The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). While molecular-based testing is used to diagnose COVID-19, serologic testing of antibodies specific to SARS-CoV-2 is used to detect past infection.
KEYWORDS: antibody, antibody testing, COVID-19, infectious disease, SARS-CoV-2, serology, virology, immunoassays
ABSTRACT
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). Molecular-based testing is used to diagnose COVID-19, and serologic testing of antibodies specific to SARS-CoV-2 is used to detect past infection. While most serologic assays are qualitative, a quantitative serologic assay was recently developed that measures antibodies against the S protein, the target of vaccines. Quantitative antibody determination may help determine antibody titer and facilitate longitudinal monitoring of the antibody response, including antibody response to vaccines. We evaluated the quantitative Roche Elecsys anti-SARS-CoV-2 S assay. Specimens from 167 PCR-positive patients and 103 control specimens were analyzed using the Elecsys anti-SARS-CoV-2 S assay on the cobas e411 (Roche Diagnostics). Analytical evaluation included assessing linearity, imprecision, and analytical sensitivity. Clinical evaluation included assessing clinical sensitivity, specificity, cross-reactivity, positive predictive value (PPV), negative predictive value (NPV), and serial sampling from the same patient. The Elecsys anti-SARS-CoV-2 S assay exhibited its highest sensitivity (84.0%) at 15 to 30 days post-PCR positivity and exhibited no cross-reactivity, a specificity and PPV of 100%, and an NPV between 98.3% and 99.8% at ≥14 days post-PCR positivity, depending on the seroprevalence estimate. Imprecision was <2% at 9.06 U/ml across 6 days, the negative quality control (QC) was consistently negative (<0.40 U/ml), the manufacturer’s claimed limit of quantitation of 0.40 U/ml was verified, and linearity across the analytical measuring range was observed, except at the low end (<20 U/ml). Lastly, antibody response showed high interindividual variation in level and time of peak antibody titer and trends over time.
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). First declared a Public Health Emergency of International Concern in January 2020 by the World Health Organization (WHO), COVID-19 has infected over 88 million people globally, causing over 1.9 million deaths as of 10 January 2021 (https://www.who.int/publications/m/item/weekly-epidemiological-update---12-january-2021). While molecular-based testing is used to diagnose COVID-19 (1), serologic testing of antibodies specific to SARS-CoV-2 is used to detect past infection (2). Serologic testing may aid in surveying asymptomatic infection, assessing past SARS-CoV-2 infection prevalence, or serving as an adjunct for COVID-19 diagnosis if used ≥15 days after symptom onset in cases with suggestive clinical presentation but negative or unavailable reverse transcriptase PCR (RT-PCR) results (3–6). Most serologic assays are qualitative and use either full-length or truncated versions of the nucleocapsid (N) or spike (S) SARS-CoV-2 protein as the target for antibody detection. Roche recently developed a quantitative serologic assay that measures antibodies against the receptor-binding domain of the S protein, the target of vaccines in development and in use (7), and thus may aid in characterizing the immune response to vaccines. The S protein facilitates viral entry to various host cells via binding to angiotensin-converting enzyme 2 (ACE2) (8), and antibodies directed against the S protein have been shown to have potent antiviral activity and correlate to potential immunity (9). Quantitative determination of anti-SARS-CoV-2 antibodies may help determine specific antibody titer, facilitate longitudinal monitoring of the antibody response in individual patients, and specifically monitor antibody response to vaccines.
In this study, we clinically and analytically evaluated the quantitative anti-SARS-CoV-2 S assay, including assessment of linearity, precision, analytical and clinical sensitivity, specificity, cross-reactivity, positive and negative predictive value, and serial sampling from the same patients.
MATERIALS AND METHODS
This work was exempt from quality improvement (QI) review and Research Ethics Board (REB) approval at the University Health Network (UHN; Toronto, Canada). Deidentified residual patient serum and plasma samples were collected from UHN and analyzed using the Elecsys anti-SARS-CoV-2 S assay on the cobas e411 (Roche Diagnostics) for the quantitative detection of antibodies to the SARS-CoV-2 spike protein receptor binding domain. This assay is a double-antigen sandwich electrochemiluminescence immunoassay, which uses streptavidin-coated microparticles to separate bound from unbound substances prior to applying a voltage to the electrode. This assay has a measuring range of 0.40 to 250 U/ml (up to 2,500 U/ml with on-board 1:10 dilution), with a concentration of <0.80 U/ml considered negative and ≥0.80 U/ml considered positive. To determine the presence or absence of SARS-CoV-2 infections, SARS-CoV-2 viral RNA was measured in nasopharyngeal swabs using the Seegene Allplex 2019-nCoV assay.
