to RNA as 1.5 × 10−4 pg per RNA copy. This ratio corresponds to about 1.8 × 103 protein molecules per RNA copy, based on a molecular weight of 49 kDa for SARS-CoV-2 nucleocapsid, which is higher than the 200 to 400 nucleocapsid molecules per genome in a typical coronavirus particle (17, 18). Using these conversion factors, virus concentrations measured at the Ag assay thresholds are around 4.5 TCID50/ml and 27,000 RNA copies/ml for the R-PLEX assay and 0.36 TCID50/ml and 2,100 RNA copies/ml for the S-PLEX assay. The analytical sensitivity of the S-PLEX Ag assay is therefore on par with the LODs of many PCR assays (2 copies/μl of sample) (19). In comparison, the detection limits for the commercial BD Veritor and Quidel Sofia SARS-CoV-2 antigen tests, based on the values in the package inserts, are roughly 140 TCID50/ml and 226 TCID50/ml, which, based on our calculated conversion factors, correspond to 121 pg/ml and 204 pg/ml of Ag, respectively, or roughly 500-fold less sensitive than the S-PLEX Ag test.
Assay specificity was confirmed by testing culture preparations of the 229E and OC43 circulating coronavirus strains. No measurable cross-reactivity was observed with these strains.
The assay was highly reproducible. The CVs for the low and high concentration controls run in duplicate on each assay plate during the study (10 plates per assay, run over several days) were 7.7 and 8.3%, respectively, for the R-PLEX assay and 7.0 and 7.7% for the S-PLEX assay.
Distribution of nucleocapsid antigen concentrations in clinical samples.
Nucleocapsid concentrations were measured in NP samples from 85 adult employees (35 PCR positive, 50 PCR negative) and 141 pediatric patients (101 PCR positive, 40 PCR negative). Each sample was the earliest available sampling time point for that employee/patient in the hospital system. The clinical characteristics of the adult and pediatric patients are summarized in Table 1. At the time of testing, 4/35 PCR-positive adult employees and 2/101 PCR-positive pediatric patients were asymptomatic.
TABLE 1.
Clinical and laboratory characteristics of adult and pediatric patients contributing samples
| Parameter | Pediatric patients, PCR positive (n = 101) | Adult patients, PCR positive (n = 35) | Pediatric patients, PCR negative (n = 40) | Adult patients, PCR negative (n = 50) |
|---|---|---|---|---|
| Age in yrs, n (%) | ||||
| <1 | 22 (22) | 0 (0) | 4 (10) | 0 (0) |
| 1 to 5 | 18 (18) | 0 (0) | 18 (45) | 0 (0) |
| 6 to 10 | 20 (20) | 0 (0) | 2 (5) | 0 (0) |
| 11 to 18 | 27 (27) | 1 (3) | 10 (25) | 0 (0) |
| 19 to 25 | 13 (13) | 5 (14) | 4 (10) | 4 (8) |
| 26 to 44 | 1 (1) | 18 (51) | 2 (5) | 34 (68) |
| 45 to 64 | 0 (0) | 11 (31) | 0 (0) | 11 (22) |
| 65 and older | 0 (0) | 0 (0) | 0 (0) | 1 (2) |
| % female | 54.5 | 71.4 | 62.5 | 84 |
| Median days (IQR)a of symptoms prior to COVID test | 3 (2–6)b | 3 (1–7)c | NAd | NA |
| Median CT (IQR) | 25.8 (19.1–36.2) | 26.4 (19–32.7) | NA | NA |
IQR, interquartile range.
Range, 0 to 40 days.
Range, 0 to 33 days (n = 33).
NA, not applicable.
