The HBV preS1 amino acid 2 to 47 region (preS1/2–47) is essential for virus binding to sodium taurocholate cotransporting polypeptide. Several MAbs targeting preS1/2–47 have been reported to neutralize HBV infection; however, which region in preS1/2–47 contains the critical neutralizing epitope(s) for HBV infection is unclear.
KEYWORDS: HBV, epitope, neutralizing antibodies, preS1
ABSTRACT
Hepatitis B virus (HBV) infection is a major public health problem. Human hepatocytes are infected with HBV via binding between the preS1 region in the large envelope protein of HBV and sodium taurocholate cotransporting polypeptide. Although several monoclonal antibodies (MAbs) that recognize the receptor binding domain in preS1 and neutralize HBV infection have been isolated, details of neutralizing epitopes are not understood. In this study, we generated 13 MAbs targeting the preS1 receptor binding domain from preS1-specific memory B cells derived from DNA-immunized mice. The MAbs were classified into three groups according to the epitope regions, designated epitopes I to III. A virus neutralization assay revealed that MAbs recognizing epitopes I and III neutralized HBV infection, suggesting that these domains are critical epitopes for viral neutralization. In addition, a neutralization assay against multiple genotypes of HBV revealed that epitope I is a semipangenotypic neutralizing epitope, whereas epitope III is a genotype-specific epitope. We also showed that neutralizing MAbs against preS1 could neutralize HBV bearing a vaccine-induced escape mutation. These findings provide insight into novel immunoprophylaxis for the prevention and treatment of HBV infection.
IMPORTANCE The HBV preS1 amino acid 2 to 47 region (preS1/2–47) is essential for virus binding to sodium taurocholate cotransporting polypeptide. Several MAbs targeting preS1/2–47 have been reported to neutralize HBV infection; however, which region in preS1/2–47 contains the critical neutralizing epitope(s) for HBV infection is unclear. Here, we generated several MAbs targeting preS1/2–47, and we found that MAbs recognizing the N or C terminus of preS1/2–47 remarkably neutralized HBV infection. We further confirmed the neutralizing activity of anti-preS1 MAbs against HBV with a vaccine escape mutation. These data clarified the relationship between the antibody epitope and the virus-neutralizing activity and also suggested the potential ability of a vaccine antigen containing the preS1 region to overcome the weakness of current hepatitis B vaccines comprising the small S protein.
INTRODUCTION
Hepatitis B virus (HBV) causes acute and chronic liver diseases and is a major global health problem. The World Health Organization estimated that, in 2015, 257 million people were living with chronic HBV infection and 887,000 deaths were caused by HBV infection-related diseases such as cirrhosis, liver failure, and hepatocellular carcinoma (1).
HBV is a small, enveloped DNA virus of the family Hepadnaviridae. The HBV envelope consists of three HBV surface antigens (HBs), small (HBs-S), middle (HBs-M), and large (HBs-L) envelope proteins. These three envelope proteins are encoded by a single open reading frame from three in-frame start codons. Human hepatocytes are infected with HBV via the sodium taurocholate cotransporting polypeptide (NTCP), which is a membrane transporter involved in bile acid uptake (2). The preS1 region of HBs-L protein binds to NTCP, and the N-terminal 47 amino acids of the preS1 domain (genotype D) are crucial for the preS1-NTCP interaction (3–8). Indeed, inhibition of the preS1-NTCP interaction effectively blocks HBV infection (9–11). Thus, the preS1-NTCP interaction is considered to be a potent target for the prevention and treatment of HBV infection. Once HBV infects human hepatocytes via NTCP binding, covalently closed circular DNA is formed in the nucleus of hepatocytes, making elimination of HBV by antiviral drug therapy difficult (12). Therefore, prevention of HBV infection by immunoprophylaxis such as vaccine therapy is important.
Anti-HBV neutralizing antibodies (NAbs) are considered to contribute to the prevention of infection as well as viral clearance. The “a” determinant, the antigenic loop polypeptide present at positions 101 to 172 in HBs-S protein, is an essential immunodominant region for HBV prevention by vaccination with current hepatitis B vaccines consisting of HBs-S protein (4, 13–15). In addition, antibodies targeting the NTCP binding domain of the preS1 region in HBs-L can also neutralize virus infection in vivo and in vitro (4, 16–18). However, neutralizing epitope analysis of the preS1 region has not been systemically performed.
Here, we report the generation of murine monoclonal antibodies (MAbs) against a 46-residue peptide corresponding to the NTCP-interacting domain of preS1 (preS1/2–47) from preS1/2–47-specific memory B cells derived from mice immunized with DNA encoding preS1 and characterization of neutralization epitopes in the receptor binding domain of HBV. Epitope analysis revealed the presence of three major epitopes, designated epitopes I to III. A virus neutralization assay demonstrated that MAbs targeting epitopes I and III have potent neutralizing activity against HBV. Importantly, epitope I is a semipangenotypic neutralizing epitope, and epitope III is a genotype-specific neutralizing epitope. In addition, all virus-neutralizing MAbs (NAbs) could neutralize infection of HBV with the G145R mutation, which is a representative vaccine escape mutation (VEM) observed in patients. These findings provide new insights into neutralizing epitopes in the preS1/2–47 region.
RESULTS
Construction and immunization of plasmids encoding amino acids 2 to 47 of HBV preS1.
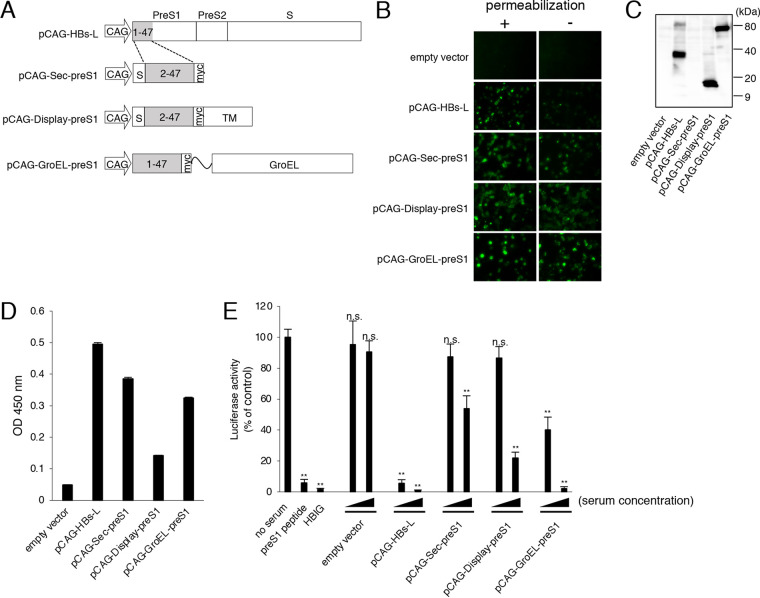
NTCP is an entry receptor of HBV, and the N-terminal myristoylated peptide corresponding to preS1/2–47 of HBs-L protein effectively binds to NTCP (2). We constructed various plasmids encoding preS1/2–47 (Fig. 1A) to obtain NAbs against preS1. Plasmid pCAG-HBs-L encodes HBs-L protein derived from genotype C. Plasmid pCAG-Sec-preS1 encodes preS1/2–47 with the secretion signal at the N terminus and a Myc tag at the C terminus. Plasmid pCAG-Display-preS1 is identical to pCAG-Sec-preS1 except for the presence of the platelet-derived growth factor receptor (PDGFR) transmembrane domain at the C terminus for expression of preS1/2–47 on the cell surface. Plasmid pCAG-GroEL-preS1 encodes preS1/2–47-Myc fused to GroEL, a chaperone protein derived from Escherichia coli, which is a molecular adjuvant for DNA immunization (19).
FIG 1.
Immunization of mice with plasmids encoding HBV preS1/2–47. (A) Schematic diagram of plasmids encoding preS1/2–47, showing the position of the CAG promoter (CAG), murine Ig κ-chain leader sequence (S), Myc tag (myc), PDGFR transmembrane domain (TM), and GroEL. The amino acid sequence of preS1/2–47 is derived from HBV genotype C. The residue numbering of the HBV preS1 domain is based on HBV genotype D. (B) Detection of preS1/2–47 in 293T cells transfected with each plasmid with the immunofluorescence assay. PreS1/2–47 in cells with (+) or without (−) permeabilization was detected using anti-preS1 antibody. (C) Expression of preS1/2–47. Lysates of 293T cells transfected with each plasmid were harvested at 3 days posttransfection and subjected to immunoblotting using anti-preS1 antibody. (D) Detection of serum antibodies binding to the preS1/2–47 peptide. Pooled sera from mice immunized with the indicated plasmids were diluted 500-fold and added to ELISA microtiter wells containing the preS1/2–47 peptide. Bound antibody was detected using HRP-conjugated anti-mouse IgG secondary antibody. Data are averages of triplicate values, with error bars showing standard deviations. OD, optical density. (E) Neutralizing activities of sera from mice immunized with preS1 expression plasmids. HBV/NL derived from genotype C was preincubated with pooled sera derived from mice immunized with the indicated plasmids (500- or 50-fold dilution) and then used to infect G2/NT-18 cells for 16 h. Luciferase activity was determined at 7 days postinfection and is expressed relative to activity without serum. Myristoylated preS1 peptide (100 nM) or HBIG (40 mIU/ml) was used as a control. Data are averages of quadruplicate values, with error bars showing standard deviations. **, P < 0.001 versus no-serum control group, Student's t test; n.s., not significant.
