The COVID-19 pandemic has created massive demand for widespread, distributed tools for detecting SARS-CoV-2 genetic material. The hurdles to scalable testing include reagent and instrument accessibility, availability of highly trained personnel, and large upfront investment.
KEYWORDS: CRISPR, Cas13, COVID-19, point of care, SARS-CoV-2, testing
ABSTRACT
The COVID-19 pandemic has created massive demand for widespread, distributed tools for detecting SARS-CoV-2 genetic material. The hurdles to scalable testing include reagent and instrument accessibility, availability of highly trained personnel, and large upfront investment. Here, we showcase an orthogonal pipeline we call CREST (Cas13-based, rugged, equitable, scalable testing) that addresses some of these hurdles. Specifically, CREST pairs commonplace and reliable biochemical methods (PCR) with low-cost instrumentation, without sacrificing detection sensitivity. By taking advantage of simple fluorescence visualizers, CREST allows a binary interpretation of results. CREST may provide a point-of-care solution to increase the distribution of COVID-19 surveillance.
INTRODUCTION
The coronavirus 2019 (COVID-19) pandemic presents the world with an unprecedented public health challenge. The lack of COVID-19 symptoms in a significant proportion (estimates range from 18% to 29%) of individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) fuels covert transmission of the virus (1, 2). Even in cases in which symptoms do occur, the virus can be transmitted before symptom onset (3, 4). The high proportion of asymptomatic or mildly symptomatic and highly infectious individuals, combined with the risk of severe disease outcomes and mortality rates ranging from 1.38% to as high as 20% in people 80 years and older, places an unprecedented burden on our health care system (5, 6).
A range of mitigation efforts have been implemented across the globe to slow the transmission and “flatten the curve,” including sheltering in place and the requirement for face covering in public. However, without widely available and accurate testing, it remains unclear exactly how effective these methods are at curbing the spread of the virus. In addition, these risk mitigation efforts have a catastrophic economic and social impact, which makes long-term compliance challenging until a vaccine or treatment is widely available (7). To reduce the socioeconomic burden, we need to implement effective containment measures, such as contact tracing, quarantine of confirmed cases, and enhanced surveillance, all of which depend on data obtained from widespread testing (8). Proactive, prevalent, and regular testing have been effective measures for the control of COVID-19 in countries such as South Korea and Taiwan and will be required for the establishment of effective strategies for the relaxation of social-distancing measures worldwide (9, 10).
A critical hurdle to deploying massively widespread, recurrent testing is the availability of reagents and specialized equipment. Suggested solutions to make sample collection scalable include laboratory-made viral transport media and 3D-printed swabs and bypassing biochemical RNA extraction through heat and chemical extraction techniques (11–14). However, there have been few end-to-end solutions to SARS-CoV-2 testing that fulfill the requirements of being immediately scalable and low in cost without sacrificing sensitivity. Here, we focused on developing a CRISPR (clustered regularly interspaced palindromic repeats)-based SARS-CoV-2 RNA detection method that is low in cost, highly sensitive, and easy to deploy at sites with minimal infrastructure.
Cas12 and Cas13 (CRISPR-based) methods have been transformative with regard to pathogen detection. They are sensitive and, when coupled to isothermal amplification methods and lateral flow immunochromatography detection, have been made field deployable. These methods are promising tools for the detection of SARS-CoV-2 (15–18). However, because of the global demand for testing, key reagents in these protocols are difficult to obtain. To lower the barrier to COVID-19 diagnostics, we devised a method we call CREST (Cas13-based, rugged, equitable, scalable testing). CREST addresses three of the main hurdles—reagent accessibility, equipment availability, and cost—that limit the scalability of Cas13-based testing, by taking advantage of widely available enzymes, low-cost thermocyclers, and easy-to-use fluorescent visualizers. Moreover, CREST is equivalent in sensitivity to the gold standard reverse transcription quantitative PCR (RT-qPCR) method most often deployed for COVID-19 testing. With these advantages, CREST has the potential to facilitate early detection of positive cases, regular monitoring of individuals at high risk, and implementation of informed containment measures for infected individuals.
METHODS AND RESULTS
To design a sensitive, low-cost, and easy-to-use SARS-CoV-2 detection method, we first identified critical steps that require limiting reagents, specific equipment, and highly trained individuals to perform them and thus present a barrier to testing at sites with limited infrastructure and resources. For CRISPR-Cas13-based methods, these steps include (i) the amplification of the target material prior to detection and (ii) the visualization of Cas13 activity by either colorimetric (immunochromatography) or fluorescent methods.
