Dengue virus (DENV), the etiological agent of dengue, a febrile and hemorrhagic disease, infects millions of people per year in tropical and subtropical countries. When infecting cells, DENV induces stress conditions known to inhibit canonical protein synthesis.
KEYWORDS: dengue fever, internal ribosome entry site, translation initiation
ABSTRACT
Dengue virus (DENV) is an enveloped, positive-sense, single-stranded RNA virus belonging to the Flaviviridae family. Translation initiation of DENV mRNA can occur by a cap-dependent or a cap-independent mechanism. Two non-mutually exclusive cap-independent mechanisms of translation initiation have been described for DENV mRNA. The first corresponds to a 5′-end-dependent, internal ribosome entry site (IRES)-independent mechanism, while the second relies on IRES-dependent initiation. In this report, we study the recently discovered DENV IRES. The results show that the DENV IRES is functional in the rabbit reticulocyte lysate (RRL). In accordance, the activity of the DENV IRES was resistant to the cleavage of eukaryotic initiation factor 4G (eIF4G) by the Foot-and-mouth disease virus leader protease in RRL. In cells, the DENV IRES exhibited only marginal activity under standard culture conditions. The DENV IRES showed weak activity in HEK 293T cells; however, DENV IRES activity was significantly enhanced in HEK 293T cells expressing Human rhinovirus 2A protease. These findings suggest that the DENV IRES enables viral protein synthesis under conditions that suppress canonical translation initiation.
IMPORTANCE Dengue virus (DENV), the etiological agent of dengue, a febrile and hemorrhagic disease, infects millions of people per year in tropical and subtropical countries. When infecting cells, DENV induces stress conditions known to inhibit canonical protein synthesis. Under these conditions, DENV mRNA thrives using noncanonical modes of translation initiation. In this study, we characterize the mechanism dependent on an internal ribosome entry site (IRES). Here, we describe the activity of the DENV IRES in vitro and cells. We show that in cells, the DENV IRES enables the viral mRNA to translate under conditions that suppress canonical translation initiation.
INTRODUCTION
Dengue virus (DENV) is an enveloped, positive-sense, single-stranded RNA virus that can lead to diverse clinical symptoms in humans, ranging from self-limiting dengue fever (DF) to life-threatening dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) (1). As with other members of the Flaviviridae, after the entry of the virus into targeted cells, DENV genomic RNA, which has at its 5′ end an (m7GpppA2′OMe)-cap structure, directly acts as the mRNA (2, 3). However, in sharp contrast to most eukaryotic mRNAs, the viral mRNA lacks a 3′ poly(A) tail (2).
Initiation of the translation of DENV mRNA is mainly cap dependent (2–4). In cap-dependent translation initiation, eukaryotic translation initiation factor 4F (eIF4F) recognizes the cap structure at the 5′ end of the mRNA and bridges it with the 40S ribosomal subunit. The eIF4F complex is formed by the cap-binding protein eIF4E, eIF4A, an ATP-dependent RNA helicase, and eIF4G, a scaffold protein with binding sites for eIF4A, eIF4E, eIF3, and the poly(A) binding protein (PABP) (5, 6). The 40S ribosomal subunit is recruited to the mRNA as part of the 43S initiation complex, composed of the subunit bound to eIF2-GTP/Met-tRNAi (ternary complex), eIF1A, eIF1, eIF3, and eIF5 (5, 6). It is eIF4G that recruits the 43S complex to the mRNA through its interaction with eIF3. For most cellular mRNAs, eIF4G also bridges the mRNA 5′–3′-end interaction through its simultaneous contact with the 5′ cap-bound eIF4E and the 3′ poly(A)-associated PABP (5, 6). The mRNA 5′–3′-end interaction [5′ cap–eIF4E–eIF4G–PABP–3′ poly(A)] enhances translation initiation, translation reinitiation through ribosome recycling, and RNA stability (7–9). The recruited 43S complex scans the 5′ UTR of the mRNA in a 5′-to-3′ direction until the initiation codon is encountered (5, 6). Recognition of the initiation codon by the anticodon of the initiator Met-tRNAi leads to 60S subunit joining, assembling an elongation-competent 80S ribosome (5, 6). In DENV mRNA, the 3′ UTR substitutes for the poly(A) tail, enhancing cap-dependent translation initiation (4, 10–12). DENV mRNA can circularize (mRNA 5′–3′-end communication) through RNA-RNA interactions in the absence of cellular proteins (12), or via 5′–3′ UTR protein-protein bridging, as PABP interacts with its 3′ UTR, favoring viral mRNA circularization in a poly(A) tail-independent fashion (3′ UTR–PABP–eIF4G–eIF4E–5′ cap) (13). In sharp contrast to most cellular mRNAs (14), DENV mRNA can also initiate translation in a cap-independent manner (15–18). Two non-mutually exclusive cap-independent translation initiation mechanisms have been described for DENV mRNA (15, 18). The first corresponds to a 5′-end-dependent, internal ribosome entry site (IRES)-independent mechanism (15), while the second relies on IRES-mediated initiation (18). Neither of these cap-independent mechanisms of translation initiation has been fully characterized.
IRESs were first identified in the naturally uncapped mRNAs of the Picornaviridae (19, 20). The discovery of IRESs was achieved by inserting the 5′ UTR of Poliovirus (PV) or Encephalomyocarditis virus (EMCV) mRNA into the intercistronic spacer of an artificial bicistronic RNA (19, 20). In this artificial bicistronic reporter system, expression of the second cistron documented the ability of the tested sequence to promote internal initiation (21, 22). The mRNAs of other members of the Picornaviridae, as well as those of viruses from other families, including members of the Flaviviridae, such as Hepatitis C virus (HCV) (23, 24), Zika virus (18), and DENV (18), also harbor IRESs.
In this study, we sought to characterize further, both in an in vitro translation system and in cultured cells, the recently discovered DENV IRES (18). The results showed that the DENV IRES was functional in the rabbit reticulocyte lysate (RRL) in vitro translation system (25). In accordance, the activity of the DENV IRES was resistant to eIF4G cleavage by the Foot-and-mouth disease virus (FMDV) leader protease (L protease) in RRL. In cells, the DENV IRES exhibited only marginal activity under standard cell culture conditions. The DENV IRES exhibited low activity, if any, in HEK 293T cells. However, we show that DENV IRES activity is significantly increased in HEK 293T cells expressing the Human rhinovirus (HRV) 2A protease, suggesting that the DENV IRES enables the viral mRNA to translate under conditions that suppress canonical translation initiation.
(This research was conducted by L. Fernández-Garcia as part of a Ph.D. thesis at the Universidad de Chile, by A. Barrera in partial fulfillment of the requirements for a Ph.D. from the Pontificia Universidad Católica de Chile, and by H. Ramos as part of an undergraduate biochemistry thesis at the Pontificia Universidad Cátolica de Chile.)
RESULTS
Monocistronic DENV-like mRNAs are capable of noncanonical translation initiation in vitro.