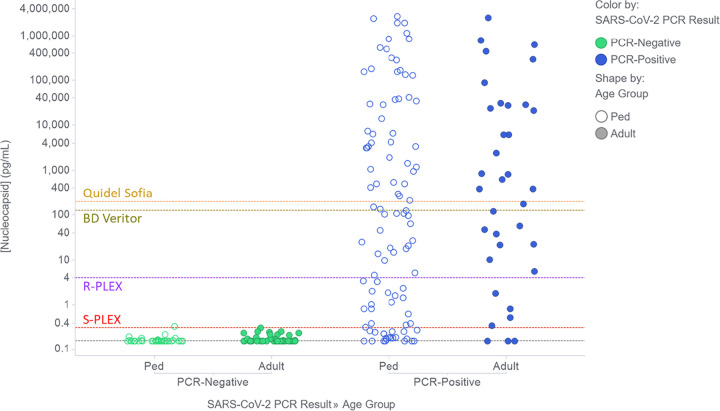
The measured concentrations of nucleocapsid in the swab samples, as measured using the R-PLEX and S-PLEX assays without any additional sample dilution, are shown in Fig. S1 in the supplemental material. Using the optimal assay format or dilution for each sample (see Materials and Methods), Fig. 1 plots the distributions of Ag concentrations in adult and pediatric samples that tested negative or positive by PCR. Figure 1 also shows the assay thresholds for the S-PLEX and R-PLEX nucleocapsid assays and, for comparison, the estimated LODs for the Quidel Sofia and BD Veritor assays. PCR-negative samples clustered tightly with measured Ag concentrations around or below the LOD for the S-PLEX assay, with only one sample providing a concentration slightly above the S-PLEX assay threshold. Ag concentrations for PCR-positive samples were evenly distributed over a wide range of concentrations spanning almost 8 orders of magnitude (from <160 fg/ml to 2.7 μg/ml) in samples from both adults and children. The sample incubation step in the assay protocol includes 0.5% Triton X-100 to release Ag from virus particles. However, testing a small set of samples without Triton generated similar calculated concentrations, suggesting that most of the Ag in the sample was not confined within intact virus particles.
FIG 1.
Nucleocapsid concentrations in clinical NP swab samples from PCR-negative (green) and PCR-positive (blue) adults (filled circles) and pediatric patients (Ped, open circles). The figure shows the concentrations measured using the optimal assay format and dilution for each sample as described in Materials and Methods. Concentrations below the LOD for the S-PLEX assay were assigned the LOD value (gray dashed line). The plots also show dashed lines to indicate the applied assay thresholds for the ultrasensitive S-PLEX and conventional R-PLEX ECL assays, as well as the estimated analogous values for the commercial BD Veritor and Quidel Sofia systems.
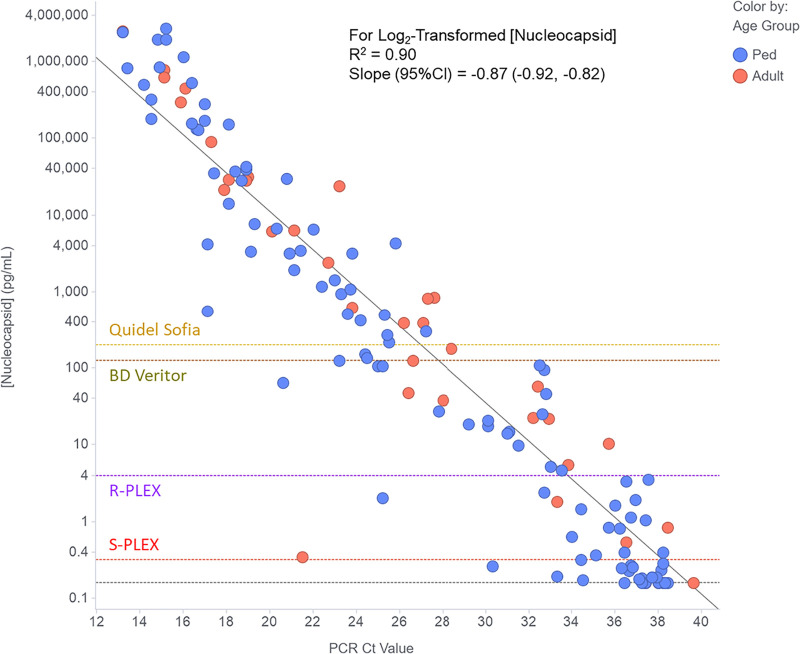
Figure 2 plots the measured Ag concentrations for the PCR-positive samples from Fig. 1 against the PCR CT values for those samples. Linear regression demonstrates a strong (R2 = 0.90) correlation of the log2-transformed concentration values (in pg/ml) with CT values across the large range of values measured with both assays. The estimates (95% confidence intervals [CI]) for the slope and y-intercept are −0.87 (−0.92 to −0.82) and 30.9 (29.5 to 32.2). Given that a one unit decrease in CT value should correspond to just under a doubling of the RNA concentration, a slope for the relationship of log2 nucleocapsid concentration with CT that is close to −1.0 is consistent with a roughly linear relationship between viral protein and RNA concentrations.
