HIV remains a major public health problem worldwide, and new therapies and preventive strategies are necessary for controlling the epidemic. Broadly neutralizing antibodies (bNAbs) have been developed in the past decade to fill this gap.
KEYWORDS: bNAb, human immunodeficiency virus
ABSTRACT
Novel therapeutic and preventive strategies are needed to contain the HIV-1 epidemic. Broadly neutralizing human antibodies (bNAbs) with exceptional activity against HIV-1 are currently being tested in HIV-1 prevention trials. The selection of anti-HIV-1 bNAbs for clinical development was primarily guided by their in vitro neutralizing activity against HIV-1 Env-pseudotyped viruses. Here, we report on the neutralizing activity of 9 anti-HIV-1 bNAbs now in clinical development against 126 clade A, C, and D peripheral blood mononuclear cell (PBMC)-derived primary African isolates. The neutralizing potency and breadth of the bNAbs tested were significantly reduced compared to those seen with pseudotyped-virus panels. The difference in sensitivity between pseudotyped viruses and primary isolates varied from 3- to nearly 100-fold depending on the bNAb and the HIV-1 clade. Thus, the neutralizing activity of bNAbs against primary African isolates differs from their activity against pseudovirus panels. The data have significant implications for interpreting the results of ongoing HIV-1 prevention trials.
IMPORTANCE HIV remains a major public health problem worldwide, and new therapies and preventive strategies are necessary for controlling the epidemic. Broadly neutralizing antibodies (bNAbs) have been developed in the past decade to fill this gap. The neutralizing activity of these antibodies against diverse HIV strains has mostly been measured using Env-pseudotyped viruses, which overestimate bNAb coverage and potency. In this study, we measured the neutralizing activity of nine bNAbs against clade A, C, and D HIV isolates derived from cells of African patients living with HIV and produced in peripheral blood mononuclear cells. We found that the coverage and potency of bNAbs were often significantly lower than what was predicted by Env-pseudotyped viruses and that this decrease was related to the bNAb binding site class. These data are important for the planning and analysis of clinical trials that seek to evaluate bNAbs for the treatment and prevention of HIV infection in Africa.
INTRODUCTION
The development of broadly neutralizing antibodies (bNAbs) against HIV has matured in the past few years as several bNAbs were evaluated in clinical trials. VRC01 (1), 3BNC117 (2) and 10-1074 (3) have been the most extensively evaluated to date, in healthy volunteers (4, 5), in viremic people living with HIV (6–9), and in the setting of analytical treatment interruption (10–13), with encouraging results. These trials were restricted to patients living in the United States and Europe, limiting the assessment of the global utility of these antibodies, since the majority of the individuals in the regions in question were infected with clade B HIV-1 (14).
One potential limitation in the development of bNAbs is that their activity has been documented primarily using panels of Env-pseudotyped viruses. However, we (15) and others (16–19) have shown that using Env-pseudotyped viruses often overestimates both the breadth and potency of bNAbs compared to peripheral blood mononuclear cell (PBMC)-derived HIV isolates.
Here, we report on the breadth and potency of nine bNAbs currently in clinical development against primary PBMC-derived HIV-1 viruses isolated from individuals living in South Africa, Uganda, and Kenya. We compared these results with data from Env-pseudotyped virus panels as well as matched Env-pseudotyped viruses derived from the African isolates.
RESULTS
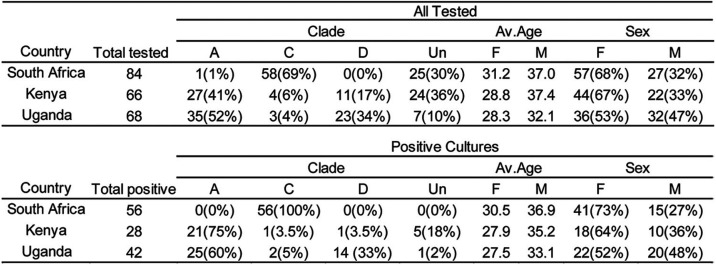
To examine the coverage of bNAbs in clinical development against HIV-1 variants circulating in Africa, we obtained 218 cryopreserved PBMC samples from people living with HIV-1 who participated in one of three studies: the Partners in Prevention HSV/HIV Transmission Study (20), the Couples Observational Study (21), or the Partners PrEP Study (22). The samples were collected from participants recruited at sites in South Africa (n = 84), Uganda (n = 68), and Kenya (n = 66). Bulk CD4+ T lymphocytes were cultured, yielding 126 (58%) HIV-1 isolates after 21 days (Table 1).
TABLE 1.
Demographical characteristics of all tested individualsa
F, female; M, male; Un, undetermined.
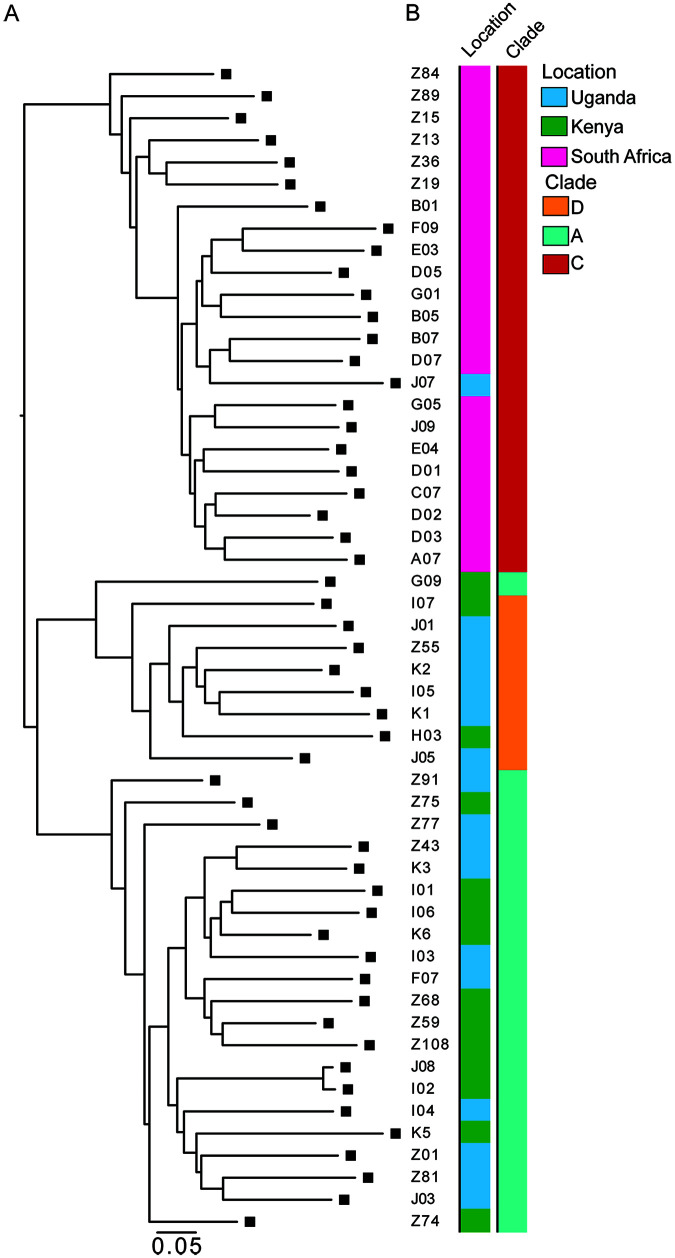
To examine the genetic diversity of the HIV-1 viruses obtained from the cultures, we performed single-genome amplification (SGA) on 53 viral supernatants and obtained 172 independent sequences representing clades A, C, and D, with 2 sequences per supernatant on average. We observed that the viruses were phylogenetically grouped in large part by their geographic origins and clades (Fig. 1A and B).