To lower the first barrier, we analyzed options for detection of specific SARS-CoV-2 genomic sequences upon enzymatic amplification (Fig. 1). The gold standard method relies on RT-qPCR (19). Quantitative detection is accomplished using specialized instruments that detect fluorescent probes which report the extent of amplification of the target sequence in real time. While sensitivity is high (on the order of tens of target molecules per microliter), the main limitation is the requirement of real-time thermocyclers, analysis software, and trained personnel for data interpretation. Limitations aside, the core of the technology—amplification of a target nucleic acid sequence by PCR—is robust and sensitive and makes use of a widely available enzyme, Taq polymerase. These advantages motivated us to pair PCR with CRISPR-based detection of viral sequences, an approach that has been successful for the detection of DNA sequences using Cas9 (20). The thermocyclers required for PCR are expensive, specialized instruments that are generally limited to professional laboratories. However, the recent “do-it-yourself biology” (DIYbio) movement has made PCR accessible through the creation of affordable, Bluetooth-enabled, field-ready thermocyclers, which can even be battery operated. These versatile thermocyclers can be used in unconventional environments and perform as well as traditional thermocyclers in moderate temperatures (21–23). We reasoned that these devices, such as the mini-PCR mini16, offer a low-cost solution for the amplification of the viral target material and can make COVID-19 testing widely available (Fig. 2).
FIG 1.
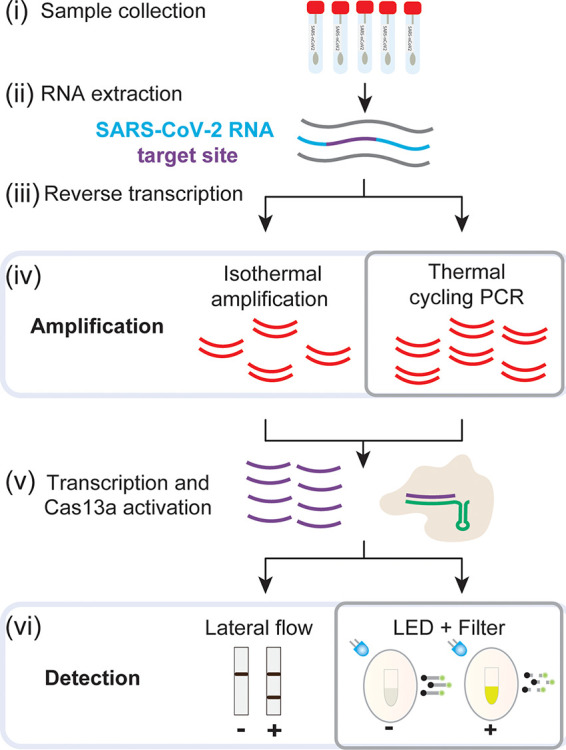
Overview of Cas13-based detection methods and CREST modifications. (i to iii) Standard sample collection, RNA extraction, and reverse transcription. (iv) Amplification using cost-effective Taq polymerase and portable thermocyclers instead of isothermal reactions. (v) Transcription and Cas13 activation are followed by fluorescence detection of dequenched poly(U) cleavage reporter visualized with a blue LED (∼495 nm) and orange filter or other fluorescence detection system.
FIG 2.