FIG 2.
Correlation of nucleocapsid concentration with PCR CT value for clinical NP swab samples from PCR-positive adults (red circles) and pediatric patients (Ped, blue circles). The figure shows the concentrations measured using the optimal assay format and dilution for each sample as described in Materials and Methods. Concentrations below the LOD for the S-PLEX assay were assigned the LOD value (gray dashed horizontal line). The diagonal dashed gray line is the linear regression fit to the data (using log-transformed nucleocapsid concentrations). To provide estimates of the expected CT value for samples at the threshold for different nucleocapsid assay formats, the plots also show horizontal dashed lines to indicate the applied assay thresholds for the ultrasensitive S-PLEX and conventional R-PLEX ECL assays, as well as the estimated analogous values for the commercial BD Veritor and Quidel Sofia systems.
Using the regression model, the expected CT values for samples at the antigen assay thresholds for the R-PLEX and S-PLEX ECL Ag assays, and the commercial BD and Quidel assays are 34, 38, 28, and 27 cycles, respectively.
Assay clinical performance.
The sensitivity and specificity of the S-PLEX Ag assay versus PCR in adult and pediatric samples are shown in Table 2. The specificities in PCR-negative samples were 100% (93 to 100%) and 98% (87 to 100%) in adult and pediatric samples, respectively. The sensitivities in PCR-positive samples were 91% (77 to 98%) and 79% (70 to 87%) in adult and pediatric samples, respectively. The correlation plot in Fig. 2 shows that the lower sensitivity in pediatric samples is largely accounted for by a number of pediatric samples with CT values between about 36 and 38 (see Table S1 in the supplemental material). Considering only samples with CT values of ≤35, the S-PLEX Ag test sensitivity increased to 100% (88 to 100%) and 96% (88 to 99%), respectively, for adult and pediatric samples. We note that these high-CT samples are also potentially on the borderline for detection by the Hologic assay, since the CT value corresponding to the Panther Fusion LOD is 35.6 according to the package insert (20).
TABLE 2.
Performance of S-PLEX Ag assay versus PCR for detection of SARS-CoV-2 in samples from adult and pediatric patients
| Age group | PCR-negative patients |
PCR-positive patients |
||||
|---|---|---|---|---|---|---|
| All CT values |
CT values ≤ 35 |
|||||
| n | % specificity (95% CI) | n | % sensitivity (95% CI) | n | % sensitivity (95% CI) | |
| Pediatric | 40 | 98 (87–100) | 101 | 79 (70–87) | 72 | 96 (88–99) |
| Adult | 50 | 100 (93–100) | 35 | 91 (77–98) | 29 | 100 (88–100) |
| Combined | 90 | 99 (94–100) | 136 | 82 (75–88) | 101 | 97 (92–99) |
This data set of measured nucleocapsid values provides a tool for estimating the clinical sensitivity that can be achieved with less-sensitive assays. Based on the S-PLEX assay measurements, the sensitivity of the R-PLEX ECL assay can be predicted based on its 4 pg/ml threshold to be 68%, which agrees well with the actual observed sensitivity for this assay of 70% (see Table S2) based mostly on measurements with the S-PLEX assay. Using the estimated LODs for the BD and Quidel assays, the predicted sensitivities of the commercial assays for this sample set would be approximately 51 and 47%, respectively; the sensitivity in samples with a CT of <30 would be 88 and 82%, and those in samples with a CT of <25 would be 97 and 92%, respectively.
Characterization of discordant samples.
Samples with discordant PCR and S-PLEX Ag assay results were analyzed further. We found that 21 of 24 false-negative samples were from pediatric patients, and 21 of 24 had CT values of >35 (see Table S1). Since the linear regression model relating nucleocapsid concentration with CT value predicted measurable nucleocapsid levels up to CT values of 38, we conducted spike recovery experiments (see Methods S1) to determine whether matrix interference may have led to under detection of antigen. Only 3 of the 24 samples exhibited significant matrix interference (spike recovery < 25%).
