Kaposi’s sarcoma and primary effusion lymphoma cells are latently infected with Kaposi’s sarcoma-associated herpesvirus (KSHV), whereas KSHV replication is frequently observed in multicentric Castleman disease. Although KSHV replication can be induced by some chemical reagents (e.g., 12-O-tetradecanoylphorbol-13-acetate), the mechanism of KSHV replication is not fully understood.
KEYWORDS: Kaposi’s sarcoma-associated herpesvirus, apoptosis, histone acetylation, histone deacetylase inhibitors, lymphoma
ABSTRACT
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi’s sarcoma as well as primary effusion lymphoma (PEL), an aggressive B-cell neoplasm that mostly arises in immunocompromised individuals. Lytic replication of KSHV is also associated with a subset of multicentric Castleman diseases. At present, there is no specific treatment available for PEL, and its prognosis is poor. In this study, we found that the histone deacetylase inhibitor suberoyl bis-hydroxamic acid (SBHA) induced KSHV reactivation in PEL cells in a dose-dependent manner. Next-generation sequencing analysis showed that >40% of all transcripts expressed in SBHA-treated PEL cells originated from the KSHV genome, compared with <1% in untreated cells. Chromatin immunoprecipitation assays demonstrated that SBHA induced histone acetylation targeting the promoter region of the KSHV replication and transcription activator gene. However, there was no significant change in the methylation status of the promoter region of this gene. In addition to its effect on KSHV reactivation, this study revealed that SBHA induces apoptosis in PEL cells in a dose-dependent manner, inducing the acetylation and phosphorylation of p53, cleavage of caspases, and expression of proapoptotic factors such as Bim and Bax. These findings suggest that SBHA reactivates KSHV from latency and induces apoptosis through the mitochondrial pathway in PEL cells. Therefore, SBHA can be considered a new tool for the induction of KSHV reactivation and could provide a novel therapeutic strategy against PEL.
IMPORTANCE Kaposi’s sarcoma and primary effusion lymphoma cells are latently infected with Kaposi’s sarcoma-associated herpesvirus (KSHV), whereas KSHV replication is frequently observed in multicentric Castleman disease. Although KSHV replication can be induced by some chemical reagents (e.g., 12-O-tetradecanoylphorbol-13-acetate), the mechanism of KSHV replication is not fully understood. We found that the histone deacetylase inhibitor suberoyl bis-hydroxamic acid (SBHA) induced KSHV reactivation with high efficiency through histone acetylation in the promoter of the replication and transcription activator gene, compared with 12-O-tetradecanoylphorbol-13-acetate. SBHA also induced apoptosis through the mitochondrial pathway in KSHV-infected cells, with a lower half-maximal effective concentration (EC50) than that measured for viral reactivation. SBHA could be used in a highly efficient replication system for KSHV in vitro and as a tool to reveal the mechanism of replication and pathogenesis of KSHV. The ability of SBHA to induce apoptosis at lower levels than those needed to stimulate KSHV reactivation indicates its therapeutic potential.
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) (also known as human herpesvirus 8 [HHV-8]) is a member of the gammaherpesviruses, first discovered in AIDS-associated Kaposi’s sarcoma (1). KSHV has also been identified as the etiological agent of several lymphoproliferative disorders, including primary effusion lymphoma (PEL) and multicentric Castleman disease (2, 3). Similar to other herpesviruses, the life cycle of KSHV consists of two distinct phases: latent and lytic infection (4). Various environmental and physiological factors trigger KSHV reactivation from latency. For instance, hypoxia and reactive oxygen species have been shown to induce lytic infection (5, 6). The lytic phase is also inducible in vitro by some chemical reagents, including 12-O-tetradecanoylphorbol-13-acetate (TPA) and histone deacetylase (HDAC) inhibitors (7, 8).
PEL is an aggressive B-cell lymphoma that usually presents as serous effusions without solid-mass formation, arising in body cavities such as the pleural, pericardial, or peritoneal cavities (9). Most cases occur in immunocompromised individuals such as patients with AIDS or severe immunodeficiency or recipients of solid-organ transplantation (9). Currently, no specific treatment is available for PEL, and its prognosis is unfavorable (10). The median survival time for PEL patients was 6.2 months under a combination of a CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone)-like regimen and antiretroviral therapy, which is typically administered to AIDS-related PEL patients (11).
HDAC inhibitors are a class of chemical compounds that exhibit various antitumor effects, including apoptosis, growth arrest, differentiation, and autophagy (12). However, the mechanism of action is complex, as HDAC inhibitors affect gene expression profiles through the induction of histone acetylation (12). To date, four HDAC inhibitors have been approved by the U.S. Food and Drug Administration (FDA) for the treatment of hematological malignancies, although none of these are clinically available for PEL treatment (13).
In a previous study, we reported that suberoyl bis-hydroxamic acid (SBHA), an HDAC inhibitor, strongly induced KSHV lytic infection and decreased cell viability in a PEL cell line (14). However, the mechanisms underlying these effects have yet to be elucidated. In this study, we demonstrate that SBHA induced histone acetylation on the promoter region of the replication and transcription activator (RTA) gene of KSHV, resulting in KSHV reactivation. We also found that SBHA induced the apoptosis of PEL cell lines through the mitochondrial pathway.
RESULTS
SBHA reactivated KSHV from latency.
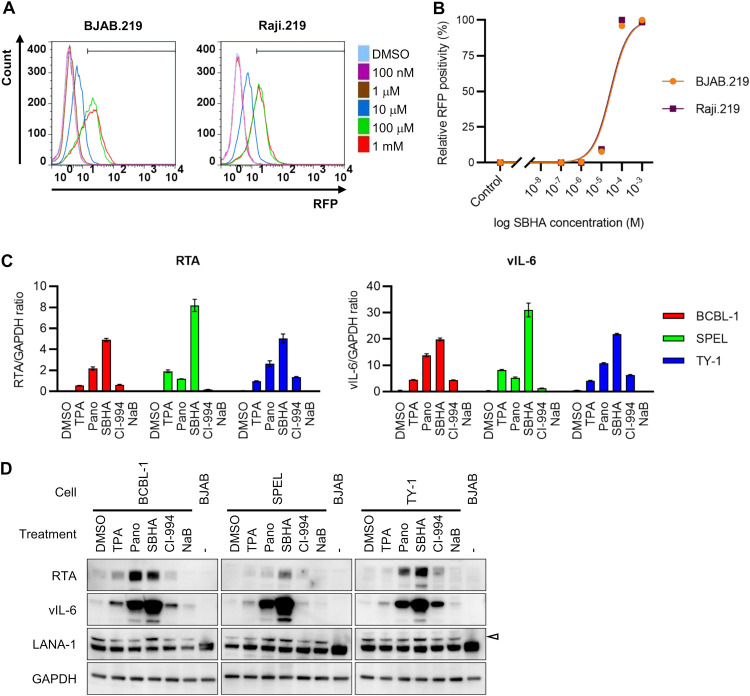
To investigate the effect of SBHA on KSHV reactivation, two rKSHV.219-infected Burkitt lymphoma cell lines (BJAB.219 and Raji.219) were exposed to various concentrations of SBHA and analyzed by flow cytometry. rKSHV.219 is a recombinant KSHV that expresses red fluorescent protein (RFP) from the KSHV lytic PAN promoter and green fluorescent protein (GFP) from the EF-1a promoter (15). The analysis revealed that SBHA reactivated rKSHV.219 from latency in a dose-dependent manner (Fig. 1A and B). The half-maximal effective concentrations (EC50s) for rKSHV.219 reactivation in BJAB.219 and Raji.219 cells were calculated to be 2.95 × 10−5 M and 2.71 × 10−5 M, respectively (Fig. 1B). To examine whether SBHA induces KSHV reactivation in PEL cells, the expression of KSHV mRNA was determined by real-time reverse transcription (RT)-PCR, and protein levels were assessed by Western blotting. Real-time RT-PCR analysis showed that the mRNA levels of two KSHV lytic genes, RTA and viral interleukin-6 (vIL-6), were higher in SBHA-treated PEL cells than in those treated with TPA or other HDAC inhibitors (Fig. 1C). Furthermore, Western blot analysis demonstrated that SBHA induced the expression of RTA and vIL-6 proteins more than did TPA and other HDAC inhibitors in PEL cell lines (Fig. 1D). These data indicate that SBHA strongly induces KSHV lytic infection in B cells, including PEL cells.
FIG 1.