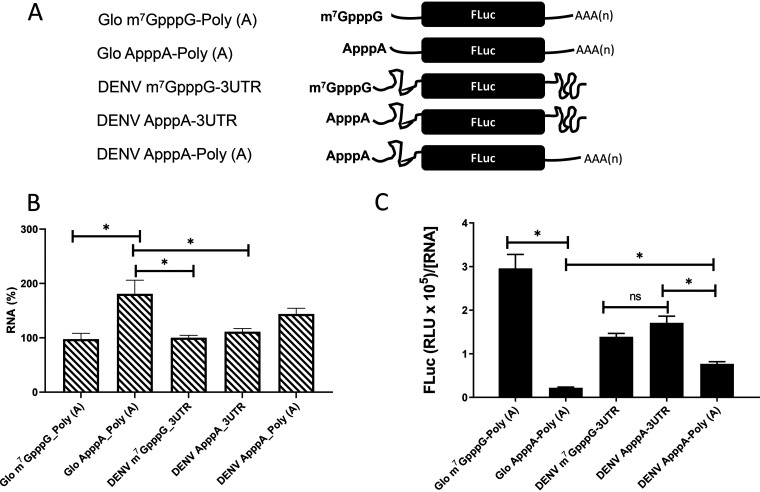
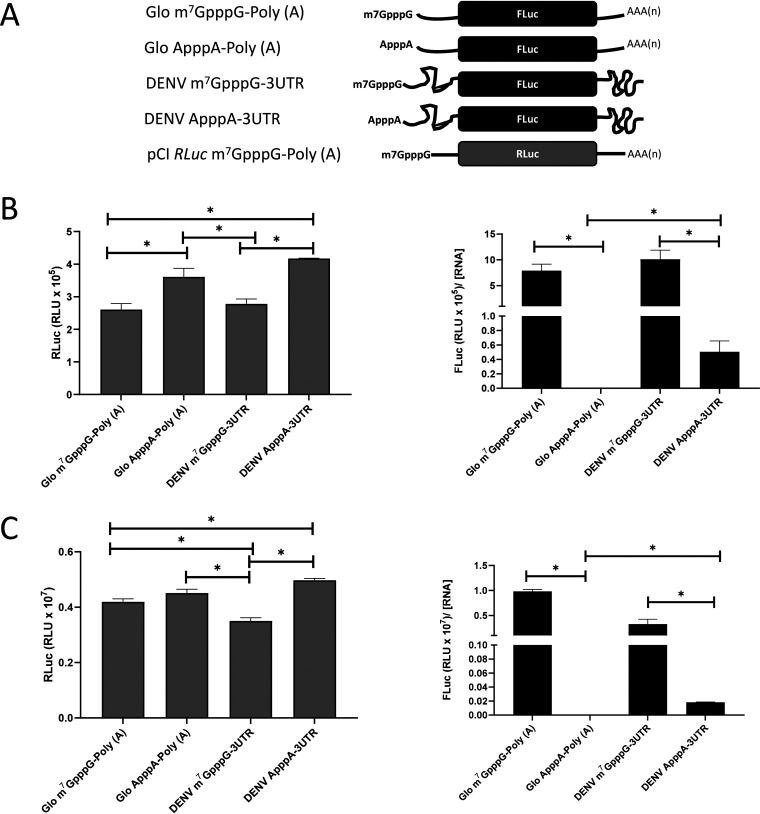
DENV mRNA can initiate translation using a cap-dependent or a cap-independent mechanism (15, 18). We wondered whether these findings could be recapitulated in the conventional nuclease-treated rabbit reticulocyte lysate (RRL) (25). For this investigation, T7 polymerase in vitro-transcribed RNA generated from plasmid pGL5′3′DV was used to program RRL. In these RNAs, the 5′ UTR-C62 (nucleotides [nt] 1 to 160) and the 3′ UTR of DENV 2 (strain 16681; GenBank accession no. NC_001474.2) mRNA flank the firefly luciferase (FLuc) open reading frame (ORF) (12). Three mRNAs—the DENV m7GpppG-3′UTR, DENV ApppA-3′UTR, and DENV ApppA-Poly(A) mRNAs—were generated using different commercially available cap analogs (depicted in Fig. 1A). Noteworthy, the ApppA cap analog is not recognized by the cap-binding protein eIF4E (25, 26). In the DENV ApppA-Poly(A) mRNA, the DENV 3′ UTR was replaced by a poly(A) tail. As controls for cap-dependent translation initiation, we used the Glo m7GpppG-Poly(A) or Glo ApppA-Poly(A) mRNA, in which the 5′ UTR of globin mRNA and a 3′ poly(A) tail flank the FLuc ORF (Fig. 1A). After in vitro translation, the luciferase activity was determined, and total RNA was recovered from the mixture and quantified by reverse transcription-quantitative PCR (RT-qPCR). The RNA concentration ([RNA]) was expressed relative to the content of the DENV m7GpppG-3′UTR mRNA, set at 100% (Fig. 1B). Relative to the DENV m7GpppG-3′UTR mRNA, significantly more (∼80%) Glo ApppA-Poly(A) mRNA, the negative control, was used and recovered in the assay (Fig. 1B). Luciferase activity, expressed in relative light units (RLU) normalized to the relative mRNA content ([RNA]), showed that replacing the m7GpppG cap with the ApppA cap reduced translation from the globin-like mRNA by 93%, confirming that the ApppA cap abrogates cap-dependent translation initiation (Fig. 1C) (25). In RRL, translation from DENV mRNA was shown to be less efficient than that of the globin control. However, replacement of the m7GpppG cap by the ApppA cap did not suppress FLuc expression (Fig. 1C). Replacement of the DENV 3′ UTR with a poly(A) tail reduced translation of the A-capped mRNA by 45%, indicating that in the context of this monocistronic RNA, the 3′ UTR also participates in cap-independent translation initiation in RRL (Fig. 1C) (4, 11). These results confirm that monocistronic mRNAs harboring the 5′ UTR of DENV mRNA support cap-independent translation initiation in RRL.
FIG 1.
The DENV 5′ UTR in the context of monocistronic mRNAs has cap-independent translation activity in RRL. (A) Schematic representation of the in vitro-transcribed RNAs used in the in vitro translation studies. The capped (m7GpppG) and A-capped (ApppA) DENV 2 RNA-like reporters were transcribed from the pGL5′3′DV plasmid (12). The DENV 2 RNA-like reporters comprise the 5′ UTR-C62 (nt 1 to 160) and the 3′ UTR of DENV 2 (strain 16681; GenBank accession no. NC_001474.2) mRNA or a poly(A) tail flanking the firefly luciferase (FLuc) open reading frame (ORF) (12). Capped or A-capped polyadenylated globin RNA reporters that harbor the 5′ UTR of the globin mRNA and a poly(A) tail flanking the FLuc ORF were also used. (B and C) In vitro-transcribed reporter RNAs were translated in RRL. (B) RNA was recovered, and the RNA levels were determined by RT-qPCR and expressed relative to the content of the DENV m7GpppG-3′UTR mRNA, set to 100%. Statistical analysis was performed by a two-tailed t test. (C) FLuc activity was determined and is expressed in relative light units (RLU) normalized by the levels of FLuc RNA relative to that of 18S rRNA present in the RRL mixture. Values are means (± standard errors of the means) for three independent experiments, each performed in duplicate.
The 5′ UTR of DENV mRNA enables IRES-mediated translation initiation in RRL.
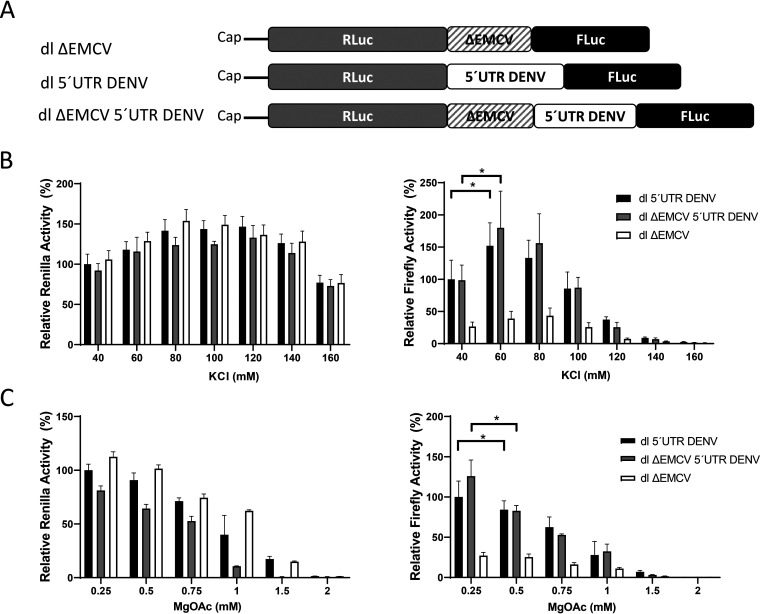
The results presented in Fig. 1 did not allow us to discriminate which mechanism of cap-independent translation initiation, IRES dependent or IRES independent, was in play (15, 18). Next, we focused exclusively on evaluating the IRES activity of the DENV 5′ UTR (nt 1 to 96 of DENV 2 mRNA; GenBank accession no. NC_001474.2) in RRL (18). For this purpose, the 5′ UTR of DENV 2 mRNA was inserted into a dual-luciferase (dl) reporter containing an upstream Renilla luciferase (RLuc) gene and a downstream FLuc gene, creating the dl 5′UTR DENV construct. Because the 3′ UTR of DENV mRNA impacted the noncanonical translation of DENV mRNA (Fig. 1C) (4, 10–12), we decided to omit its use in our constructs. The omission of the DENV mRNA 3′ UTR from the dl constructs allowed us to focus exclusively on the study of the IRES present within the 5′ UTR of DENV mRNA in the absence of any other viral sequence (18). In the context of the dl-RNA, translation driven by the DENV 5′ UTR was monitored by using FLuc activity as the readout. At the same time, the RLuc reporter gene was translated by a cap-dependent mechanism and served as an internal control (21). As an additional control, plasmid dl ΔEMCV 5′UTR DENV RNA was constructed. In this reporter, the highly structured defective EMCV IRES sequence (ΔEMCV), known to impede ribosome reinitiation and readthrough (27), was inserted upstream of the DENV 5′ UTR (depicted in Fig. 2A).
FIG 2.
Effects of potassium and magnesium on the translation of the Renilla or firefly luciferase (RLuc or FLuc) open reading frame. (A) Schematic representation of the in vitro-transcribed dual-luciferase (dl) RNAs used in the in vitro translation studies. In these dl-RNAs, the first cistron corresponds to the RLuc ORF, while the second cistron corresponds to the FLuc ORF. The dl ΔEMCV construct harboring a defective encephalomyocarditis virus (ΔEMCV) 5′ UTR that lacks IRES activity in the intercistronic region has been described previously (27). The dl 5′UTR DENV construct harbors the 5′ UTR (nt 1 to 96; GenBank accession no. U87411) of DENV 2 (strain 16681) mRNA within the intercistronic region. In the dl ΔEMCV 5′UTR DENV RNA, the structured ΔEMCV region was upstream of the DENV 5′ UTR. (B and C) The dl 5′UTR DENV (filled bars), dl ΔEMCV 5′UTR DENV (shaded bars), and dl ΔEMCV (open bars) in vitro transcripts were translated in RRL in the presence of various concentrations of KCl (40 to 160 mM) (B) or MgOAc (0.25 to 2 mM) (C). RLuc (left) and FLuc (right) activities (± standard errors of the means) are presented relative to the luciferase activity obtained in RRL without additional salt supplementation (40 mM KCl, 0.25 mM MgOAc), which was set to 100%. Statistical analysis was performed by one-way ANOVA, followed by Tukey’s multiple-comparison test. Asterisks indicate significant differences (P < 0.05) from the negative control.
In vitro, IRES-mediated translation initiation is dependent on the concentration of potassium (K+) and magnesium (Mg2+) ions present within the translation mixture (28–30). So, as a first approach in evaluating DENV IRES activity, RRL was programmed with either the dl ΔEMCV 5′UTR DENV or the dl 5′UTR DENV bicistronic in vitro-transcribed RNA using a range of potassium chloride (KCl) (Fig. 2B) or magnesium acetate (MgOAc) (Fig. 2C) concentrations. In these assays, the dl ΔEMCV RNA (27), which lacks IRES activity, was used as a negative control. The results are presented relative to the RLuc and FLuc activities obtained with the dl 5′UTR DENV RNA in RRL (40 mM KCl and 0.25 mM MgOAc) without additional salt supplementation, set to 100%. When RRL was programmed with the dl 5′UTR DENV or the dl ΔEMCV 5′UTR DENV bicistronic mRNA, FLuc expression was significantly enhanced (P < 0.05) at 60 mM KCl (Fig. 2B, right). Increasing the concentrations of MgOAc over 0.75 mM reduced translation driven from the 5′ UTR of DENV and the ΔEMCV 5′UTR DENV RNA (Fig. 2C, right). From these data, we concluded that the optimal salt concentrations for the in vitro activity of the DENV IRES in RRL were 60 mM KCl and 0.25 mM MgOAc (no additional MgOAc added to the RRL) (Fig. 2B and C, right). The two cistrons, RLuc (left) and FLuc (right), showed different responses to varying K+ and Mg2+ concentrations (Fig. 2B and C), suggesting that in RRL, translation of the RLuc cistron and translation of the FLuc cistron are independent of one another.