FIG 1.
(A) Maximum-likelihood phylogenetic tree of env sequences of viruses isolated from outgrowth cultures by SGA. (B) The first bar to the right of the phylogenetic tree represents the country of origin of the sample; the second bar represents the HIV-1 clade of every sample.
The following bNAbs were tested for neutralizing activity against the PBMC isolates in TZM-bl assays: 3BNC117-LS, VRC01, VRC07-523LS, and 1-18, all of which are CD4 binding site specific (CD4bs), (1, 2, 23, 24); 10-1074-LS, and BG18, which target the base of the V3 glycan and surrounding glycans (3, 25); and PGDM1400 and CAP256-VRC25.26, which are specific for the V2 loop (26, 27). We also tested the combination of 3BNC117-LS and 10-1074-LS, which is currently in clinical development (9, 12) (clinicaltrials.gov/ct2/show/NCT03254277, clinicaltrials.gov/ct2/show/NCT03554408, clinicaltrials.gov/ct2/show/NCT04250636, and clinicaltrials.gov/ct2/show/NCT04173819).
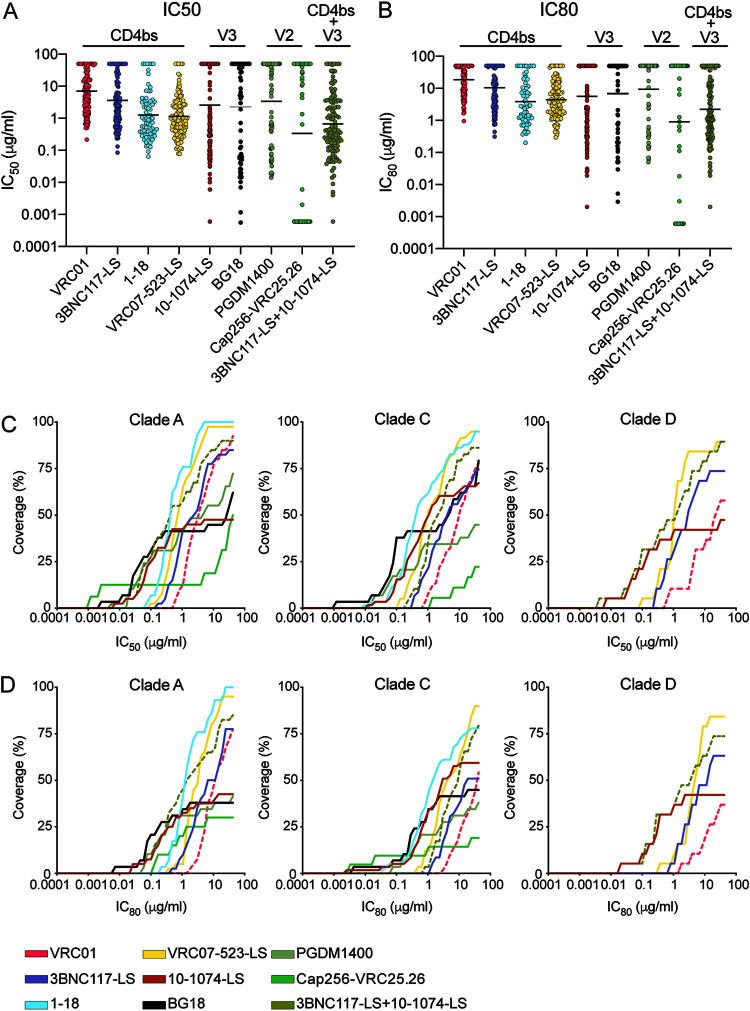
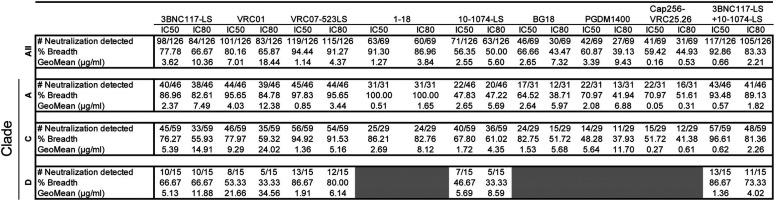
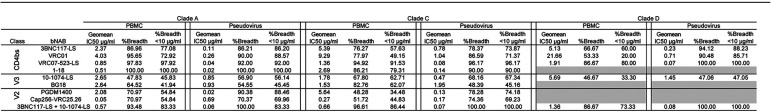
The geometric mean 50% inhibitory concentration (IC50) for VRC01, which is now being tested in two large efficacy prevention trials (clinicaltrials.gov/ct2/show/NCT02568215 and clinicaltrials.gov/ct2/show/NCT02716675), was 7.01 µg/ml for all viral isolates (Fig. 2A; Table 2). Only 57% of the viruses tested were sensitive to VRC01 at concentrations below 10 µg/ml (Fig. 2A to D; Table 2; also, see Data Set S1 in the supplemental material). Other CD4bs antibodies were substantially more potent than VRC01, including VRC07-523LS and 1-18, with geometric mean IC50s of 1.14 µg/ml and 1.27 µg/ml, respectively. These two antibodies alone covered 92% and 87% of the viruses tested at concentrations below 10 µg/ml (Fig. 2A to D; Table 2; Data Set S1). 10-1074-LS and BG18, which target the base of the V3 loop, demonstrated geometric mean IC50s of 2.55 µg/ml and 2.64 µg/ml, respectively (Fig. 2A to D; Table 2; Data Set S1). PGDM1400 and CAP256-VRC25.26, which target the V2 loop, demonstrated geometric mean IC50s of 3.38 µg/ml and 0.16 µg/ml, respectively. However, 10-1074-LS, BG18, PGDM1400, and CAP256-VRC25.26 covered only 52%, 49%, 46%, and 47% of the viruses, respectively, at concentrations below 10 µg/ml (Fig. 2A to D; Table 2; Data Set S1). The combination of 3BNC117-LS and 10-1074-LS performed better than any single antibody alone in terms of potency. The geometric mean IC50 for the combination was 0.65 µg/ml, and 84% of the viruses were sensitive at concentrations below 10 µg/ml (Fig. 2A to D; Table 2; Data Set S1).