PreS1/2–47 expression in cells transfected with all plasmids was confirmed with an immunofluorescence assay following Triton X-100 treatment (Fig. 1B). Immunofluorescence signals were also observed in the cells transfected with pCAG-Sec-preS1, pCAG-Display-preS1, and pCAG-GroEL-preS1 without Triton X-100 treatment (Fig. 1B), suggesting that these preS1/2–47 peptides were expressed on the cell surface. Major single bands with predicted molecular weights were also observed with Western blotting of proteins derived from cells transfected with pCAG-HBs-L, pCAG-Display-preS1, and pCAG-GroEL-preS1 (Fig. 1C). We failed to detect the preS1 peptide in cells transfected with pCAG-Sec-preS1 with Western blotting, possibly due to the small molecular size.
To induce anti-preS1 antibodies, mice were immunized with each constructed plasmid four times, followed by intraperitoneal injection of 293T cells transfected with each plasmid as a final booster immunization. Immunized mouse sera were subjected to an enzyme-linked immunosorbent assay (ELISA) to detect specific antibodies against the preS1/2–47 peptide. Pooled sera of mice immunized with pCAG-HBs-L, pCAG-Sec-preS1, and pCAG-GroEL-preS1 clearly showed positivity in the ELISA (Fig. 1D). Mouse sera were further subjected to an HBV neutralizing assay using the nanoluciferase reporter virus of HBV (HBV/NL) from genotype C (20). HBV/NL infection was inhibited by myristoylated preS1/2–47 peptide (preS1 peptide) or hepatitis B immunoglobulin (HBIG) (Fig. 1E). Sera from mice immunized with plasmids encoding preS1/2–47 showed dose-dependent neutralization of HBV/NL infection. Among them, sera from mice immunized with pCAG-HBs-L and pCAG-GroEL-preS1 showed higher neutralizing activity, although pCAG-HBs-L could induce not only anti-preS1 antibodies but also anti-preS2 and anti-HBs-S antibodies.
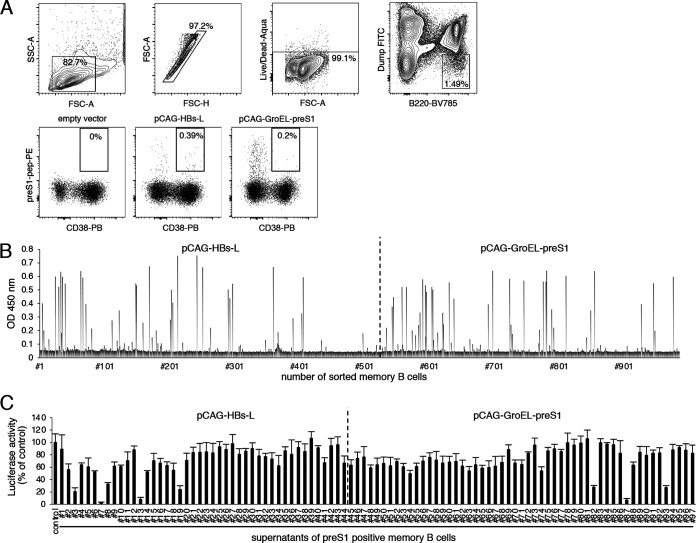
Isolation of preS1/2–47-binding memory B cells.
To obtain preS1/2–47-binding memory B cells, splenocytes were extracted from mice immunized with pCAG-HBs-L or pCAG-GroEL-preS1, due to the results of the ELISA as well as neutralizing activity. B220+/CD38+ memory B cells that bound to the preS1 peptide were isolated using a magnetically activated cell sorting system and flow cytometry. Flow cytometry analysis revealed the presence of preS1-specific memory B cells in mice immunized with pCAG-HBs-L or pCAG-GroEL-preS1 (Fig. 2A). Single sorted B cells were cultured for 10 days, and culture supernatants containing clonal IgG were screened by ELISA. Of 987 isolated memory B cells, 411 were positive for anti-mouse IgG Fab antibody in ELISA (data not shown) and 97 were positive for preS1/2–47 in ELISA (Fig. 2B). These 97 samples were further evaluated for virus-neutralizing activities using HBV/NL. Several clones showed more than 70% reduction in HBV/NL infection (clones 3, 7, 13, 82, and 87) (Fig. 2C).
FIG 2.
Isolation and screening of preS1-specific memory B cells. (A) Splenocytes derived from mice immunized with the indicated plasmids were subjected to flow cytometric analysis. Upper panels show representative data for the gating strategy. Lower panels show representative flow data for preS1/2–47-binding memory B cells. (B) Detection of antibodies to the preS1/2–47 peptide in memory B cell culture supernatants. Culture supernatants of isolated memory B cells were diluted 5-fold and added to ELISA microtiter wells containing preS1/2–47. Antibody binding was detected using HRP-conjugated anti-mouse IgG secondary antibody. OD, optical density. (C) Neutralizing activities of memory B cell culture supernatants. HBV/NL derived from genotype C was preincubated with culture supernatants (1:1) for 1 h and then used to infect G2/NT-18 cells. Luciferase activity of the cells was determined 7 days postinfection and is expressed relative to activity without supernatant. Data are averages of quadruplicate values, with error bars showing standard deviations.
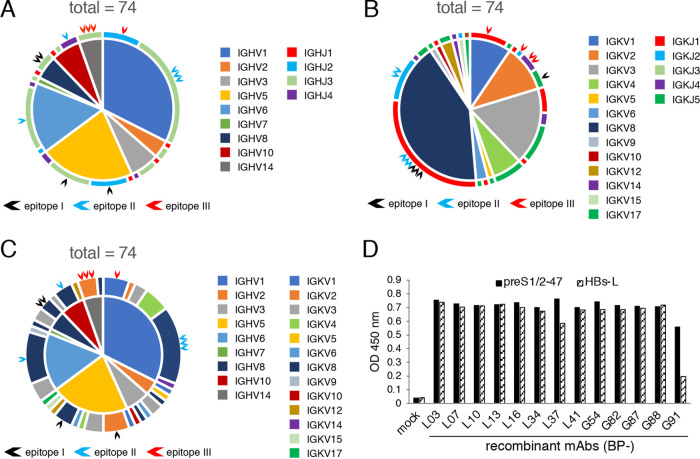
Sequence analysis, expression, and epitope mapping of MAbs against preS1/2–47.
To generate recombinant MAbs against preS1/2–47, cDNAs of the variable region were amplified from RNA extracted from memory B cells. Ig gene sequences of heavy and light chain variable regions (VH and VL, respectively) were analyzed by using IgBLAST, the web tool for sequence analysis of the Ig variable domain. We determined sequences of 74 sets of VH and VL from 97 memory B cell samples that were positive for preS1 in ELISA. Amplification was unsuccessful for either or both of the VH and VL regions in the remaining 23 samples. Sequence analysis revealed that 20 of 74 MAbs belonged to the subgroup IGHV1 with IGHJ3 (Fig. 3A) and 21 MAbs belonged to the subgroup IGKV8 with IGKJ1 (Fig. 3B). Approximately 43% of MAbs (32 of 74 MAbs) belonged to the IGKV8 subgroup. Sequence analysis also revealed 33 different MAbs in the IGHV/IGKV subgroup combination (Fig. 3C), indicating that anti-preS1/2–47 MAbs obtained in this study represent a diversity of clones. Among the 74 MAb sequences, 13 MAbs were selected for generation of recombinant MAbs, due to the remarkable neutralizing activity (Fig. 2C) or representative sequence combination (Fig. 3C). Selected MAbs are indicated by arrowheads in Fig. 3A to C. Generated recombinant MAbs are listed in Table 1. The complementarity-determining region 3 (CDR3) amino acid sequences of VH and VL are shown in Table 2, implying that some MAbs were derived from clonally expanded B cells (e.g., BP-L03, L07 and L13, or BP-G82 and G88). After expression and purification of the 13 recombinant MAbs, binding of MAbs against preS1/2–47 and HBs-L was confirmed by ELISA (Fig. 3D). Epitopes of these MAbs were further analyzed using overlapping peptides, as shown in Fig. 3E. Each epitope of the MAbs obtained in this study is summarized in Fig. 3F. Interestingly, 13 MAbs obtained in our study were classified in three groups according to the major region of the epitopes, i.e., amino acids 8 to 16 of preS1/2–47 (designated epitope I), amino acids 23 to 35 of preS1/2–47 (designated epitope II), and amino acids 35 to 41 of preS1/2–47 (designated epitope III).
FIG 3.
