Accommodating large increases in sample workloads has presented a major challenge to clinical laboratories during the coronavirus disease 2019 (COVID-19) pandemic. Despite the implementation of automated detection systems and previous efficiencies, including barcoding, electronic data transfer, and extensive robotics, capacities have struggled to meet the demand.
KEYWORDS: COVID-19, enhanced throughput, laboratory capacity, pooling
ABSTRACT
Accommodating large increases in sample workloads has presented a major challenge to clinical laboratories during the coronavirus disease 2019 (COVID-19) pandemic. Despite the implementation of automated detection systems and previous efficiencies, including barcoding, electronic data transfer, and extensive robotics, capacities have struggled to meet the demand. Sample pooling has been suggested as an additional strategy to address this need. The greatest concern with this approach in clinical settings is the potential for reduced sensitivity, particularly detection failures with weakly positive samples. To investigate this possibility, detection rates in pooled samples were evaluated, with a focus on pools containing weakly positive specimens. Additionally, the frequencies of occurrence of weakly positive samples during the pandemic were reviewed. Weakly positive specimens, with threshold cycle (CT) values of 33 or higher, were detected in 95% of 60 five-sample pools but only 87% of 39 nine-sample pools. The proportion of positive samples with very low viral loads rose markedly during the first few months of the pandemic, peaking in June, decreasing thereafter, and remaining level since August. At all times, weakly positive specimens comprised a significant component of the sample population, ranging from 29% to >80% for CT values above 31. In assessing the benefits of pooling strategies, however, other aspects of the testing process must be considered. Accessioning, result data management, electronic data transfer, reporting, and billing are not streamlined and may be complicated by pooling procedures. Therefore, the impact on the entire laboratory process needs to be carefully assessed prior to implementing such a strategy.
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic has presented numerous challenges to the health care industry in general and laboratory testing specifically. Not least among the latter has been a dramatic increase in the sample testing load (1). Efforts to meet the demand have included the increased use of automated instrumentation, multiplexing of molecular detection assays, streamlined testing protocols, as well as increasingly varied acceptable sample types, collection devices, and transport media (2–4). Accommodating the testing workload and reagent shortage during the symptomatic pandemic wave was a significant undertaking (1). However, more recent tasks, testing returning health care workers and patients returning for elective services and other clinical procedures as well as mass screening of numerous asymptomatic populations, have generated an even greater challenge. One proposed solution has been sample pooling (5–8), a method previously used for numerous other situations where large-scale testing was needed (9–12).
When used as an epidemiological surveillance tool, acceptable parameters and limits may be quite different from those when pooled testing is applied in a clinical testing setting (6, 13–16). In the latter case, the issue of detection sensitivity for every individual specimen becomes critical. Methods for pooling vary widely, and testing large numbers of pooled samples is not possible without an inherent loss of sensitivity. However, there is the potential for more limited pooling without a significant loss of sensitivity. We sought to investigate the extent to which this was possible while maintaining the detection of weakly positive samples.
MATERIALS AND METHODS
The CDC 2019 novel coronavirus (nCoV) real-time reverse transcription-PCR (RT-PCR) diagnostic panel (17–19) was used throughout, with extraction on the bioMérieux eMAG system (bioMérieux Inc., Durham, NC). For individual specimens, 110 μl of viral transport medium (VTM; Regeneron, Rensselaer, NY) from an upper respiratory swab was extracted into 110 μl of the eluate.
The bioMérieux eMAG extraction system accommodates a maximum volume of 3 ml. Therefore, a maximum of nine 110-μl samples can be added to the 2-ml lysis buffer tube. Assuming that the same extraction efficiency is maintained when nine samples are loaded as when one is loaded, and the eluate is still 110 μl, the same total nucleic acid from each sample should be extracted. Provided that there is no increase in the PCR inhibition of the pooled eluate, the detection sensitivity should therefore also still be the same for each sample.
Pool sizes of five and nine samples were tested, with each pool containing a single severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-positive specimen and the remaining four (five-sample pool) or eight (nine-sample pool) specimens being negative specimens. In an initial experiment, strongly, moderately, and weakly positive samples in VTM were tested for each pool size. In subsequent experiments, 20 weakly positive specimens were tested in five-sample pools, and 13 weakly positive specimens were tested in nine-sample pools. All pools were prepared and tested in triplicate.