FIG 2.
(A) Dot plot showing IC50s of unique PBMC-derived viruses for each bNAb tested. (B) Dot plot showing IC80s of unique PBMC-derived viruses for each bNAb tested. Each dot represents a single virus. Black bars represent geometric mean IC50s and IC80s. (C and D) Coverage curves. For each antibody, the graph shows the percentage of viruses neutralized in the TZM-bl assay at a given IC50 (C) or IC80 (D) for the PBMC-derived primary isolates for HIV-1 clades A, C, and D. All bNAbs are represented with the same color scheme as in panel A. The pink dotted line represents VRC01, and the olive green represents the combination of 3BNC117-LS and 10-1074-LS.
TABLE 2.
Breadth, IC50s, and IC80s in TZM-bl cells for PBMC-derived isolates
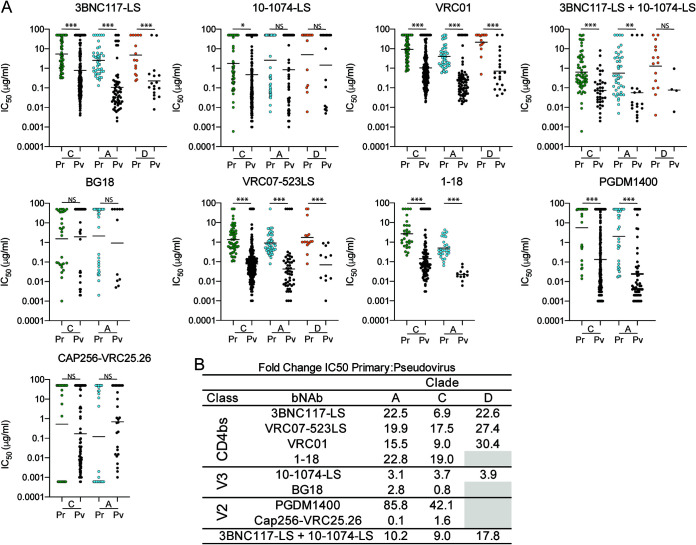
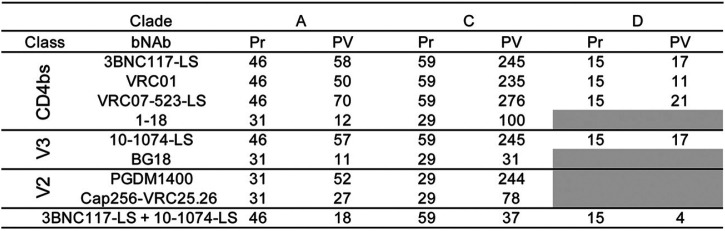
To determine whether the sensitivity of the primary African isolates to bNAbs differs from that of standard pseudovirus panels, we compared the data obtained from the outgrowth cultures with those from well-characterized clade A, C, and D pseudoviruses (Fig. 3A and B; Tables 3 and 4). All of the bNAbs tested were more potent and showed increased breadth against the pseudoviruses compared to the primary isolates. The difference between pseudovirus and primary isolates varied between antibodies. For example, CD4 binding site-specific bNAbs showed significant average decreases in potency of 20-, 13-, and 27-fold for primary isolates from clades A, C, and D, respectively (P < 0.0001 for all clades tested for CD4 binding site-specific antibodies). Moreover, these antibodies neutralized an average of 4.2%, 13.5%, and 28.1% fewer clade A, C, and D primary isolates, respectively, when tested against primary isolates than against pseudoviruses at concentrations below 10 µg/ml.
FIG 3.
(A) IC50s of unique PBMC-derived viruses (Pr) shown in color, and corresponding clade pseudovirus panel in black (Pv). The viruses are organized by HIV-1 clade. Each dot represents a single virus. Black bars represent geometric mean IC50s. (B) Fold change between the geometric mean IC50s (in micrograms per milliliter) of PBMC-derived viruses and the corresponding pseudovirus panel. Statistical analysis was done using the Mann-Whitney test. Statistical significance was defined as a P value of <0.05 unless stated otherwise. P values smaller than 0.05 were considered statistically significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
TABLE 3.
Breadth, IC50s, and IC80s in TZM-bl cells for PBMC-derived isolates and pseudovirus panels
TABLE 4.
Number of tested samples for PBMC derived isolates (Pr) and pseudovirus panels (PV)
The difference in potency and breadth between pseudoviruses and primary isolates was less dramatic for bNAbs targeting the V3 glycan. On average, there was only a 3-fold difference in IC50 between primary isolates and pseudovirus panels for clades A, C, and D (P = 0.002 for 10-1074-LS on clade C; the difference was not significant for clades A and D for both 10-1074-LS and BG18). V3 glycan antibodies also retained most of their breadth, as shown by the numbers of strains reaching IC50s at concentrations below 10 µg/ml (Fig. 3A and B; Tables 3 and 4).