Sequence analysis, expression, and epitope mapping of MAbs against preS1/2–47. cDNA sequences of VH and VL genes derived from preS1/2–47-specific memory B cells were analyzed by using IgBLAST. (A) Combinations of heavy chain V-J subgroups from 74 preS1/2–47 MAbs. The sections inside the circle represent the IGHV subgroups, and the outer circumference shows IGHJ subgroups. Black, blue, and red arrows indicate generated recombinant MAbs recognizing epitopes I, II, and III, respectively. (B) Combinations of light chain V-J combinations from 74 MAbs. The sections inside the circle represent the IGKV subgroups, and the outer circumference shows IGKJ subgroups. Black, blue, and red arrows indicate generated recombinant MAbs recognizing epitopes I, II, and III, respectively. (C) IGHV-IGKV combinations of 74 preS1/2–47 MAbs. The sections inside the circle represent the IGHV subgroup, and the outer circumference shows the IGKV subgroup. Black, blue, and red arrows indicate generated recombinant MAbs recognizing epitopes I, II, and III, respectively. (D) ELISA for binding of recombinant MAbs (1 μg/ml in PBS) to preS1/2–47 or HBs-L protein. OD, optical density. (E) Epitope mapping of preS1-specific recombinant MAbs. Forty-seven synthetic overlapping peptides of 10 to 20 amino acids corresponding to preS1/2–47 were used. The horizontal axis for each MAb shows absorbance units. The peptides showing antibody binding are marked by an asterisk. (F) Summary of minimum epitopes recognized by each MAb. The preS1/2–47 amino acid sequences used for immunization are shown. Yellow numbers in black bars represent the amino acid positions.
TABLE 1.
Summary of recombinant MAbs targeting the preS1 receptor binding domain
| Memory B cell clone |
Recombinant MAb |
Immunized plasmid |
Type of: |
|||
|---|---|---|---|---|---|---|
| IGHV | IGHJ | IGKV | IGKJ | |||
| 3 | BP-L03 | pCAG-HBs-L | 14-3 | 3 | 2-137 | 4 |
| 7 | BP-L07 | pCAG-HBs-L | 14-3 | 3 | 2-137 | 4 |
| 10 | BP-L10 | pCAG-HBs-L | 1-14 | 3 | 8-30 | 1 |
| 13 | BP-L13 | pCAG-HBs-L | 14-3 | 3 | 2-137 | 2 |
| 16 | BP-L16 | pCAG-HBs-L | 6-6 | 4 | 8-30 | 1 |
| 34 | BP-L34 | pCAG-HBs-L | 5-6-3 | 2 | 8-27 | 1 |
| 37 | BP-L37 | pCAG-HBs-L | 1-87 | 3 | 8-30 | 1 |
| 41 | BP-L41 | pCAG-HBs-L | 5-4 | 3 | 2-137 | 5 |
| 54 | BP-G54 | pCAG-GroEL-preS1 | 10-3 | 4 | 8-30 | 2 |
| 82 | BP-G82 | pCAG-GroEL-preS1 | 8-12 | 3 | 8-30 | 1 |
| 87 | BP-G87 | pCAG-GroEL-preS1 | 1S29 | 2 | 1-110 | 1 |
| 88 | BP-G88 | pCAG-GroEL-preS1 | 8-12 | 3 | 8-30 | 1 |
| 91 | BP-G91 | pCAG-GroEL-preS1 | 1S135 | 3 | 8-30 | 2 |
TABLE 2.
CDR3 sequences of preS1/2–47-specific MAbs
| Recombinant MAb |
CDR3 sequence of: |
|
|---|---|---|
| Heavy chain | Light chain | |
| BP-L03 | ALNWDGAWFAY | MQHLEYPFT |
| BP-L07 | ALNWDGAWFAY | MQHLEYPFT |
| BP-L10 | VRSPYYDYPWFAY | QQYYTYPWT |
| BP-L13 | ALNWDGAWFSY | MQHLEYPFT |
| BP-L16 | TSPTTATPFAY | QQYYSYPWT |
| BP-L34 | ARFDY | HQYLSSWT |
| BP-L37 | ARRGREGGFAY | QQYYTYPWT |
| BP-L41 | VRDLDY | MQHLESPFT |
| BP-G54 | VREGRAMDY | QQYYTYPYT |
| BP-G82 | ARVHSYDDPVFAY | QQYYSYGT |
| BP-G87 | ANGDL | SQSTHVPWT |
| BP-G88 | ARVYDYDDPVFAY | QQYYSFGT |
| BP-G91 | AGLGQAY | QQYYSYPYT |
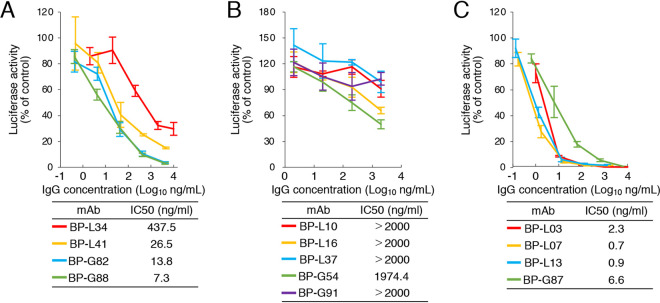
Neutralizing activities of recombinant MAbs against HBV infection.
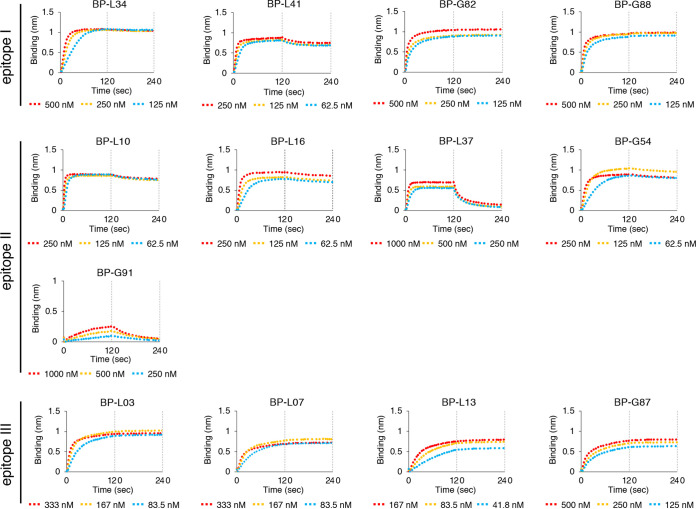
We performed the virus neutralization assay with recombinant MAbs using HBV/NL derived from genotype C. MAbs recognizing epitope I (BP-L34, L41, G82, and G88) neutralized 50% of HBV/NL infection at 7 to 438 ng/ml (Fig. 4A). MAbs recognizing epitope III (BP-L03, L07, L13, and G87) showed higher neutralizing activity (50% inhibitory concentration [IC50] values of <7 ng/ml), compared to that of epitope I-recognizing MAbs. MAbs recognizing epitope II (BP-L10, L16, L37, G54, and G91) showed low or no neutralizing activity against HBV/NL infection (IC50 values of >1,974 ng/ml) (Fig. 4B). Kinetic analysis of these MAbs using the preS1/2–47 peptide was also performed (Fig. 5 and Table 3). Although BP-L37 and G91 showed low affinity against the preS1/2–47 peptide, three other MAbs that recognize epitope II (BP-L10, L16, and G54) showed high affinity against the preS1/2–47 peptide, indicating that the low or absent neutralizing activity of MAbs recognizing epitope II is not due to low affinity for the preS1/2–47 peptide. These data suggest that epitopes I and III of preS1/2–47 are potential neutralizing epitopes for HBV infection.
FIG 4.
Neutralization assay of recombinant MAbs using HBV/NL derived from genotype C. MAbs targeting epitopes I (A), II (B), and III (C) were incubated with HBV/NL (20 GEq/cell) for 1 h and then infected into G2/NT-18 cells for 16 h. The luciferase activity of cells was determined at 7 days postinfection and is expressed relative to activity without antibodies. The concentration of antibody that results in 50% neutralization of HBV/NL infection (IC50) was calculated and is shown. Data are averages of quadruplicate values, with error bars showing standard deviations.
FIG 5.
Binding kinetics of MAbs recognizing epitopes I to III against preS1/2–47 peptide derived from HBV genotype C, based on biolayer interferometry. Association and dissociation measurements for 2-fold serially diluted MAbs were carried out for 2 min each. Kinetic parameters (kon and koff) and affinities (KD) were calculated from a nonlinear fit of BLItz instrument data using BLItz software.
TABLE 3.