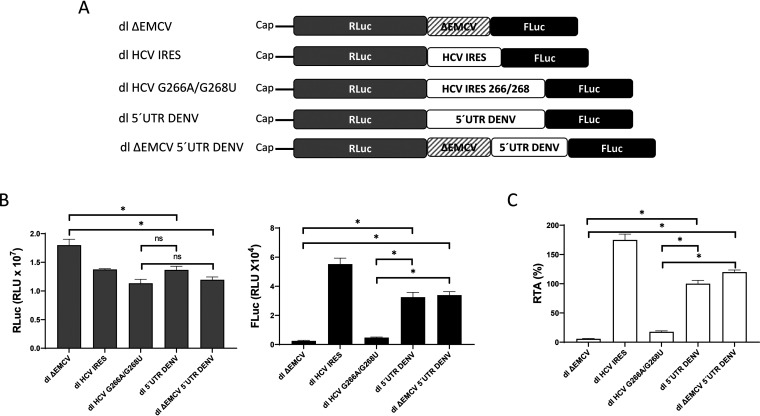
Next, we decided to compare the translational activity of the DENV 5′ UTR with that of the IRES of the HCV 1b genotype (31). For this purpose, we used salt-optimized RRL programmed with in vitro-transcribed dl HCV IRES (designated dl HCV 1b in reference 31), dl 5′UTR DENV, or dl ΔEMCV 5′UTR DENV RNA (depicted in Fig. 3A). As negative controls, we used the dl ΔEMCV and dl HCV G266A/G268U mRNAs (27, 31, 32). The dl HCV G266A/G268U mRNA harbors a mutant HCV genotype 1b 5′ UTR (G266A G268U), which lacks IRES activity (31, 32). As expected, all RNAs expressed RLuc, indicating that cap-dependent translation was functional (Fig. 3B, left). RLuc activity was evidenced for the mRNAs included in the study (Fig. 3B, left). FLuc activity was detected only in RRL programmed with the dl HCV IRES, dl 5′UTR DENV, or dl ΔEMCV 5′UTR DENV mRNA (Fig. 3B, right). The absence of FLuc activity confirms that the ΔEMCV and HCV 5′UTR G266A/G268U mRNAs (27, 31, 32) lack IRES activity in RRL (Fig. 3B, right). The results were also expressed as relative translation activity (RTA), which corresponds to the FLuc/RLuc ratio, a direct index of IRES-mediated translation initiation (Fig. 3C). The RTA value obtained for the dl 5′UTR DENV RNA was arbitrarily set to 100%. The results showed that translation driven by the DENV 5′ UTR or ΔEMCV-DENV 5′UTR RNA was, on average, ∼82% or ∼102% higher than that of the dl HCV266/268 control, or ∼95% and ∼115% higher than that of the dl ΔEMCV RNA, respectively (Fig. 3C). The fusion of the ΔEMCV RNA upstream of the DENV 5′ UTR did not reduce translation driven by the DENV 5′ UTR, suggesting that a reinitiation or readthrough mechanism from the first cistron was not responsible for FLuc expression from the dl 5′UTR DENV RNA (Fig. 3C). These findings support those of a previous report (18) and show that in RRL, the short nucleotide sequence of the DENV 2 5′ UTR (96 nt) is a competent IRES. However, in contrast to the results that Song et al. obtained in cells (18), in RRL, translation driven by the DENV 5′ UTR or ΔEMCV-DENV 5′UTR RNA was weaker than that of the HCV IRES RNA by an average of ∼77% or ∼57%, respectively. From these observations, we conclude that the 5′ UTR of DENV mRNA harbors an IRES, although it displays weaker activity than that of the HCV IRES in RRL.
FIG 3.
The DENV 5′ UTR in the context of a bicistronic mRNA enables IRES-dependent translation in RRL. (A) Schematic representation of the in vitro-transcribed dl-RNAs used in the in vitro translation studies. The dl HCV IRES and dl HCV G266A/G268U constructs, harboring an active or inactive genotype 1b HCV IRES, respectively, have been described previously (31, 32). (B) The dl ΔEMCV, dl HCV G266A/G268U, dl HCV IRES, dl 5′UTR DENV, and dl ΔEMCV 5′UTR DENV in vitro transcripts were translated in RRL. RLuc (left) and FLuc (right) activities (expressed in relative light units [RLU]) were measured. (C) Relative translation activity (RTA), corresponding to the FLuc/RLuc ratio relative to that of the dl 5′UTR DENV construct, which was arbitrarily set at 100%. The values shown are means (± standard errors of the means) for three independent experiments, each conducted in duplicate. Statistical analysis was performed by one-way ANOVA, followed by Tukey’s multiple-comparison test (*, P < 0.05).
The DENV 5′ UTR directs translation in RRL in the presence of the FMDV L protease.
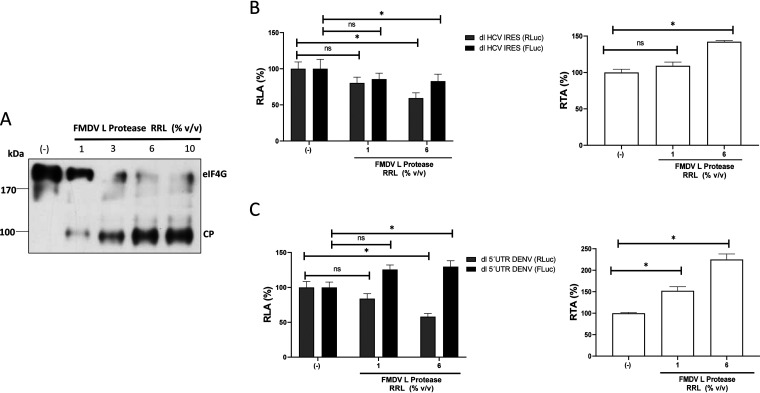
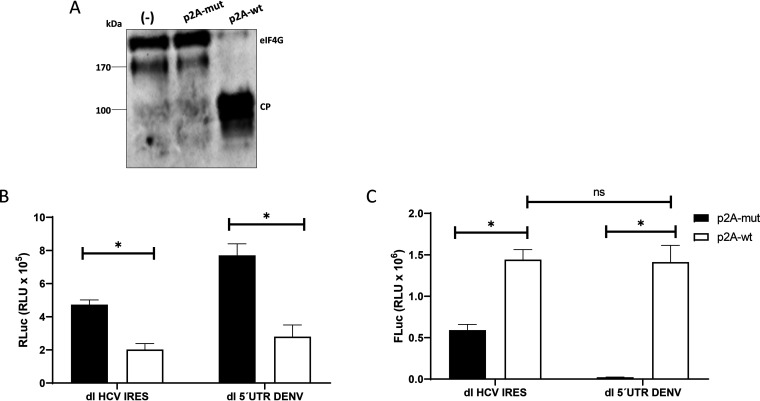
To further validate our conclusions, next, we evaluated the translational activity of the DENV 5′ UTR in RRL treated with the FMDV L protease. The FMDV L protease cleaves eIF4G in such a way that it impedes the interaction of cap-bound eIF4E with the rest of the eIF4F complex, inhibiting cap-dependent, but not IRES-dependent, translation initiation (33, 34). As described previously (35), the FMDV L protease was synthesized in RRL and was added to fresh RRL (1% to 10% [vol/vol]). The cleavage of eIF4G in the total mixture was confirmed by Western blot analysis (Fig. 4A). Next, in vitro-transcribed dl HCV IRES and dl 5′UTR DENV RNAs were translated in RRL in the absence or the presence (1% or 6% [vol/vol]) of the FMDV L protease. The results are presented as relative luciferase activities (RLAs), where the RLuc and FLuc activities obtained in RRL alone were set to 100%. Cap-dependent translation (RLuc) from the dl HCV IRES and dl 5′UTR DENV RNAs was reduced in the presence of the FMDV L protease by averages of ∼18% (1% [vol/vol] FMDV L protease in RRL) and ∼42% (6% [vol/vol] FMDV L protease in RRL), respectively (Fig. 4B and C, left). In the case of the HCV IRES, and in agreement with previous reports (33, 36), FLuc activity was not significantly altered by the presence of the FMDV L protease (Fig. 4B, left). Strikingly, translation driven by the DENV 5′ UTR increased by ∼27% (1% [vol/vol] FMDV L protease) or ∼31% (6% [vol/vol] FMDV L protease) in the presence of FMDV L protease in RRL (Fig. 4C, left). To better evidence IRES activity, results are also presented as the RTA (expressed as a percentage), where the values obtained in the absence of FMDV L protease were set to 100% (Fig. 4B and C, right). Treatment with the FMDV L protease increased HCV IRES activity in RRL by ∼10% (with 1% [vol/vol]) and ∼43% (with 6% [vol/vol]) (Fig. 4B, right). In the case of the DENV 5′ UTR, FLuc activity increased by ∼52% (1% [vol/vol]) and ∼124% (6% [vol/vol]) in RRL treated with the FMDV L protease (Fig. 4C, right). The concurrent decrease in RLuc activity and increase in FLuc activity in the presence of FMDV L protease confirm that the cistrons were independently translated. This observation also indicated that FLuc expression mediated by the DENV 5′ UTR was not due to 40S ribosomal subunits being recruited at, and tethered from, the 5′ capped end of the dl RNA. Collectively, these data strongly suggest that in vitro, the 5′ UTR of DENV mRNA can drive IRES-dependent translation initiation in RRL. Also, the results indicated that in RRL, the activity of the DENV IRES is enhanced when cap-dependent translation is suppressed by the FMDV L protease.