The two V2-loop bNAbs were unusual in that they had very different relative potencies against primary and pseudotyped clade A and C viruses. Whereas CAP256-VRC25.26 showed no significant difference in activity, PGDM1400 was 85- and 42-fold less active against primary clade A and C viruses than pseudotyped viruses, respectively (P < 0.0001) (Fig. 3A and B; Tables 3 and 4). These antibodies neutralized 24% and 32% fewer clade A and C primary isolates, respectively, than pseudoviruses at concentrations below 10 µg/ml. Finally, the 3BNC117-LS/10-1074-LS combination was on average 12-fold less active against the primary isolates than pseudoviruses and showed no decrease in breadth for clade A but did show 13.5% and 26.5% decreases in breadth with regard to the numbers of strains reaching IC50s at concentrations below 10 µg/ml for clades C and D, respectively (Fig. 3A and B; Tables 3 and 4).
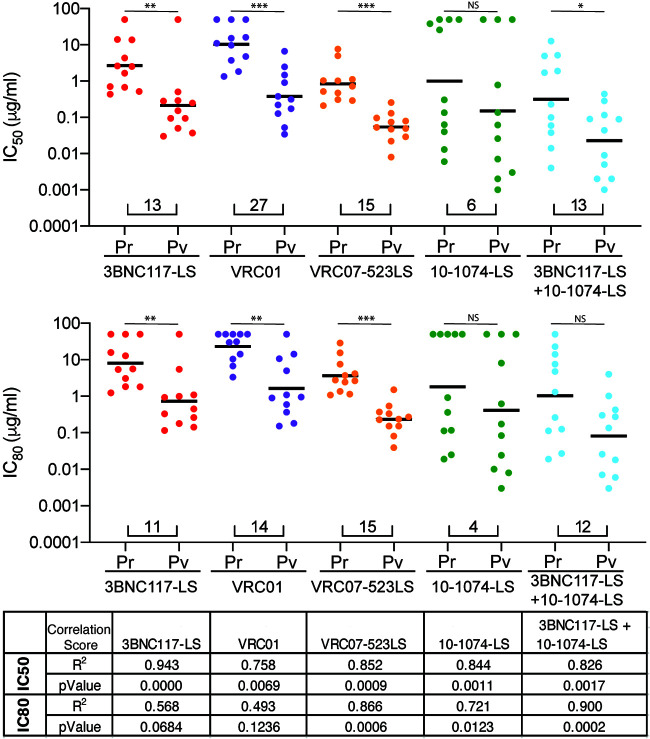
To determine whether the differences between primary isolates and pseudovirus panels were attributable to sequence differences between the viruses being tested, we cloned HIV-1 env genes from 11 different primary cultures, expressed them as pseudotyped viruses, and tested them against a panel of 5 bNAbs in the TZM-bl neutralization assay. IC50s and IC80s for the PBMC-derived viruses and the matched pseudoviruses showed similar fold differences than those found between primary isolates and pseudovirus panels for all bNAbs tested (Fig. 4). Besides the underestimation of the resistance levels of to bNAbs presented in the pseudovirus experiments, we observed a significant correlation between the results of both experiments (Fig. 4). The data suggest that there are significant differences in bNAb potency and breadth between primary clade A, C, and D isolates and pseudotyped viruses and that the magnitude of these differences is bNAb specific.
FIG 4.
(Top and middle) IC50s and IC80s of unique PBMC-derived clonal viruses (Pr) and corresponding pseudoviruses (Pv) for each antibody. Each dot represents a single virus. Black bars represent geometric mean IC50s. Numbers under the dots indicate the fold change in geometric mean IC50s between the 2 groups. Statistical analysis was done using the Mann-Whitney test. Statistical significance was defined as a P value of <0.05 unless stated otherwise. P values smaller than 0.05 were considered statistically significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. (Bottom) Correlation analysis between Pr and Pv. r2 and P values were obtained from the Pearson correlation coefficient.
DISCUSSION
We measured the neutralization profile of nine bNAbs currently in clinical development on 126 primary isolates obtained from PBMC cultures from individuals infected with HIV-1 clades A, C, and D. VRC01, the most advanced clinical candidate, is nearly 15 times less active against primary isolates than pseudotyped viruses. Similar results were obtained with other CD4 binding site antibodies. In contrast, the two V2-directed antibodies tested varied widely in their ability to neutralize pseudotyped viruses and primary isolates. Thus, the results obtained with pseudotyped virus panels cannot be translated directly to bNAb activity on primary isolates.
Our results extend earlier work with less potent antibodies (16–18) and with bNAbs against clade B viruses (15) to clades A, C, and D. In all cases, primary isolates were less sensitive to bNAbs than pseudotyped viruses. However, the relative reduction in activity differed between antibodies that target different epitopes on the envelope spike, with V3 glycan bNAbs 10-1074-LS and BG18 being least affected and PGDM1400 the most affected. In addition, the magnitude of the differences varies among viral clades. Combinations of bNAbs, as exemplified by 3BNC117-LS and 10-1074-LS, are advantageous in this respect, as also suggested by in vitro and in silico analysis using Env- pseudotyped panels (28).
A number of non-mutually exclusive hypotheses have been suggested to explain the enhanced susceptibility of 293T-derived pseudotyped viruses to neutralization by bNAbs. For example, sensitivity to neutralization could be dependent on the number of envelope protein spikes, with fewer spikes bound on the surface of 293T-derived pseudotyped viruses than PBMC-derived primary isolates (16, 18). Another possibility involves differential glycosylation by different packaging cell types. bNAbs frequently target glycan-dependent epitopes; therefore, the differential glycosylation profile of the envelope spike produced in different cell types could also alter their neutralization profile (49, 50). However, V3 glycan bNAbs and CAP256-VRC25.26, which target highly glycan-dependent epitopes, were the least affected. Similarly, PG9, a V2 peptide glycan-specific bNAb (29, 30), showed only small changes in its neutralization profile between clade B pseudotyped viruses and PBMC-derived viruses (18). Still another possibility is that most of the pseudoviruses tested in the standard panels were isolated between 1998 and 2010, whereas our samples were collected between 2007 and 2012 (31), and there appears to be increased bNAb resistance over time (32–35).