Relationship between nucleocapsid concentration and symptom duration.
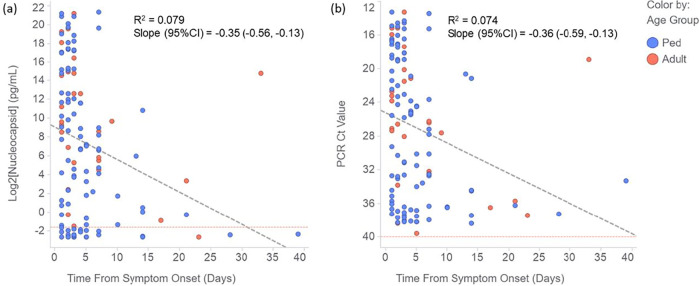
For samples taken from symptomatic PCR-positive patients, Fig. 3 plots the nucleocapsid concentration (Fig. 3a) and PCR CT value (Fig. 3b) as a function of the time from onset of symptoms to collection of the sample. The correlations of antigen concentration and CT value with time from symptom onset were evaluated by linear regression (Fig. 3). The correlations were relatively weak (R2 values were 0.079 and 0.074 for nucleocapsid [after log2 transformation] and PCR CT values, respectively), due to the large between-patient variation in Ag concentrations and CT values. However, as expected, both nucleocapsid and RNA concentrations decreased on average over time. Notably, the slopes of decline were similar for the two analytes, suggesting that they may follow similar clearance kinetics (Fig. 3).
FIG 3.
(a and b) Correlation of nucleocapsid concentration (log2 tranformed) (a) and PCR CT value (b) with time from symptom onset for clinical NP swab samples from PCR-positive adults (red circles) and pediatric patients (Ped, blue circles). The plots also show the linear regression fit to the data (diagonal dashed gray lines).
DISCUSSION
This is the first characterization of SARS-CoV-2 nucleocapsid antigen concentration distributions in clinical nasopharyngeal swab samples. We compared measured Ag concentrations and qualitative (positive/negative) results to PCR results and CT values generated using the Hologic Panther Fusion test, which has one of the lowest LODs among molecular assays tested with a U.S. Food and Drug Administration (FDA) reference panel (19). Our work demonstrated that Ag concentrations in clinical samples span 8 orders of magnitude and that there is a close correlation between Ag and RNA concentrations (as reflected by CT values) throughout the range of viral loads in adult and pediatric individuals with a range of symptom duration prior to testing. The S-PLEX Ag assay had nearly perfect specificity and high sensitivity compared to PCR, with nearly 100% sensitivity in NP swab samples with CT values of ≤35, offering the potential for a new approach to COVID diagnosis.
The strong correlation of Ag concentrations with CT values and the high sensitivity of the S-PLEX Ag test in samples covering the range of viral loads observed in newly symptomatic individuals with COVID-19 suggests that the high false-negative rates that have been observed with some Ag RDT tests (versus PCR) in similar populations are due to the relative analytical insensitivity of the RDT tests rather than to any major difference in the kinetics of expression of Ag and RNA in the nose or nasopharynx. A highly sensitive assay could detect individuals earlier in infection than an Ag RDT or potentially support Ag testing in pooled samples.
Studies such as ours that use a high sensitivity quantitative assay to measure an infectious disease biomarker provide a useful data set for predicting the clinical performance of less-sensitive methods or for setting analytical sensitivity targets to achieve specific clinical performance targets. To this point, our estimated clinical sensitivities for the BD and Quidel RDTs, based on our measured distributions of Ag concentrations, are consistent with the field performance of Ag RDTs in a recent European prospective study (3). The report evaluated Ag RDT test performance at POC versus PCR and found that the best-performing visually read Ag RDT was 76.6% sensitive and 99.3% specific, with high sensitivity in samples with CT values <25, moderate sensitivity in samples with CT <30, and poor sensitivity above a CT of 30 (3).