FIG 4.
Proteolytic cleavage of eIF4G by the FMDV L protease negatively impacts cap-dependent translation initiation but not translation driven by the DENV 5′ UTR. (A) In vitro translation reaction mixtures in RRL were supplemented with 1, 3, 6, or 10% (vol/vol) RRL that was or was not (–) programmed with an FMDV L protease RNA template. Western blot analysis was performed using polyclonal antibodies against eIF4GI. The positions of molecular mass standards are shown. The eIF4G cleavage product (CP) has been characterized previously (34). (B and C) In vitro-transcribed dl HCV IRES (B) and dl 5′UTR DENV (C) RNAs were translated in salt-optimized RRL in the absence (–) or presence of 1 or 6% (vol/vol) RRL programmed with FMDV L protease. (Left) RLuc and FLuc activities are shown as the relative luciferase activity (RLA) for each RNA, where the activity obtained by the translation reaction conducted in the absence of FMDV L protease was set to 100%. (Right) The results are also shown as the RTA (expressed as a percentage). Statistical analysis was performed with two-tailed t tests (*, P < 0.05). The values shown are means (± standard errors of the means) for five independent experiments, each conducted in duplicate.
The DENV 5′ UTR is unable to support noncanonical translation initiation in cells efficiently.
To further examine the function of the DENV IRES, we next decided to evaluate its activity in cells. However, first, we sought to determine if, as observed in RRL (Fig. 1), a noncanonical mode of initiation could occur in cells. For this purpose, in vitro-transcribed monocistronic virus-like mRNAs DENV m7GpppG-3′UTR and DENV ApppA-3′UTR (Fig. 5A) were transfected into Vero E6 (Fig. 5B) or BHK-21 (Fig. 5C) cells, together with an m7GpppG-Poly(A) monocistronic RNA encoding RLuc, pCI RLuc (a control for the efficiency of transfection) (Fig. 5A). The Glo m7GpppG-Poly(A) and Glo ApppA-Poly(A) RNAs (Fig. 5A) were incorporated into the assay as supplementary controls for translation. Four hours posttransfection, cells were lysed, luciferase activity was determined, and total RNA was extracted from the cell lysates. Total RNA was used as a template in an RT-qPCR assay to determine the relative amount of FLuc-encoding mRNA in cells. When analyzed, RLuc activities were similar in cell extracts, suggesting that equivalent amounts of RNAs were transfected in Vero E6 (Fig. 5B, left) and BHK-21 (Fig. 5C, left) cells. RLuc activity was higher in BHK-21 cells (Fig. 5C, left) than in Vero E6 cells (Fig. 5B, left). In accord with this observation, FLuc activity, normalized to the relative amount of FLuc-expressing RNA ([RNA]), also was 2 log units higher in BHK-21 cells (Fig. 5C, right) than in Vero E6 cells (Fig. 5B, right). In cells, replacement of the m7GpppG cap with an ApppA cap abrogated translation from the Glo m7GpppG-Poly(A) reporter in both cell lines (Fig. 5B and C, right). In agreement with the findings of Edgil et al. in 2006 (15), replacement of the m7GpppG cap with an ApppA cap in the DENV m7GpppG-3′UTR RNA led to a ∼95% reduction in translation in both cell lines (Fig. 5B and C, right). Noteworthy, the remaining ∼5% of FLuc activity of the DENV ApppA-3′UTR was significantly higher than what was obtained with the Glo ApppA-Poly(A) RNA, used as a negative control. However, in agreement with Edgil et al. (15), we conclude that the 5′ UTR of DENV 2 mRNA cannot drive efficient cap-independent translation initiation in the context of a monocistronic mRNA either in Vero E6 (Fig. 5B) or BHK-21 (Fig. 5C) cells.
FIG 5.
In the context of monocistronic RNAs, the DENV 5′ UTR exhibits negligible IRES activity in cells. (A) Schematic representation of in vitro-transcribed RNAs transfected into mammalian cells. (B and C) The capped or A-capped DENV 2-like RNAs (encoding FLuc) and the globin-like RNA (encoding FLuc) were transfected together with the capped and polyadenylated pCI RLuc RNA (encoding RLuc) into Vero E6 (B) or BHK-21 (C) cells. RNA was extracted and was used to determine the relative amount of FLuc-encoding RNA. RLuc (left) and FLuc (right) activities were determined. RLuc activity is expressed in relative light units, while FLuc activity was normalized to the level of FLuc-encoding RNA (FLuc/[RNA]). Values are means (± standard errors of the means) for three independent experiments, each performed in duplicate. Statistical analysis was performed using a two-tailed t test. Asterisks indicate P values of <0.05.
Next, we evaluated the translational activity of the DENV 5′ UTR, but in the context of a dl-RNA. We decided to examine DENV IRES activity in different cell lines, assuming that its activity could be cell type dependent (24, 37). In this study, we included the Vero E6 (African green monkey cells), HEK 293T (human cells, kidney), BHK-21 (hamster cells, kidney), A549 (human cells, lung carcinoma), Jurkat (human cells, T cells), Huh-7 (human cells, hepatocarcinoma), HeLa (human cells, cervical tumor), MDCK (canine cells, kidney), and C6/36 (mosquito cells) cell lines. Cells were transfected with the capped and polyadenylated versions of the dl 5′UTR DENV RNA, the dl HCV IRES RNA (a positive control for IRES activity), or the dl HCV G266A/G268U RNA (a negative control for IRES activity). Four hours posttransfection, RLuc and FLuc activities were measured. The results, summarized in Table 1, showed that luciferase activities differed in the different cell lines; however, the activities of RLuc were comparable within the same cell line (Table 1). Therefore, within each cell line, cap-dependent translation initiation did not significantly vary among the reporter dl-RNAs. The HCV IRES was active in all cell lines, except for the mosquito cell line C6/36, as reported previously by others (18, 38) (Table 1). Due to this observation and the lack of appropriate positive and negative controls for IRES activity, the mosquito cells were not considered further in this study. As reported elsewhere (31, 32), the IRES mutant, dl HCV G266A/G268U, exhibited background levels of FLuc activity compared to that with the HCV 5′ IRES RNA (Table 1). FLuc expression from the DENV 5′ UTR was significantly lower than that of the HCV IRES. The RTA data, presented relative to DENV IRES activity (arbitrarily set at 1), showed that the activity of the DENV IRES was 5 to 15 times (depending on the cell line) weaker than that of the HCV IRES (Table 1). However, DENV IRES activity, though marginal, was statistically higher than that of the HCV G266A/G268U 5′ UTR control in BHK-21, Vero E6, HEK 293T, HeLa, Huh-7, and A549 cells. In contrast, in Jurkat and MDCK cells, the 5′ UTR of DENV mRNA exhibited no evident IRES activity (Table 1). Thus, in agreement with Edgil et al. (15), we find that in the context of a bicistronic RNA, the 5′ UTR of DENV mRNA exhibits a negligible (statistically significant but weakly above background) ability to drive IRES activity in cells.
TABLE 1.
Transfection of the dl-reporter RNAs in cell lines
| Cell line | Transfected RNA | Luciferase activity (RLU) (mean ± SEM) |
RTA (FLuc/RLuc) | Normalized RTA | |
|---|---|---|---|---|---|
| RLuc | FLuc | ||||
| Vero E6 | dl HCV IRES | 8.60 × 106 ± 247,909 | 4.50 × 104 ± 6,676 | 5.14 × 10−3 | 7.5 |
| dl HCV G266A/G268U | 6.70 × 106 ± 38,358 | 1.753 × 103 ± 204 | 2.89 × 10−4 | 0.4 | |
| dl 5′UTR DENV | 5.89 × 106 ± 381,428 | 4.10 × 103 ± 928 | 6.84 × 10−4 | 1 | |
| HEK 293T | dl HCV IRES | 1.63 × 107 ± 231,008 | 4.79 × 105 ± 77,190 | 2.93 × 10−2 | 12 |
| dl HCV G266A/G268U | 1.27 × 107 ± 342,424 | 1.17 × 104 ± 2,498 | 9.12 × 10−4 | 0.4 | |
| dl 5′UTR DENV | 1.31 × 107 ± 159,300 | 3.19 × 104 ± 4,958 | 2.44 × 10−3 | 1 | |
| BHK-21 | dl HCV IRES | 1.68 × 107 ± 384,921 | 2.54 × 105 ± 20,248 | 1.5 × 10−2 | 13 |
| dl HCV G266A/G268U | 1.40 × 107 ± 397,720 | 4.65 × 103 ± 128 | 3.32 × 10−4 | 0.3 | |
| dl 5′UTR DENV | 1.42 × 107 ± 298,458 | 1.62 × 104 ± 1,078 | 1.14 × 10−3 | 1 | |
| A549 | dl HCV IRES | 1.46 × 106 ± 44,044 | 1.76 × 104 ± 1,360 | 1.21 × 10−2 | 11 |
| dl HCV G266A/G268U | 9.32 × 105 ± 71,568 | 5.17 × 102 ± 50 | 5.55 × 10−4 | 0.5 | |
| dl 5′UTR DENV | 1.07 × 106 ± 47,434 | 1.2 × 103 ± 47 | 1.12 × 10−3 | 1 | |
| Jurkat | dl HCV IRES | 1.44 × 105 ± 11,224 | 1.19 × 103 ± 135 | 8.22 × 10−3 | 5 |
| dl HCV G266A/G268U | 8.6 × 104 ± 13,317 | 1.65 × 102 ± 12 | 1.97 × 10−3 | 1.2 | |
| dl 5′UTR DENV | 1.45 × 105 ± 12,437 | 2.39 × 102 ± 17 | 1.65 × 10−3 | 1 | |
| Huh-7 | dl HCV IRES | 1.07 × 107 ± 938,542 | 7.87 × 104 ± 8,174 | 7.5 × 10−3 | 10 |
| dl HCV G266A/G268U | 8.10 × 106 ± 831,655 | 2.43 × 103 ± 212 | 3.11 × 10−4 | 0.4 | |
| dl 5′UTR DENV | 8.82 × 106 ± 960,521 | 6.58 × 103 ± 696 | 7.66 × 10−4 | 1 | |
| HeLa | dl HCV IRES | 6 × 106 ± 95,609 | 2.42 × 105 ± 12,578 | 4.04 × 10−2 | 15 |
| dl HCV G266A/G268U | 4.11 × 106 ± 143,745 | 5.02 × 103 ± 202 | 1.22 × 10−3 | 0.4 | |
| dl 5′UTR DENV | 4.4 × 106 ± 113,433 | 1.2 × 104 ± 667 | 2.73 × 10−3 | 1 | |
| MDCK | dl HCV IRES | 1.52 × 106 ± 71,888 | 4.84 × 103 ± 1,035 | 3.14 × 10−3 | 6 |
| dl HCV G266A/G268U | 1.16 × 106 ± 52,019 | 6.29 × 102 ± 139 | 5.36 × 10−4 | 1 | |
| dl 5′UTR DENV | 9.84 × 105 ± 54,237 | 5.08 × 102 ± 31 | 5.22 × 10−4 | 1 | |
| C6/36 | dl HCV IRES | 4.53 × 106 ± 211,645 | 9.94 × 102 ± 157 | 2.23 × 10−4 | 0.9 |
| dl HCV G266A/G268U | 2.83 × 106 ± 87,714 | 5.75 × 102 ± 35 | 2.04 × 10−4 | 0.8 | |
| dl 5′UTR DENV | 4.58 × 106 ± 131,534 | 1.14 × 103 ± 122 | 2.5 × 10−4 | 1 | |
DENV IRES activity in BHK-21 and Vero E6 cells.