Clinical trials testing bNAbs for HIV-1 prevention are now being conducted in Africa and other parts of the world. The largest of these trials is testing VRC01 at several sites in Africa (Botswana, Kenya, Malawi, Mozambique, South Africa, Tanzania, and Zimbabwe), where the majority of the HIV-1 infections are caused by clade A, C, and D viruses (14). Although the results of those trials are not yet known, data are available from smaller trials where bNAbs were administered to individuals undergoing analytical treatment interruption (ATI). In the absence of antiretroviral therapy, nearly all participants experience viral rebound in 2 to 3 weeks, and it is believed that recrudescence of viremia is due to reactivation of HIV-1 from latently infected CD4+ T cells (36). Single antibodies were able to decrease viremia levels or delay the return of quantifiable viremia, but their ability to do so correlated with their neutralizing activity against primary isolates and not pseudotyped viruses (6, 7, 10, 11, 37, 38). For example, VRC01 had little measurable effect on delaying HIV-1 rebound when administered during ATI (13, 39). In contrast, antibody combinations maintain suppression of viremia during ATI in individuals harboring bNAb-sensitive viruses for as long as antibody concentrations remain above 10 µg/ml (12). Should the clinical outcomes in the ongoing VRC01 prevention trials track with bNAb activity against primary isolates as opposed to pseudotyped virus panels, there could be up to a 15-fold difference between the predicted and observed outcomes of the trial. Nevertheless, by analogy with the ATI trials, if the AMP trials demonstrate even a smaller-than-projected effect with VRC01, it provides a proof of concept that passive immunization can prevent sexual transmission of sensitive HIV-1 strains and indicates that combinations should be highly effective.
MATERIALS AND METHODS
Samples.
The study was conducted with the approval of The Rockefeller University Institutional Review Board. Samples were collected during the course of three studies in sub-Saharan Africa. (i) The first is the Partners in Prevention HSV/HIV Transmission Study. Between November 2004 and April 2007, 3,408 HIV-serodiscordant heterosexual couples were enrolled from 14 study sites in sub-Saharan Africa into this phase III clinical trial evaluating the efficacy of herpes simplex virus 2 (HSV-2) suppressive therapy (acyclovir 400 mg orally twice daily versus matching placebo) provided to persons infected with both HIV-1 and HSV-2 who had CD4 counts of ≥250 at enrollment to prevent HIV transmission to their HIV-uninfected heterosexual partner (20). (ii) The second was the Couples Observational Study. A total of 485 HIV-serodiscordant heterosexual couples were recruited at two of the same sites as the Partners in Prevention HSV-2/HIV Transmission Study (Kampala, Uganda, and Soweto, South Africa) for a prospective, observational study of biologic correlates of HIV protection; there was no HSV-2 coinfection or CD4 count enrollment requirement (21). (iii) The third was the Partners PrEP Study. This was a randomized, phase III clinical trial of antiretroviral pre-exposure chemoprophylaxis (300 mg tenofovir once daily versus 300 mg tenofovir plus 200 mg emtricitabine once daily versus matching placebo) conducted at nine sites in Kenya and Uganda (22).
CD4+ T cell outgrowth culture.
Bulk outgrowth cultures were performed as previously described (11). Briefly, PBMCs were obtained from HIV-1-infected individuals, and CD4+ T lymphocytes were isolated by negative selection with magnetic beads (Miltenyi). A total of 2 × 106 CD4+ T lymphocytes were activated using anti-CD3/CD2/CD28 beads (Miltenyi) and cultured in the presence of 100 U/ml interleukin 2 (IL-2) (Peprotech) at 37°C and 5% CO2. CD4+ T lymphocytes were cocultured with irradiated heterologous PBMCs from healthy donors (1 × 106). After 24 h of activation, 1 × 105 Molt 4 CCR5 cells were added. The medium was replaced twice a week, and the presence of p24 in the culture supernatant was quantified by the Lenti-X p24 Rapid Titer kit (Clontech) after 7, 14, and 21 days of culture. The infectivity of viral cultures was confirmed by a 50% tissue culture infective dose assay with TZM-bl cells (40). We performed a single outgrowth culture for each tested individual (n = 218) and further analyzed the ones with a positive enzyme-linked immunosorbent assay (ELISA) signal (n = 126).
Neutralization assays.
TZM-bl cell neutralization assays were performed as previously described (40, 41). Neutralization assays were conducted in laboratories meeting good clinical laboratory practice quality assurance criteria. All bulk outgrowth culture primary isolates were tested against 3BNC117-LS, VRC01, 10-1074-LS, VRC07-523LS and the combination of 3BNC117-LS and 10-1074-LS. Sixty-nine were also tested against PGDM1400 (provided by Dennis Burton, Scripps Research Institute), BG18, 1-18 (provided by Florian Klein, University of Cologne), and CAP256-VRC25.36 (provided by John Mascola, NIH Vaccine Research Center). The maximum antibody concentration tested was 50 µg/ml. All the experiments were performed in triplicate. Neutralization values used in Fig. 3 for the pseudovirus panel were obtained from the Antibody Database software (42), the CATNAP database (43), or the original reports for antibodies 1-18 (25), BG18 (24), and the combination of 3BNC117-LS and 10-1074-LS (44).
Virus sequence analysis.
HIV env sequences from p24-positive supernatants were obtained and analyzed as previously described (45). Sequences derived from each bulk culture that had double peaks (cutoff consensus identity for any residue, <75%) or stop codons or were shorter than the expected envelope size were omitted from downstream analysis. Phylogenetic analysis was performed by generating nucleotide alignments using MAFFT (46) and posterior phylogenetic trees using PhyML v3.1 (47), using the GTR model with 1,000 bootstraps. Clade determination was performed using the NCBI subtyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi). For samples not sequenced in this study, the clade was determined by sequencing a 514-bp region of the env gene (C2-V3-C3 region) from plasma samples as previously described (20–22).
Pseudotyped-virus production.