Dissociation constants of MAbs recognizing epitopes I to III
| Epitope and MAb | KD (nM) | kon (M−1 s−1) | koff (s−1) |
|---|---|---|---|
| Epitope I | |||
| BP-L34 | 0.45 | 2.0E+05 | 8.7E−05 |
| BP-L41 | 2.2 | 4.5E+05 | 9.9E−04 |
| BP-G82 | <1.0E−03 | 4.0E+05 | <1.0E−07 |
| BP-G88 | <1.0E−03 | 2.4E+05 | <1.0E−07 |
| Epitope II | |||
| BP-L10 | 1.6 | 7.5E+05 | 1.2E−03 |
| BP-L16 | 1.5 | 5.3E+05 | 7.9E−04 |
| BP-L37 | 2.7E+02 | 1.6E+05 | 4.2E−02 |
| BP-G54 | 1.8 | 3.4E+05 | 6.1E−04 |
| BP-G91 | 1.3E+06 | 11 | 1.4E−02 |
| Epitope III | |||
| BP-L03 | <1.0E−03 | 3.66E+05 | <1.0E−07 |
| BP-L07 | <1.0E−03 | 1.36E+05 | <1.0E−07 |
| BP-L13 | <1.0E−03 | 1.43E+05 | <1.0E−07 |
| BP-G87 | <1.0E−03 | 7.33E+04 | <1.0E−07 |
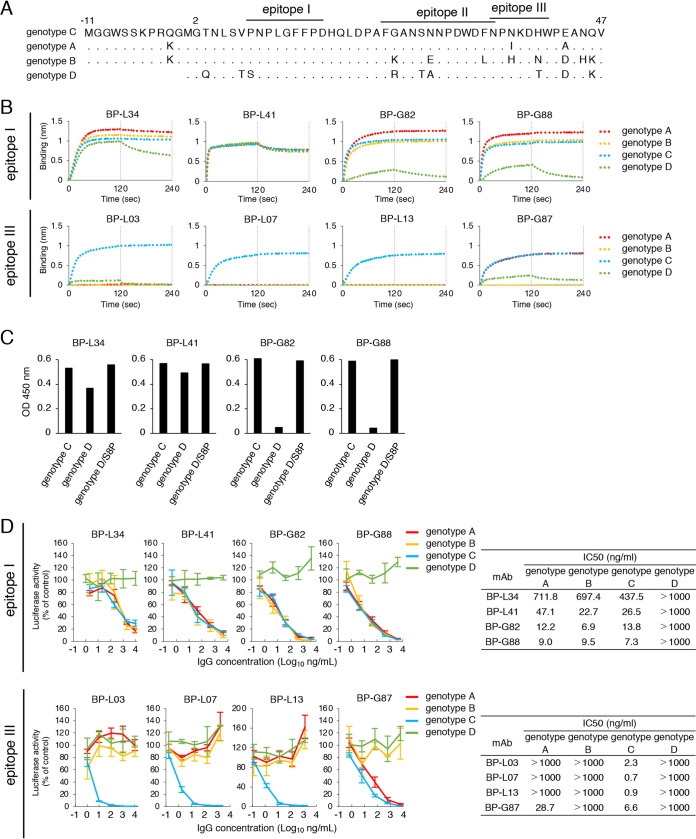
We further examined the effect of selected MAbs against HBV derived from genotype A, B, or D. MAbs recognizing epitope I bound to the preS1/2–47 peptide from genotypes A to C, although these MAbs except BP-L41 showed lower affinity against the genotype D peptide (Fig. 6B and Table 4). Because a 1-amino acid difference is present in genotype D at position 8 (Fig. 6A), we examined the effect of this amino acid on the binding of these MAbs. In accordance with Fig. 6B, BP-L34, G82, and G88 bound to the amino acid 3 to 22 peptide of genotype C and bound less to the peptide of genotype D (Fig. 6C). The lower binding was completely restored when the peptide of genotype D with a serine-to-proline mutation at position 8 was used, suggesting that the proline residue at position 8 was crucial for binding of BP-L34, BP-G82, and BP-G88 to preS1. BP-L03, L07, and L13 recognizing epitope III bound to the genotype C peptide only, whereas BP-G87 bound to the preS1/2–47 peptide from genotypes A and C (Fig. 6B and Table 4). These data could be explained by the fact that the amino acid sequences of epitope I were highly conserved among genotypes A to D, with a 1-amino acid difference at position 8 in genotype D, whereas epitope III contains a few amino acid differences among genotypes (Fig. 6A). A neutralization assay using HBV/NL derived from different genotypes revealed that MAbs recognizing epitope I neutralized infection by genotype A and B HBV/NL as well as genotype C in a dose-dependent manner (Fig. 6D), whereas these MAbs did not neutralize HBV/NL derived from genotype D. Among the four MAbs that recognize epitope III, three MAbs (BP-L03, L07, and L13) did not neutralize infection by HBV genotypes A, B, and D (Fig. 6D). BP-G87 neutralized genotype A HBV/NL in addition to genotype C HBV/NL but not genotype B and D HBV/NL, suggesting that the virus-neutralizing activity of MAbs was dependent on the binding activity to the preS1/2–47 peptide. We also evaluated the neutralizing activity of MAbs using cell culture-generated HBV (HBVcc) derived from genotypes C and D. As shown in Fig. 6E, MAbs recognizing epitope I (BP-L34, L41, G82, and G88) neutralized infection by HBV genotype C virus. However, infection by HBV genotype D was not inhibited by these MAbs, in accordance with the result of the neutralization assay using reporter virus (Fig. 6D). Taken together, MAbs recognizing epitope I are NAbs that broadly bind to HBV genotypes A to C, and MAbs recognizing epitope III are genotype-specific NAbs. Notably, BP-L41 recognizing epitope I bound to the genotype D peptide, similar to other genotypes, regardless of not having neutralizing activity against HBV genotype D (Fig. 6D and E).
FIG 6.
(Continued)
TABLE 4.
Dissociation constants of MAbs recognizing epitopes I and III against HBV genotypes A to D
| Epitope and MAb |
KD (nM) for: |
|||
|---|---|---|---|---|
| Genotype A | Genotype B | Genotype C | Genotype D | |
| Epitope I | ||||
| BP-L34 | 3.4 | 1.4 | 0.45 | 2.7E+02 |
| BP-L41 | 2.8 | 2.4 | 2.2 | 12 |
| BP-G82 | <1.0E−03 | <1.0E−03 | <1.0E−03 | 1.5E+05 |
| BP-G88 | <1.0E−03 | <1.0E−03 | <1.0E−03 | 2.3E+03 |
| Epitope III | ||||
| BP-L03 | —a | — | <1.0E−03 | 1.6E+05 |
| BP-L07 | — | — | <1.0E−03 | — |
| BP-L13 | — | — | <1.0E−03 | — |
| BP-G87 | <1.0E−03 | — | <1.0E−03 | 1.9E+02 |
—, The parameter was not determined because no binding curve could be obtained.
Evaluation of neutralizing activities against HBV with VEM.
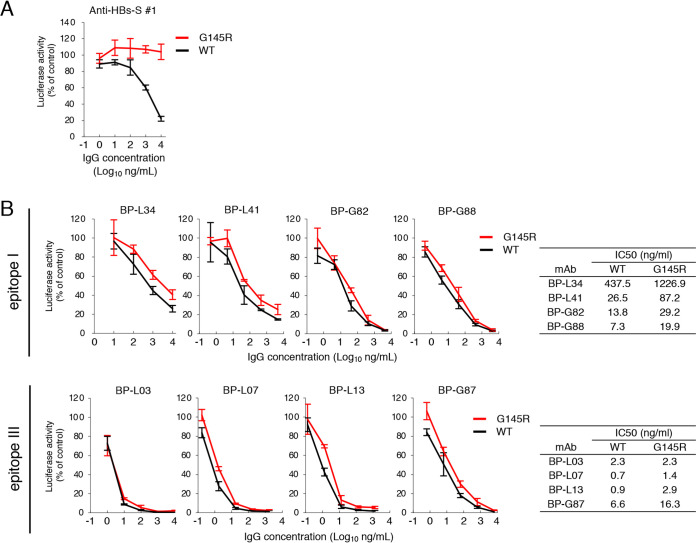
NAbs induced by HBV vaccination are targeted largely toward the conformational epitope of the “a” determinant (15). Mutations within this region cause conformational changes that can allow infection of mutated viruses in vaccinated people (21). Although no reports have indicated that anti-preS1 antibodies neutralize HBV with such mutations, we hypothesized that anti-preS1 antibodies would neutralize HBV with VEM. To assess whether MAbs against preS1 could neutralize infection caused by HBV with VEM, we generated HBV/NL with a G145R mutation, which is representative of VEM observed in patients (14, 21). A MAb against HBs-S neutralized wild-type genotype C HBV/NL but not HBV/NL with VEM (Fig. 7A). In contrast, MAbs against preS1 showed equivalent neutralizing activity against HBV/NL with or without mutation (Fig. 7B). Thus, MAbs targeting preS1/2–47 could neutralize HBV as well as HBV with VEM.
FIG 7.
Neutralization assay of MAbs against HBV/NL genotype C with VEM. Anti-HBs-S MAb 1 (A) or MAbs recognizing epitope I or III (B) were incubated with 20 GEq/cell of HBV/NL for 1 h and then infected into G2/NT-18 cells for 16 h. WT, wild-type. The luciferase activity of cells was determined at 7 days postinfection and is expressed relative to activity without antibodies. The concentration of antibody that results in 50% neutralization of HBV/NL infection (IC50) was calculated and is shown. Data are averages of triplicate values, with error bars showing standard deviations.