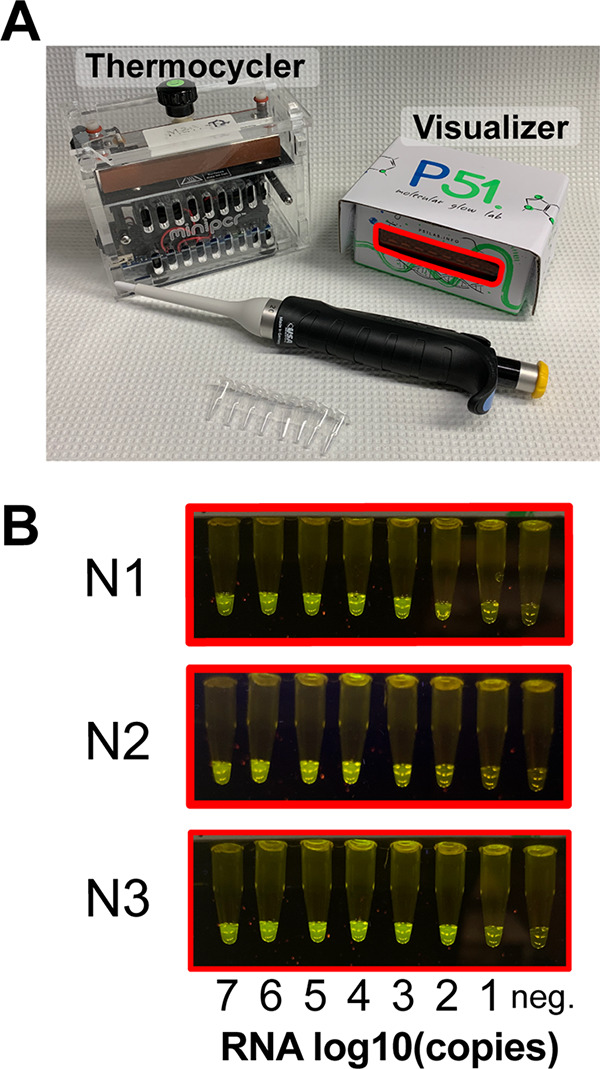
Detection of SARS-CoV-2 RNA using CREST. (A) The mini-PCR mini16 thermocycler and P51 molecular fluorescence visualizer used in this study. Both are portable, can be operated with batteries, and have minimal footprint. (B) Fluorescence visualization of N1, N2, and N3 synthetic targets using the P51 visualizer.
To reduce the second hurdle, the visualization of test outcomes, we explored options available for the detection of Cas13 activity. When bound to its target, Cas13 catalyzes the nonspecific cleavage of RNAs. This target-specific recognition can be detected in many ways, for example, either by lateral flow immunochromatography or by fluorescence visualization through the use of a fluorescein- and quencher-conjugated poly(U) RNA cleavage reporter. Lateral-flow test strips are a promising detection method. These strips utilize capillary action to move analytes through a solid support material striped with antibodies that can detect them, resulting in binary readouts. While they are simple to use and read—they may eventually enable in-home testing—their current availability is limited, and they are expensive relative to the cost per test (Fig. 3B; see also Data Set S1 in the supplemental material). For this reason, we sought an affordable, scalable, easy-to-interpret, solution for visualization of positive results. In the CREST protocol, we use a P51 cardboard fluorescence visualizer, powered by a 9-V battery, for the detection of Cas13 activity instead of immunochromatography (Fig. 2).
FIG 3.
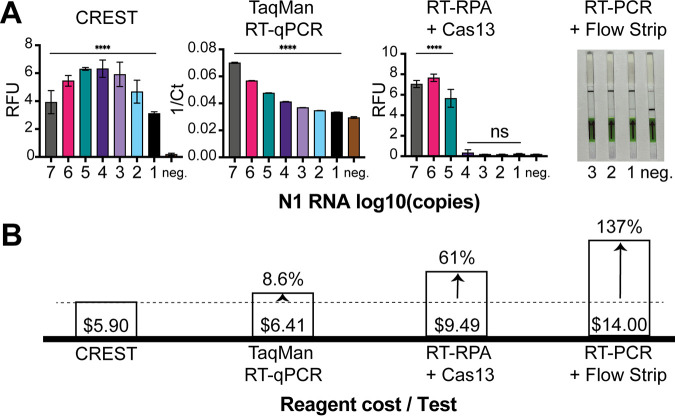
Comparative analysis of method sensitivity and reagent cost per test. (A) (Left) Comparison of method sensitivity using a quantitative fluorescence detection instrument. (Right) RT-PCR + Cas13 detection visualized with lateral flow strips. (B) Associated costs of reagents per test of each testing method (excluding up-front instrumentation costs). A test is defined as a single sample run in triplicate.
To validate a streamlined workflow with these devices, we measured the presence of the three viral sequences that correspond to the CDC RT-qPCR test (19). Briefly, we PAGE purified annealed synthetic DNA oligonucleotides flanked by an upstream T7 RNA polymerase promoter that encode sequences corresponding to the N1, N2, and N3 sites in the SARS-CoV-2 nucleocapsid gene (Table S3). Next, we transcribed the DNA in vitro to obtain target RNAs. After Cas13 purification and extensive optimization of the reaction conditions (Fig. S1 and S2), we used these targets to determine the detection limit of the CREST protocol and found that we could detect as few as 10 copies of a target RNA molecule per μl (Fig. 2B). This result shows that CREST has sensitivity comparable to that of the corresponding RT-qPCR in our hands, demonstrating the power of CREST for pairing a thermal cycling amplification step (PCR) with a linear amplification step (transcription), combined with enzymatic signal amplification through fluorescence detection. In addition, we calculated CREST’s limit of detection in negative human nasopharyngeal (NP) swabs with spiked heat-inactivated SARS-CoV-2 virus (ATCC VR-1986HK) to be 200 copies per μl (Fig. S3).