KSHV reactivation by SBHA. (A) Flow cytometry analysis of RFP expression in rKSHV.219-infected Burkitt lymphoma cell lines BJAB.219 and Raji.219. The cells were analyzed by flow cytometry 36 h after the addition of various concentrations of suberoyl bis-hydroxamic acid (SBHA) to the culture medium. (B) Dose-response relationship between the SBHA concentration and RFP positivity in BJAB.219 and Raji.219 cells. (C) Real-time RT-PCR analysis of KSHV-encoded RTA (left) and vIL-6 (right) mRNA expression in three KSHV-positive PEL cell lines treated with each drug for 48 h. The averages from three independent experiments are shown, and error bars indicate the standard deviations. (D) Western blot analysis of KSHV-encoded proteins RTA, vIL-6, and LANA-1. The cells were treated with each drug for 48 h. The arrowhead indicates LANA-1. DMSO, dimethyl sulfoxide; TPA, 12-O-tetradecanoylphorbol-13-acetate; Pano, panobinostat; NaB, sodium butyrate.
SBHA dramatically altered the gene expression profile of KSHV-infected PEL cells.
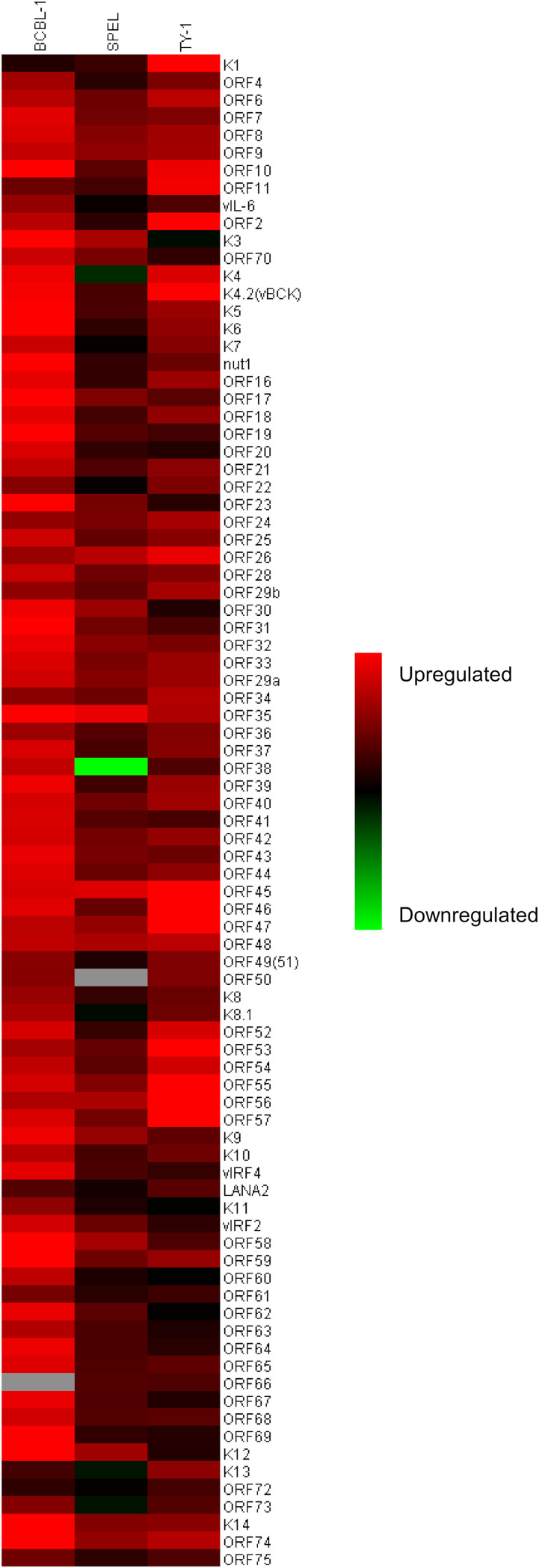
A real-time PCR array was carried out on KSHV gene products to evaluate the effect of SBHA on KSHV gene expression. The KSHV real-time PCR array revealed the global activation of KSHV gene transcription in SBHA-treated PEL cells (Fig. 2; see also Fig. S1 in the supplemental material). SBHA increased almost all KSHV gene transcripts in three different PEL cell lines, although the pattern and degree of change were very different between cell lines.
FIG 2.
KSHV gene expression profile of SBHA-treated PEL cell lines. The heat map was generated from the results of the KSHV real-time PCR array. Red and green indicate upregulation and downregulation of gene transcription, respectively. Gray indicates missing values.
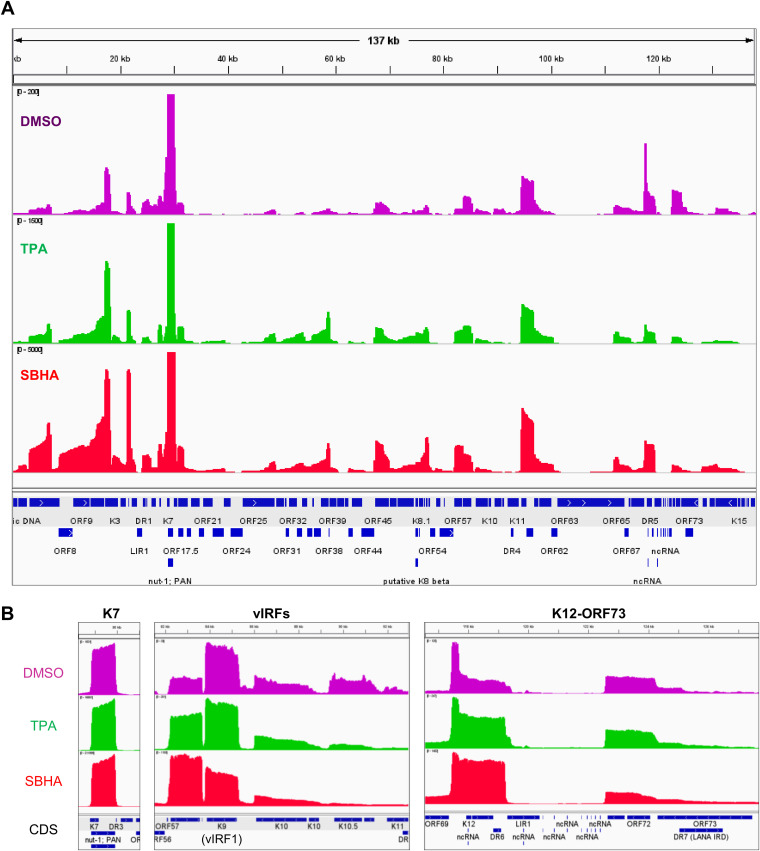
Next, KSHV transcriptome analysis by next-generation sequencing (NGS) was performed. As shown in Table 1, <1% of total reads were mapped to the KSHV genome in untreated BCBL-1 and SPEL cells, and ∼3% were mapped to the KSHV genome in TPA-treated cells. In contrast, more than 40% of the total reads originated from KSHV in SBHA-treated PEL cells (Table 1). Read coverage demonstrated the differences in KSHV gene expression profiles between untreated (dimethyl sulfoxide [DMSO] only) and TPA- and SBHA-stimulated cells (Fig. 3A and Fig. S2A). The number of KSHV-mapped reads was dramatically increased by SBHA stimulation, which increased the expression of almost all lytic genes, including vIL-6 (K2), K4, open reading frame 45 (ORF45), ORF46, ORF59, and ORF65 (Fig. 3A and B and Fig. S2A and B). Notably, the expression profiles of viral interferon regulatory factors (vIRFs) (including K9 [vIRF1], K10 [vIRF4], K10.5 [vIRF3], and K11 [vIRF2]) and latent genes (including K12, ORF71, ORF72, and ORF73) changed after SBHA treatment (Fig. 3B and Fig. S2B). Taken together, these data show that SBHA intensely induces the transcription of KSHV lytic genes and modifies the gene expression profile in PEL cells.
TABLE 1.
Summary of transcriptome reads mapping to the KSHV genome
| Cell type | Treatment | Total no. of reads | No. of KSHV mapped reads | % of total reads |
|---|---|---|---|---|
| BCBL-1 | DMSO | 1,304,979 | 8,522 | 0.65 |
| TPA | 904,733 | 27,672 | 3.06 | |
| SBHA | 657,182 | 296,867 | 45.17 | |
| SPEL | DMSO | 756,210 | 3,076 | 0.41 |
| TPA | 912,306 | 25,383 | 2.78 | |
| SBHA | 628,538 | 275,229 | 43.79 | |
FIG 3.
Transcriptome analysis of KSHV genes in SPEL cells. (A) Overview of the coverage of reads mapped to the KSHV genome. Read coverages of DMSO (violet)-, TPA (green)-, or SBHA (red)-treated SPEL cells mapped to the KSHV genome (GenBank accession no. AP017458) are shown. Maximum coverages in the images of DMSO, TPA, and SBHA are 200, 1,500, and 5,000, respectively. Coverage of the K7 gene is over the maximum reads in each sample. The bottom row indicates coding sequences (CDS) of open reading frames. (B) Read coverage in SPEL cells of K7 (left), vIRFs (middle), and latent gene clusters (K12-ORF73) (right). DR, direct repeat; IRD, internal repeat domain; LIR, long interspersed repeats; ncRNA, noncoding RNA; ORF, open reading frame.