Linearity across the claimed analytical measuring range (AMR; 0.40 to 250 U/ml) was assessed by various protocols. In the first linearity assessment, linearity was assessed by mixing high (1,097 U/ml) and low (<0.40 U/ml) patient plasma sample pools to create 14 samples, including the neat high and low sample pools. Secondly, linearity was assessed by mixing high (1,012 U/ml) and low (<0.40 U/ml) single patient plasma samples to create eight samples, including the neat high and low samples. Next, a high patient plasma sample (1,012 U/ml) was diluted with manufacturer provided diluent to create eight samples, including the neat high sample and neat diluent. Lastly, the lower end of the measuring range was assessed by similarly diluting a patient sample (64.2 U/ml) with manufacturer provided diluent to create nine samples, including the neat high sample and neat diluent.
Imprecision was assessed using two levels of quality control (QC) material (i.e., <0.40 U/ml and 9.06 U/ml) across 6 days. Analytical sensitivity was assessed by verifying the manufacturer’s claimed limit of quantitation (LoQ) of 0.40 U/ml by analyzing five samples with low concentrations (i.e., 1.05 U/ml, 0.58 U/ml, 0.51 U/ml, <0.40 U/ml, <0.40 U/ml) in replicates of five.
Clinical sensitivity was determined using serum or plasma samples collected from 167 patients that were confirmed positive for SARS-CoV-2 infection by PCR testing within the previous 0 to 73 days. Total sensitivity and sensitivity in different categories of days post-PCR positivity were determined.
Cross-reactivity was determined using serum or plasma samples collected from 103 patients that were positive for viruses other than SARS-CoV-2 (e.g., hepatitis A, hepatitis B, hepatitis C, human immunodeficiency virus, rubella, Epstein-Barr virus, cytomegalovirus), had autoantibodies or a known autoimmune condition, had elevations of other analytes (e.g., C-reactive protein, IgA, IgG, IgM), or had the influenza vaccine in 2019. Specificity was assessed using 32 of these samples that were collected from patients in 2019, before SARS-CoV-2 was thought to be circulating in Ontario, Canada.
Positive predictive value (PPV) and negative predictive value (NPV) at seroprevalence values of 1%, 5%, and 10% were calculated using sensitivity for <14 days and ≥14 days post-PCR positivity as well as specificity as determined in this study.
Lastly, antibody titers were examined over time since PCR positivity using serial serum and plasma samples (n = 6 to 20) collected from five patients.
RESULTS
Linearity of the Elecsys anti-SARS-CoV-2 S assay was assessed using various protocols, which all similarly showed a linear response across the AMR, except for the lower end, particularly <20 U/ml (see Fig. S1 in the supplemental material). Assessing linearity by mixing high and low patient plasma sample pools exhibited an average percent difference of 5.60% from the expected linear relationship, except for values at the lower end with expected concentrations of 11.0 U/ml and 5.49 U/ml exhibiting percent difference values of 67.8% and 69.8%, respectively (Table 1). Mixing a high and low patient sample and mixing a high patient sample with diluent produced similar results, with a greater deviation from a linear response at lower concentrations (see Table S1 in the supplemental material).
TABLE 1.
Linearity assessment of the quantitative Roche SARS-CoV-2 S assaya
| Sample | Rep 1 (U/ml) | Rep 2 (U/ml) | Avg (U/ml) | Expected (U/ml) | Dilution factor | Difference (U/ml) | % difference |
|---|---|---|---|---|---|---|---|
| Linearity assessment 1 | |||||||
| Neat high sample | 1,102 | 1,092 | 1,097 | 1,097 | N/Ab | N/A | N/A |
| 1 | 1,021 | 1,030 | 1,026 | 987 | 1.11 | 38.2 | 3.87 |
| 2 | 917 | 932 | 925 | 878 | 1.25 | 47.2 | 5.38 |
| 3 | 767 | 780 | 773 | 768 | 1.43 | 5.40 | 0.70 |
| 4 | 691 | 705 | 698 | 658 | 1.67 | 39.8 | 6.04 |
| 5 | 568 | 569 | 568 | 549 | 2 | 19.7 | 3.59 |
| 6 | 407 | 415 | 411 | 439 | 2.5 | −27.8 | −6.32 |
| 7 | 319 | 321 | 320 | 329 | 3.33 | −9.05 | −2.75 |
| 8 | 218 | 229 | 224 | 219 | 5 | 4.40 | 2.01 |
| 9 | 126 | 129 | 127 | 110 | 10 | 17.7 | 16.1 |
| 10 | 24.7 | 25.1 | 24.9 | 27.4 | 40 | −2.53 | −9.21 |
| 11 | 18.2 | 18.6 | 18.4 | 11.0 | 100 | 7.44 | 67.8 |
| 12 | 9.18 | 9.45 | 9.32 | 5.49 | 200 | 3.83 | 69.8 |
| Neat low sample | <0.40 | <0.40 | <0.40 | <0.40 | N/A | N/A | N/A |
All values are reported up to 3 significant figures.