Next, we quantitively compared CREST’s sensitivity and cost to those of established methods. First, we compared it to RT-qPCR (one-step TaqMan assay) and found that, while being similarly sensitive, CREST’s reagents cost less than RT-qPCR’s even at the low scale of our pilot experiment (Fig. 3; Data Set S1). In addition, the up-front cost of CREST instrumentation is 30 to 50 times lower. Second, we compared the RT-PCR amplification step of CREST to Cas13-based protocols that utilize RT-recombinase polymerase amplification (RT-RPA). We found thermal cycling amplification (20 cycles) to be substantially more efficient with comparable amplification reaction times (Fig. 3A; Fig. S4). Moreover, in stark contrast to the proprietary, high-cost, relatively small-batch production rates of reagents required for RPA, Taq polymerase, which has been in high-volume production for decades and is a workhorse of modern molecular biology, is readily accessible and stable at room temperature and lowers costs significantly (Fig. 3B). Last, we compared lateral-flow test strip visualization to CREST, and while we found them as sensitive as fluorescent detection methods (Fig. 3A), their high cost and difficulty to obtain can limit their distribution and scaled use in this pandemic.
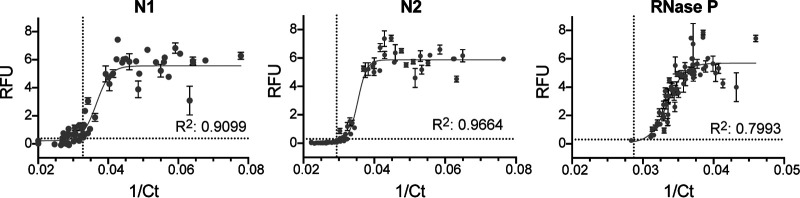
To test the efficacy of CREST on human samples, we obtained 64 de-identified nasopharyngeal (NP) swabs from individuals collected through the Santa Barbara County Department of Public Health. We purified RNA from these samples, which were stored in viral transport medium, using a commercially available RNA extraction kit (QIAamp Mini Elute virus spin kit; Qiagen). We used this RNA as input for a parallel comparison between CREST and the CDC-recommended one-step TaqMan assay (our end-to-end CREST protocol is described in the supplemental material). Considering that CREST was designed to provide a binary outcome, we fit the CREST-to-TaqMan comparison to a sigmoid. We then calculated a goodness-of-fit R value between assays for detection of N1, N2, and RNase P (Fig. 4). These analyses revealed high concordance between CREST and TaqMan assays (R2 > 0.9 for SARS-CoV-2 genes and R2 > 0.79 for RNase P). Of note, CREST appears to be more sensitive than TaqMan for detection of N1, whereas the converse appears to be true for N2. We conducted two additional comparisons, one on 95 asymptomatic individuals at University of California, Santa Barbara (UCSB), where oropharyngeal (OP) self-sampling was carried out, and a second one on 30 positive and 30 negative NP/OP samples obtained from University of California, San Francisco (UCSF) (Fig. S6; Data Set S2). Taken together, the results show that CREST had a sensitivity of 97% and a specificity of 98% (Fig. 5).
FIG 4.
Comparative analysis between CREST and TaqMan for SARS-CoV-2 N1 and N2 sites and RNase P. De-identified human samples are shown as gray circles. Error bars show standard deviations (n = 4). Dotted lines indicate the detection threshold for each assay.
FIG 5.