Additionally, the positivity rate and the percentage of positive samples at different viral loads, as assessed by threshold cycle (CT) values, were reviewed across the entire duration of the pandemic to determine when pooling might have been feasible if weakly positive specimens comprise a significant component of the total tested specimens and whether this proportion has changed over the course of the pandemic.
This study was approved by the New York State Institutional Review Board under study number 07-022.
RESULTS
In initial experiments, when pooled specimens with high, medium, or low viral loads were compared, the detection of samples with lower CT values (high viral loads) was not impaired in either five- or nine-sample pools (Table 1). However, in one of the nine-sample pools, the detection of one of the weak specimens (specimen D) shifted from positive to inconclusive, with an N1 CT value of 32.25 and an N2 CT value of 40.74. The CT values of positive samples were increased by 0.4 to 1.51 CTs when pooled with four negative samples. In contrast, pooling with 8 negative samples caused CT increases ranging from 0.94 to 8.49 when comparing the CT of the single specimen to that of the pool.
TABLE 1.
Detection of strongly, moderately, and weakly SARS-CoV-2-positive nasopharyngeal swab specimens in five- and nine-sample pools
| Pool size (no. of samples) | Positive specimenb | N1a |
N2a |
||||
|---|---|---|---|---|---|---|---|
| Sample CT | Pool CT | ΔCT | Sample CT | Pool CT | ΔCT | ||
| 5 | A | 18.60 | 19.00 | 0.40 | 19.17 | 19.59 | 0.42 |
| B | 24.27 | 25.18 | 0.90 | 24.40 | 25.41 | 1.01 | |
| C | 29.61 | 30.44 | 0.83 | 29.85 | 30.46 | 0.61 | |
| D | 33.09 | 32.59 | 0.50 | 32.25 | 33.76 | 1.51 | |
| 9 | A | 18.60 | 22.71 | 4.11 | 19.17 | 22.79 | 3.62 |
| B | 24.27 | 25.33 | 1.06 | 24.40 | 25.34 | 0.94 | |
| C | 29.61 | 30.57 | 0.95 | 29.85 | 30.85 | 0.99 | |
| D | 33.09 | 36.20 | 3.11 | 32.25 | 40.74 | 8.49 | |
Real-time RT-PCR assay virus target.
bIn VTM.
The effect of pooling was then assessed more stringently on weakly positive specimens, all of which had CT values of 33 or higher when tested individually. A total of 20 such specimens were retrieved for these experiments and used to produce 60 five-sample pools and 39 nine-sample pools for comparative testing. When combined with negative specimens, these weakly positive specimens were still detected in 95% of the five-specimen pools (Table 2), failing detection in one of the three replicate pools for specimens with original CT values of 34.92, 35.38, and 36.01. However, in the nine-specimen pools, detection dropped to 87% (Table 3), with 5 of the 39 pools being undetected, from corresponding specimens with original CT values ranging from 35.38 to 36.13. Of note, the changes in CT values between the N1 and N2 targets across the five- and nine-sample pools are not consistent with this overall picture: the N1 CT changes are slightly greater for nine-sample pools than for five-sample pools, while the reverse is true for N2 CT changes (Tables 2 and 3).
TABLE 2.