DISCUSSION
In this study, we performed single-cell sorting of high-affinity memory B cells using fluorescent labeling of the preS1/2–47 peptide and in vitro cell culture of mouse IgG memory B cells and acquired 97 clones of preS1/2–47-specific memory B cells. The method based on antigen-specific memory B cell sorting allows for rapid generation of large numbers of functional MAbs. Furthermore, this method is an efficient tool to generate MAbs from single memory B cells at any maturation or differentiation stage, compared to conventional hybridoma technology (22).
We determined the sequences of 74 murine MAbs against preS1/2–47 and found a diverse preS1/2–47-specific memory B cell receptor repertoire; >20 combinations of the V-J subgroup were found in both VH and VL. The VH gene allele is biased for specific antigens. For example, NAbs that broadly target the CD4 binding site on the HIV envelope spike preferentially use either IGHV1-2 or IGHV1-46 germline alleles (23). In HIV-infected individuals, the IGHV1-69 gene is dominant in NAbs that target the V2 loop of the GP120 trimer (24). The IGHV1-69 gene is also frequently observed among NAbs that broadly target the highly conserved stem region of influenza virus hemagglutinin (25). The VH1-69 gene is the only human heavy chain gene that encodes two hydrophobic residues at the tip of the CDR H2 loop (26), which is considered to provide the ability to interact with similarly charged residues in antigens (25, 26). Profiles of VH and VL genes of anti-HBsAg antibodies were analyzed (27, 28). However, a comprehensive analysis of immunoglobulin usage of anti-preS1 antibodies has not yet been reported. In this study, we determined 74 sequences of MAbs derived from preS1/2–47-specific memory B cells (Fig. 3A to C). The top 10 VDJ rearrangement frequencies of these 74 IGHV sequences are shown in Table 5. Several IGHV sequences with no/low frequencies in naive BALB/c mice (29) (e.g., IGHV1-87, IGHV1S135, and IGHV1S29) were observed at the highest frequencies in this study. The major subgroups of κ light chain in 74 MAbs were IGKV2, IGKV3, and IGKV8 genes, which are also different from the B cell receptor in naive BALB/c mice (30). These data suggest that IGHV and IGKV repertoires of antibodies against preS1/2–47 seem to be biased, although the detailed mechanism is not yet clear.
TABLE 5.
IGHV rearrangement frequencies in naive BALB/c mice reported by Collins et al. (29) and in preS1/2–47-specific memory B cell genes in this study
| IMGT description | Rearrangement frequency (%) in: |
|
|---|---|---|
| Collins et al. (29) | This study (total of 74 samples) | |
| IGHV1-87 | —a | 13.5 |
| IGHV6-6 | 1.96 | 13.5 |
| IGHV1S135 | 0.06 | 6.8 |
| IGHV5-4 | 2.65 | 6.8 |
| IGHV5-6-3 | 0.23 | 5.4 |
| IGHV14-3 | 2.08 | 5.4 |
| IGHV3-2 | 5.84 | 4.1 |
| IGHV1-14 | 1.25 | 2.7 |
| IGHV1S29 | — | 2.7 |
| IGHV3-6 | 1.88 | 2.7 |
| IGHV5-15 | 0.25 | 2.7 |
| IGHV6-3 | — | 2.7 |
| IGHV8-12 | 0.88 | 2.7 |
| IGHV10-3 | 1.06 | 2.7 |
| IGHV10S3 | 0.64 | 2.7 |
—, Not applicable in data.
We generated 13 anti-preS1 MAbs targeting the receptor binding site derived from preS1/2–47. Eight of 13 MAbs inhibited HBV infection. Two neutralizing epitopes at the N and C termini of preS1/2–47 were identified. MAbs recognizing epitope I at the N terminus of preS1/2–47 showed semipangenotypic neutralization against HBV infection. A previous report using a synthetic preS1 peptide showed that the preS1/2–21 peptide could competitively inhibit HBV infection, whereas the preS1/9–48 peptide could not inhibit HBV infection (7). These findings suggested that the N-terminal region of preS1/2–47 is important for direct binding between HBV and NTCP. In fact, the amino acid sequence of the N-terminal half of preS1/2–47 is highly conserved among multiple genotypes. These findings imply that antibodies targeting epitope I are effective for broad neutralization of HBV infection. Conversely, MAbs recognizing epitope III at the C terminus of preS1/2–47 showed high neutralizing activity in a genotype-specific manner. As a previous report showed that the preS1/2-33 peptide inhibits HBV infection less efficiently than the preS1/2–47 peptide (7), epitope III may support efficient preS1 binding to the host receptor, although the region is not essential for binding between preS1 and NTCP.
Although MAbs recognizing epitope I neutralized viral infection of genotypes A to C, these MAbs could not neutralize genotype D reporter virus or HBVcc. A 1-amino acid difference is present in epitope I between genotypes C and D. This difference affects the affinity and neutralizing activity of BP-L34, G82, and G88 against the genotype D sequence. Interestingly, BP-L41 did not show neutralizing activity against HBV/NL genotype D, although the binding activity for the peptide derived from genotype D was the same as that for peptides from genotypes A to C (Fig. 6B and C). HBV genotype D has an 11-amino acid deletion (d11) in the preS1 region. This deletion is associated with efficient HBV infection in vitro, possibly due to increased glycosylation of HBs-L protein (20). The glycosylation status of the viral envelope may affect the interaction between preS1 and NTCP. Therefore, d11 in preS1 of HBV genotype D may also affect the binding of BP-L41 to the epitope of viral particles. Further study is needed to understand the mechanisms.
The G145R mutation in the HBs-S region was first identified 30 years ago in infants born to HBsAg-carrier mothers who developed breakthrough infections despite the presence of protective antibody levels produced in response to the administration of HBIG plus vaccine received at birth (31). Other HBs-S gene mutations in the “a” determinant that can lead to increased viral evasion in vaccinated people have been identified (32, 33). Because most of the current hepatitis B vaccines consist of HBs-S protein, VEM is a concern (31). To our knowledge, we demonstrated for the first time that anti-preS1 antibody neutralizes HBV with or without VEM. In addition, about 10% of adults are vaccine nonresponders or low responders, resulting in a lack of immunity against HBV. Immune responses induced by vaccination with the hepatitis B vaccine containing HBs-S derived from genotype C are associated with human leukocyte antigen class II alleles (34), implying that a novel or additional vaccine antigen is needed to elicit NAbs in nonresponders/low responders.
A previous report showed that murine BX-182 MAb and humanized KR127 MAb targeting preS1 protected chimpanzees from HBV infection (18, 35). The epitope of BX-182 MAb was mapped between positions 5 and 9 in the preS1/2–47 sequence, which partially overlaps with epitope I. The epitope of KR127 MAb was mapped between positions 26 and 34 in the preS1/2–47 sequence, which is located within epitope II (9), whereas our MAbs recognizing epitope II did not show strong neutralizing activity. Recently, anti-preS1 MAb 2H5-A14, the epitope of which was mapped between positions 19 and 27 in the preS1/2–47 sequence and partially overlaps with epitope II, also neutralizes HBV infection in vitro and acts therapeutically by eliciting Fc-dependent immunological effector functions that effectively suppress viral infection in HBV-infected mice (17). Interestingly, the neutralizing activity of 2H5-A14 was 200-fold higher than that of KR127 MAb. Considering the preferential presence of the HBs-L protein on infectious HBV particles (36), NAbs targeting preS1 may be a novel clinical candidate for prevention of HBV infection.
In summary, we identified two critical neutralizing epitopes in preS1/2–47 by systematically characterizing 13 preS1/2–47-specific MAbs. Our data provide evidence for a potential novel vaccine containing the preS1 region, to overcome the weakness of current hepatitis B vaccines containing HBs-S, and new insights into neutralizing epitopes in the preS1/2–47 region, thus enhancing our understanding of novel immunoprophylaxis for the prevention and treatment of HBV infection.
MATERIALS AND METHODS
Cell cultures.
The human embryonic kidney 293T cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with nonessential amino acids (NEAA), 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum. The HepG2 cell line expressing NTCP, G2/NT-18C (37), was maintained in DMEM supplemented with NEAA, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 1 μg/ml puromycin, and 10% fetal bovine serum. HepG2-NTCPsec+ cells were maintained as described (38). Cells were cultured at 37°C in a 5% CO2 incubator. The Expi293F cell line (Thermo Fisher Scientific) was cultured in accordance with the manufacturer’s instructions.