N/A, not available.
Imprecision was unable to be calculated for the low QC material (mean, <0.40 U/ml), as all results were <0.40 U/ml. For the high QC material (mean, 9.06 U/ml), the imprecision was 1.26% across 6 days and two reagent lots. The manufacturer’s claimed LoQ of 0.40 U/ml was verified with a coefficient of variation (CV) of <4% (see Table S2 in the supplemental material).
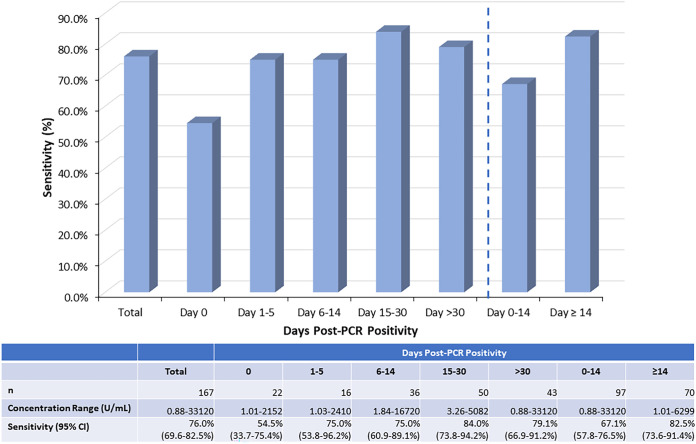
Total sensitivity was 76.0% (95% confidence interval [CI], 69.6% to 82.5%), with the highest sensitivity observed 15 to 30 days post-PCR positivity (84.0% [95% CI, 73.8% to 94.2%]) and the lowest sensitivity observed on the same day as PCR positivity (54.5% [95% CI, 33.7% to 75.4%]), although the CIs slightly overlapped (Fig. 1). Sensitivity was higher at ≥14 days post-PCR positivity (82.5% [95% CI, 73.6% to 91.4%]) compared to that at <14 days post-PCR positivity (67.1% [95% CI, 57.8% to 76.5%]), although the CIs slightly overlapped.
FIG 1.
Total sensitivity and sensitivity for 0, 1 to 5, 6 to 14, 15 to 30, >30, 0 to 14, and ≥14 days post-PCR positivity for the quantitative Roche Elecsys anti-SARS-CoV-2 S assay using serum or plasma samples collected from 167 patients confirmed SARS-CoV-2 positive within the previous 0 to 73 days.
We previously determined the sensitivity of qualitative anti-SARS-CoV-2 serology assays, including the qualitative Roche anti-SARS-CoV-2 assay, using the same patient samples (10). No samples that were positive by the qualitative Roche anti-SARS-CoV-2 assay were negative by the quantitative Roche anti-SARS-CoV 2 S assay. However, seven samples (five samples <14 days post-PCR positivity and two samples ≥14 days) were positive by the quantitative Roche anti-SARS-CoV 2 S assay but negative by the qualitative Roche anti-SARS-CoV-2 assay. The overall sensitivity of the quantitative assay was slightly higher, although not significantly different, than the qualitative assay (total sensitivity of 76.0% [95% CI, 69.6% to 82.5%] compared to 73.6% [95% CI, 67.0% to 80.1%], respectively).
The antibody concentration ranges for all positive samples and positive samples for each category of days post-PCR positivity are provided in Fig. 1. An extensively wide total concentration range of positive samples was observed, ranging from 0.88 to 33,120 U/ml, with 59.1% of positive samples having a concentration of >250 U/ml. While the upper AMR is 250 U/ml, it is increased to 2,500 U/ml with the on-board 1:10 dilution. This upper limit could then be further increased for the 11.0% of positive samples with concentrations >2,500 U/ml by performing manual dilutions with manufacturer-recommended universal diluent in addition to on-board dilution. For example, to obtain the highest concentration of 33,120 U/ml, a 1:100 manual dilution was performed in addition to the on-board 1:10 dilution to ultimately perform a 1:1,000 dilution.