Low-titer SARS-CoV-2-positive nasopharyngeal swab specimens in five-specimen poolsa
| Positive sample | No. of PCR-positive pools/no. of pool replicates | N1 |
N2 |
||||
|---|---|---|---|---|---|---|---|
| Sample CT | Pool CTb | ΔCTb | Sample CT | Pool CTb | ΔCT (range)b | ||
| A | 3/3 | 35.56 | 35.89 | 0.33 | 35.63 | 36.40 | 0.81 |
| B | 3/3 | 35.15 | 34.70 | −0.45 | 34.29 | 36.09 | 1.80 |
| C | 3/3 | 36.13 | 36.52 | 0.39 | 38.05 | 36.81 | −1.24 |
| D | 3/3 | 36.23 | 35.31 | −0.92 | 39.07 | 36.83 | −2.24 |
| E | 3/3d | 33.36 | 34.80 | 1.44 | 33.98 | 38.49 | 4.51 |
| F | 3/3 | 33.49 | 35.24 | 1.75 | 34.78 | 36.33 | 1.55 |
| G | 3/3 | 33.33 | 33.28 | −0.05 | 35.10 | 34.42 | −0.68 |
| H | 3/3 | 33.09 | 35.03 | 1.94 | 34.06 | 36.78 | 2.72 |
| I | 3/3 | 33.03 | 34.04 | 1.01 | 33.86 | 35.23 | 1.37 |
| J | 3/3 | 33.34 | 33.53 | 0.19 | 34.00 | 35.89 | 1.89 |
| K | 2/3c,d | 34.92 | 36.88 | 1.96 | 37.69 | 38.56 | 0.90 |
| L | 3/3 | 34.63 | 36.43 | 1.80 | 36.36 | 39.31 | 2.95 |
| M | 3/3d | 34.84 | 35.90 | 1.06 | 36.11 | 36.29 | 0.18 |
| N | 3/3d | 34.37 | 36.70 | 2.33 | 36.59 | 38.14 | 1.55 |
| O | 2/3c,d | 35.38 | 35.32 | −0.06 | 36.29 | 36.53 | 0.24 |
| P | 3/3 | 34.18 | 35.70 | 1.52 | 34.84 | 35.23 | 0.39 |
| Q | 2/3c,e | 36.01 | Undetected | NA | 35.61 | 38.90 | 3.29 |
| R | 3/3 | 33.65 | 35.05 | 1.40 | 35.01 | 37.74 | 2.73 |
| S | 3/3 | 33.72 | 35.40 | 1.68 | 35.45 | 39.67 | 4.22 |
| T | 3/3f | 35.55 | 37.04 | 1.49 | 37.21 | 38.61 | 1.40 |
| Aggregate datag | 57/60 (95%) | 34.50 (33.01–36.01) | 35.20 (33.64–37.34) | 0.97 (−0.92–2.63), 0.67 | 35.70 (33.86–37.21) | 36.95 (34.87–38.90) | 1.58 (−2.24–3.29), 1.80 |
Each positive specimen (n = 20) was pooled with four negative specimens. Each pool was prepared three times such that pools were prepared and tested in triplicate. NA, not applicable.
Values represent the means from three replicates unless otherwise noted.
N1 and N2 were undetected in one of three replicates.
N1 was undetected in one of three replicates. The pool would be deconvoluted for individual sample testing.
N1 was undetected in two of three replicates. Pools would be deconvoluted for individual sample testing.
N2 was undetected in two of three replicates. Pools would be deconvoluted for individual sample testing.
Aggregate data are shown as the total number of PCR-positive pools/total number of pools tested (percent); mean (range); or mean (range), standard deviation.
TABLE 3.
Low-titer SARS-CoV-2-positive nasopharyngeal swab specimens in nine-specimen poolsa
| Positive sample | No. of PCR-positive pools/no. of pool replicates | N1 |
N2 |
||||
|---|---|---|---|---|---|---|---|
| Sample CT | Pool CTb | ΔCTb | Sample CT | Pool CTb | ΔCTb | ||
| A | 1/3d,e | 35.56 | 36.12 | 0.56 | 35.63 | 39.51 | 3.95 |
| B | 3/3 | 35.15 | 35.24 | 0.09 | 34.29 | 36.00 | 1.71 |
| C | 2/3c | 36.13 | 36.52 | 0.38 | 38.05 | 37.59 | −0.46 |
| D | 3/3 | 36.23 | 36.11 | −0.12 | 39.07 | 37.07 | −2.00 |
| E | 3/3 | 33.36 | 34.19 | 0.83 | 33.98 | 34.43 | 0.45 |
| F | 3/3 | 33.49 | 35.65 | 2.16 | 34.78 | 36.71 | 1.93 |
| G | 3/3 | 33.33 | 35.71 | 2.38 | 35.10 | 35.80 | 0.70 |
| H | 3/3f,g | 34.92 | 36.30 | 1.38 | 37.69 | 37.97 | 0.28 |
| L | 3/3e,g | 34.63 | 36.92 | 2.29 | 36.36 | 37.46 | 1.10 |
| O | 1/3d,e | 35.38 | Undetected | NA | 36.29 | 37.26 | 0.97 |
| P | 3/3 | 34.18 | 35.77 | 1.59 | 34.84 | 36.02 | 1.18 |
| Q | 3/3f,g | 36.01 | 36.51 | 0.50 | 35.61 | 37.22 | 1.61 |
| R | 3/3e,g | 33.65 | 36.63 | 2.98 | 35.01 | 38.63 | 3.62 |
| Aggregate datah | 34/39 (87%) | 34.77 (33.33–36.23) | 35.97 (34.19–36.92) | 1.25 (−0.12–2.98), 1.02 | 35.90 (33.98–38.05) | 37.05 (34.43–39.51) | 1.16 (−2.00–3.95), 1.56 |
Each positive specimen (n = 13) was pooled with eight negative specimens. Each pool was prepared three times such that pools were prepared and tested in triplicate. NA, not applicable.