Reports show that IRESs exhibit higher activities when the dl-vectors are delivered into cells as DNA (22, 39). Even though DENV is an RNA virus, we sought to explore this possibility. For this purpose, the dl 5′UTR DENV plasmid DNA was transfected into BHK-21, Vero E6, and HEK 293T cells. The dl ΔEMCV plasmid was used as a negative control for IRES activity. RLuc and FLuc activities were measured 24 h posttransfection. As before (Table 1), RLuc activity was readily detected in the BHK-21, Vero E6, and HEK 293T cell lines transfected with either plasmid, confirming the adequate transfection and expression of the reporter RNAs (Table 2). The RLuc activities of the two reporter constructs were comparable (in the same cell line), indicating that equivalent amounts of DNA were initially transfected, and they were similarly expressed (Table 2). Similarly to what was observed when RNA was transfected (Table 1), RLuc activity differed from cell line to cell line (Table 2). As expected, the dl ΔEMCV reporter exhibited no IRES activity (Table 2). Strikingly, in BHK-21 and Vero E6 cells, FLuc activity was significantly above the background level given by the dl ΔEMCV control (Table 2). Relative to the RLuc and FLuc activities obtained by transfecting RNA (Table 1), despite similar RLuc activities (Table 2), FLuc activity was increased by an average of 2 log units when the dl 5′UTR DENV reporter was delivered as DNA (Table 2). The results expressed as RTA, relative to the activity of the “DENV IRES” (set at 1), showed that IRES-mediated translation initiation driven by the 5′ UTR of DENV mRNA was significantly above background levels (Table 2). In contrast to what was observed in BHK-21 and Vero E6 cells, or when the dl 5′UTR DENV plasmid was delivered as RNA (Table 1), no IRES activity was evident in HEK 293T cells transfected with the dl 5′UTR DENV plasmid delivered as DNA (Table 2).
TABLE 2.
Transfection of the dl-reporter plasmids in cell lines
| Cell line | Transfected DNA | Luciferase activity (RLU) (mean ± SEM) |
RTA (FLuc/RLuc) | Normalized RTA | |
|---|---|---|---|---|---|
| RLuc | FLuc | ||||
| BHK-21 | dl ΔEMCV | 2.070 × 107 ± 264,626 | 0.3 × 105 ± 11,633 | 1.58 × 10−2 | 0.13 |
| dl 5′UTR DENV | 1.5 × 107 ± 398,903 | 1.8 × 105 ± 45,206 | 1.19 × 10−1 | 1 | |
| Vero E6 | dl ΔEMCV | 5.2 × 106 ± 1,347,204 | 0.02 × 105 ± 1,568 | 4.78 × 10−4 | 0.06 |
| dl 5′UTR DENV | 5.3 × 106 ± 1,449,812 | 0.44 × 105 ± 37,428 | 8.24 × 10−3 | 1 | |
| HEK 293T | dl ΔEMCV | 5.7 × 106 ± 996,239 | 0.018 × 106 ± 4,222 | 3.16 × 10−3 | 0.96 |
| dl 5′UTR DENV | 2.88 × 106 ± 930,362 | 0.09 × 106 ± 3,569 | 3.28 × 10−3 | 1 | |
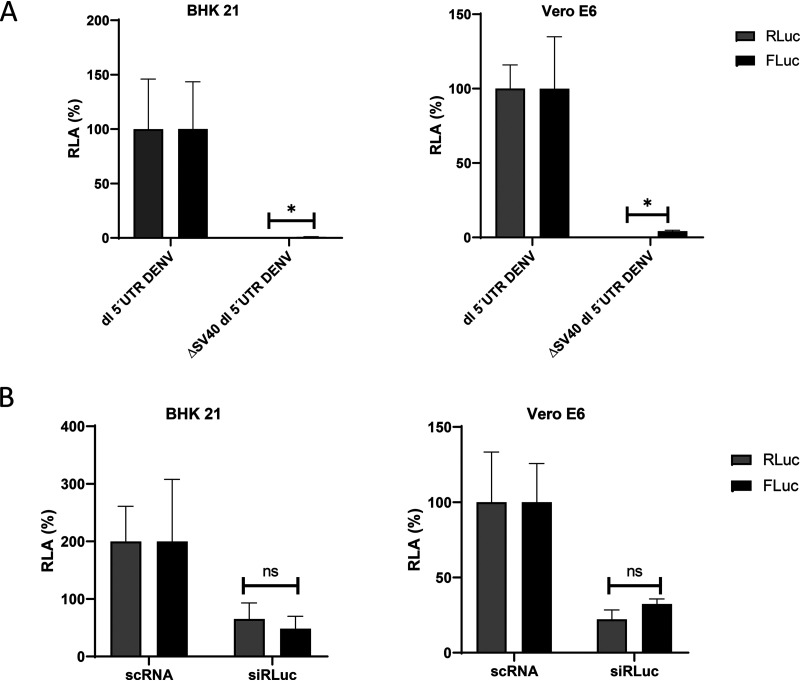
A caveat with regard to the delivery of the dl-vectors to cells as DNA is the potential for false-positive results or artifactual “non-IRES” activity caused by alternative splicing or by cryptic promoter activity (21, 22, 40, 41). Given our data (Table 2), a critical issue was to determine whether the low FLuc expression detected in BHK-21 and Vero E6 cells transfected with the dl 5′UTR DENV plasmid was due to the activity of a cryptic promoter or to an alternative splicing event. To evaluate the existence of a possible cryptic promoter within the dl-DNA, the simian virus 40 (SV40) promoter was removed from the dl 5′UTR DENV plasmid, generating the ΔSV40 dl 5′UTR DENV vector. The dl 5′UTR DENV plasmid or the promoterless ΔSV40 dl 5′UTR DENV plasmid was transfected into BHK-21 or Vero E6 cells. In the absence of the SV40 promoter, both the RLuc and FLuc activities were diminished in both cell lines (Fig. 6A), suggesting that the expression of FLuc from the dl 5′UTR DENV DNA cannot be linked to a cryptic promoter that is active. To evaluate if alternative splicing was occurring, the Renilla luciferase ORF of the dl 5′UTR DENV mRNA was targeted with a small interfering RNA (siRNA) for RLuc (siRLuc) as described previously (35, 42, 43). The siRLuc, or a commercially available scrambled control RNA (scRNA), was cotransfected with the dl 5′UTR DENV plasmid into BHK-21 or Vero E6 cells, and the RLuc and FLuc activities were determined. In the presence of the siRLuc RNA, the RLuc and FLuc activities were reduced in equivalent proportions, strongly suggesting that both reporter proteins were synthesized from the same transcript (Fig. 6B). Together, these findings indicate that when transfected as DNA, the DENV 5′ UTR is capable of weak but genuine internal initiation.
FIG 6.
FLuc expression from the dual-luciferase reporter is due to IRES activity. (A) BHK21 (left) and Vero E6 (right) cells were transfected with the dl 5′UTR DENV and ΔSV40 dl 5′UTR DENV plasmids (200 ng). The latter plasmid shares the full backbone of the dl 5′UTR DENV and lacks only the SV40 promoter. The RLuc (shaded bars) and FLuc (filled bars) activities obtained for the dl 5′UTR DENV DNA were set to 100%. (B) A 50 nM concentration of a scrambled control RNA (scRNA) or the siRLuc siRNA, which targets the RLuc ORF, was cotransfected with the dl 5′UTR DENV plasmid (200 ng) into BHK-21 (left) or Vero E6 (right) cells. RLuc (shaded bars) and FLuc (filled bars) activities were measured and are expressed relative to the luciferase activities obtained when cells were transfected with the scRNA, set at 100%. Statistical analysis was performed with a two-tailed t test. Asterisks indicate P values of <0.05; ns, no significant difference. The values shown are means (± standard errors of the means) for three independent experiments, each performed in duplicate.