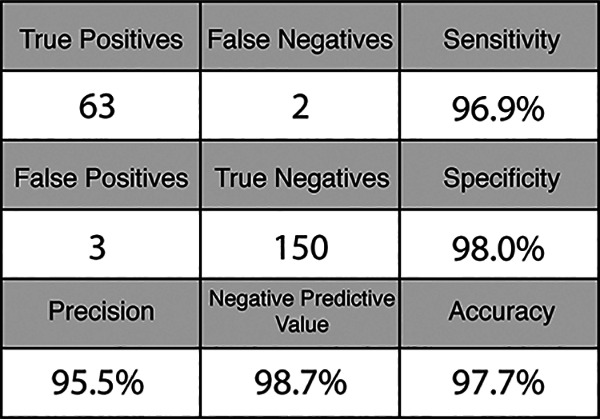
Confusion matrix of combined results from Santa Barbara County Department of Public Health, UCSF, and UCSB asymptomatic clinical sample sets.
Finally, while we designed CREST to be an accessible and scalable assay for detecting SARS-CoV-2, it still requires RNA extraction using commercial kits, which limits its widespread adoption. In a companion paper, we present a method called PEARL (precipitation-enhanced analyte retrieval) which uses common laboratory reagents to bypass this limitation (24). To lift the final obstacle to CREST’s accessibility, we coupled PEARL with CREST and found that commercial RNA extraction could be omitted (Fig. S7).
DISCUSSION
The unparalleled spread of SARS-CoV-2 demands orthogonal solutions to expand testing capacities. Here, we describe an optimized Cas13-based protocol that uses accessible reagents and affordable equipment while maintaining sensitivity that is comparable to that of established testing methods. Considering the need for a rugged, equitable, and scalable test, we coupled spartan, Bluetooth-enabled thermocyclers that can be plugged into a battery and run using mobile device applications, with simple plastic-filter-based LED visualizers. Results can be simply documented with a smartphone camera and uploaded to the cloud, enabling distributed point-of-care (POC) testing. Operational costs could be further lowered by multiplexing the amplification of viral targets.
Current methods to detect SARS-CoV-2 presence or prior exposure in patient samples rely on detection of the viral genome or antibodies in nucleic acid and serological tests, respectively (24). The detection of viral genome sequences by RT-qPCR is recommended by public health organizations worldwide. However, these recommendations fail to account for the massive need for widespread testing. Implementing this method alone places a single large burden on the manufacturing capacity for RT-qPCR testing reagents and consumables, specialized instrumentation, and trained laboratory personnel, thus hampering widespread testing. Other POC diagnostic platforms still require specialized instrumentation and are low throughput (e.g., Abbott ID NOW).
These limitations underscore the need for alternative methods, such as CRISPR-based detection, to relieve some of the strain on the global supply chain for testing reagents. These state-of-the-art methods are scalable, do not entail specialized instrumentation, and require very little specialized training. CREST can be run, from RNA sample to result, with no need for AC power or a dedicated facility and with minimal handling, in ∼2 h. It also has a throughput comparable to that of RT-qPCR tests. Indeed, in a companion study, we processed ∼300 CREST samples per day with two operators and two thermocyclers (25).
We found CREST to have sensitivity comparable to that of RT-qPCR. Of the 218 clinical samples we tested with both of these methods, we identified 2 false-negative and 3 false-positive results by CREST. The 3 false-positive samples were reported as negative by RT-qPCR because the levels of the N1 target were below the detection threshold, although N2 detection by this method was positive. The 2 false-negative samples were above CREST’s detection threshold for N1 but not N2. Indeed, N1 CREST target detection was consistently slightly more sensitive than that for N2. In a clinical testing scenario, discordant results between N1 and N2 may be resolved by retesting. Finally, because CREST’s signal saturates even at low input levels (Fig. 2B), CREST offers the added advantage of binary result interpretation, similar to lateral-flow test strips, but at a fraction of the price, as it does not require the costly antibodies or antibody conjugates needed for lateral-flow immunochromatography.
By utilizing off-the-shelf components and an efficient purification protocol of Cas13, CREST’s costs remain close to those of RT-qPCR, one of the least expensive testing modalities currently available. We chose to compare CREST’s cost to that of Thermo Fisher’s general-purpose TaqPath reagent because of its validation by the CDC (19) and EUA status. However, purpose-built reaction mixes could make RT-qPCR costs closer to, or even lower than, that of CREST. Though Cas13 is not yet commercially available, we do not foresee its commercial-scale production to be a bottleneck. Indeed, we were able to purify enough protein from a 1-liter bacterial culture for more than 500,000 reactions, which leads us to conclude its cost per reaction is minimal. We expect Cas13 protein cost to be in the range of costs of other enzymes used in this study, between $0.05 and $0.50 per reaction. Other cost considerations include the labor involved in handling samples and setting up reactions.