Pseudotyped-virus production was performed as previously described (48). The cytomegalovirus (CMV) promoter was amplified by PCR from the pcDNA 3.1D/V5-His-TOPO plasmid (Life Technologies) with forward primer 5′-AGTAATCAATTACGGGGTCATTAGTTCAT-3′ and reverse primer 5′-CATAGGAGATGCCTAAGCCGGTGGAGCTCTGCTTATATAGACCTC-3′. A 1-µl volume of the first-round PCR product from each individual env gene obtained from bulk cultures was amplified with primers 5′-CACCGGCTTAGGCATCTCCTATGGCAGGAAGAA-3′ and 5′-ACTTTTTGACCACTTGCCACCCAT-3′. PCR products were purified with the Macherey-Nagel gel and PCR purification kit. The CMV promoter amplicon was fused to individual env genes by overlap PCR with 10 ng of env and 0.5 ng of CMV with forward primer 5′-AGTAATCAATTACGGGGTCATTAGTTCAT-3′ and reverse primer 5′-ACTTTTTGACCACTTGCCACCCAT-3′. Resulting amplicons were analyzed by gel electrophoresis, purified with the Macherey-Nagel gel and PCR purification kit, and cotransfected with pSG3Δenv backbone vector (NIH AIDS Reagent Program) into HEK293T cells to produce pseudoviruses as previously described (48).
Statistical analysis.
Statistical analyses were performed with GraphPad Prism 8.0 software. Statistical analysis presented in Fig. 3 and 4 were analyzed using the Mann-Whitney test. Correlations were tested by Pearson correlation coefficient. Statistical significance was defined as a P value of <0.05 unless stated otherwise. P values smaller than 0.05 were considered statistically significant. The data are shown as means and individual data points.
Supplementary Material
ACKNOWLEDGMENTS
We thank the study participants who devoted time to our research and all members of the Nussenzweig lab for helpful discussions.
This work was supported in part by Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD) grants OPP1146996 (M.S.S.), OPP1092074, and OPP1168933 (M.C.N.) and by the NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-06 (M.C.N.). M.C.N. is a Howard Hughes Medical Investigator.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen YZ, Butler AL, Millard K, Witmer-Pack M, Levin R, Unson-O'Brien C, Patel R, Shimeliovich I, Lorenzi JCC, Horowitz J, Walsh SR, Lin S, Weiner JA, Tse A, Sato A, Bennett C, Mayer B, Seaton KE, Yates NL, Baden LR, deCamp AC, Ackerman ME, Seaman MS, Tomaras GD, Nussenzweig MC, Caskey M. 2019. Safety, pharmacokinetics, and immunogenicity of the combination of the broadly neutralizing anti-HIV-1 antibodies 3BNC117 and 10-1074 in healthy adults: a randomized, phase 1 study. PLoS One 14:e0219142. doi: 10.1371/journal.pone.0219142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer KH, Seaton KE, Huang Y, Grunenberg N, Isaacs A, Allen M, Ledgerwood JE, Frank I, Sobieszczyk ME, Baden LR, Rodriguez B, Van Tieu H, Tomaras GD, Deal A, Goodman D, Bailer RT, Ferrari G, Jensen R, Hural J, Graham BS, Mascola JR, Corey L, Montefiori DC, HVTN 104 Protocol Team, and the NIAID HIV Vaccine Trials Network . 2017. Safety, pharmacokinetics, and immunological activities of multiple intravenous or subcutaneous doses of an anti-HIV monoclonal antibody, VRC01, administered to HIV-uninfected adults: results of a phase 1 randomized trial. PLoS Med 14:e1002435. doi: 10.1371/journal.pmed.1002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O'Brien C, Weiland D, Robles A, Kümmerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP, Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F. 2017. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O’Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun T-W, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE, VRC 601 Study Team . 2015. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-On Y, Gruell H, Schoofs T, Pai JA, Nogueira L, Butler AL, Millard K, Lehmann C, Suárez I, Oliveira TY, Karagounis T, Cohen YZ, Wyen C, Scholten S, Handl L, Belblidia S, Dizon JP, Vehreschild JJ, Witmer-Pack M, Shimeliovich I, Jain K, Fiddike K, Seaton KE, Yates NL, Horowitz J, Gulick RM, Pfeifer N, Tomaras GD, Seaman MS, Fätkenheuer G, Caskey M, Klein F, Nussenzweig MC. 2018. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat Med 24:1701–1707. doi: 10.1038/s41591-018-0186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu C-L, Lorenzi JCC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T, Shimeliovich I, Patel R, Burke L, Cohen YZ, Hadrigan S, Settler A, Witmer-Pack M, West AP, Jr, Juelg B, Keler T, Hawthorne T, Zingman B, Gulick RM, Pfeifer N, Learn GH, Seaman MS, Bjorkman PJ, Klein F, Schlesinger SJ, Walker BD, Hahn BH, Nussenzweig MC, Caskey M. 2016. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535:556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen YZ, Lorenzi JCC, Krassnig L, Barton JP, Burke L, Pai J, Lu C-L, Mendoza P, Oliveira TY, Sleckman C, Millard K, Butler AL, Dizon JP, Belblidia SA, Witmer-Pack M, Shimeliovich I, Gulick RM, Seaman MS, Jankovic M, Caskey M, Nussenzweig MC. 2018. Relationship between latent and rebound viruses in a clinical trial of anti-HIV-1 antibody 3BNC117. J Exp Med 215:2311–2324. doi: 10.1084/jem.20180936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suárez I, Oliveira TY, Lorenzi JCC, Cohen YZ, Wyen C, Kümmerle T, Karagounis T, Lu C-L, Handl L, Unson-O'Brien C, Patel R, Ruping C, Schlotz M, Witmer-Pack M, Shimeliovich I, Kremer G, Thomas E, Seaton KE, Horowitz J, West AP, Bjorkman PJ, Tomaras GD, Gulick RM, Pfeifer N, Fätkenheuer G, Seaman MS, Klein F, Caskey M, Nussenzweig MC. 2018. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561:479–484. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge KE, Tustin NB, Madden PJ, Oden K, O’Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun T-W. 2016. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 375:2037–2050. doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bbosa N, Kaleebu P, Ssemwanga D. 2019. HIV subtype diversity worldwide. Curr Opin HIV AIDS 14:153–160. doi: 10.1097/COH.