The DENV IRES was activated in HEK 293T cells expressing the HRV 2A protease.
DENV mRNA can support translation when cap-dependent translation is pharmacologically suppressed (15). We wondered if, in the experiments conducted with HEK 293T cells (Table 2), the DENV IRES was in an inactive state. This notion agrees with a transition of the viral mRNA from a cap-dependent to a cap-independent translation initiation mechanism during DENV infection (15–17). In their study, Edgil et al. unsuccessfully attempted to stimulate DENV IRES activity by using drugs that target the availability of eIF4E (15). However, from our observations, treatment of RRL with FMDV L protease stimulated DENV IRES activity (Fig. 4). Because we lack a plasmid for expressing the FMDV L protease in cells, we decided to use the Human rhinovirus (HRV) 2A protease (p2A) to target eIF4G (43). A plasmid expressing either a wild-type (wt) HRV p2A (p2A-wt) or an inactive mutant protease (p2A-mut) was transfected into HEK 293T cells together with the dl HCV IRES or dl 5′UTR DENV plasmid by following a previously described strategy (43). Cleavage of eIF4G in the presence of HRV p2A-wt was confirmed by Western blot analysis (Fig. 7A). No cleavage of eIF4G was observed in cells transfected with the p2A-mut plasmid (Fig. 7A). RLuc activities in the two plasmids were dissimilar, yet despite this difference, the presence of HRV p2A-wt significantly reduced RLuc activity by an average of ∼64% for either vector from that with its HRV p2A-mut control (Fig. 7B). In the presence of HRV p2A-wt, FLuc activity was significantly increased, by ∼140%, for the dl HCV IRES over that with its HRV p2A-mut control (Fig. 7C). Strikingly, FLuc activity driven by the DENV 5′ UTR increased from background levels (no detectable activity) to levels comparable to the activity exhibited by the HCV IRES (Fig. 7C). These results indicate that in the presence of the HRV 2A protease, the activity of the DENV IRES is switched on in HEK 293T cells. This result supports in full the notion proposed by Edgil et al. (15), indicating that suppression of cap-dependent protein synthesis activates DENV mRNA cap-independent translation initiation.
FIG 7.
Translation promoted by the DENV 5′ UTR is activated for the proteolytic cleavage of eIF4G by HRV 2A protease. The dl HCV IRES or dl 5′UTR DENV plasmid (200 ng) was transfected into HEK 293T cells together with a plasmid expressing either wild-type HRV 2A protease (p2A-wt) or an inactive mutant (p2A-mut) (500 ng). (A) The cleavage of eIF4G was monitored by Western blotting using polyclonal antibodies against eIF4GI. The eIF4G cleavage product (CP) has been characterized previously (34). (B and C) RLuc (B) and FLuc (C) activities were measured and are expressed in relative light units (RLU). Statistical analysis was performed with a two-tailed t test. Asterisks indicate P values of <0.05; ns, no significant difference. Values represent means (± standard errors of the means) for three independent experiments, each conducted in duplicate.
DISCUSSION
DENV mRNA translation is resistant to the pharmacological suppression of cap-dependent translation initiation (15). In accordance, early after infection, DENV induces dramatic repression of host cell translation (17). Interestingly, the inhibition of cellular mRNA translation in DENV-infected cells is at the stage of mRNA translation initiation (17). In parallel, DENV infection induces cellular endoplasmic reticulum (ER) stress, an event required for viral replication (44). In general, ER stress leads to oligomerization and activation of PERK, which mediates the phosphorylation of eIF2α, resulting in the shutdown of global translation (45). Based on these results, several authors have suggested that DENV mRNA translation initiation can transition from a cap-dependent to a cap-independent mode (15–17). Two different, non-mutually exclusive mechanisms have been proposed to explain how DENV mRNA translation thrives when cap-dependent translation initiation is shut down (15, 18). In this study, we focused exclusively on one of these mechanisms and evaluated the function of the DENV IRES both in vitro and in cells. Translation in the conventional RRL system has proven useful for deciphering the general features of mRNA translation (25). It is clear that, like all in vitro systems, the RRL system has limitations (28, 46). Yet in vitro translation in RRL has proven useful in the characterization of multiple IRESs, including the identification of the IRES of HCV (47), which, like DENV, is a member of the Flaviviridae family of viruses. Based on this, we sought to determine if the DENV IRES was also functional in RRL. Interestingly, we were successful in reproducing in the RRL results reported when DENV-like RNAs were translated in BHK-based cell-free translational extracts or when RNA was directly transfected into cells (15, 18). So we confirmed in RRL that the DENV-like RNA was capable of driving noncanonical translation initiation (Fig. 1). Next, we geared our study toward the DENV IRES by using in vitro-transcribed bicistronic RNAs. In agreement with the report of Song et al. (18), but using RRL, we confirmed that the DENV 5′ UTR has IRES activity (Fig. 2 and 3). However, in contrast to what was reported previously (18), in RRL, DENV IRES activity was less robust than HCV IRES activity (Fig. 3). Though different, this result was not unexpected, since translation initiation of several viral IRESs is inefficient in RRL unless the in vitro translation system is supplemented with cell extracts (36, 48, 49). The presence of an IRES within the DENV 5′ UTR was further demonstrated by showing that the 5′ UTR of DENV mRNA enables translation of the second cistron of a bicistronic RNA under experimental conditions that inhibit cap-dependent translation (Fig. 4). Interestingly, treatment of RRL with the FMDV L protease stimulated DENV IRES activity. In agreement with previous reports, minor stimulation of the HCV IRES was observed in FMDV L protease-treated RRL (Fig. 4) (33, 36). Noteworthy, proteinase-mediated stimulation of IRES-mediated translation initiation is observed mainly under conditions where the basal level of translation is relatively low (36). These results suggest that DENV IRES activity, though functional in RRL, is not optimal in this in vitro system. Despite this, we consider that the DENV IRES can be studied in RRL. For us, this is a relevant finding, because preparing in-house eukaryotic cell-free protein synthesis systems can be tricky and time-consuming, and the RRL is a well-characterized commercially available system (25).
Having established that RRL is suitable for characterizing the DENV IRES, we decided to transfect RNAs into cells. In agreement with the findings of Edgil et al. (15), when monocistronic or bicistronic RNAs were transfected into cells, only marginal, if any, noncanonical translation could be documented (Fig. 5; Table 1). These results contrast with those of Song et al. (18), who described robust DENV IRES activity in BHK-21 and Vero E6 cells. These contradictory observations cannot be readily explained. We presume that the difference between these studies could be associated with the reporter used. Like Edgil et al. (15), we used FLuc as a reporter, while Song et al. (18) used the luciferase from Gaussia prínceps, Gaussia luciferase (GLuc). GLuc is secreted from cells, so during the experiment, the reporter can be found in the medium and within cells (50). The mean values, combining both intracellular and extracellular activities in mammalian cells, have been documented to be >1,000-fold higher for GLuc than for FLuc (50). When one is comparing intracellular luciferase activity only in mammalian cell lysates, thus eliminating the contribution of the secreted form of the reporter, the activity of GLuc is 100-fold higher than that of FLuc (50). Thus, at least in part, the difference between the studies could be due to the sensitivity of the assay used. This possibility prompted us to evaluate the same bicistronic mRNAs when delivered as DNAs, considering reports indicating that the efficiency of the IRES is enhanced when it is delivered as DNA (22, 39). When the bicistronic mRNAs were delivered as DNAs, low IRES activity was detected in BHK-21 and Vero E6 cells, while no FLuc activity was detected in HEK 293T cells (Table 2). We confirmed that the IRES activity obtained in BHK-21 and Vero E6 cells, though low, was genuine (Fig. 6). Following the report of Edgil et al. (15) showing that noncanonical translation initiation of DENV mRNA is observed when cap-dependent translation is suppressed, and considering that in RRL, DENV IRES activity was stimulated by eIF4G cleavage (Fig. 4), we decided to use a similar approach in cells. In agreement with our in vitro findings (Fig. 4), when cap-dependent translation initiation was reduced in cells, DENV IRES activity was significantly enhanced in HEK 293T cells (Fig. 7). Surprisingly, the activity of the DENV IRES increased to levels similar to those obtained for the HCV IRES. The increase in DENV IRES activity obtained when cells were treated with the HRV 2A protease cannot, at this stage, be ascribed exclusively to the cleavage of eIF4G (Fig. 7). The stimulation of IRES activity could also be explained by a combined effect of the protease acting on the expression or stability of several different proteins within the cells (51). The latter possibility was not evaluated in this study. Despite the unknowns, our findings suggest that under normal growth conditions in HEK 293T cells, translation initiation from DENV mRNA is mainly, if not exclusively, cap dependent. Even though negligible FLuc activity was detected when the reporters were transfected as RNA (Table 1), no IRES activity at all was observed when the vector was delivered as DNA (Table 2). However, suppression of cap-dependent translation, in our case by the HRV 2A protease, led translation to switch to an IRES mode (Fig. 7). These findings fully support reports proposing that during the DENV-induced shutoff of cap-dependent translation, viral mRNA translation thrives by transitioning from a cap-dependent to a cap-independent mechanism (16, 17). Thus, we provide evidence indicating that the DENV IRES enables viral protein synthesis under conditions that suppress canonical translation initiation.
MATERIALS AND METHODS
Plasmids.