CREST requires innovations before it can be fully POC capable. CREST entails two pipetting steps, one of which involves opening the reaction tube postamplification. This step increases the possibility of contaminating other samples (26). Removing these steps by either employing thermostable Cas variants that can survive the amplification reaction (27) or using disposable microfluidic cartridges with specialized reaction vessels to separate compartments (28) will be essential for establishing CREST’s full POC capability. Other requirements for field deployment of Cas13 include optimizing long-term enzyme storage conditions. Indeed, our Cas13 master mix is not affected significantly by undergoing numerous freeze-thaw cycles (Fig. S5), which increases CREST’s potential for field deployment by overcoming the need for professional laboratories.
Regular testing may be a key determinant impacting the ability to ascertain foci of infection and reduce false-negative rates. Recent studies document that in many individuals, even those with active symptoms, initial tests can be negative, while subsequent tests are positive, and intermittent detection of the virus in samples can also occur (29–32). Despite the biochemical reliability of currently available diagnostics, the initial quality of a sample and the viral load in the patient can affect the likelihood of a false result. Because of its low costs and ease of use, CREST could be employed for regular testing as well as for disambiguation of results obtained with established methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank Feng Zhang and Michael Springer for the kind gift of Cas13a purification plasmids. We thank Radu Gogoana for his helpful comments on scalability and Cameron Myhrvold for his thoughts on assay design. We thank Sebastian Kraves and Ezequiel Alvarez-Saavedra for the P51 visualization devices. We are grateful to Stewart W. Comer of the Santa Barbara County Public Health Department for testing advice and for granting access to SARS-CoV-2 positive samples. We thank Steve Miller at the UCSF Clinical Microbiology Laboratory for providing SARS-CoV-2 clinical samples. We thank Keith Blaha for designing a Web database to store CREST data. We thank Jennifer Smith and the Biological Nanostructures Laboratory within the California NanoSystems Institute for their equipment. We thank Monte Radeke for use of his fast protein liquid chromatograph (FPLC), Apeel Sciences for use of their FPLC columns, and Kevin Brackett for technical assistance.
We thank UC Santa Barbara Office of Research and IEX for their generous support.
REFERENCES
- 1.Mizumoto K, Kagaya K, Zarebski A, medRxiv GC. 2020. Estimating the asymptomatic ratio of 2019 novel coronavirus onboard the Princess Cruises ship, 2020. medrxiv.org.
- 2.Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, Ma H, Chen W, Lin Y, Zheng Y, Wang J, Hu Z, Yi Y, Shen H. 2020. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 4.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 5.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, Cuomo-Dannenburg G, Thompson H, Walker PGT, Fu H, Dighe A, Griffin JT, Baguelin M, Bhatia S, Boonyasiri A, Cori A, Cucunubá Z, FitzJohn R, Gaythorpe K, Green W, Hamlet A, Hinsley W, Laydon D, Nedjati-Gilani G, Riley S, van Elsland S, Volz E, Wang H, Wang Y, Xi X, Donnelly CA, Ghani AC, Ferguson NM. 2020. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis 20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onder G, Rezza G, Brusaferro S. 2020. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA 323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 7.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. 2020. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson NM, Laydon D, Nedjati-Gilani G, Imai N, Ainslie K, Baguelin M, Bhatia S, Boonyasiri A, Cucunubá Z, Cuomo-Dannenburg G, Dighe A, Dorigatti I, Fu H, Gaythorpe K, Green W, Hamlet A, Hinsley W, Okell LC, van Eisland S, Thompson H, Verity R, Volz E, Wang H, Wang Y, Walker PGT, Walters C, Winskill P, Whittaker C, Donnelly CA, Riley S, Ghani ACH. 2020. Report 9: Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Imperial College, London, United Kingdom.
- 9.Wang CJ, Ng CY, Brook RH. 2020. Response to COVID-19 in Taiwan: big data analytics, new technology, and proactive testing. JAMA 323:1341–1342. doi: 10.1001/jama.2020.3151. [DOI] [PubMed] [Google Scholar]
- 10.Normile D. 17 March 2020. Coronavirus cases have dropped sharply in South Korea. What’s the secret to its success? Science. doi: 10.1126/science.abb7566. [DOI] [Google Scholar]
- 11.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF, Barnes KG, Chak B, Mondini A, Nogueira ML, Isern S, Michael SF, Lorenzana I, Yozwiak NL, MacInnis BL, Bosch I, Gehrke L, Zhang F, Sabeti PC. 2018. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sentmanat M, Kouranova E, Cui X. 2020. One-step RNA extraction for RT-qPCR detection of 2019-nCoV. bioRxiv.