For the cross-reactivity assessment, all 103 samples were negative for anti-SARS-CoV-2 S, with only one value above the LoQ (0.40 U/ml) at 0.56 U/ml but still below the positivity cutoff of ≥0.80 U/ml. When assessing 32 of these samples for specificity (collected from patients in 2019), all samples were below the LoQ, resulting in a specificity of 100%.
PPV was 100% for <14 days and ≥14 days post-PCR positivity across all seroprevalence estimates because of 100% specificity (Table 2). NPV ranged from 96.8% (95% CI, 95.7% to 97.9%) to 99.7% (95% CI, 99.3% to 100%) for <14 days post-PCR positivity and 98.3% (95% CI, 97.5% to 99.1%) to 99.8% (95% CI, 99.6% to 100%) for ≥14 days post-PCR positivity, depending on the seroprevalence estimate.
TABLE 2.
Positive and negative predictive values at 1%, 5%, and 10% seroprevalence for the quantitative Roche SARS-CoV-2 S assay
| Parameter | Estimate (95% CI) |
|---|---|
| <14 Days Post-PCR Positivity | |
| Sensitivity | 67.1 (57.8–76.5) |
| Specificity | 100 (100–100) |
| Seroprevalence | |
| 1% | |
| PPV | 100 (100–100) |
| NPV | 99.7 (99.3–100) |
| 5% | |
| PPV | 100 (100–100) |
| NPV | 98.4 (97.6–99.2) |
| 10% | |
| PPV | 100 (100–100) |
| NPV | 96.8 (95.7–97.9) |
| ≥14 Days Post-PCR Positivity | |
| Sensitivity | 82.5 (73.6–91.4) |
| Specificity | 100 (100–100) |
| Seroprevalence | |
| 1% | |
| PPV | 100 (100–100) |
| NPV | 99.8 (99.6–100) |
| 5% | |
| PPV | 100 (100–100) |
| NPV | 99.1 (98.6–99.7) |
| 10% | |
| PPV | 100 (100–100) |
| NPV | 98.3 (97.5–99.1) |
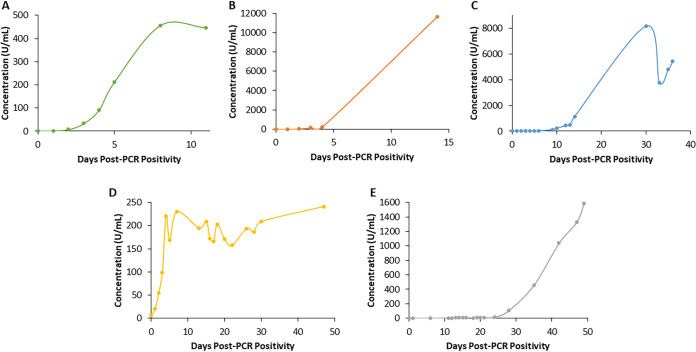
The antibody titers examined over time since PCR positivity for the first patient are shown in Fig. 2A. This patient was monitored up to 11 days and exhibited an exponential increase in antibody titer that peaked at 8 days post-PCR positivity and subsequently plateaued. The second patient (Fig. 2B), monitored up to 14 days, exhibited a gradual increase in antibody titer up to 4 days, and on day 14, a value over 50-fold higher was obtained. The third patient (Fig. 2C), monitored up to 36 days, exhibited peak titers on day 30, followed by a trough, with levels subsequently rising again. The fourth patient (Fig. 2D), monitored up to 47 days, exhibited an exponential increase in antibody titer up to day 4, with levels subsequently plateauing, although with variation. Lastly, the fifth patient (Fig. 2E), monitored up to 49 days, exhibited a much later increase in antibody titers, with levels beginning to rise at 28 days and not yet plateauing or declining by day 49. Overall, a very heterogenous antibody response between patients was observed, varying in level and time of peak antibody titer and trends over time.
FIG 2.
Anti-SARS-CoV-2 antibody response by days post-PCR positivity in five patients as measured by the quantitative Roche Elecsys anti-SARS-CoV-2 S assay.