Plasmids pGL5′3′DV (12), dl ΔEMCV (27), dl HCV 1b (referred to here as dl HCV IRES) (31), dl HCV G266A/G268U (31), and dl PV IRES (52) have been described previously. The pGL5′3′DV and Glo-FLuc vectors were kindly provided by Andrea Gamarnik (Fundación Instituto Leloir, Argentina) and Ricardo Soto-Rifo (Universidad de Chile, Chile), respectively. Plasmids expressing the functional wild-type (genotype A16) human rhinovirus (HRV) 2A protease or an inactive mutant were generously supplied by Ann C. Palmenberg and Kelly Watters (University of Wisconsin—Madison, USA) and have been validated previously (43). To generate the dual-luciferase (dl) bicistronic vectors, dl Δ EMCV 5´UTR DENV and dl 5´UTR DENV, the DENV 5′ UTR was recovered from the pGL5′3′DV vector by PCR using primers DENV 5UTR F Eco RI (5′-CTCAAAGAATTCAGTTGTTAGTCTACGTGG-3′) and DENV 5UTR R NcoI (5′-CTCAAACCATGGCAGAGATCTGCTCTCTAA-3′). The amplicons were digested using EcoRI and NcoI (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were cloned into the intercistronic space of the dl HIV IRES described in reference 53, which had been digested previously with EcoRI/NcoI and NcoI/XbaI as described in reference 35. The promoterless vector ΔSV40 dl 5′UTR DENV was generated by digesting the dl 5′UTR DENV plasmid with MluI and Eco147I (StuI) (Thermo Fisher Scientific), followed by a ligation reaction.
Cell culture.
Huh-7 cells were provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany) and have been used in the laboratory previously (31). HEK 293T (ATCC CRL-11268), HeLa (ATCC CRM-CCL-2), Vero E6 (ATCC CRL-1586), A549 (ATCC CCL-185), MDCK (ATCC PTA-6500), and BHK-21 (ATCC CCL-10) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (catalog no. SH30022; HyClone, GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (catalog no. SH30910; HyClone, GE Healthcare Life Sciences), 1% penicillin-streptomycin (1,000 U/ml) (catalog no. SV30010; HyClone, GE Healthcare Life Sciences), and 1% amphotericin B (25 mg/ml) (catalog no. SV30078.01; HyClone). Huh-7 cell culture medium was supplemented with MEM nonessential amino acid solution (catalog no. 11140050; Thermo Fisher Scientific). Aedes albopictus clone C6/36 cells were kindly provided by Andrea Gamarnik (Fundación Instituto Leloir, Argentina). Jurkat (ATCC TIB-152) and C6/36 cells were grown in RPMI 1640 medium (catalog no. 12633020; Thermo Fisher Scientific). All cells were cultured at 37°C under a 5% CO2 atmosphere, with the exception of C6/36 cells, which were kept at 28°C.
In vitro transcription.
For in vitro transcription, plasmids were digested with BamHI, except for the dl PV IRES, which was treated with XhoI, and the Glo-FLuc vector, which was treated with EcoRI. To obtain monocistronic DENV mRNA reporters, pGL5′3′DV was digested with BamHI and XbaI for DENV 3′ UTR deletion. The RNAs were synthesized using the mMESSAGE mMACHINE T7 transcription kit (catalog no. AM1344; Thermo Fisher Scientific) according to the producer’s recommendations. mRNAs with a nonfunctional cap (Acap) were synthesized in the presence of the A(5′)ppp(5′)A cap analog (catalog no. NU-506-5; Jena Bioscience, Jena, Thuringia, Germany). The poly(A) tail was added using the Poly(A) tailing kit (catalog no. AM1350; Thermo Fisher Scientific). After transcription, the RNA was treated with 2 U of Turbo DNase (catalog no. AM2238; Thermo Fisher Scientific) for 30 min at 37°C, precipitated for 1 h at −20°C in the presence of 2.5 M LiCl, centrifuged at 16,000 × g for 30 min at 4°C, washed with 70% ethanol, and resuspended in 50 µl of nuclease-free water. RNA concentrations were determined spectrophotometrically using nanospectrophotometry (NanoPhotometer N60; Implen, Westlake Village, CA, USA). RNA integrity was assessed by electrophoresis in a 1% agarose gel using formamide as a denaturing agent in the loading buffer.
In vitro translation.
In vitro translation reactions were carried out in nuclease-treated rabbit reticulocyte lysate (RRL) (catalog no. L4960; Promega Corporation) as described previously using 0.01 pmol RNA (43). Optimal salt concentrations were used to translate the dl HCV IRES (100 mM KCl and 0.75 mM MgOAc2) and dl PV IRES in vitro as described previously (32, 35). The L protease–RRL was diluted 1:5 or 1:10 in nuclease-free water (IDT), added (to a final concentration of 1, 3, 6, or 10% [vol/vol]) to the fresh nuclease-treated 35% (vol/vol) RRL, and preincubated for 15 min at 30°C prior to the addition of the bicistronic mRNA.
DNA transfection.
Cells were seeded (5 × 104 BHK-21 cells/ml, 1.2 × 105 HEK 293T cells/ml, and 8 × 104 Vero E6 cells/ml) 24 h prior to transfection in 24-well culture plates. DNA transfection experiments were performed at 70 to 80% confluence using polyethyleneimine (PEI; Gibco, Thermo Fisher Scientific, Inc.). The activity of the DENV IRES was evaluated by cotransfecting 200 ng of bicistronic vectors with 50 ng of the pcDNA 3.1-LacZ plasmid. pSP64 Poly(A) was used as a negative control and also as an irrelevant DNA to reach equal quantities of transfected material. In all transfection assays, equivalent concentrations of total DNA were used. For the HRV 2A protease experiments in HEK 293T cells, the cells were cotransfected with 200 ng of the dl HCV IRES or dl 5´UTR DENV plasmid and 500 ng of HRV p2A-wt or HRV p2A-mut per well in a 24-well plate. In all experiments, at 24 h posttransfection, the culture medium was removed, and the cells were harvested using the Passive Lysis buffer supplied with the Dual-Luciferase reporter assay system (catalog no. E1960; Promega Corporation) according to the manufacturer’s protocols.
siRNA and RNA transfection.
The cells described above were seeded (5 × 104 BHK-21 cells/ml, 1.2 × 105 HEK 293T cells/ml, 1 × 105 Jurkat cells/ml, 8 × 104 Vero E6 cells/ml, 6 × 104 Huh-7 cells/ml, 1 × 105 HeLa cells/ml, 8 × 104 MDCK cells/ml, 2 × 105 C6/36 cells/ml, and 1 × 105 A549 cells/ml) 24 h before RNA transfection to obtain 90% confluence per well in a 24-well culture plate. Then 0.5 pmol of the in vitro-transcribed RNA was transfected. The transfection was performed using the Lipofectamine 2000 system (catalog no. 11668019; Thermo Fisher Scientific). Four hours posttransfection, cells were harvested. For RLuc silencing, DNA cotransfection was performed at 60 to 70% confluence using the Lipofectamine 2000 system (Thermo Fisher Scientific). For transfection, 200 ng of the dl 5′UTR DENV plasmid was cotransfected with a 50 nM concentration of either Silencer Select Negative Control no. 1 siRNA (catalog no. 4390844; Ambion, Thermo Fisher Scientific) or a siRNA that targets the RLuc ORF (Renilla siRNA [5′-UAUAAGAACCAUUACCAGAUUUGCCUG-3′]; IDT) (42). The RLuc and FLuc activities were measured after 24 h posttransfection (42).
Luciferase assays.
The activities of firefly luciferase (FLuc) and Renilla luciferase (RLuc) were measured using the dual-luciferase reporter (DLR) assay system (catalog no. E1960; Promega Corporation), according to the manufacturer’s instructions, on 20 μl of cell lysates using a Sirius Single Tube luminometer (Berthold Detection Systems GmbH). Data are expressed as the relative luciferase activity (RLA) or relative translation activity (RTA) (35, 42, 43). For in vitro translations, FLuc activity was normalized to FLuc RNA abundance (FLuc RNA was quantified relative to the amount of 18S rRNA by the ΔΔCT method, as described below). When monocistronic RNA reporters were employed in cells, FLuc activity was normalized to FLuc RNA abundance (FLuc RNA was quantified relative to the amount of glyceraldehyde-3-phosphate dehydrogenase [GAPDH] RNA by the ΔΔCT method, as described below).
RNA extraction and RT-qPCR.
RNA was extracted as described previously (42). Cells were washed twice with 1× phosphate-buffered saline (PBS) (catalog no. SH30256; HyClone, GE Healthcare Life Sciences) and incubated on ice. A 500-μl volume of TRIzol reagent (catalog no. 15596018; Life Technologies Corporation, Thermo Fisher Scientific, Inc.) was added to the supernatant, and the RNA was recovered according to the manufacturer’s protocol. The RNA was resuspended in 20 μl of nuclease-free water, treated with DNase (catalog no. AM1907; Ambion, Thermo Fisher Scientific, Inc.), and purified. The RNA concentration was determined by nanospectrophotometry (NanoPhotometer N60; Implen). Real-time RT-qPCR experiments were carried out using the Brilliant II SYBR green RT-qPCR one-step master mix (catalog no. 600835; Agilent Technologies, Santa Clara, CA, USA). qPCRs without reverse transcriptase were carried out to control for contaminant DNA. FLuc RNA was detected with firefly sense (5′-ACTTCGAAATGTCCGTTCGG-3′) and firefly antisense (5′-GCAACTCCGATAAATAACGCG-3′) primers, and GAPDH mRNA was amplified using primers GAPDH sense (5′-TCCACCACCCTGTTGCTGTAG-3′) and GAPDH antisense (5′-ACCCACTCCTCCACCTTTGAC-3′) as described previously (54). 18S rRNA, used as a reference gene in RRL, was amplified using primers 18S sense (5′-GTGGAGCGATTTGTCTGGTT-3′) and 18S antisense (5′-CGCTGAGCCAGTCAGTGTAG-3′) as described in reference 43. Data were analyzed using the previously described ΔΔCT method (55, 56).