Plasmid construction for DNA immunization.
pCAG-HBs-L was constructed by insertion of cDNA encoding the genotype C HBV L protein from pHBV/C-AT (39) into pCAGGS. To generate pCAG-Sec-preS1, cDNA encoding amino acids 2 to 47 of preS1 with the Myc tag sequence (EQKLISEEDL) at the C terminus was amplified from pCAG-HBs-L, and cDNA encoding the murine Ig κ-chain secretion signal was amplified from pDisplay (Thermo Fisher Scientific). The residue numbering of the HBV preS1 domain is based on HBV genotype D. The resultant fragments were inserted into pCAGGS by using the In-Fusion HD cloning kit (TaKaRa). To generate pCAG-Display-preS1, cDNA encoding the PDGFR transmembrane domain from pDisplay was inserted into the C terminus of the Myc tag sequence of pCAG-Sec-preS1. pCAG-GroEL-preS1 was constructed by insertion of cDNA fragments encoding amino acids 2 to 4 of preS1 with the Myc tag and cDNA fragments encoding GroEL from pCADEST1-ETAR-GroEL (19) into pCAGGS.
IgG expression plasmids were described previously (22). The constant region of the heavy chain expression plasmid was replaced with the murine IgG 2a constant region (GenBank accession number V00825).
DNA transfection.
For Western blotting and the immunofluorescence assay, 293T cell monolayers were transfected with plasmid DNA using FuGENE 6 transfection reagent (Promega) in accordance with the manufacturer’s instructions. To produce MAbs, Expi293F cells were transfected with H and L chain expression plasmids in accordance with the manufacturer’s instructions.
Antibodies.
Anti-preS1 antibody (LT-1) was obtained from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-coupled anti-mouse IgG was obtained from Cell Signaling. Anti-HBc antibody (Ab-1) and Alexa Fluor 555-conjugated anti-rabbit IgG antibody were obtained from Thermo Fisher Scientific. Anti-mouse IgM (II/41), IgD (11-26c), CD4 (GK1.5), CD5 (53-7.3), Gr1 (RB6-8C5), and CD93 (AA4.1) were obtained from eBioScience. Anti-mouse CD43 (S7) was obtained from BD Biosciences. Anti-mouse CD11b (M1/70), B220/CD45R (RA3-6B2), and CD38 (90) were obtained from BioLegend. HBIG was obtained from Japan Blood Products Organization. Anti-HBs-S MAbs 1 and 6 were described previously (40). Antiflavivirus MAb (D1-4G2) was obtained from Millipore and used as a negative control.
Immunoblotting.
Cells on the culture plate were washed with phosphate-buffered saline (PBS) and suspended in lysis buffer (50 mM Tris-HCl [pH 7.6], 300 mM NaCl, 1% Triton X-100). Cell lysates were sonicated with a Bioruptor UCD-250 (Tosho Denki) and boiled for 10 min at 99°C after mixing with SDS sample buffer. Samples were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membrane was blocked for 1 h in Tween 20/Tris-buffered saline (TTBS) (0.05% Tween 20, 50 mM Tris-HCl, 150 mM NaCl) with 1% skim milk (Becton, Dickinson). Then, the membrane was washed three times with TTBS for 5 min and incubated for 1.5 h at room temperature with anti-preS1 antibody (LT-1) diluted in Can Get Signal solution 1 (Toyobo). The membrane was further washed three times with TTBS for 5 min and incubated for 1 h at room temperature with HRP-conjugated anti-mouse IgG secondary antibody (Cell Signaling) in Can Get Signal solution 2 (Toyobo). After washing three times with TTBS, immunoreactive signals were visualized using an enhanced chemiluminescence detection system (SuperSignal West Femto maximum sensitivity substrate; Thermo Fisher Scientific) and an LAS-3000 imager (GE Healthcare).
Immunofluorescence.
Cells on the culture plate were washed with PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. Fixed cells were permeabilized for 30 min with 0.2% Triton X-100 in PBS and washed with PBS. Cells were then blocked for 1 h with Block Ace (Snow Brand Milk Products Co.). After washing with PBS, cells were incubated for 1.5 h at room temperature with primary antibody diluted in PBS. Cells were subsequently washed three times with PBS and incubated for 1 h at room temperature with Alexa Fluor 488-goat anti-mouse IgG secondary antibody (Invitrogen) in PBS. Cells were then washed three times with PBS, and immunoreactive signals were visualized by using the IX81 inverted fluorescence microscope (Olympus).
ELISA.
Approximately 0.5 μM amino acids 2 to 47 of the HBV preS1 peptide (genotype C) or 1 μg/ml HBs-L protein (BCL-AG-001; Beacle) in Tris-buffered saline (TBS) (50 mM Tris-HCl, 150 mM NaCl) was adsorbed onto MaxiSorp 96-well plates (Nunc) by overnight incubation at 4°C. Wells were washed, blocked with 1:5 diluted Block Ace (Snow Brand Milk Products Co.), and then washed with PBS containing 0.05% Tween 20 (TPBS). Diluted sera or antibodies were added to each well, and plates were incubated for 1.5 h at room temperature. The supernatant was then discarded, and each well was washed three times with TPBS. HRP-coupled anti-mouse IgG (Cell Signaling) was added to each well, and plates were incubated for 1 h at room temperature prior to addition of substrate (peroxidase detection kit; Sumitomo Bakelite). The absorbance at 450 nm was determined with a plate reader (Bio-Rad).
Production and infection of HBV reporter virus encoding nanoluciferase.
Production of an HBV reporter virus encoding nanoluciferase (HBV/NL) was described previously (20, 41). Briefly, to generate HBV/NL, in the plasmid containing 1.2-fold HBV genome HBe/c was replaced with NL (designated pUC1.2xHBV/NL), and the plasmid bearing a packaging-defective 1.2-fold HBV genome (designated pUC1.2xHBV-DdelS) was used. pUC1.2xHBV/NL encodes the NL gene instead of HBe/c antigens, and the pregenome RNA generated from this plasmid is incorporated into the virion. On the other hand, pUC1.2xHBV-DdelS has mutations in the encapsidation signal and thus is not incorporated into the virion. This plasmid produces all HBV proteins except for HBs antigens. The HBV genome in these plasmids was derived from the isolate C_JPNAT, genotype C (GenBank accession number AB246345). To generate HBV/NL containing all HBs species (HBs-L, -M, and -S) of genotypes A, B, and D, the coding region of these proteins in pUC1.2xHBV/NL was replaced with that of genotype A (GenBank accession number LC488828), genotype B (GenBank accession number AB246341), and genotype D (GenBank accession number LC488706) strains. The plasmid of HBV-dEdelS was also prepared by deleting the expression of all HBs species of HBV-dE (originally designated pUC1.2xHBV-D, encoding the 1.2-fold HBV genome lacking the encapsidation signal). To generate the HBV/NL containing the VEM, the representative VEM of glycine to arginine at amino acid 145 (G145R) was introduced into the plasmids of pUC1.2xHBV/NL. To obtain HBV/NL, these plasmids were transfected into HepG2 cells with Lipofectamine 3000 reagent. The secreted HBV/NL was harvested 1 week after transfection, and the viral titer was measured by real-time PCR with the primer and probe set targeting the Nano Luc gene after treatment with DNase (RQ1 RNase-Free DNase). Twenty genome equivalents (GEq)/cell of HBV/NL was used for infection of G2/NT-18 cells.
Production and infection of HBVcc.
To obtain HBVcc of genotype C, a replication-competent HBV genotype C clone (GenBank accession number AB246345) with a 1.38-fold genome length was transfected into HepG2-NTCPsec+ cells with Lipofectamine 3000 reagent (Thermo Fisher Scientific). The culture supernatant was harvested 1 week posttransfection. To obtain HBV of genotype D, HepG2.2.15 cells were cultured without G418 (42). The generated HBVs in the culture medium were passed through a 0.45-μm filter to remove cell debris, concentrated with Amicon Ultra centrifugal filters (100 kDa; Merck Millipore, Billerica, MA), purified with iodixanol gradients, and centrifuged at 38,000 rpm for 16 h at 4°C in an SW 41 Ti rotor (20). The DNA titer of HBVcc was measured by real-time PCR with the primer and probe set targeting HBs, and HBVcc was inoculated into HepG2-NTCPsec+ cells at 1,000 GEq/ml (genotype C) or 1 GEq/ml (genotype D) after incubation with MAbs or HBIG for 1 h at 37°C. After 12 days of culture, HBV-infected cells were fixed with 4% paraformaldehyde and permeabilized. The HBV core protein-positive cells were visualized by staining with rabbit polyclonal anti-HBc antibody (Ab-1) and Alexa Fluor 555-conjugated anti-rabbit IgG (Thermo Fisher Scientific). The nuclei were stained with 4′,6- diamidino-2-phenylindole (DAPI) (42).
Virus neutralization assay.
For the HBV neutralization assay, HBV/NL was incubated with diluted sera or antibodies in the presence of 4% polyethylene glycol (PEG) 8000 and 2% dimethyl sulfoxide (DMSO) at room temperature for 1 h and then inoculated into G2/NT-18 cells for 16 h at 37°C. Following infection, the supernatant was removed, cells were washed three times with PBS, and cells were then incubated with culture medium containing 2% DMSO. Luciferase activity of cells was determined at 7 days postinfection using the Nano-Glo luciferase assay system (Promega).
Immunization of mice.