DISCUSSION
We performed an analytical and clinical evaluation of the quantitative anti-SARS-CoV-2 S assay. Analytically, we assessed linearity, precision, and analytical sensitivity. The poor linearity observed below 20 U/ml by mixing patient pools, mixing single patient samples, or using diluent may not be particularly concerning, as samples should not be diluted down to concentrations this low in clinical practice. Indeed, the Roche package insert states that the concentration of the diluted sample must be ≥20 U/ml. Our linearity assessments involved creating and measuring dilution series, which determines whether the measured concentration changes as expected according to the proportional relationship between samples created with different dilution factors. Poor linearity below 20 U/ml would be concerning if inaccuracy was observed in this concentration range, perhaps if materials of known concentration (not created by dilution) were analyzed across the measuring range. The concentrations of both QC materials used for imprecision assessment were <20 U/ml and were within the manufacturer’s stated ranges, suggesting accuracy at low concentrations. Imprecision was minimal (i.e., 1.26% at 9.06 U/ml), and the manufacturer’s claimed LoQ of 0.40 U/ml was verified.
Clinically, we assessed clinical sensitivity, specificity, cross-reactivity, positive and negative predictive values, and serial sampling from the same patients. While we report the same observation as the manufacturer of higher sensitivity at ≥14 days post-PCR positivity compared to that at <14 days, we obtained lower estimates than the manufacturer’s claimed sensitivity of 98.8% at ≥14 days after diagnosis with positive PCR and 86.1% at <14 days. This may be due to the patient population characteristics, particularly the large immunocompromised population at UHN. Overall, 90.0% of patients positive for SARS-CoV-2 by PCR testing but negative by the Elecsys anti-SARS-CoV-2 S assay were also negative on four other qualitative serologic assays (10), supporting the inability of these patients to produce antibodies against SARS-CoV-2. Another potential reason that our sensitivity estimates were lower than those claimed by the manufacturer could be the potential preferential inclusion or requirement of individuals with low cycle threshold values on PCR by the manufacturer, therefore including subjects with a relatively higher viral load. To ensure that the reason for our lower sensitivity was not simply having antibody titers just below the positivity cutoff (i.e., between the LoQ of 0.40 U/ml and the positivity cutoff of 0.80 U/ml), we recalculated the total sensitivity and sensitivity for different categories based on days since PCR positivity using 0.40 U/ml as the positivity cutoff. No sensitivity estimates significantly changed, as only two samples had antibody concentration between 0.40 U/ml and 0.80 U/ml. It is important to note that negative results cannot rule out current or past SARS-CoV-2 infection because results are negative in the preseroconversion phase of infection, some patients with confirmed SARS-CoV-2 infection do not develop antibodies, and antibody titers may wane in individuals within months of infection (11, 12).
Our specificity estimate of 100% is in accordance with the manufacturer’s claimed specificity of 99.98%. We also recalculated the specificity using 0.40 U/ml as the positivity cutoff, and the specificity did not significantly change (i.e., decreased from 100% [95% CI, 100% to 100%] to 99.0% [95% CI, 97.1% to 100%]). We observed no cross-reactivity in 103 samples tested that contained potentially cross-reacting substances (e.g., samples from patients with autoimmune disease or other viral infections), similar to the manufacturer (i.e., no cross-reactivity in 1,100 samples tested). Due to the estimated specificity of 100%, PPV was also 100% for <14 days and ≥14 days post-PCR positivity across all seroprevalence estimates. NPV was slightly lower than PPV, was higher at ≥14 days post-PCR positivity compared to that at <14 days, and decreased with higher seroprevalence estimates.
We previously reported on heterogeneity in antibody response observed with four qualitative serologic assays (i.e., Abbott SARS-CoV-2 IgG and SARS-CoV-2 IgM, DiaSorin SARS-CoV-2 S1/S2 IgG, and Roche Elecsys anti-SARS-CoV-2 total) by representing antibody levels relative to the manufacturer supplied cutoff value (10). Others have also reported on interindividual difference in SARS-CoV-2 antibody responses (4, 13), yet this is the first report of SARS-CoV-2 antibody trends using a quantitative assay. As mentioned previously, quantitative determination of anti-SARS-CoV-2 antibodies may help facilitate longitudinal monitoring of the antibody response in individual patients and specifically monitor antibody response to vaccines. As observed from the results of antibody titer in serial samples from PCR-positive patients, this assay was useful in delineating various trends over time in antibody response. However, we did not specifically examine the ability of this assay to monitor antibody response to vaccines. While this test may have utility in this area due to the antigen used to capture the antibodies in the assay exhibiting similarity to that used in the vaccines, monitoring the response to vaccines with antibody neutralization assays would likely be more clinically useful to ensure subjects develop a functional antibody response and develop immunity (14). Lastly, in terms of using this quantitative anti-SARS-CoV-2 antibody assay to determine specific antibody titer, it is important to note that the majority of positive samples had a concentration above the upper AMR (>250 U/ml). However, with the on-board 1:10 dilution, the upper limit in increased to 2,500 U/ml, and only 11.0% of samples had to be manually diluted in addition to performing the on-board dilution to obtain a quantitative result.