Western blotting.
Cells were lysed using the 5× Passive Lysis buffer (catalog no. E1941; Promega Corporation), and the concentration of total protein was determined by the Bradford assay using the Bio-Rad Protein Assay (catalog no. 5000006; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Thirty micrograms of total protein was resolved in a 15% Tricine-SDS-polyacrylamide gel and was transferred to a 0.45-µm-pore-size polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific, MA, USA). A rabbit polyclonal anti-eIF4G antibody (H-300; catalog no. sc-11373; Santa Cruz Biotechnology) at a 1:1,000 dilution, followed by a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (catalog no. AP132P; Merck, Darmstadt, Germany) at a 1:10,000 dilution, was used to develop the blot. Western blots were visualized by enhanced luminescence using a chemiluminescence reaction with 4-hydroxycinnamic acid (catalog no. 800237; Merck) and luminol (catalog no. 09253; Fluka, Sigma-Aldrich).
Sequence and statistical analysis.
Graphics were created, and statistical analysis was carried out, by using GraphPad Prism, version 8.0.0 for Windows (GraphPad Software, San Diego, CA, USA) and performing t tests for individual comparisons or ordinary one-way analysis of variance (ANOVA) for group comparisons. Serial Cloner, version 2.6.1, was used for primer design and sequence alignments.
ACKNOWLEDGMENTS
We thank Andrea Gamarnik, Ann C. Palmenberg, Kelly Watters, and Ricardo Soto Rifo for kindly providing plasmids or reagents used in this study.
This work was supported by the Comision Nacional de Investigación Cientifica y Tecnologica (CONICYT), CONICYT-Programa de Investigación Asociativa (PIA) ACT1408, and the Proyecto P09/016-F de la Iniciativa Científica Milenio (ICM) del Ministerio de Economía, Fomento y Turismo (to M.L.-L.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. L.F.-G. (fellowship 21160121) and A.B. (fellowship 21161331) were supported by CONICYT doctoral fellowships.
M.L.-L. and L.F.-G. designed the study. L.F.-G. performed the cloning and plasmid construction. L.F.-G., J.A., and K.P. performed the in vitro experiments. L.F.-G., J.A., K.P., A.B., H.R., and J.V.-O. performed the experiments in cells. M.L.-L. wrote the manuscript. All authors read and approved the final manuscript.
We declare that no conflict of interest exists.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paranjape SM, Harris E. 2010. Control of dengue virus translation and replication. Curr Top Microbiol Immunol 338:15–34. doi: 10.1007/978-3-642-02215-9_2. [DOI] [PubMed] [Google Scholar]
- 3.Barrows NJ, Campos RK, Liao KC, Prasanth KR, Soto-Acosta R, Yeh SC, Schott-Lerner G, Pompon J, Sessions OM, Bradrick SS, Garcia-Blanco MA. 2018. Biochemistry and molecular biology of flaviviruses. Chem Rev 118:4448–4482. doi: 10.1021/acs.chemrev.7b00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holden KL, Harris E. 2004. Enhancement of dengue virus translation: role of the 3' untranslated region and the terminal 3' stem-loop domain. Virology 329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Pelletier J, Sonenberg N. 2019. The organizing principles of eukaryotic ribosome recruitment. Annu Rev Biochem 88:307–335. doi: 10.1146/annurev-biochem-013118-111042. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alekhina OM, Terenin IM, Dmitriev SE, Vassilenko KS. 2020. Functional cyclization of eukaryotic mRNAs. Int J Mol Sci 21:1677. doi: 10.3390/ijms21051677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson AL, Pasquinelli AE. 2019. Tales of detailed poly(A) tails. Trends Cell Biol 29:191–200. doi: 10.1016/j.tcb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu WW, Kinney RM, Dreher TW. 2005. Control of translation by the 5'- and 3'-terminal regions of the dengue virus genome. J Virol 79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgil D, Diamond MS, Holden KL, Paranjape SM, Harris E. 2003. Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology 317:275–290. doi: 10.1016/j.virol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polacek C, Friebe P, Harris E. 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol 90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- 14.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351:aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 15.Edgil D, Polacek C, Harris E. 2006. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol 80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou JN, Chen TH, Chiang YH, Peng JY, Yang TH, Cheng CC, Sofiyatun E, Chiu CH, Chiang-Ni C, Chen WJ. 2017. PERK signal-modulated protein translation promotes the survivability of dengue 2 virus-infected mosquito cells and extends viral replication. Viruses 9:262. doi: 10.3390/v9090262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, Stoecklin G, Ruggieri A. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. mBio 8:e02150-16. doi: 10.1128/mBio.02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Mugavero J, Stauft CB, Wimmer E. 2019. Dengue and Zika virus 5′ untranslated regions harbor internal ribosomal entry site functions. mBio 10:e00459-19. doi: 10.1128/mBio.00459-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62:2636–2643. doi: 10.1128/JVI.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 21.Thompson SR. 2012. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA 3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. 2004. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwan T, Thompson SR. 2019. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 11:a032672. doi: 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailliot J, Martin F. 2018. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip Rev RNA 9. doi: 10.1002/wrna.1458. [DOI] [PubMed] [Google Scholar]
- 25.Merrick WC, Barth-Baus D. 2007. Use of reticulocyte lysates for mechanistic studies of eukaryotic translation initiation. Methods Enzymol 429:1–21. doi: 10.1016/S0076-6879(07)29001-9. [DOI] [PubMed] [Google Scholar]
- 26.Bergamini G, Preiss T, Hentze MW. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson JE, Powell MJ, Hoover SE, Sarnow P. 2000. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol 20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulous S, Malnou CE, Michel YM, Kean KM, Borman AM. 2003. Comparison of the capacity of different viral internal ribosome entry segments to direct translation initiation in poly(A)-dependent reticulocyte lysates. Nucleic Acids Res 31:722–733. doi: 10.1093/nar/gkf695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasso MC, Jackson RJ. 1989. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res 17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson RJ. 1991. Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: unique features of encephalomyocarditis virus RNA translation. Biochim Biophys Acta 1088:345–358. doi: 10.1016/0167-4781(91)90124-5. [DOI] [PubMed] [Google Scholar]
- 31.Barria MI, Gonzalez A, Vera-Otarola J, Leon U, Vollrath V, Marsac D, Monasterio O, Perez-Acle T, Soza A, Lopez-Lastra M. 2009. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res 37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angulo J, Ulryck N, Deforges J, Chamond N, Lopez-Lastra M, Masquida B, Sargueil B. 2016. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res 44:1309–1325. doi: 10.1093/nar/gkv1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlmann T, Rau M, Pain VM, Morley SJ. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J 15:1371–1382. doi: 10.1002/j.1460-2075.1996.tb00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 35.Olivares E, Landry DM, Caceres CJ, Pino K, Rossi F, Navarrete C, Huidobro-Toro JP, Thompson SR, Lopez-Lastra M. 2014. The 5' untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J Virol 88:5936–5955. doi: 10.1128/JVI.00279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borman AM, Bailly JL, Girard M, Kean KM. 1995. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res 23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plank TD, Whitehurst JT, Kieft JS. 2013. Cell type specificity and structural determinants of IRES activity from the 5' leaders of different HIV-1 transcripts. Nucleic Acids Res 41:6698–6714. doi: 10.1093/nar/gkt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter JR, Fraser TS, Fraser MJ. 2008. Examining the relative activity of several dicistrovirus intergenic internal ribosome entry site elements in uninfected insect and mammalian cell lines. J Gen Virol 89:3150–3155. doi: 10.1099/vir.0.2008/003921-0. [DOI] [PubMed] [Google Scholar]
- 39.Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res 28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han B, Zhang JT. 2002. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol Cell Biol 22:7372–7384. doi: 10.1128/mcb.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. 2003. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene 318:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- 42.Carvajal F, Vallejos M, Walters B, Contreras N, Hertz MI, Olivares E, Caceres CJ, Pino K, Letelier A, Thompson SR, Lopez-Lastra M. 2016. Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J 283:2508–2527. doi: 10.1111/febs.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caceres CJ, Angulo J, Lowy F, Contreras N, Walters B, Olivares E, Allouche D, Merviel A, Pino K, Sargueil B, Thompson SR, Lopez-Lastra M. 2018. Non-canonical translation initiation of the spliced mRNA encoding the human T-cell leukemia virus type 1 basic leucine zipper protein. Nucleic Acids Res 46:11030–11047. doi: 10.1093/nar/gky802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YR, Kuo SH, Lin CY, Fu PJ, Lin YS, Yeh TM, Liu HS. 2018. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci Rep 8:489. doi: 10.1038/s41598-017-18909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 46.Soto Rifo R, Ricci EP, Decimo D, Moncorge O, Ohlmann T. 2007. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res 35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J Virol 66:1476–1483. doi: 10.1128/JVI.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J Virol 50:507–514. doi: 10.1128/JVI.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallejos M, Deforges J, Plank TD, Letelier A, Ramdohr P, Abraham CG, Valiente-Echeverria F, Kieft JS, Sargueil B, Lopez-Lastra M. 2011. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res 39:6186–6200. doi: 10.1093/nar/gkr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 51.Watters K, Inankur B, Gardiner JC, Warrick J, Sherer NM, Yin J, Palmenberg AC. 2017. Differential disruption of nucleocytoplasmic trafficking pathways by rhinovirus 2A proteases. J Virol 91:e02472-16. doi: 10.1128/JVI.02472-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem 273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 53.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol 77:3939–3949. doi: 10.1128/jvi.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vera-Otarola J, Solis L, Soto-Rifo R, Ricci EP, Pino K, Tischler ND, Ohlmann T, Darlix JL, Lopez-Lastra M. 2012. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J Virol 86:2176–2187. doi: 10.1128/JVI.06223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Yuan JS, Reed A, Chen F, Stewart CN, Jr. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]