Values represent the means from three replicates unless otherwise noted.
N1 and N2 were undetected in one of three replicates.
N1 and N2 were undetected in two of three replicates.
N1 was undetected in one of three replicates. The pool would be deconvoluted for individual sample testing.
N1 was undetected in two of three replicates. Pools would be deconvoluted for individual sample testing.
N2 was undetected in one of three replicates. The pool would be deconvoluted for individual sample testing.
Aggregate data are shown as the total number of PCR-positive pools/total number of pools tested (percent); mean (range); or mean (range), standard deviation.
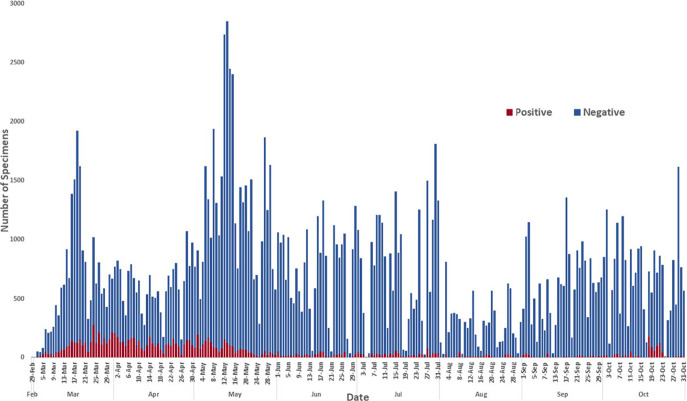
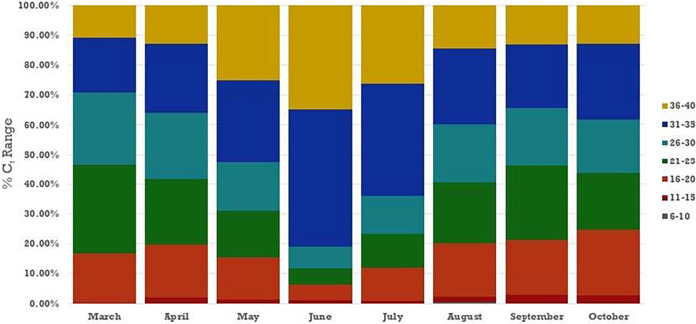
We then sought to assess the positivity rate during the months since the onset of the pandemic in New York because pooling strategies are not efficient unless the sample positivity rate is low. Despite the large range and fluctuations in the number of specimens tested per day from late February to the end of October (Fig. 1), the positivity rate remained consistently low after late May, except for a brief surge in late October. The potential impact of pooling on the detection sensitivity of positive samples during the pandemic was also assessed by determining the percentage of positive samples tested that were comprised of weakly positive samples. The distribution of viral loads in specimens across time showed an interesting picture. From March through June, the percentage of samples with very low viral loads (CT values of 36 to 40) and low viral loads (CT values of 31 to 35) continually increased. In March, these two groups combined accounted for 30% of positive specimens, and by June, the two groups comprised 80% of positive samples. By July, the trend reversed, and the distribution has remained relatively consistent for the last few months (Fig. 2).
FIG 1.
Number of specimens tested per day at the Wadsworth Center for SARS-CoV-2, from 29 February to 31 October 2020.
FIG 2.
Percentage of specimens in each category of viral load, as approximated by CT range, by month, from March through October 2020. CT values are the averages of the N1 and N2 values for each specimen.