One important consideration for COVID-19 diagnostics is their ability to identify patients with high loads of active virus that are more at risk of transmitting disease. Several investigators have reported difficulty in culturing virus from patient samples with measured viral loads below approximately 1 × 105 RNA copies/ml (21–24). However, virus has been recovered from samples with RNA levels as low as 1.2 × 104 copies/ml (25) and from samples with a CT value of 34 to 35 on a range of PCR assays (26–28). Using our conversion factor from the correlation of Ag and RNA levels, 1 × 104 copies/ml translates to 1.5 pg/ml, which is about five times higher than the S-PLEX Ag assay cutoff (0.32 pg/ml), suggesting that the S-PLEX Ag assay should be able to detect Ag in most if not all samples from which virus is culturable (though we note that cultures themselves have variable sensitivity [29] and that lack of ability to culture virus does not preclude transmission).
If converted to a clinical diagnostic, the S-PLEX Ag assay offers some potential advantages over existing diagnostic approaches. The assay’s analytical sensitivity is similar to that of PCR-based methods, and the assay was able to detect nearly all samples with a CT of ≤35. The S-PLEX Ag assay is ∼500-fold more sensitive than existing Ag RDTs with electronic readers, with near perfect specificity in clinical samples in UTM and normal saline. We found a higher rate of false-negative results for samples with CT values between 36 and 38 than would be expected based on the correlation of CT values with Ag concentration. These false-negative samples were largely pediatric samples eluted in saline. It is possible that there may be some loss of Ag by adsorption from saline compared to samples eluted in protein-containing UTM, warranting further study. Depending on the use case, the inconsistent detection of samples with CT values of >35 may or may not be a disadvantage (if the goal is to detect all individuals with evidence of infection, e.g., to optimize hospital safety and inform contact tracing, versus to identify those most likely to transmit [29]). The high specificity of the S-PLEX Ag assay, if confirmed in a clinical diagnostic setting, should be an advantage over Ag RDTs, some of which have generated concerning specificity data in actual practice (8) despite having high specificities in EUA validation studies (5, 6).
While the S-PLEX Ag test offers analytical advantages over Ag RDTs, given that the Ag RDTs can be performed at the POC, the operational characteristics of the S-PLEX Ag assay (cost, speed, and operator time) must be carefully considered (30). The turnaround time of the assay is 4 h, and 78 samples can be run on each plate. A single operator can process as many as five plates (390 samples) in a run, with higher throughputs available with automation. The MSD plate reader has a cost in the typical range for luminescence or multimode plate readers. The assay utilizes entirely different reagents than used for molecular testing, which might make it less susceptible to current supply chain issues.
Our study has some limitations. First, this was a retrospective study using frozen samples previously tested by PCR; future studies will test fresh samples prospectively, but the tight correlation between Ag and CT values suggests that freezing was not a significant issue. We note that the sensitivity in the pediatric sample set was lower than in the adult sample set, but we ascribe this to a higher CT value distribution in the pediatric samples and, possibly, to a higher proportion extracted in saline relative to UTM. For some of the patients who provided samples, the time between onset of symptoms and testing was long, but this reflects real life clinical test use. The majority of the patients whose samples were included in our study were symptomatic, so the results will need confirmation in asymptomatic individuals (31). Finally, the performance of the test in an independent clinical laboratory setting remains to be demonstrated.
In summary, we have shown that an ultrasensitive Ag assay is able to detect Ag throughout the range of viral load in clinical samples in adults and children infected with SARS-CoV-2. The assay has high sensitivity and specificity, offering an alternative to PCR and a clear analytical advantage over Ag RDT. Future prospective studies evaluating test performance in programmatic screening of symptomatic and asymptomatic individuals will demonstrate whether the S-PLEX Ag assay can offer a diagnostic alternative that is inexpensive, rapid, and high throughput, thus contributing a novel diagnostic tool for management of this pandemic.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Rossi for assistance with PCR CT data capture, and we thank the virology laboratory staff at Boston Children’s Hospital (BCH) for their assistance with sample aliquoting. We thank Pradeepthi Bathala, Shraddha S. Kale, Salvia Misaghian, Nikhil Padmanabhan, Daniel Romero, and Navaratnam Manjula for conducting the assay measurements.
The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate USA-WA1/2020, gamma-irradiated, NR-52287.
The work at Meso Scale Diagnostics (MSD) was funded by MSD. The BCH team did not have dedicated funding for this study.