SBHA altered histone modification but not CpG methylation of promoters of KSHV lytic genes.
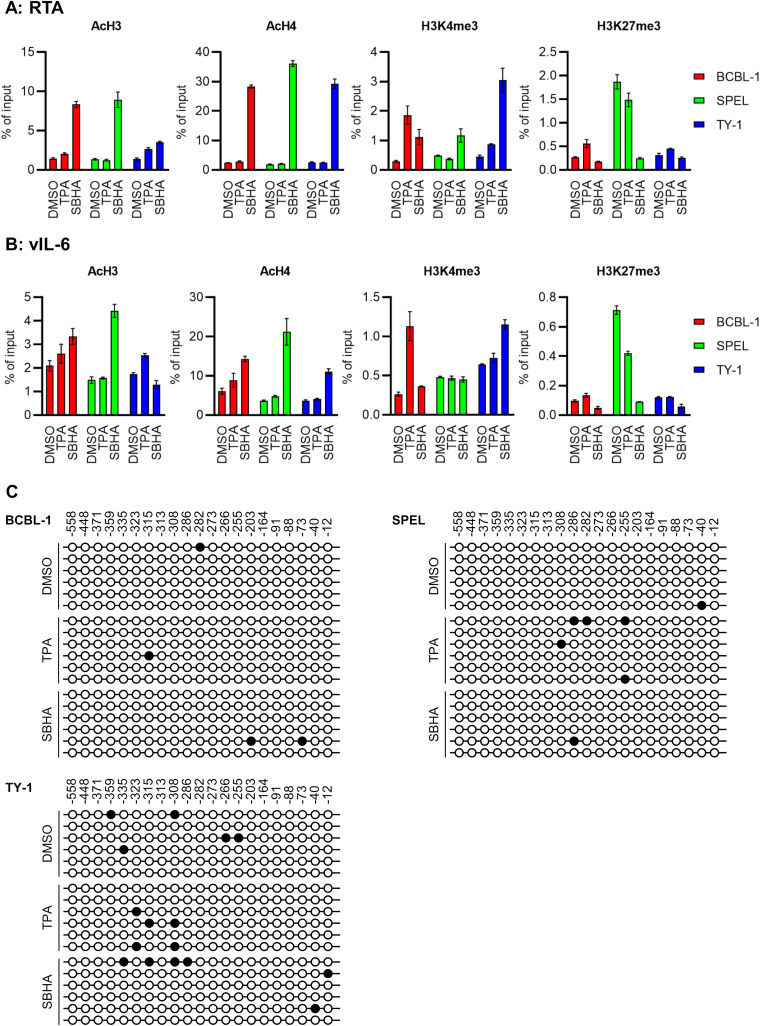
PEL cells were treated with TPA or SBHA for 12 h, and histone modification of the KSHV lytic gene promoter was analyzed. A chromatin immunoprecipitation (ChIP) assay revealed that activating histone modifications, acetyl-histone H3 (AcH3) and acetyl-histone H4 (AcH4), on RTA and vIL-6 promoters were strongly induced by SBHA, while minimal change was induced by TPA (Fig. 4A and B). The level of another activating histone mark, H3K4me3, was increased by SBHA in TY-1 and by TPA in BCBL-1 cells. In contrast, the repressive histone mark H3K27me3 was reduced by SBHA in SPEL cells (Fig. 4A and B).
FIG 4.
Epigenetic effects of SBHA on PEL cells. (A and B) ChIP assays for RTA (A) and vIL-6 (B) promoters. Chromatin samples were reacted with the antibody against modified histones shown at the top of each panel. The averages from three independent experiments are shown, and error bars indicate standard deviations. (C) Bisulfite sequencing of the RTA promoter. Six clones from each cell were subjected to analysis. Closed circles indicate methylated CpG sites, while open ones indicate unmethylated CpG sites. The position relative to the transcription start site of RTA is indicated above the panel.
The methylation pattern of the RTA promoter was also investigated since HDAC inhibitors reverse CpG methylation (16). Bisulfite sequencing analysis revealed that no or only a few CpG sites were methylated at the RTA promoter in PEL cells, regardless of SBHA or TPA treatment (Fig. 4C).
Taken together, these results indicate that the reactivation of KSHV by SBHA is mainly associated with an alteration of histone modifications in the promoter of KSHV lytic genes.
SBHA induced apoptosis and inhibited the growth of PEL cells.
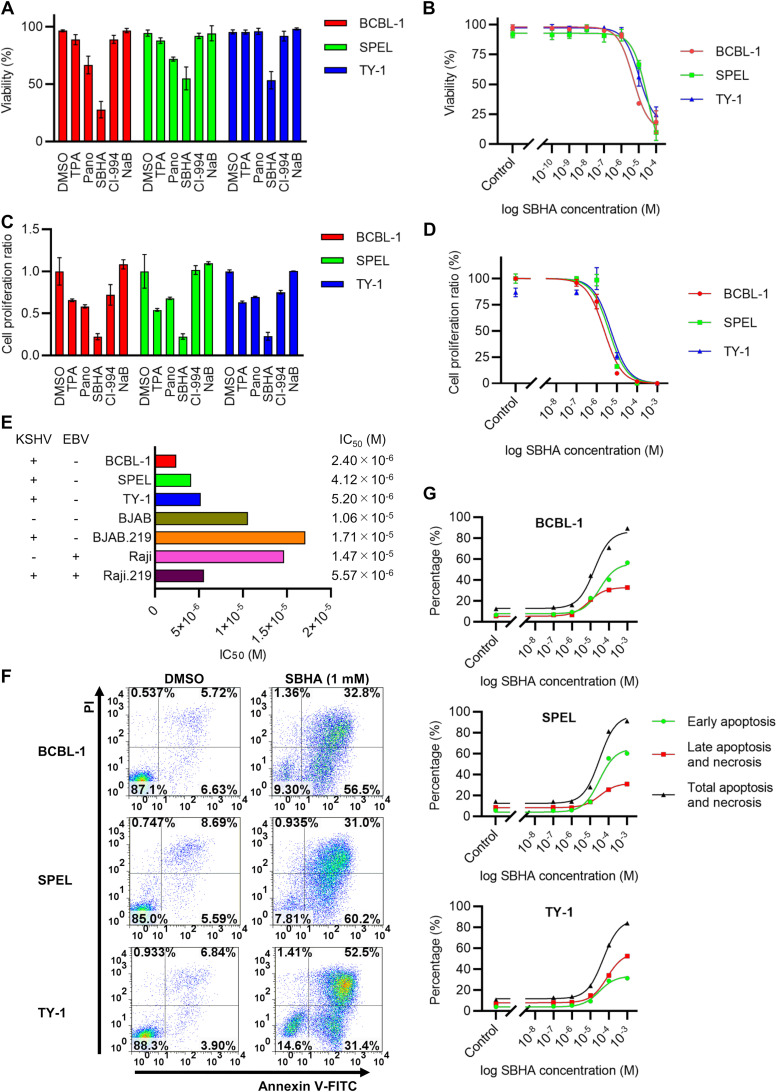
Next, the effect of SBHA on cell growth was investigated. SBHA consistently decreased the viability of PEL cell lines compared with TPA and other drugs following 48 h of drug treatment, as demonstrated by trypan blue staining (Fig. 5A). Notably, SBHA inhibited the growth of PEL cells in a dose-dependent manner (Fig. 5B). The growth-inhibitory effects of SBHA were confirmed by a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt (XTT) assay with results similar to the trypan blue staining data (Fig. 5C and D). Each half-maximal inhibitory concentration (IC50) for cell growth inhibition by SBHA in BCBL-1, SPEL, and TY-1 cells was calculated to be between 2.40 × 10−6 and 5.20 × 10−6 M (Fig. 5D and E). The growth of Burkitt lymphoma cells (BJAB and Raji) and their recombinant KSHV-infected derivatives (BJAB.219 and Raji.219) was also inhibited by SBHA in a dose-dependent manner. However, each associated IC50 was higher than that found in PEL cell lines (Fig. 5E). Finally, annexin V assays demonstrated that SBHA induced apoptosis in PEL cells in a dose-dependent manner (Fig. 5F and G). These data suggest that SBHA inhibits the growth of PEL cells by inducing apoptosis.
FIG 5.