DISCUSSION
As the pandemic has evolved, case counts, hospitalization rates, and fatalities have risen and fallen with repeated waves, and laboratories continue to face new challenges. Workloads have increased with requirements to perform surveillance screening of asymptomatic health care workers, residents in congregate settings, students, and patients undergoing elective procedures, in addition to symptomatic patients. These policies have pushed test numbers beyond those encountered even at the height of the pandemic wave. Suggestions to help manage the load have included the pooling of samples to enhance throughput capacity. When being considered for application in a clinical testing environment, of greatest concern is the potential for this strategy to increase the false-negative rate. With that, the greatest risk of false-negative results is with pooling of weakly positive samples with negative samples. To investigate the potential limits of pooling in this situation, this study focused on pooling weakly positive samples in relatively small pool sizes.
There are multiple methods for pooling samples, some of which carry an inherent risk of reducing test sensitivity and some of which do not. In the method described in this paper, the same volumes of each sample as those that would normally be added if the samples were being tested individually are added to lysis buffer and eluted. Therefore, theoretically, the pooling strategy should not adversely affect sensitivity as long as the extraction efficiency is maintained and the level of PCR inhibition is not increased by the additional load through the extraction device. When the data were analyzed, despite some minor shifts in CT values, a minimal loss of detection was observed in the five-pool experiments. However, when the same weakly positive samples were pooled with 8 negative samples rather than 4, to create pools of 9, more detection failures were observed, with failure rates increasing from 5% to 13%.
The use of pooling as a throughput enhancement strategy is efficient only if the positivity rate of the samples being tested is low enough such that a minimal number of pools will test positive; otherwise, multiple pools will have to be deconvoluted for retesting of individual samples. The optimal or maximum positivity level at which pooling starts to become efficient depends on the pool size being used. For pool sizes as small as 5, this maximum positivity level is considerably higher than that for very large pool sizes, which are sometimes suggested for large-scale epidemiological screening studies. However, large pools incur additional issues such as the potential adverse impact of overloading nucleic acid into the test system and disruption to optimal ratios and concentrations of magnesium and other key reaction components. At a positivity rate of 1% and a pool size of 5, on average, only 1 in every 20 pools will be positive and need to be deconvoluted for retesting. Therefore, for every 100 samples, testing could be achieved with a total of 25 tests (20 pools and 1 deconvoluted pool). The positivity rate in our laboratory was generally <1.5% from May to October, and such a strategy may have been efficient for extraction and detection procedures then. However, a positivity surge in October caused by two large outbreaks would have been disruptive, and more recent rises would have made the strategy inefficient.
Importantly, as the pandemic progressed from February through June, there was an increasing proportion of samples in the weakly positive range, and major consideration must be given to the importance of detection sensitivity for weak samples during this peak time of a pandemic. The subsequent decrease in the proportion of these high-CT-value specimens from July onward is interesting, although the reasons are unclear. It may be associated with late maturation of a pandemic, altered patient populations for which the laboratory was receiving samples for testing, or the pandemic entering its second or third wave. Perhaps the subsequent leveling out of the distribution of viral loads reflects the community entering an endemic epidemic rather than a pandemic.
It must be noted that extraction and detection are not the only components of laboratory operation affected by pooling, and other aspects of processing may be complicated rather than enhanced. Processes for specimen receiving and accessioning, as well as those for result data management, reporting, and billing, are not reduced by pooling, and it may be especially difficult to adjust electronic data transfer and laboratory information systems to accommodate these strategies. A large increase in throughput facilitated by a pooling strategy may in fact create serious bottlenecks for these areas of the operation. Therefore, the global implications need to be carefully assessed, especially in the clinical testing environment, before implementation.
ACKNOWLEDGMENTS
We thank Kamran Zamani for assistance with data management and the preparation of figures. We also thank Jennifer Laplante, Rene Hull, and Steven Zink for specimen selection and retrieval. We thank the Wadsworth COVID response team for months of dedicated, skilled testing and careful specimen archiving that provided the well-characterized samples accessed for this work.
S.B.G. and G.V.S. declare no conflict of interest. K.S.G. receives research support from Thermo Fisher for the evaluation of new assays for the diagnosis and characterization of viruses. She also has a royalty-generating collaborative agreement with Zeptometrix.