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 15.Cohen YZ, Lorenzi JCC, Seaman MS, Nogueira L, Schoofs T, Krassnig L, Butler A, Millard K, Fitzsimons T, Daniell X, Dizon JP, Shimeliovich I, Montefiori DC, Caskey M, Nussenzweig MC. 2017. Neutralizing activity of broadly neutralizing anti-HIV-1 antibodies against clade B clinical isolates produced in peripheral blood mononuclear cells. J Virol 92:e01883-17. doi: 10.1128/JVI.01883-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louder MK, Sambor A, Chertova E, Hunte T, Barrett S, Ojong F, Sanders-Buell E, Zolla-Pazner S, McCutchan FE, Roser JD, Gabuzda D, Lifson JD, Mascola JR. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339:226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Mann AM, Rusert P, Berlinger L, Kuster H, Günthard HF, Trkola A. 2009. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS 23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- 18.Provine NM, Cortez V, Chohan V, Overbaugh J. 2012. The neutralization sensitivity of viruses representing human immunodeficiency virus type 1 variants of diverse subtypes from early in infection is dependent on producer cell, as well as characteristics of the specific antibody and envelope variant. Virology 427:25–33. doi: 10.1016/j.virol.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etemad B, Ghulam-Smith M, Gonzalez O, White LF, Sagar M. 2015. Single genome amplification and standard bulk PCR yield HIV-1 envelope products with similar genotypic and phenotypic characteristics. J Virol Methods 214:46–53. doi: 10.1016/j.jviromet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celum C, Wald A, Lingappa JR, Magaret AS, Wang RS, Mugo N, Mujugira A, Baeten JM, Mullins JI, Hughes JP, Bukusi EA, Cohen CR, Katabira E, Ronald A, Kiarie J, Farquhar C, Stewart GJ, Makhema J, Essex M, Were E, Fife KH, de Bruyn G, Gray GE, McIntyre JA, Manongi R, Kapiga S, Coetzee D, Allen S, Inambao M, Kayitenkore K, Karita E, Kanweka W, Delany S, Rees H, Vwalika B, Stevens W, Campbell MS, Thomas KK, Coombs RW, Morrow R, Whittington WLH, McElrath MJ, Barnes L, Ridzon R, Corey L, Partners in Prevention HSV/HIV Transmission Study Team . 2010. Acyclovir and transmission of HIV-1 from persons infected with HIV-1 and HSV-2. N Engl J Med 362:427–439. doi: 10.1056/NEJMoa0904849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lingappa JR, Petrovski S, Kahle E, Fellay J, Shianna K, McElrath MJ, Thomas KK, Baeten JM, Celum C, Wald A, de Bruyn G, Mullins JI, Nakku-Joloba E, Farquhar C, Essex M, Donnell D, Kiarie J, Haynes B, Goldstein D, Partners in Prevention HSV/HIV Transmission Study Team . 2011. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLoS One 6:e28632. doi: 10.1371/journal.pone.0028632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baeten JM, Donnell D, Ndase P, Mugo NR, Campbell JD, Wangisi J, Tappero JW, Bukusi EA, Cohen CR, Katabira E, Ronald A, Tumwesigye E, Were E, Fife KH, Kiarie J, Farquhar C, John-Stewart G, Kakia A, Odoyo J, Mucunguzi A, Nakku-Joloba E, Twesigye R, Ngure K, Apaka C, Tamooh H, Gabona F, Mujugira A, Panteleeff D, Thomas KK, Kidoguchi L, Krows M, Revall J, Morrison S, Haugen H, Emmanuel-Ogier M, Ondrejcek L, Coombs RW, Frenkel L, Hendrix C, Bumpus NN, Bangsberg D, Haberer JE, Stevens WS, Lingappa JR, Celum C, Partners PrEP Study Team . 2012. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N Engl J Med 367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudicell RS, Kwon YD, Ko S-Y, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, Shi W, Yang Z-Y, Doria-Rose NA, McKee K, O'Dell S, Schmidt SD, Chuang G-Y, Druz A, Soto C, Yang Y, Zhang B, Zhou T, Todd J-P, Lloyd KE, Eudailey J, Roberts KE, Donald BR, Bailer RT, Ledgerwood J, Mullikin JC, Shapiro L, Koup RA, Graham BS, Nason MC, Connors M, Haynes BF, Rao SS, Roederer M, Kwong PD, Mascola JR, Nabel GJ, NISC Comparative Sequencing Program . 2014. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol 88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schommers P, Gruell H, Abernathy ME, Tran M-K, Dingens AS, Gristick HB, Barnes CO, Schoofs T, Schlotz M, Vanshylla K, Kreer C, Weiland D, Holtick U, Scheid C, Valter MM, van Gils MJ, Sanders RW, Vehreschild JJ, Cornely OA, Lehmann C, Fätkenheuer G, Seaman MS, Bloom JD, Bjorkman PJ, Klein F. 2020. Restriction of HIV-1 escape by a highly broad and potent neutralizing antibody. Cell 180:471–489.E22. doi: 10.1016/j.cell.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freund NT, Wang H, Scharf L, Nogueira L, Horwitz JA, Bar-On Y, Golijanin J, Sievers SA, Sok D, Cai H, Cesar Lorenzi JC, Halper-Stromberg A, Toth I, Piechocka-Trocha A, Gristick HB, van Gils MJ, Sanders RW, Wang L-X, Seaman MS, Burton DR, Gazumyan A, Walker BD, West AP, Jr, Bjorkman PJ, Nussenzweig MC. 2017. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci Transl Med 9:eaal2144. doi: 10.1126/scitranslmed.aal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sok D, van Gils MJ, Pauthner M, Julien J-P, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, Hua Y, Seaman MS, Moore JP, Ward AB, Wilson IA, Sanders RW, Burton DR. 2014. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A 111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang G-Y, Pancera M, Cale EM, Ernandes MJ, Louder MK, Asokan M, Bailer RT, Druz A, Fraschilla IR, Garrett NJ, Jarosinski M, Lynch RM, McKee K, O'Dell S, Pegu A, Schmidt SD, Staupe RP, Sutton MS, Wang K, Wibmer CK, Haynes BF, Abdool-Karim S, Shapiro L, Kwong PD, Moore PL, Morris L, Mascola JR. 2016. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol 90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagh K, Seaman MS, Zingg M, Fitzsimons T. 2018. Potential of conventional & bispecific broadly neutralizing antibodies for prevention of HIV-1 subtype A, C & D infections. PLoS 14:e1006860. doi: 10.1371/journal.ppat.1006860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR, Protocol G Principal Investigators . 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol 84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hraber P, Rademeyer C, Williamson C, Seaman MS, Gottardo R, Tang H, Greene K, Gao H, LaBranche C, Mascola JR, Morris L, Montefiori DC, Korber B. 2017. Panels of HIV-1 subtype C Env reference strains for standardized neutralization assessments. J Virol 91:e00991-17. doi: 10.1128/JVI.00991-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouvin-Pley M, Morgand M, Moreau A, Jestin P, Simonnet C, Tran L, Goujard C, Meyer L, Barin F, Braibant M. 2013. Evidence for a continuous drift of the HIV-1 species towards higher resistance to neutralizing antibodies over the course of the epidemic. PLoS Pathog 9:e1003477. doi: 10.1371/journal.ppat.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouvin-Pley M, Morgand M, Meyer L, Goujard C, Moreau A, Mouquet H, Nussenzweig M, Pace C, Ho D, Bjorkman PJ, Baty D, Chames P, Pancera M, Kwong PD, Poignard P, Barin F, Braibant M. 2014. Drift of the HIV-1 envelope glycoprotein gp120 toward increased neutralization resistance over the course of the epidemic: a comprehensive study using the most potent and broadly neutralizing monoclonal antibodies. J Virol 88:13910–13917. doi: 10.1128/JVI.02083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hake A, Pfeifer N. 2017. Prediction of HIV-1 sensitivity to broadly neutralizing antibodies shows a trend towards resistance over time. PLoS Comput Biol 13:e1005789. doi: 10.1371/journal.pcbi.1005789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouvin-Pley M, Beretta M, Moreau A, Roch E, Essat A, Goujard C, Chaix M-L, Moiré N, Martin L, Meyer L, Barin F, Braibant M. 2018. Evolution of the envelope glycoprotein of HIV-1 clade B toward higher infectious properties over the course of the epidemic. J Virol 93:e01171-18. doi: 10.1128/JVI.01171-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengupta S, Siliciano RF. 2018. Targeting the latent reservoir for HIV-1. Immunity 48:872–895. doi: 10.1016/j.immuni.2018.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephenson KE, Julg B, Ansel J, Walsh SR, Tan CS, Maxfield L, Abbink P, Gelderblom HC, Priddy F, deCamp AC. 2019. Therapeutic activity of PGT121 monoclonal antibody in HIV infected adults, abstr 145. In Conference on Retroviruses and Opportunistic Infections, Seattle, WA. [Google Scholar]
- 38.Chen G, Coates E, Fichtenbaum C, Koletar S, Landovitz R, Presti R, Overton T, Santana J, Rothwel RS, Roa J, Donaghy E, Holman L, Novik L, Berkowitz N, Larkin B, Conan-Cibotti M, Tressler R, Wang J, Hu Z, Capparelli E, Arnold F, Bailer R, McDermott A, Gama L, Graham B, Koup R, Mascola J, Ledgerwood J, Tebas P, VRC 607/ACTG A5378 study team . 2019. Safety and virologic effect of the HIV-1 broadly neutralizing antibodies, VRC01LS or VRC07-523LS, administered to HIV-infected adults in a phase 1 clinical trial, abstr 4941. In 10th IAS Conference on HIV Science. [Google Scholar]
- 39.Crowell TA, Colby DJ, Pinyakorn S, Sacdalan C, Pagliuzza A, Intasan J, Benjapornpong K, Tangnaree K, Chomchey N, Kroon E, de Souza MS, Tovanabutra S, Rolland M, Eller MA, Paquin-Proulx D, Bolton DL, Tokarev A, Thomas R, Takata H, Trautmann L, Krebs SJ, Modjarrad K, McDermott AB, Bailer RT, Doria-Rose N, Patel B, Gorelick RJ, Fullmer BA, Schuetz A, Grandin PV, O'Connell RJ, Ledgerwood JE, Graham BS, Tressler R, Mascola JR, Chomont N, Michael NL, Robb ML, Phanuphak N, Ananworanich J, RV397 Study Group . 2019. Safety and efficacy of VRC01 broadly neutralising antibodies in adults with acutely treated HIV (RV397): a phase 2, randomised, double-blind, placebo-controlled trial. Lancet HIV 6:e297–e306. doi: 10.1016/S2352-3018(19)30053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12:Unit 12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 42.West AP, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. 2013. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Natl Acad Sci U S A 110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon H, Macke J, West AP, Jr, Foley B, Bjorkman PJ, Korber B, Yusim K. 2015. CATNAP: a tool to compile, analyze and tally neutralizing antibody panels. Nucleic Acids Res 43:W213–W219. doi: 10.1093/nar/gkv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong R, Louder MK, Wagh K, Bailer RT, deCamp A, Greene K, Gao H, Taft JD, Gazumyan A, Liu C, Nussenzweig MC, Korber B, Montefiori DC, Mascola JR. 2015. Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol 89:2659–2671. doi: 10.1128/JVI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenzi JCC, Cohen YZ, Cohn LB, Kreider EF, Barton JP, Learn GH, Oliveira T, Lavine CL, Horwitz JA, Settler A, Jankovic M, Seaman MS, Chakraborty AK, Hahn BH, Caskey M, Nussenzweig MC. 2016. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci U S A 113:E7908–E7916. doi: 10.1073/pnas.1617789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New Algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 48.Kirchherr JL, Lu X, Kasongo W, Chalwe V, Mwananyanda L, Musonda RM, Xia S-M, Scearce RM, Liao H-X, Montefiori DC, Haynes BF, Gao F. 2007. High throughput functional analysis of HIV-1 env genes without cloning. J Virol Methods 143:104–111. doi: 10.1016/j.jviromet.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J Virol 82:5807–5814. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utachee P, Nakamura S, Isarangkura-Na-Ayuthaya P, Tokunaga K, Sawanpanyalert P, Ikuta K, Auwanit W, Kameoka M. 2010. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. J Virol 84:4311–4320. doi: 10.1128/JVI.02619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.