Groups (n = 4 each) of specific-pathogen-free female BALB/c mice (9 weeks old) were purchased from SLC Japan. Plasmid injection via the intramuscular route followed by in vivo electroporation was performed according to a previously described method (43). Briefly, hyaluronidase (50 U) was injected into the quadriceps muscles. After 15 min, mice were anesthetized with isoflurane, and 20 μg of each plasmid was injected into the quadriceps muscles. The same procedure was performed on the other quadriceps muscle. Electrode needles were then inserted into the muscle, and electric pulses were delivered using an electric pulse generator (NEPA21; Nepa Gene). Three poring pulses (50 V for 30 ms), followed by three transfer pulses (20 V for 50 ms), were administered three times to each injection site. Booster DNA immunizations were given 2, 4, and 8 weeks after the primary immunization. Additionally, 1 × 107 cells/head 293T cells transfected with each plasmid were intraperitoneally injected at week 13 as a final booster immunization. Sera were collected 1 week after the last immunization. Blood samples were collected in Bloodsepar vials (Immuno-Biological Laboratories) and centrifuged at 2,500 × g for 2 min at room temperature. Supernatants were collected and heat inactivated at 56°C for 30 min for use in neutralization assays. To obtain preS1-specific memory B cells, pCAG-HBs-L- and pCAG-GroEL-preS1-immunized mice were further immunized at week 29 with 293T cells transfected with each construct, followed by intraperitoneal injection at week 32 of 20 μg/head of preS1/2–47 peptide conjugated to keyhole limpet hemocyanin. Murine spleens were collected 6 days after the last boost. All animal experiments were approved by the Animal Care and Use Committee of the National Institute of Infectious Diseases and were carried out in accordance with the approved guidelines.
Isolation of memory B cells from splenocytes of mice immunized with preS1 plasmids.
Single-cell suspensions from two mouse spleens for each construct were incubated with anti-mouse IgM, IgD, CD43, CD4, CD5, CD11b, Gr1, and CD93 directly coupled to fluorescein isothiocyanate (FITC). Cells were further incubated with anti-FITC MicroBeads (Miltenyi Biotec), and stained cells were removed by magnetically activated cell sorting to negatively select class-switched memory B cells. The resulting cells were further stained with BV785 anti-mouse B220/CD45R, Pacific Blue-coupled anti-mouse CD38, LIVE/DEAD Fixable Aqua stain (Thermo Fisher), and phycoerythrin-coupled preS1/2–47 peptide. Single memory B cells binding to the preS1 peptide were sorted using FACSAria (Becton, Dickinson) according to surface marker expression patterns (FITC negative, B220 positive, and CD38 positive).
Single-cell culture of memory B cells.
Single-cell culture of memory B cells was described previously (44). Briefly, sorted memory B cells were cultured at a single cell per well in 96-well plates with NB-21.2D9 cells in B cell medium (RPMI 1640 medium supplemented with 10% fetal bovine serum, 55 μM 2-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 1 mM sodium pyruvate, and 1% MEM NEAA). After 10 days of culture in B cell medium with 2 ng/ml recombinant mouse interleukin-4, all culture supernatants were harvested, and plates were stored at −80°C. Screening with the preS1 ELISA and HBV/NL neutralization assay were performed by using 5-fold diluted culture supernatants.
Immunoglobulin cDNA cloning from memory B cells.
Total RNA from memory B cells was reverse transcribed using final amounts/concentrations of 150 ng random hexamer primer, 1 mM each deoxynucleoside triphosphate, 10 mM dithiothreitol, 10 U Prime RNase inhibitor, and 50 U SuperScript III reverse transcriptase (Invitrogen). Reverse transcription reactions were performed at 25°C for 10 min, 50°C for 50 min, and 94°C for 5 min. Mouse Igh and Igk V gene transcripts were independently amplified by two rounds of PCR starting with 1 μl cDNA as the template. The first round of PCR was performed at 95°C for 15 min, followed by 43 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final incubation at 72°C for 5 min. Nested PCR was performed using 1 μl unpurified first-round PCR product at 95°C for 15 min, followed by 43 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final incubation at 72°C for 5 min. PCR products were analyzed on 1% agarose gels. After identification of germline Ig V and J genes by IgBLAST (http://www.ncbi.nlm.nih.gov/igblast), PCR was repeated using 1 μl unpurified second-round PCR product as the template and combinations of single gene-specific V and J gene primers. All primers used in Ig gene-specific PCRs included restriction sites (AgeI and SalI for Igh and AgeI and BsiWI for Igk) that allowed direct cloning into expression vectors encoding mouse Igγ2a or Igκ constant regions (22). PCR products were purified with the Monarch PCR and DNA cleanup kit (New England BioLabs), followed by digestion with AgeI and SalI (Igh) or BsiWI (Igk). Ligation reactions were performed using the DNA ligation kit (TaKaRa Bio).
Expression and purification of recombinant antibodies.
Plasmids encoding Ig heavy and light chains were cotransfected into Expi293F cells in accordance with the manufacturer’s instructions. The culture supernatant containing recombinant MAbs was harvested 4 days after transfection. Recombinant MAb in the supernatant was concentrated with Amicon Ultra centrifugal filters (Merck) and purified using PureSpeed 20-μl ProG resin tips (Rainin). The IgG concentration was determined by the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific).
Epitope mapping of preS1 MAbs.
Epitope mapping was performed by ELISA using 10- to 20-amino acid overlapping peptides of the preS1/2–47 sequence. Peptides (20 μM) in TBS (50 mM Tris-HCl, 150 mM NaCl) were adsorbed onto MaxiSorp 96-well plates (Nunc) by overnight incubation at 4°C. The wells were washed, blocked with 1:5 diluted Block Ace (Snow Brand Milk Products Co.), and then washed with TPBS. Recombinant MAbs were added to each well, and plates were incubated for 16 h at 4°C. MAbs then were discarded, and each well was washed three times with TPBS. HRP-coupled anti-mouse IgG (GE Healthcare) was added to each well, and plates were incubated at room temperature for 1 h prior to the addition of substrate (peroxidase detection kit; Sumitomo Bakelite). The absorbance at 450 nm was determined with a plate reader (Bio-Rad).
Kinetic analysis of antibody binding to preS1/2–47.
Binding kinetics of MAbs targeting preS1/2–47 were measured by biolayer interferometry on the BLItz instrument (Primetech). Streptavidin biosensors were loaded with 1.5-μg/ml solutions of biotinylated preS1/2–47 peptides derived from HBV genotype A to D consensus sequences. Loaded sensors were washed with BLI buffer (PBS containing 0.02% Tween 20 and 0.1% bovine serum albumin), and association and dissociation measurements for 2-fold serially diluted MAbs were carried out for 2 min each. Kinetic parameters (kon and koff) and affinities (KD) were calculated from a nonlinear fit of BLItz instrument data using BLItz software.
ACKNOWLEDGMENTS
We are grateful to M. Sasaki for technical assistance and Y. Hirama for secretarial assistance. Anti-HBs-S MAbs 1 and 6 were kindly provided by T. Kiyohara. pCADEST1-ETAR-GroEL was kindly provided by J. Chiba (Tokyo University of Science). The HepG2-NTCPsec+ cell line was kindly provided by Marc Peter Windisch (Institute Pasteur Korea, Seoul, South Korea). We also thank S. Yamagoe for allowing us to conduct the kinetic analyses of antibodies using the BLItz instrument.
This research was supported by the Japan Agency for Medical Research and Development (AMED) under grant JP19fk0310120.
REFERENCES
- 1.World Health Organization. 2019. Hepatitis B. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. Accessed 1 May 2020.