REFERENCES
- 1.Patel R, Babady E, Theel ES, Storch GA, Pinsky BA, St George K, Smith TC, Bertuzzi S. 2020. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: value of diagnostic testing for SARS-CoV-2/COVID-19. mBio 11:e00722-20. doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. 2020. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pettit SD, Jerome KR, Rouquie D, Mari B, Barbry P, Kanda Y, Matsumoto M, Hester S, Wehmas L, Botten JW, Bruce EA. 2020. ‘All in’: a pragmatic framework for COVID-19 testing and action on a global scale. EMBO Mol Med 12:e12634. doi: 10.15252/emmm.202012634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y, Chang L, Wang L. 2020. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev Med Virol 30:e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, Iwen PC. 2020. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol 153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogan CA, Sahoo MK, Pinsky BA. 2020. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 323:1967–1969. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres I, Albert E, Navarro D. 2020. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J Med Virol 92:2306–2307. doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yelin I, Aharony N, Tamar ES, Argoetti A, Messer E, Berenbaum D, Shafran E, Kuzli A, Gandali N, Shkedi O, Hashimshony T, Mandel-Gutfreund Y, Halberthal M, Geffen Y, Szwarcwort-Cohen M, Kishony R. 2020. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin Infect Dis 71:2073–2078. doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dwyre DM, Fernando LP, Holland PV. 2011. Hepatitis B, hepatitis C and HIV transfusion-transmitted infections in the 21st century. Vox Sang 100:92–98. doi: 10.1111/j.1423-0410.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- 10.Van TT, Miller J, Warshauer DM, Reisdorf E, Jernigan D, Humes R, Shult PA. 2012. Pooling nasopharyngeal/throat swab specimens to increase testing capacity for influenza viruses by PCR. J Clin Microbiol 50:891–896. doi: 10.1128/JCM.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keys JR, Leone PA, Eron JJ, Alexander K, Brinson M, Swanstrom R. 2014. Large scale screening of human sera for HCV RNA and GBV-C RNA. J Med Virol 86:473–477. doi: 10.1002/jmv.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lima GFMC, Levi JE, Geraldi MP, Sanchez MCA, Segurado AAC, Hristov AD, Inoue J, Costa-Nascimento MDJ, Di Santi SM. 2011. Malaria diagnosis from pooled blood samples: comparative analysis of real-time PCR, nested PCR and immunoassay as a platform for the molecular and serological diagnosis of malaria on a large-scale. Mem Inst Oswaldo Cruz 106:691–700. doi: 10.1590/s0074-02762011000600008. [DOI] [PubMed] [Google Scholar]
- 13.Ray KJ, Zhou Z, Cevallos V, Chin S, Enanoria W, Lui F, Lietman TM, Porco TC. 2014. Estimating community prevalence of ocular Chlamydia trachomatis infection using pooled polymerase chain reaction testing. Ophthalmic Epidemiol 21:86–91. doi: 10.3109/09286586.2014.884600. [DOI] [PubMed] [Google Scholar]
- 14.Aragon-Caqueo D, Fernandez-Salinas J, Laroze D. 2020. Optimization of group size in pool testing strategy for SARS-CoV-2: a simple mathematical model. J Med Virol 92:1988–1994. doi: 10.1002/jmv.25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kacena KA, Quinn SB, Howell MR, Madico GE, Quinn TC, Gaydos CA. 1998. Pooling urine samples for ligase chain reaction screening for genital Chlamydia trachomatis infection in asymptomatic women. J Clin Microbiol 36:481–485. doi: 10.1128/JCM.36.2.481-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaiakovou M, Wright J, Pilotte N, Chooneea D, Schar F, Truscott JE, Dunn JC, Gardiner I, Walson JL, Williams SA, Littlewood DTJ. 2019. Pooling as a strategy for the timely diagnosis of soil-transmitted helminths in stool: value and reproducibility. Parasit Vectors 12:443. doi: 10.1186/s13071-019-3693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention RVB, Division of Viral Diseases. 2020. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. [Google Scholar]
- 18.Centers for Disease Control and Prevention RVB, Division of Viral Diseases. 2021. CDC 2019-nCoV real-time RT-PCR diagnostic panel, acceptable alternative primer and probe sets. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/downloads/List-of-Acceptable-Commercial-Primers-Probes.pdf. [Google Scholar]
- 19.Centers for Disease Control and Prevention RVB, Division of Viral Diseases. 2020. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel, instructions for use. Centers for Disease Control and Prevention, Atlanta, GA. https://www.fda.gov/media/134922/download. [Google Scholar]