SBHA induces apoptosis in PEL cells. (A) Cell viability after drug treatment for 48 h determined by trypan blue stain. (B) Dose-response relationship between the SBHA concentration and cell viability. (C and D) XTT assay after drug treatment for 48 h. Cell proliferation ratios were compared among TPA and several HDAC inhibitors (C), and the dose-response relationship between the SBHA concentration and cell growth was determined (D). The averages from three independent experiments are shown, and error bars indicate the standard deviations. (E) IC50s of each drug against various cell lines. KSHV and EBV statuses are shown to the left. (F) Flow cytometry of annexin V. The fluorescence of FITC-labeled annexin V and PI was measured following 24 h of exposure to various concentrations of SBHA. The data from control (DMSO-treated) cells and SBHA-treated (1 mM) cells are shown. (G) Dose-response relationship between the SBHA concentration and apoptosis of PEL cells determined by an annexin V assay.
SBHA triggered the mitochondrial pathway of apoptosis by inducing p53 acetylation and phosphorylation.
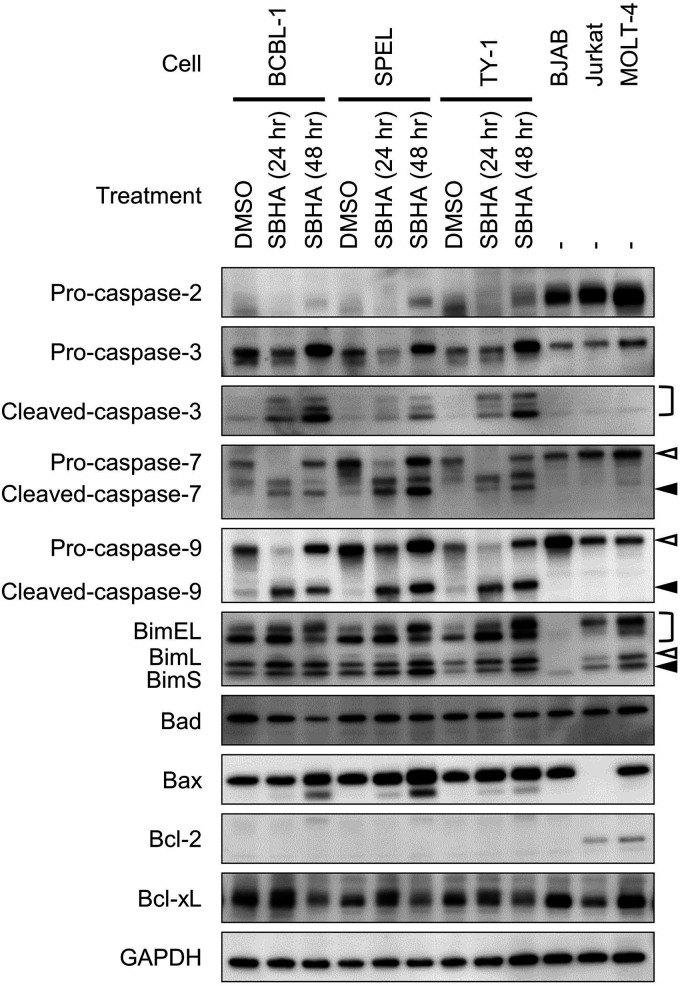
The effect of SBHA on apoptosis-related factors was investigated. Western blot analysis demonstrated that the expression levels of proapoptotic factors of the Bcl-2 family, including Bim and Bax, were upregulated by SBHA in PEL cells (Fig. 6). The cleavage of caspases downstream of Bim and Bax, including caspase-3, caspase-7, and caspase-9, was also stimulated (Fig. 6). Furthermore, SBHA downregulated Bcl-xL, an antiapoptotic factor from the Bcl-2 family (Fig. 6).
FIG 6.
Activation of the mitochondrial pathway of apoptosis induced by SBHA in PEL cells. PEL cells were treated with SBHA for 24 or 48 h. Protein samples from BJAB, Jurkat, and MOLT-4 cells were applied as positive controls, with GAPDH as a loading control. White arrowheads indicate uncleaved caspases or BimL, and black arrowheads indicate cleaved caspases or BimS.
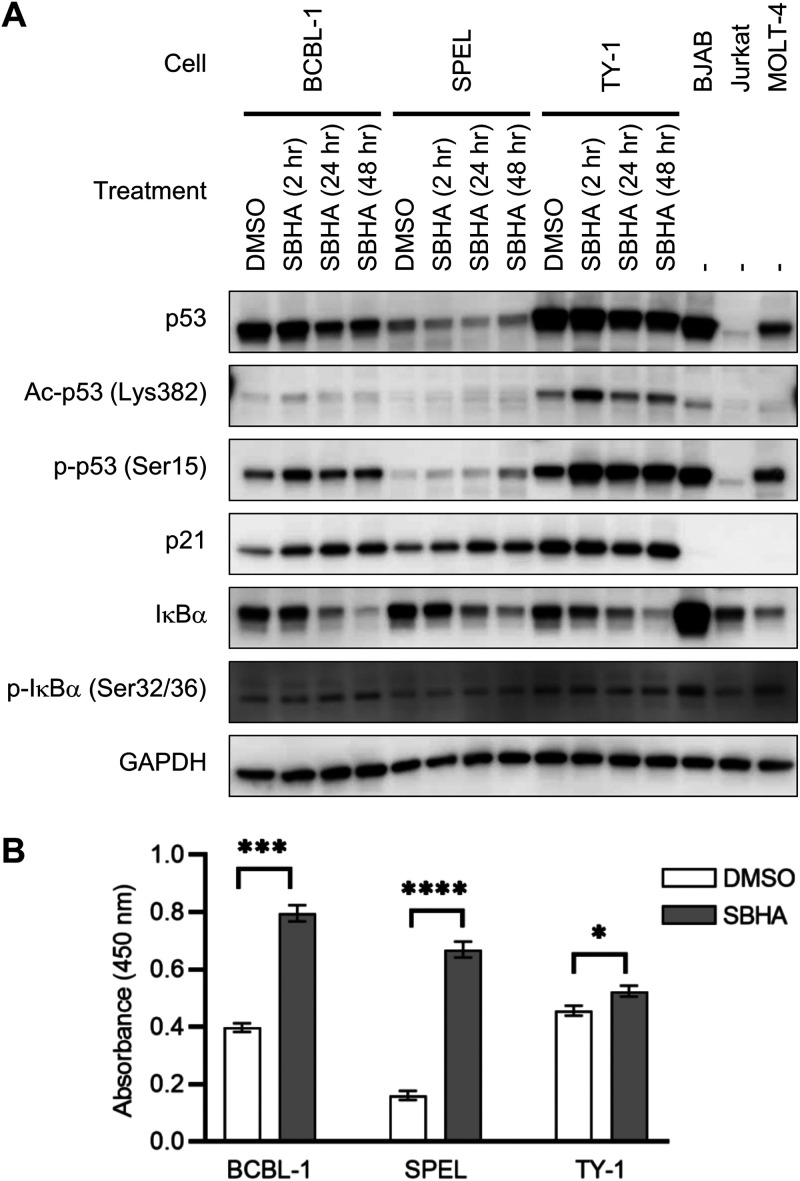
To further elucidate the molecular mechanism underlying the induction of apoptosis in PEL cells, the contribution of p53, a transcriptional activator of Bax, was evaluated (17–20). Western blot analysis demonstrated that acetylation of p53 at lysine 382 (Lys382) was transiently induced 2 h after SBHA stimulation in BCBL-1 and TY-1 cells, while phosphorylation of p53 at serine 15 (Ser15) was induced by SBHA in all three PEL cell lines (Fig. 7A). Furthermore, the expression of p21, a target gene of p53, was induced by SBHA at least in BCBL-1 and SPEL cells (Fig. 7A).
FIG 7.
Activation of signaling pathways of p53 and NF-κB by SBHA. (A) Western blot analysis of p53- and NF-κB-associated proteins. BJAB, Jurkat, and MOLT-4 cells were applied as positive controls. Ac-p53, acetyl-p53; p-p53, phospho-p53; p-IκBα, phospho-IκBα. (B) DNA binding activity of NF-κB p65 determined by an ELISA. SBHA augmented p65 DNA binding activity in nuclear extracts of PEL cells. Data represent the averages from three independent experiments, and error bars indicate standard deviations. Student’s t test was performed to estimate statistical significance (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001).
Next, the role of the NF-κB pathway in apoptosis was examined because NF-κB activation upregulates the expression of antiapoptotic factors such as Bcl-xL (21) and promotes the survival of PEL cells (22, 23). Western blot analysis demonstrated that SBHA induced the phosphorylation of IκBα in BCBL-1 and SPEL cells and decreased IκBα in all PEL cells (Fig. 7A). In addition, an enzyme-linked immunosorbent assay (ELISA) detecting the DNA binding activity of NF-κB p65 revealed that SBHA potentiated the DNA binding activity of p65 in nuclear extracts of PEL cells (Fig. 7B), suggesting the activation of the NF-κB pathway.