Overall, the Elecsys anti-SARS-CoV-2 S assay exhibited a highest sensitivity of 84.0% 15 to 30 days post-PCR positivity, no cross-reactivity, specificity and PPV of 100%, and NPV of between 98.3% and 99.8% at ≥14 days post-PCR positivity, depending on the seroprevalence estimate. Linearity across the AMR was observed, except at the low end, particularly for antibody titers of <20 U/ml. Imprecision was minimal, and the LoQ of 0.40 U/ml was verified. Lastly antibody response showed high interindividual variation in level and time of peak antibody titer as well as trends over time.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UHN Core Lab Specimen Management team, Tech IVs, and biochemists for their help with sample retrieval and input.
Reagent kits were provided by Roche Diagnostics.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso JM, Gregg AC, Soares DJ, Beskid TR, Jervey SR, Liu C. 2020. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci 6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Caeseele P, Bailey D, Forgie SE, Dingle TC, Krajden M, COVID-19 Immunity Task Force. 2020. SARS-CoV-2 (COVID-19) serology: implications for clinical practice, laboratory medicine and public health. CMAJ 192:E973–E979. doi: 10.1503/cmaj.201588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, Adriano A, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Dittrich S, Emperador D, Hooft L, Leeflang MM, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group. 2020. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev 6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, Liao P, Qiu J-F, Lin Y, Cai X-F, Wang D-Q, Hu Y, Ren J-H, Tang N, Xu Y-Y, Yu L-H, Mo Z, Gong F, Zhang X-L, Tian W-G, Hu L, Zhang X-X, Xiang J-L, Du H-X, Liu H-W, Lang C-H, Luo X-H, Wu S-B, Cui X-P, Zhou Z, Zhu M-M, Wang J, Xue C-J, Li X-F, Wang L, Li Z-J, Wang K, Niu C-C, Yang Q-J, Tang X-J, Zhang Y, Liu X-M, Li J-J, Zhang D-C, Zhang F, Liu P, Yuan J, Li Q, Hu J-L, Chen J, et al. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 5.Kofler N, Baylis F. 2020. Ten reasons why immunity passports are a bad idea. Nature 581:379–381. doi: 10.1038/d41586-020-01451-0. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein MC, Freedberg KA, Hyle EP, Paltiel AD. 2020. Waiting for certainty on Covid-19 antibody tests—at what cost? N Engl J Med 383:e37. doi: 10.1056/NEJMp2017739. [DOI] [PubMed] [Google Scholar]
- 7.Zhu F-C, Guan X-H, Li Y-H, Huang J-Y, Jiang T, Hou L-H, Li J-X, Yang B-F, Wang L, Wang W-J, Wu S-P, Wang Z, Wu X-H, Xu J-J, Zhang S, Jia S-Y, Wang B-S, Hu Y, Liu J-J, Zhang J, Qian X-A, Qiong L, Pan H-X, Jiang H-D, Deng P, Gou J-B, Wang X-W, Wang X-H, Chen W. 2020. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. 2020. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci 63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins V, Fabros A, Wang XY, Bhandari M, Daghfal DJ, Kulasingam V. 2020. Analytical and clinical evaluation of four anti-SARS-CoV-2 serologic (IgM, IgG, and total) immunoassays. medRxiv doi: 10.1101/2020.10.23.20217810. [DOI] [Google Scholar]
- 11.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, Elliott J, Hofmann C, Hausner MA, Ferbas KG, Tobin NH, Aldrovandi GM, Yan OO. 2020. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med 383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu A, Wang W, Zhao X, Zhou X, Yang D, Lu M, Lv Y. 2020. Disappearance of antibodies to SARS-CoV-2 in a -COVID-19 patient after recovery. Clin Microbiol Infect 26:1703–1705. doi: 10.1016/j.cmi.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 2020. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis 71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muruato AE, Fontes-Garfias CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, Shi P-Y. 2020. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun 11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.