N.R.P., H.W., T.J.S., and R.A.L. do not have any conflicts of interest to declare. A.M., M.S., and G.B.S. are employees of MSD.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Center for Systems Science and Engineering. 2020. COVID-19 dashboard by the Center for Systems Science and Engineering (CCSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html. Accessed 31 October 2020.
- 2.Centers for Disease Control. 2020. COVID data tracker. Centers for Disease Control and Prevention, Atlanta, GA. https://covid.cdc.gov/covid-data-tracker/#testing_testsperformed. Accessed 31 October 2020. [Google Scholar]
- 3.Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, Schnitzler P, Kräusslich HG, Lindner AK, Nikolai O, Mockenhaupt FP, Seybold J, Corman VM, Drosten C, Pollock NR, Cubas-Atienzar AI, Kontogianni K, Collins A, Wright AH, Knorr B, Welker A, de Vos M, Sacks JA, Adams ER, Denkinger CM. 2020. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv doi: 10.1101/2020.10.01.20203836:2020.10.01.20203836. [DOI] [Google Scholar]
- 4.Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, Krüger L, Gaeddert M, Tobian F, Lainati F, Köppel L, Seybold J, Corman VM, Drosten C, Hofmann J, Sacks J, Mockenhaupt F, Denkinger CM. 2020. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected anterior nasal swab versus professional-collected nasopharyngeal swab. medRxiv doi: 10.1101/2020.10.26.20219600:2020.10.26.20219600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FDA. 2020. Package insert for the BD VeritorTM system for rapid detection of SARS-CoV-2. Food and Drug Administration, Washington, DC. https://www.fda.gov/media/139755/download. Accessed 2 November 2020. [Google Scholar]
- 6.FDA. 2020. Package insert for the Quidel Sofia SARS antigen FIA test. Food and Drug Administration, Washington, DC. https://www.fda.gov/media/137885/download. Accessed 2 November 2020. [Google Scholar]
- 7.FDA. 2020. Package insert for the Abbott BinaxNOW Ag card. Food and Drug Administration, Washington, DC. https://www.fda.gov/media/141570/download. Accessed 2 November 2020. [Google Scholar]
- 8.Rubin R. 2020. The challenges of expanding rapid tests to curb COVID-19. JAMA 324:1813. doi: 10.1001/jama.2020.21106. [DOI] [PubMed] [Google Scholar]
- 9.Sethuraman N, Jeremiah SS, Ryo A. 2020. Interpreting diagnostic tests for SARS-CoV-2. JAMA 323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 10.Kissler SM, Fauver JR, Mack C, Tai C, Shiue KY, Kalinich C, Jednak S, Ott I, Vogels C, Wohlgemuth J, Weisberger J, DiFiori J, Anderson DJ, Mancell J, Ho D, Grubaugh ND, Grad YH. 2020. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. medRxiv doi: 10.1101/2020.10.21.20217042:2020.10.21.20217042. [DOI] [Google Scholar]
- 11.Debad JD, Glezer EN, Wohlstadter JN, Sigal GB. 2004. Clinical and biological applications of ECL. In Electrogenerated chemiluminescence, p 43–78. Marcel Dekker, New York, NY. [Google Scholar]
- 12.Sigal GB, Pinter A, Lowary TL, Kawasaki M, Li A, Mathew A, Tsionsky M, Zheng RB, Plisova T, Shen K, Katsuragi K, Choudhary A, Honnen WJ, Nahid P, Denkinger CM, Broger T. 2018. A novel sensitive immunoassay targeting the 5-methylthio-d-xylofuranose-lipoarabinomannan epitope meets the WHO’s performance target for tuberculosis diagnosis. J Clin Microbiol 56:e01338-18. doi: 10.1128/JCM.01338-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broger T, Tsionksy M, Mathew A, Lowary TL, Pinter A, Plisova T, Bartlett D, Barbero S, Denkinger CM, Moreau E, Katsuragi K, Kawasaki M, Nahid P, Sigal GB. 2019. Sensitive electrochemiluminescence (ECL) immunoassays for detecting lipoarabinomannan (LAM) and ESAT-6 in urine and serum from tuberculosis patients. PLoS One 14:e0215443. doi: 10.1371/journal.pone.0215443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poorbaugh J, Samanta T, Bright SW, Sissons SE, Chang CY, Oberoi P, MacDonald AJ, Martin AP, Cox KL, Benschop RJ. 2019. Measurement of IL-21 in human serum and plasma using ultrasensitive MSD S-PLEX® and Quanterix SiMoA methodologies. J Immunol Methods 466:9–16. doi: 10.1016/j.jim.