Taken together, these data suggest that SBHA induces apoptosis through the activation of p53 and the mitochondrial pathway in PEL cells.
SBHA did not stimulate the Notch1 signaling pathway in PEL cells.
SBHA stimulates the cleavage of the full-length Notch1 protein, leading to the release of the active form of Notch1, the Notch1 intracellular domain (NICD), in thyroid carcinoma and neuroendocrine tumors (24–28). However, such an activation of the Notch1 signaling pathway by SBHA in PEL cells was not detected by Western blot analysis (Fig. S3).
DISCUSSION
HDAC inhibitors are a group of chemical compounds that exhibit antitumor effects. Since suberoylanilide hydroxamic acid (SAHA) (also known as vorinostat) was first approved for clinical use, four HDAC inhibitors have been approved by the FDA for hematological malignancies (13). SBHA, an analogue of SAHA, has been proven to demonstrate antitumor effects in vitro in various types of tumors, including lung, breast, and thyroid carcinomas; melanoma; acute leukemia; and myeloma (24–32). We previously showed that SBHA robustly decreased the viability of SPEL cells compared with other HDAC inhibitors. To the best of our knowledge, this is the first report showing an antitumor effect of SBHA against a virus-associated malignancy (14). Here, we demonstrate that SBHA induces apoptosis in PEL cells through the mitochondrial pathway of apoptosis and reactivates KSHV from latency by altering histone modification in the promoter region of KSHV lytic genes.
According to a previous report by Bhatt et al. (20), SAHA induces massive apoptosis in PEL cells combined with a proteasome inhibitor, bortezomib (Btz). SAHA induces the early acetylation of p53 and its dissociation from MDM2, resulting in the stabilization of p53, whereas Btz induces the phosphorylation of p53 and the activation of the mitochondrial pathway of apoptosis; thus, SAHA and Btz synergistically induce apoptosis in PEL cells (20). Although the combination of SAHA and Btz significantly prolonged the survival of PEL-bearing mice (20), SAHA has no benefit on the efficacy of chemotherapy clinically (33). In contrast to SAHA, we demonstrated that SBHA intensely induced apoptosis in PEL cells without Btz (Fig. 5). This difference could be explained by the effect on p53 activation; SBHA was able to induce the acetylation and phosphorylation of p53 by itself, resulting in the activation of the mitochondrial pathway of apoptosis (Fig. 6 and 7).
The mechanism underlying the antitumor effect of SBHA is not yet fully understood. However, two signaling pathways have been linked to SBHA: the mitochondrial pathway of apoptosis and Notch signaling. In the current study, we demonstrate that the mitochondrial pathway of apoptosis is activated by SBHA in PEL cells (Fig. 6). This effect is likely based on transcriptional regulation mediated by SBHA since the expression of the proapoptotic factors Bim and Bax was upregulated (Fig. 6). In addition, the antiapoptotic factor Bcl-xL was downregulated by SBHA, although its transcriptional regulator NF-κB was activated (Fig. 6 and Fig. 7B), suggesting that the antiapoptotic role of NF-κB activation by SBHA was limited. This result agrees with the hypothesis that SBHA might downregulate antiapoptotic factors through posttranscriptional regulation such as microRNAs (miRNAs) (34). Moreover, similar upregulation of Bim and downregulation of Bcl-xL have been observed in melanoma, suggesting that the activation of the mitochondrial pathway of apoptosis is the major mechanism underlying the antitumor effect of SBHA (29).
Notch signaling is an evolutionarily conserved signaling pathway that plays a crucial role in diverse developmental and physiological processes as well as tumorigenesis (35). Notch signaling is also important for the pathogenesis of KSHV-associated malignancies as it induces the proliferation of PEL cells and the reactivation of KSHV (36, 37). Several reports have demonstrated that SBHA exhibited antitumor activity in neuroendocrine tumors such as carcinoid tumor, pheochromocytoma, and thyroid carcinoma through the upregulation of Notch1 signaling (24–28). However, in this study, SBHA did not activate Notch1 in PEL cells, even though trace levels of inactive Notch1 protein were detectable by Western blotting (see Fig. S3 in the supplemental material). This variation in the response to SBHA may be due to differences in cell type or KSHV infection.
SBHA induced histone acetylation on the promoters of KSHV lytic genes, although no significant change was observed in CpG methylation of the RTA promoter (Fig. 4). At present, two different groups have reported the methylation pattern of the RTA promoter, with opposite results. Chen et al. reported that CpG sites in the RTA promoter are highly methylated in BCBL-1 cells during latent infection and that demethylation is induced by TPA treatment (38). However, Günther and Grundhoff conclude that the RTA promoter is highly methylated in HBL6 cells but that very little methylation is observed in BCBL-1 and AP3 cells in the latent phase (39). These contrasting results suggest that CpG methylation of the RTA promoter is not a key regulator for maintaining KSHV latency.
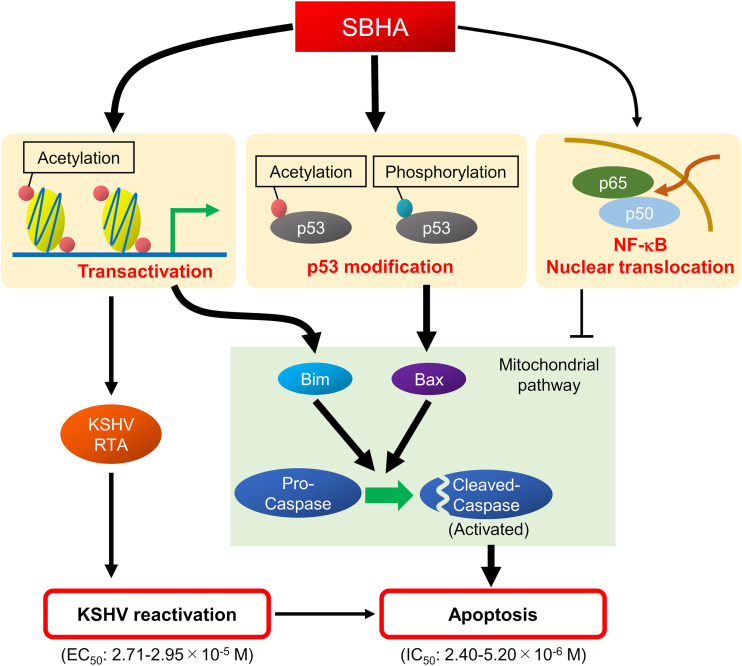
In general, lytic replication of herpesvirus leads to host cell death (40). Therefore, a combination of antiviral drugs with certain drugs or stimuli that reactivate herpesvirus is regarded as a potent therapeutic tool for herpesvirus-associated malignancies, a strategy called lytic induction therapy (41). In the present study, we demonstrate that SBHA not only reactivates KSHV but also initiates the mitochondrial pathway of apoptosis (Fig. 1 and Fig. 6). Importantly, each IC50 for cell growth inhibition in PEL cells (2.40 × 10−6 to 5.20 × 10−6 M) (Fig. 5E) is lower than the EC50 for KSHV reactivation (2.71 × 10−5 to 2.95 × 10−5 M) (Fig. 1B). This suggests that the cytotoxicity of SBHA to PEL cells principally depends on the direct activation of the mitochondrial pathway of apoptosis rather than reactivation of KSHV (Fig. 8).
FIG 8.
Schematic diagram of the effect of SBHA in PEL cells. SBHA induces histone acetylation on the promoters of various genes such as KSHV-encoded RTA and proapoptotic factors. The transactivation of target genes results in KSHV reactivation and apoptosis. In addition, SBHA induces modification of p53, resulting in its activation of the mitochondrial pathway of apoptosis. SBHA induces NF-κB activation in PEL cells, but its antiapoptotic role is limited.
The effect of SBHA on gene expression in KSHV-infected cells was striking in that more than 40% of total transcripts expressed in SBHA-treated PEL cells were of KSHV origin (Table 1). It is noteworthy that the expression profile of vIRF genes changed after SBHA treatment (Fig. 3B and Fig. S2B). vIRFs interact with cellular proteins and interfere with gene transcription and signaling pathways associated with various cellular functions such as cell death, cell cycle regulation, proliferation, and the immune response (42). According to a previous report by Muñoz-Fontela et al., vIRF3 inhibited the activation of the Bim promoter by the FOXO3a transcription factor (43). Therefore, the change in the expression profiles of vIRFs, including vIRF3, by SBHA may contribute to the induction of Bim expression (Fig. 6).
In this study, there is little investigation into the effect of SBHA on host gene expression, including miRNAs, that could contribute to the establishment of KSHV infection or tumorigenesis of KSHV-associated malignancies. However, the findings presented here provide new insight into the pathophysiology of PEL. In light of these results, SBHA should be considered a novel therapeutic strategy against PEL.