- 2.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neurath AR, Kent SB, Strick N, Parker K. 1986. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell 46:429–436. doi: 10.1016/0092-8674(86)90663-X. [DOI] [PubMed] [Google Scholar]
- 4.Glebe D, Aliakbari M, Krass P, Knoop EV, Valerius KP, Gerlich WH. 2003. Pre-S1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J Virol 77:9511–9521. doi: 10.1128/JVI.77.17.9511-9521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grun S, Bulavaite A, Sasnauskas K, Gerlich WH. 2005. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and Tupaia hepatocytes. Gastroenterology 129:234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 6.Engelke M, Mills K, Seitz S, Simon P, Gripon P, Schnolzer M, Urban S. 2006. Characterization of a hepatitis B and hepatitis delta virus receptor binding site. Hepatology 43:750–760. doi: 10.1002/hep.21112. [DOI] [PubMed] [Google Scholar]
- 7.Schulze A, Schieck A, Ni Y, Mier W, Urban S. 2010. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J Virol 84:1989–2000. doi: 10.1128/JVI.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier A, Mehrle S, Weiss TS, Mier W, Urban S. 2013. Myristoylated preS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology 58:31–42. doi: 10.1002/hep.26181. [DOI] [PubMed] [Google Scholar]
- 9.Maeng CY, Ryu CJ, Gripon P, Guguen-Guillouzo C, Hong HJ. 2000. Fine mapping of virus-neutralizing epitopes on hepatitis B virus PreS1. Virology 270:9–16. doi: 10.1006/viro.2000.0250. [DOI] [PubMed] [Google Scholar]
- 10.Yu Y, Li S, Liang W. 2018. Bona fide receptor for hepatitis B and D viral infections: mechanism, research models and molecular drug targets. Emerg Microbes Infect 7:134. doi: 10.1038/s41426-018-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watashi K, Urban S, Li W, Wakita T. 2014. NTCP and beyond: opening the door to unveil hepatitis B virus entry. Int J Mol Sci 15:2892–2905. doi: 10.3390/ijms15022892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassal M. 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 13.Zanetti AR, Van Damme P, Shouval D. 2008. The global impact of vaccination against hepatitis B: a historical overview. Vaccine 26:6266–6273. doi: 10.1016/j.vaccine.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 14.Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325–329. doi: 10.1016/0140-6736(90)91874-A. [DOI] [PubMed] [Google Scholar]
- 15.Salisse J, Sureau C. 2009. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J Virol 83:9321–9328. doi: 10.1128/JVI.00678-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobotta D, Sominskaya I, Jansons J, Meisel H, Schmitt S, Heermann KH, Kaluza G, Pumpens P, Gerlich WH. 2000. Mapping of immunodominant B-cell epitopes and the human serum albumin-binding site in natural hepatitis B virus surface antigen of defined genosubtype. J Gen Virol 81:369–378. doi: 10.1099/0022-1317-81-2-369. [DOI] [PubMed] [Google Scholar]
- 17.Li D, He W, Liu X, Zheng S, Qi Y, Li H, Mao F, Liu J, Sun Y, Pan L, Du K, Ye K, Li W, Sui J. 2017. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. eLife 6:e26738. doi: 10.7554/eLife.26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong HJ, Ryu CJ, Hur H, Kim S, Oh HK, Oh MS, Park SY. 2004. In vivo neutralization of hepatitis B virus infection by an anti-preS1 humanized antibody in chimpanzees. Virology 318:134–141. doi: 10.1016/j.virol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto A, Kosaka N, Hasegawa H, Suzuki H, Sugano S, Chiba J. 2012. Enhancement of antibody responses to native G protein-coupled receptors using E. coli GroEL as a molecular adjuvant in DNA immunization. J Immunol Methods 375:243–251. doi: 10.1016/j.jim.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Murayama A, Yamada N, Osaki Y, Shiina M, Aly HH, Iwamoto M, Tsukuda S, Watashi K, Matsuda M, Suzuki R, Tanaka T, Moriishi K, Suzuki T, Nishitsuji H, Sugiyama M, Mizokami M, Shimotohno K, Wakita T, Muramatsu M, Liang TJ, Kato T. 2020. N-terminal preS1 sequence regulates efficient infection of cell culture-generated hepatitis B virus. Hepatology doi: 10.1002/hep.31308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Deng W, Deng L, Cao L, Qin B, Li S, Wang Y, Pei R, Yang D, Lu M, Chen X. 2012. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol 86:4658–4669. doi: 10.1128/JVI.06353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiller T, Busse CE, Wardemann H. 2009. Cloning and expression of murine Ig genes from single B cells. J Immunol Methods 350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 23.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. 2012. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A 109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorny MK, Pan R, Williams C, Wang XH, Volsky B, O'Neal T, Spurrier B, Sampson JM, Li L, Seaman MS, Kong XP, Zolla-Pazner S. 2012. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals. Virology 427:198–207. doi: 10.1016/j.virol.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, Weiss RA, Sallusto F, Subbarao K, Lanzavecchia A. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest 120:1663–1673. doi: 10.1172/jci41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golsaz-Shirazi F, Amiri MM, Bahadori M, Bayat AA, Mohammadi H, Farid S, Maddah M, Khoshnoodi J, Zarnani AH, Jeddi-Tehrani M, Shokri F. 2015. Molecular characterization of murine monoclonal antibody variable regions specific for hepatitis B surface antigen. Viral Immunol 28:425–433. doi: 10.1089/vim.2015.0023. [DOI] [PubMed] [Google Scholar]
- 28.Hehle V, Beretta M, Bourgine M, Ait-Goughoulte M, Planchais C, Morisse S, Vesin B, Lorin V, Hieu T, Stauffer A, Fiquet O, Dimitrov JD, Michel ML, Ungeheuer MN, Sureau C, Pol S, Di Santo JP, Strick-Marchand H, Pelletier N, Mouquet H. 2020. Potent human broadly neutralizing antibodies to hepatitis B virus from natural controllers. J Exp Med 217:e20200840. doi: 10.1084/jem.20200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins AM, Wang Y, Roskin KM, Marquis CP, Jackson KJ. 2015. The mouse antibody heavy chain repertoire is germline-focused and highly variable between inbred strains. Philos Trans R Soc Lond B Biol Sci 370:20140236. doi: 10.1098/rstb.2014.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaaya N, Shahsavarian MA, Maffucci I, Friboulet A, Offmann B, Leger JB, Rousseau S, Avalle B, Padiolleau-Lefevre S. 2019. Genetic background and immunological status influence B cell repertoire diversity in mice. Sci Rep 9:14261. doi: 10.1038/s41598-019-50714-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanò L, Paladini S, Galli C, Raimondo G, Pollicino T, Zanetti AR. 2015. Hepatitis B vaccination. Hum Vaccin Immunother 11:53–57. doi: 10.4161/hv.34306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu HY, Chang MH, Ni YH, Lin HH, Wang SM, Chen DS. 1997. Surface gene mutants of hepatitis B virus in infants who develop acute or chronic infections despite immunoprophylaxis. Hepatology 26:786–791. doi: 10.1002/hep.510260336. [DOI] [PubMed] [Google Scholar]
- 33.Hsu HY, Chang MH, Ni YH, Chen HL. 2004. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut 53:1499–1503. doi: 10.1136/gut.2003.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida N, Sugiyama M, Sawai H, Nishina S, Sakai A, Ohashi J, Khor SS, Kakisaka K, Tsuchiura T, Hino K, Sumazaki R, Takikawa Y, Murata K, Kanda T, Yokosuka O, Tokunaga K, Mizokami M. 2018. Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology 68:848–858. doi: 10.1002/hep.29876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, Yu MY, Venable R, Alter HJ, Shih JW. 2006. Neutralization epitope responsible for the hepatitis B virus subtype-specific protection in chimpanzees. Proc Natl Acad Sci U S A 103:9214–9219. doi: 10.1073/pnas.0603316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho JK, Jeevan-Raj B, Netter HJ. 2020. Hepatitis B virus (HBV) subviral particles as protective vaccines and vaccine platforms. Viruses 12:126. doi: 10.3390/v12020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otoguro T, Tanaka T, Kasai H, Kobayashi N, Yamashita A, Fukuhara T, Ryo A, Fukai M, Taketomi A, Matsuura Y, Moriishi K. Establishment of a cell culture model permissive for infection by hepatitis B and C viruses. Hepatol Commun, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.König A, Yang J, Jo E, Park KHP, Kim H, Than TT, Song X, Qi X, Dai X, Park S, Shum D, Ryu WS, Kim JH, Yoon SK, Park JY, Ahn SH, Han KH, Gerlich WH, Windisch MP. 2019. Efficient long-term amplification of hepatitis B virus isolates after infection of slow proliferating HepG2-NTCP cells. J Hepatol 71:289–300. doi: 10.1016/j.jhep.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N, Kaito M, Miyakawa Y, Mizokami M. 2006. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 44:915–924. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto H, Satoh T, Kiyohara T, Totsuka A, Moritsugu Y. 1997. Quantitation of group-specific a antigen in hepatitis B vaccines by anti-HBs/a monoclonal antibody. Biologicals 25:373–380. doi: 10.1006/biol.1997.0109. [DOI] [PubMed] [Google Scholar]
- 41.Nishitsuji H, Harada K, Ujino S, Zhang J, Kohara M, Sugiyama M, Mizokami M, Shimotohno K. 2018. Investigating the hepatitis B virus life cycle using engineered reporter hepatitis B viruses. Cancer Sci 109:241–249. doi: 10.1111/cas.13440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada N, Murayama A, Shiina M, Aly HH, Iwamoto M, Tsukuda S, Watashi K, Tanaka T, Moriishi K, Nishitsuji H, Sugiyama M, Mizokami M, Shimotohno K, Muramatsu M, Murata K, Kato T. 2020. Anti-viral effects of interferon-λ3 on hepatitis B virus infection in cell culture. Hepatol Res 50:283–291. doi: 10.1111/hepr.13449. [DOI] [PubMed] [Google Scholar]
- 43.Matsuda M, Yamanaka A, Yato K, Yoshii K, Watashi K, Aizaki H, Konishi E, Takasaki T, Kato T, Muramatsu M, Wakita T, Suzuki R. 2018. High-throughput neutralization assay for multiple flaviviruses based on single-round infectious particles using dengue virus type 1 reporter replicon. Sci Rep 8:16624. doi: 10.1038/s41598-018-34865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. 2016. Complex antigens drive permissive clonal selection in germinal centers. Immunity 44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]