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Diamandis EP, Stanczyk FZ, Wheeler S, Mathew A, Stengelin M, Nikolenko G, Glezer EN, Brown MD, Zheng Y, Chen YH, Wu HL, Azziz R. 2017. Serum complexed and free prostate-specific antigen (PSA) for the diagnosis of the polycystic ovarian syndrome (PCOS). Clin Chem Lab Med 55:1789–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FDA. 2020. Childrens-Altona SARS-CoV-2 assay. Food and Drug Administration, Washington, DC. https://www.fda.gov/media/136971/download. Accessed 2 November 2020. [Google Scholar]
- 17.Bar-On YM, Flamholz A, Phillips R, Milo R. 2020. SARS-CoV-2 (COVID-19) by the numbers. Elife 9:e53709. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ. 2006. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol 80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FDA. 2020. Sensitivity mean estimates of the EUA authorized SARS-CoV-2 molecular diagnostic tests using the FDA SARS CoV-2 reference panel. Food and Drug Administration, Washington, DC. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data. Accessed 3 November 2020. [Google Scholar]
- 20.FDA. 2020. Package insert for the Hologic Panther Fusion SARS-CoV-2 test. Food and Drug Administration, Washington, DC. https://www.fda.gov/media/136156/download. Accessed 10 November 2020. [Google Scholar]
- 21.Jones TC, Mühlemann B, Veith T, Biele G, Zuchowski M, Hoffmann J, Stein A, Edelmann A, Corman VM, Drosten C. 2020. An analysis of SARS-CoV-2 viral load by patient age. medRxiv doi: 10.1101/2020.06.08.20125484:2020.06.08.20125484. [DOI] [Google Scholar]
- 22.Perera R, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, Chin AWH, Chu DKW, Cheng SMS, Poon LLM, Chuang VWM, Peiris M. 2020. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallichote E, Quicke K, Sexton N, Young M, Janich A, Gahm G, Carlton EJ, Ehrhart N, Ebel GD. 2020. Longitudinal surveillance for SARS-CoV-2 RNA among asymptomatic staff in five Colorado skilled nursing facilities: epidemiologic, virologic and sequence analysis. medRxiv doi: 10.1101/2020.06.08.20125989. [DOI] [Google Scholar]
- 24.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 25.L’Huillier AG, Torriani G, Pigny F, Kaiser L, Eckerle I. 2020. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg Infect Dis 26:2494–2497. doi: 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA, Public Health–Seattle and King County and CDC COVID-19 Investigation Team. 2020. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill 25:2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaafar R, Aherfi S, Wurtz N, Grimaldier C, Hoang VT, Colson P, Raoult D, La Scola B. 2020. Correlation between 3790 qPCR positives samples and positive cell cultures including 1941 SARS-CoV-2 isolates. Clin Infect Dis doi: 10.1093/cid/ciaa1491. [DOI] [Google Scholar]
- 29.Pekosz A, Cooper CK, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S, Leitch J, Gary DS, Roger-Dalbert C. 2020. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture. medRxiv doi: 10.1101/2020.10.02.20205708:2020.10.02.20205708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paltiel AD, Zheng A, Walensky RP. 2020. Assessment of SARS-CoV-2 screening strategies to permit the safe reopening of college campuses in the United States. JAMA Netw Open 3:e2016818. doi: 10.1001/jamanetworkopen.2020.16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kociolek LK, Muller WJ, Yee R, Dien Bard J, Brown CA, Revell P, Wardell H, Savage TJ, Jung S, Dominguez S, Parikh BA, Jerris RC, Kehl SC, Campigotto A, Bender JM, Zheng X, Muscat E, Linam M, Abuogi L, Smith C, Graff K, Hernandez-Leyva A, Williams D, Pollock NR. 2020. Comparison of upper respiratory viral load distributions in asymptomatic and symptomatic children diagnosed with SARS-CoV-2 infection in pediatric hospital testing programs. J Clin Microbiol 59:e02593-20. doi: 10.1128/JCM.02593-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.