MATERIALS AND METHODS
Cell culture and drug treatment.
KSHV-positive PEL cell lines (BCBL-1, SPEL, and TY-1), Burkitt lymphoma cell lines (BJAB and Raji), and T-cell leukemia cell lines (Jurkat and MOLT-4) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C with 5% CO2. KSHV-infected Burkitt lymphoma cell lines (BJAB.219 and Raji.219) were established as reported previously (44): BJAB and Raji cells were infected with rKSHV.219 (kindly provided by Jeffrey Vieira, University of Washington) at a multiplicity of infection of 0.1 and selected in RPMI 1640 medium containing 5 μg/ml puromycin. Selected cells were maintained under conditions with 0.5 μg/ml puromycin. BJAB.219 and Raji.219 cells were positive for GFP expression, and the KSHV genome was detected in both cell types by PCR after more than 10 passages. The Raji.219 cell line is also positive for Epstein-Barr virus (EBV).
Cells were treated with TPA (Sigma-Aldrich, St. Louis, MO) or one of the HDAC inhibitors panobinostat (Pano; Sigma-Aldrich), SBHA (Sigma-Aldrich), CI-994 (Sigma-Aldrich), and sodium butyrate (NaB; Sigma-Aldrich). All the reagents mentioned above were dissolved in DMSO as 1,000× stock solutions. The final concentrations used for each drug are as follows unless otherwise noted: 20 ng/ml for TPA (45), 3.6 nM for Pano, 10 μM for SBHA (14), 100 nM for CI-994, and 1.25 μM for NaB (14). These concentrations were determined individually according to previous reports showing KSHV induction in PEL cells. Concentrations of Pano and CI-994 were determined by the results of real-time RT-PCR for RTA and vIL-6 mRNAs and cell viability as described below. The viability of cells was measured by using a TC10 automated cell counter (Bio-Rad, Hercules, CA) with trypan blue staining.
Flow cytometry.
To detect reactivation of KSHV by flow cytometry, red fluorescence was measured in rKSHV.219-infected cells, as reported previously (15). The rKSHV.219-infected Burkitt lymphoma cell lines BJAB.219 and Raji.219 were seeded at densities of 2.5 × 105 cells/ml and incubated with SBHA at various concentrations for 36 h. Cells were washed twice with phosphate-buffered saline (PBS) and then analyzed by using a CyFlow SL flow cytometer (Partec, Görlitz, Germany). Data were analyzed using FlowJo software (FlowJo, Ashland, OR).
RNA isolation and real-time RT-PCR.
Total RNA was extracted from cultured cells using an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol and subsequently treated with Turbo DNase (Thermo Fisher Scientific, Waltham, MA). RNA samples were subjected to TaqMan-based real-time RT-PCR analysis to detect the mRNAs of RTA, vIL-6, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using QuantiTect multiplex RT-PCR kits (Qiagen) as described previously (46, 47).
Western blot analysis.
Protein extraction and immunoblotting were performed as described previously, with some modifications (48). Briefly, 1 × 106 cells were lysed in 50 μl of M-PER (mammalian protein extraction reagent) (Thermo Fisher Scientific). Subsequently, 7.5 μg per lane of total protein samples was separated on 4 to 12% Bolt Bis-Tris Plus gels and blotted onto a polyvinylidene difluoride membrane (Bio-Rad) using the Trans-Blot Turbo transfer system (Bio-Rad). The membranes were blocked with Block Ace solution (KAC, Kyoto, Japan) and probed with primary antibodies followed by horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies diluted with immunoreaction enhancer solution (Can Get Signal; Toyobo, Osaka, Japan). Blots were visualized by using the SuperSignal West Dura extended-duration substrate (Thermo Fisher Scientific), and signals were detected with the LAS-3000 imaging system (Fujifilm, Tokyo, Japan).
The following antibodies were used for Western blotting. GAPDH antibody (catalogue no. sc-25778) was obtained from Santa Cruz Biotechnology (Dallas, TX). Bim antibody (catalogue no. ab32158) was obtained from Abcam (Cambridge, UK). Cleaved caspase-3 (Asp175) (catalogue no. 9661), caspase-7 (catalogue no. 9492), caspase-9 (catalogue no. 9502), Bcl-xL (catalogue no. 2764), Notch1 (catalogue no. 3608), cleaved Notch1 (Val1744) (catalogue no. 4147), acetyl-p53 (Lys382) (catalogue no. 2525), phospho-p53 (Ser15) (catalogue no. 9284), p21 (catalogue no. 2947), IκBα (catalogue no. 4812), and phospho-IκBα (Ser32/36) (catalogue no. 9246) antibodies were obtained from Cell Signaling Technology (Danvers, MA). p53 antibody (catalogue no. M7001) was obtained from Dako (Copenhagen, Denmark). Apoptosis Sampler kit I (for Bad, Bax, and Bcl-2) and Apoptosis Sampler kit II (for caspase-2 and caspase-3) were obtained from BD Transduction Laboratories (Franklin Lakes, NJ). The mouse monoclonal antibody against RTA (49) and rabbit polyclonal antibodies against latency-associated nuclear antigen 1 (LANA-1) (50) and vIL-6 (51) were generated as previously described.
KSHV real-time PCR array.
To determine the expression profiles of KSHV gene transcripts, we used a KSHV real-time PCR array, a TaqMan-based real-time RT-PCR system that can detect all 87 KSHV gene transcripts simultaneously (48). RNA samples were obtained from BCBL-1, SPEL, and TY-1 cells cultured with or without SBHA for 48 h according to the procedure for RNA preparation described above. Subsequently, RNA samples were subjected to a KSHV real-time PCR array using the QuantiTect Probe RT-PCR kit (Qiagen). The copy number of each transcript was normalized to that of the GAPDH transcript, and the ratio of treated versus untreated cells was calculated.
KSHV transcriptome analysis.
BCBL-1 and SPEL cells were seeded at a density of 1 × 105 cells/ml and stimulated with TPA or SBHA for 36 h. Total RNA was extracted from the cells using Isogen (Nippon Gene, Tokyo, Japan), and mRNA was then purified from total RNA by using an Oligotex-dT30 Super mRNA purification kit (TaKaRa Bio, Kusatsu, Japan). Double-stranded cDNA was prepared from 1 μg of each mRNA sample using the PrimeScript double-strand cDNA synthesis kit (TaKaRa), and a cDNA library was established with a ligation sequencing kit (catalogue no. SQK-LSK109; Oxford Nanopore Technologies, Oxford, UK) according to the manufacturers’ instructions. Barcoded samples were pooled and ligated to a sequencing adaptor. Sequencing was performed with a MinION device using an R9.4.1 flow cell (Oxford Nanopore Technologies).
Bioinformatics.
Guppy version 3.6.0 was used for base calling. Fastq files were mapped to the KSHV genome (GenBank accession no. AP017458 for SPEL and NC_003409 for BCBL-1) by using CLC Genome Workbench (version 14; Qiagen), and the coverage data were obtained using Integrative Genomics Viewer (version 2.50 [http://software.broadinstitute.org/software/igv/home]).
ChIP assay.
BCBL-1, SPEL, and TY-1 cells were seeded at a density of 5.0 × 105 cells/ml with TPA or SBHA. After 12 h of incubation, cells were treated with 1% formaldehyde to cross-link protein and DNA, and unreacted formaldehyde was then quenched with 0.15 M glycine. Subsequently, cells were washed three times with PBS and lysed in SDS lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS). Cell lysates were sonicated using a Bioruptor II instrument (Cosmo Bio, Tokyo, Japan) to shear cross-linked DNA to approximately 200 to 700 bp in length. Chromatin samples derived from 1.0 × 106 cells were precleared with protein G Sepharose (GE Healthcare, Chicago, IL), and 1 to 5 μg of each antibody was then added to form an immune complex. After overnight incubation, immune complexes were precipitated with protein G Sepharose and sequentially washed with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA [pH 8.0], 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA [pH 8.0], 20 mM Tris-HCl [pH 8.0], 500 mM NaCl), LiCl immune complex wash buffer (250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA [pH 8.0], 10 mM Tris-HCl [pH 8.0]), and Tris-EDTA (TE) buffer. Finally, protein-DNA complexes were eluted with elution buffer (1% SDS, 100 mM NaHCO3), incubated with 0.2 M NaCl at 65°C overnight to reverse the cross-link, and then treated with RNase A and proteinase K. For the preparation of input DNA, chromatin samples from 1.0 × 104 cells were sequentially treated with 0.2 M NaCl, RNase A, and proteinase K. All DNA samples were subjected to real-time PCR analysis using a QuantiTect SYBR green PCR kit (Qiagen) with the primers described below.