- 13.Ford J, Goldstein T, Trahan S, Neuwirth A, Tatoris K, Decker S. 2020. A 3D-printed nasopharyngeal swab for COVID-19 diagnostic testing. 3D Print Med 6:21. doi: 10.1186/s41205-020-00076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control. 2020. Preparation of viral transport medium.
- 15.Metsky HC, Freije CA, Kosoko-Thoroddsen T-SF, Sabeti PC, Myhrvold C. 2020. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. bioRxiv.
- 16.Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR diagnostics.
- 17.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan C-Y, Guevara H, Wadford DA, Chen JS, Chiu CY. 2020. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding X, Yin K, Li Z, Liu C. 2020. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. [DOI] [PMC free article] [PubMed]
- 19.Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J, Tamin A, Thornburg NJ, Villanueva JM, Lindstrom S. 2020. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Zhang B, Xu X, Long F, Wang J. 2018. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci Rep 8:14126. doi: 10.1038/s41598-018-32329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Gonzalez E, Lizeth Mendoza-Ramos J, Cristina Pedroza S, Alexandra Cuellar-Monterrubio A, Roberto Marquez-Ipina A, Lira-Serhan D, Trujillo-de Santiago G, Moises Alvarez M. 2019. Validation of use of the miniPCR thermocycler for Ebola and Zika virus detection. PLoS One 14:e0215642. doi: 10.1371/journal.pone.0215642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boguraev A-S, Christensen HC, Bonneau AR, Pezza JA, Nichols NM, Giraldez AJ, Gray MM, Wagner BM, Aken JT, Foley KD, Copeland DS, Kraves S, Alvarez Saavedra E. 2017. Successful amplification of DNA aboard the International Space Station. NPJ Microgravity 3:26. doi: 10.1038/s41526-017-0033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomerantz A, Penafiel N, Arteaga A, Bustamante L, Pichardo F, Coloma LA, Barrio-Amoros CL, Salazar-Valenzuela D, Prost S. 2018. Real-time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. Gigascience 7:giy033. doi: 10.1093/gigascience/giy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponce-Rojas JC, Costello MS, Proctor DA, Kosik KS, Wilson MZ, Arias C, Acosta-Alvear D. 2021. A fast and accessible method for the isolation of RNA, DNA, and protein to facilitate the detection of SARS-CoV-2. J Clin Microbiol 59:e02403-20. doi: 10.1128/JCM.02403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauch JN, Valois E, Ponce-Rojas JC, Aralis Z, Lach RS, Zappa F, Audouard M, Solley SC, Vaidya C, Costello M, Smith H, Javanbakht A, Malear B, Polito L, Comer S, Arn K, Kosik KS, Acosta-Alvear D, Wilson MZ, Fitzgibbons L, Arias C. 2020. Rapid CRISPR-based surveillance of SARS-CoV-2 in asymptomatic college students captures the leading edge of a community-wide outbreak. medRxiv. [Google Scholar]
- 26.Aslanzadeh J. 2004. Preventing PCR amplification carryover contamination in a clinical laboratory. Ann Clin Lab Sci 34:389–396. [PubMed] [Google Scholar]
- 27.Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong S, Makarova KS, Koonin EV, Cheng DR, Scott DA. 2019. Functionally diverse type V CRISPR-Cas systems. Science 363:88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauk MG, Song J, Liu C, Bau HH. 2018. Simple approaches to minimally-instrumented, microfluidic-based point-of-care nucleic acid amplification tests. Biosensors (Basel) 8:17. doi: 10.3390/bios8010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. 2020. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. 2020. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology 296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D, Xu W, Zhang C, Yu J, Jiang B, Cao H, Li L. 2020. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID-19) in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis 71:706–712. doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, Ng O-T, Marimuthu K, Ang LW, Mak TM, Lau SK, Anderson DE, Chan KS, Tan TY, Ng TY, Cui L, Said Z, Kurupatham L, Chen MI-C, Chan M, Vasoo S, Wang L-F, Tan BH, Lin RTP, Lee VJM, Leo Y-S, Lye DC, Coronavirus S2N. 2020. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.