The following antibodies were used for chromatin immunoprecipitation. Normal rabbit IgG (catalogue no. PM035) was obtained from Medical & Biological Laboratories (Nagoya, Japan). Acetyl-histone H3 (catalogue no. 06-599), acetyl-histone H4 (catalogue no. 06-866), trimethyl-histone H3 (Lys4) (catalogue no. 04-745), and trimethyl-histone H3 (Lys27) (catalogue no. 07-449) antibodies were obtained from Millipore (Burlington, MA). The sequences of the primers were as follows: RTA promoter forward primer 5′-GGTACCGAATGCCACAATCTGTGCCCT-3′ and reverse primer 5′-ATGGTTTGTGGCTGCCTGGACAGTATTC-3′ (52), and vIL-6 promoter forward primer 5′-GCGCCTCCCGGTACAAGTCC-3′ and reverse primer 5′-GACCATTGGCGGGTAGAATC-3′ (53).
Bisulfite sequencing.
BCBL-1, SPEL, and TY-1 cells were cultured with TPA or SBHA for 24 h. DNA samples were prepared from the cells using a DNeasy blood and tissue kit (Qiagen). Before bisulfite conversion, 5 μg of each DNA sample was digested with EcoRI. Digested DNA was denatured in 0.3 M NaOH and subjected to bisulfite conversion with 4.0 M sodium bisulfite–0.4 mM hydroquinone (pH 5.0) at 55°C for 3 h. Bisulfite-converted DNA was sequentially treated with 0.3 M NaOH and 0.45 M ammonium acetate for deamination and desulfonation, respectively. Purified DNA was subjected to PCR to amplify the promoter region of the RTA gene using KOD-Multi & Epi-enzyme (Toyobo, Osaka, Japan) with the following primers: forward primer 5′-GTGTTTTATTATTTTTATAG-3′ and reverse primer 5′-CATCTAACATAACTTTAATC-3′ (39). PCR products were cloned into the pCR2.1 vector using the TA cloning kit (Invitrogen, Carlsbad, CA) to isolate single clones. The DNA sequence of each clone was determined by Sanger sequencing using M13 forward and reverse primers.
XTT assay.
XTT assays were performed using cell proliferation kit II (XTT) (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s protocol. Briefly, cells were seeded into 96-well plates at a density of 1 × 105 cells/ml with TPA or an HDAC inhibitor. After 48 h of culture, an XTT labeling mixture was added to each well. Following a further 4-h incubation, the absorbance at 450 nm was measured by using an iMARK microplate reader (Bio-Rad).
Annexin V assay.
An annexin V assay was carried out using a Mebcyto apoptosis kit (annexin V-fluorescein isothiocyanate [FITC] kit) (Medical & Biological Laboratories) according to the manufacturer’s instructions. In brief, cells were seeded at a density of 5.0 × 105 cells/ml and incubated with various concentrations of SBHA for 24 h to induce apoptosis. After washing with PBS, the cells were stained with FITC-labeled annexin V and propidium iodide (PI). Finally, cells were analyzed by using a flow cytometer as described above.
Nuclear extraction and NF-κB p65 transcription factor assay.
Nuclear extraction was performed as previously described, with slight modifications (54). Briefly, PEL cells were seeded at a density of 5.0 × 105 cells/ml and treated with or without SBHA for 24 h. After washing three times with cold PBS, 1.0 × 107 cells were lysed in hypotonic buffer (20 mM HEPES [pH 7.0], 10 mM KCl, 1 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Triton X-100) followed by centrifugation to precipitate nuclei. The pellets were resuspended in extraction buffer (20 mM HEPES [pH 7.0], 10 mM KCl, 1 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Triton X-100, 20% glycerol, 420 mM NaCl) to obtain the nuclear fraction. For the determination of the DNA binding activity of NF-κB p65, nuclear extracts containing 15 μg/μl of protein were subjected to an ELISA using an NF-κB p65 transcription factor assay kit (Abcam). The experiment was carried out according to the manufacturer’s protocol, and the absorbance at 450 nm was measured by using an iMARK microplate reader (Bio-Rad).
Data analysis.
The generation of dose-response curves, calculation of the EC50 and IC50, and two-tailed unpaired Student’s t test were carried out using GraphPad Prism 8.3.0 software (GraphPad Software, San Diego, CA). Data obtained from KSHV real-time PCR arrays were analyzed using Cluster 3.0 software (55), and heat maps were generated using Java Treeview software (56).
Data availability.
The sequence reads determined by NGS in this study were registered in the Sequence Read Archive database as accession no. DRA010668 in the DNA Data Bank of Japan Sequence Read Archive.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeffrey Vieira, Department of Laboratory Medicine, University of Washington, for providing the recombinant KSHV.
This work was partly supported by the Japan Society for the Promotion of Science (grant no. 19K07600 to H.K.) and the Japan Agency for Medical Research and Development (AMED) (no. JP20fk0410016 to H.K. and JP20fk0108104 to T.S. and H.K.).
We declare that no competing interests exist.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86:1276–1280. doi: 10.1182/blood.V86.4.1276.bloodjournal8641276. [DOI] [PubMed] [Google Scholar]
- 4.Ye F, Lei X, Gao S-J. 2011. Mechanisms of Kaposi’s sarcoma-associated herpesvirus latency and reactivation. Adv Virol 2011:193860. doi: 10.1155/2011/193860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis DA, Rinderknecht AS, Zoeteweij JP, Aoki Y, Read-Connole EL, Tosato G, Blauvelt A, Yarchoan R. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244–3250. doi: 10.1182/blood.v97.10.3244. [DOI] [PubMed] [Google Scholar]
- 6.Ye F, Zhou F, Bedolla RG, Jones T, Lei X, Kang T, Guadalupe M, Gao SJ. 2011. Reactive oxygen species hydrogen peroxide mediates Kaposi’s sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog 7:e1002054. doi: 10.1371/journal.ppat.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 8.Lu F, Day L, Gao SJ, Lieberman PM. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi’s sarcoma-associated herpesvirus lytic transcription. J Virol 80:5273–5282. doi: 10.1128/JVI.02541-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Said J, Cesarman E. 2017. Primary effusion lymphoma, p 323–324. In Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J (ed), WHO classification of tumours of haematopoietic and lymphoid tissues, revised 4th ed. International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- 10.Simonelli C, Spina M, Cinelli R, Talamini R, Tedeschi R, Gloghini A, Vaccher E, Carbone A, Tirelli U. 2003. Clinical features and outcome of primary effusion lymphoma in HIV-infected patients: a single-institution study. J Clin Oncol 21:3948–3954. doi: 10.1200/JCO.2003.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Boulanger E, Gerard L, Gabarre J, Molina JM, Rapp C, Abino JF, Cadranel J, Chevret S, Oksenhendler E. 2005. Prognostic factors and outcome of human herpesvirus 8-associated primary effusion lymphoma in patients with AIDS. J Clin Oncol 23:4372–4380. doi: 10.1200/JCO.2005.07.084. [DOI] [PubMed] [Google Scholar]
- 12.Chueh AC, Tse JW, Togel L, Mariadason JM. 2015. Mechanisms of histone deacetylase inhibitor-regulated gene expression in cancer cells. Antioxid Redox Signal 23:66–84. doi: 10.1089/ars.2014.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Waschke BC, Woolaver RA, Chen Z, Zhang G, Piscopio AD, Liu X, Wang JH. 2019. Histone deacetylase inhibition sensitizes PD1 blockade-resistant B-cell lymphomas. Cancer Immunol Res 7:1318–1331. doi: 10.1158/2326-6066.CIR-18-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osawa M, Mine S, Ota S, Kato K, Sekizuka T, Kuroda M, Kataoka M, Fukumoto H, Sato Y, Kanno T, Hasegawa H, Ueda K, Fukayama M, Maeda T, Kanoh S, Kawana A, Fujikura Y, Katano H. 2016. Establishing and characterizing a new primary effusion lymphoma cell line harboring Kaposi’s sarcoma-associated herpesvirus. Infect Agents Cancer 11:37. doi: 10.1186/s13027-016-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vieira J, O’Hearn PM. 2004. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 325:225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar S, Abujamra AL, Loew JE, Forman LW, Perrine SP, Faller DV. 2011. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res 31:2723–2732. [PubMed] [Google Scholar]
- 17.Miyashita T, Reed JC. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Roeder RG. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Shieh SY, Ikeda M, Taya Y, Prives C. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 20.Bhatt S, Ashlock BM, Toomey NL, Diaz LA, Mesri EA, Lossos IS, Ramos JC. 2013. Efficacious proteasome/HDAC inhibitor combination therapy for primary effusion lymphoma. J Clin Invest 123:2616–2628. doi: 10.1172/JCI64503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Edelstein LC, Gelinas C. 2000. The Rel/NF-kappaB family directly activates expression of the apoptosis inhibitor Bcl-x(L). Mol Cell Biol 20:2687–2695. doi: 10.1128/mcb.20.8.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller SA, Schattner EJ, Cesarman E. 2000. Inhibition of NF-kappaB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood 96:2537–2542. doi: 10.1182/blood.V96.7.2537. [DOI] [PubMed] [Google Scholar]
- 23.Keller SA, Hernandez-Hopkins D, Vider J, Ponomarev V, Hyjek E, Schattner EJ, Cesarman E. 2006. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood 107:3295–3302. doi: 10.1182/blood-2005-07-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenblatt DY, Cayo M, Ning L, Jaskula-Sztul R, Haymart M, Kunnimalaiyaan M, Chen H. 2007. Suberoyl bishydroxamic acid inhibits cellular proliferation by inducing cell cycle arrest in carcinoid cancer cells. J Gastrointest Surg 11:1515–1520; discussion, 1520. doi: 10.1007/s11605-007-0249-1. [DOI] [PubMed] [Google Scholar]
- 25.Adler JT, Hottinger DG, Kunnimalaiyaan M, Chen H. 2008. Histone deacetylase inhibitors upregulate Notch-1 and inhibit growth in pheochromocytoma cells. Surgery 144:956–961; discussion, 961–962. doi: 10.1016/j.surg.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning L, Greenblatt DY, Kunnimalaiyaan M, Chen H. 2008. Suberoyl bis-hydroxamic acid activates Notch-1 signaling and induces apoptosis in medullary thyroid carcinoma cells. Oncologist 13:98–104. doi: 10.1634/theoncologist.2007-0190. [DOI] [PubMed] [Google Scholar]
- 27.Xiao X, Ning L, Chen H. 2009. Notch1 mediates growth suppression of papillary and follicular thyroid cancer cells by histone deacetylase inhibitors. Mol Cancer Ther 8:350–356. doi: 10.1158/1535-7163.MCT-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Zheng X, Gao M, Zhao J, Li Y, Meng X, Qian B, Li J. 2017. Suberoyl bis-hydroxamic acid activates Notch1 signaling and induces apoptosis in anaplastic thyroid carcinoma through p53. Oncol Rep 37:458–464. doi: 10.3892/or.2016.5281. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XD, Gillespie SK, Borrow JM, Hersey P. 2004. The histone deacetylase inhibitor suberic bishydroxamate regulates the expression of multiple apoptotic mediators and induces mitochondria-dependent apoptosis of melanoma cells. Mol Cancer Ther 3:425–435. [PubMed] [Google Scholar]
- 30.Chen S, Dai Y, Pei XY, Grant S. 2009. Bim upregulation by histone deacetylase inhibitors mediates interactions with the Bcl-2 antagonist ABT-737: evidence for distinct roles for Bcl-2, Bcl-xL, and Mcl-1. Mol Cell Biol 29:6149–6169. doi: 10.1128/MCB.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You BR, Park WH. 2010. Suberoyl bishydroxamic acid inhibits the growth of A549 lung cancer cells via caspase-dependent apoptosis. Mol Cell Biochem 344:203–210. doi: 10.1007/s11010-010-0543-1. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Zhang N, Shi Z, Yang Z, Hu X. 2015. Histone deacetylase inhibitor suberoyl bis-hydroxamic acid suppresses cell proliferation and induces apoptosis in breast cancer cells. Mol Med Rep 11:2908–2912. doi: 10.3892/mmr.2014.3076. [DOI] [PubMed] [Google Scholar]
- 33.Ramos JC, Sparano JA, Chadburn A, Reid EG, Ambinder RF, Siegel ER, Moore PC, Rubinstein PG, Durand CM, Cesarman E, Aboulafia D, Baiocchi R, Ratner L, Kaplan L, Capoferri AA, Lee JY, Mitsuyasu R, Noy A. 2020. Impact of Myc in HIV-associated non-Hodgkin lymphomas treated with EPOCH and outcomes with vorinostat (AMC-075 trial). Blood 136:1284–1297. doi: 10.1182/blood.2019003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Autin P, Blanquart C, Fradin D. 2019. Epigenetic drugs for cancer and microRNAs: a focus on histone deacetylase inhibitors. Cancers (Basel) 11:1530. doi: 10.3390/cancers11101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bray SJ. 2006. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 36.Lan K, Choudhuri T, Murakami M, Kuppers DA, Robertson ES. 2006. Intracellular activated Notch1 is critical for proliferation of Kaposi’s sarcoma-associated herpesvirus-associated B-lymphoma cell lines in vitro. J Virol 80:6411–6419. doi: 10.1128/JVI.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan K, Murakami M, Choudhuri T, Kuppers DA, Robertson ES. 2006. Intracellular-activated Notch1 can reactivate Kaposi’s sarcoma-associated herpesvirus from latency. Virology 351:393–403. doi: 10.1016/j.virol.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Ueda K, Sakakibara S, Okuno T, Parravicini C, Corbellino M, Yamanishi K. 2001. Activation of latent Kaposi’s sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc Natl Acad Sci U S A 98:4119–4124. doi: 10.1073/pnas.051004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Günther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenney SC. 2007. Reactivation and lytic replication of EBV, p 403–433. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 41.Feng WH, Hong G, Delecluse HJ, Kenney SC. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J Virol 78:1893–1902. doi: 10.1128/jvi.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs SR, Damania B. 2011. The viral interferon regulatory factors of KSHV: immunosuppressors or oncogenes? Front Immunol 2:19. doi: 10.3389/fimmu.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muñoz-Fontela C, Marcos-Villar L, Gallego P, Arroyo J, Da Costa M, Pomeranz KM, Lam EW, Rivas C. 2007. Latent protein LANA2 from Kaposi’s sarcoma-associated herpesvirus interacts with 14-3-3 proteins and inhibits FOXO3a transcription factor. J Virol 81:1511–1516. doi: 10.1128/JVI.01816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kati S, Hage E, Mynarek M, Ganzenmueller T, Indenbirken D, Grundhoff A, Schulz TF. 2015. Generation of high-titre virus stocks using BrK.219, a B-cell line infected stably with recombinant Kaposi’s sarcoma-associated herpesvirus. J Virol Methods 217:79–86. doi: 10.1016/j.jviromet.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 45.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S, Chang Y. 1997. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol 71:314–324. doi: 10.1128/JVI.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fakhari FD, Dittmer DP. 2002. Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J Virol 76:6213–6223. doi: 10.1128/jvi.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanagisawa Y, Sato Y, Asahi-Ozaki Y, Ito E, Honma R, Imai J, Kanno T, Kano M, Akiyama H, Sata T, Shinkai-Ouchi F, Yamakawa Y, Watanabe S, Katano H. 2006. Effusion and solid lymphomas have distinctive gene and protein expression profiles in an animal model of primary effusion lymphoma. J Pathol 209:464–473. doi: 10.1002/path.2012. [DOI] [PubMed] [Google Scholar]
- 48.Kanno T, Uehara T, Osawa M, Fukumoto H, Mine S, Ueda K, Hasegawa H, Katano H. 2015. Fumagillin, a potent angiogenesis inhibitor, induces Kaposi sarcoma-associated herpesvirus replication in primary effusion lymphoma cells. Biochem Biophys Res Commun 463:1267–1272. doi: 10.1016/j.bbrc.2015.06.100. [DOI] [PubMed] [Google Scholar]
- 49.Okuno T, Jiang YB, Ueda K, Nishimura K, Tamura T, Yamanishi K. 2002. Activation of human herpesvirus 8 open reading frame K5 independent of ORF50 expression. Virus Res 90:77–89. doi: 10.1016/s0168-1702(02)00142-9. [DOI] [PubMed] [Google Scholar]
- 50.Katano H, Sato Y, Kurata T, Mori S, Sata T. 1999. High expression of HHV-8-encoded ORF73 protein in spindle-shaped cells of Kaposi’s sarcoma. Am J Pathol 155:47–52. doi: 10.1016/S0002-9440(10)65097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katano H, Sato Y, Kurata T, Mori S, Sata T. 2000. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi’s sarcoma, and multicentric Castleman’s disease. Virology 269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 52.Lu F, Zhou J, Wiedmer A, Madden K, Yuan Y, Lieberman PM. 2003. Chromatin remodeling of the Kaposi’s sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J Virol 77:11425–11435. doi: 10.1128/jvi.77.21.11425-11435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Ye F, Xie J, Kuhne K, Gao SJ. 2009. Genome-wide identification of binding sites for Kaposi’s sarcoma-associated herpesvirus lytic switch protein, RTA. Virology 386:290–302. doi: 10.1016/j.virol.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dignam JD, Martin PL, Shastry BS, Roeder RG. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol 101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 55.de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 56.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence reads determined by NGS in this study were registered in the Sequence Read Archive database as accession no. DRA010668 in the DNA Data Bank of Japan Sequence Read Archive.