For the development of vaccines based on SARS-CoV-2-specific cytotoxic T lymphocytes (CTLs), we attempted to identify HLA-A*02:01-restricted CTL epitopes derived from the nonstructural polyprotein 1a of SARS-CoV-2. Out of 82 peptides predicted by bioinformatics, 54 peptides showed good binding affinities to HLA-A*02:01.
KEYWORDS: SARS-CoV-2, COVID-19, CTL epitope, HLA-A*02:01, pp1a, vaccines
ABSTRACT
COVID-19 vaccines are being rapidly developed and human trials are under way. Almost all of these vaccines have been designed to induce antibodies targeting the spike protein of SARS-CoV-2 in expectation of neutralizing activities. However, nonneutralizing antibodies are at risk of causing antibody-dependent enhancement. Further, the longevity of SARS-CoV-2-specific antibodies is very short. Therefore, in addition to antibody-inducing vaccines, novel vaccines developed on the basis of SARS-CoV-2-specific cytotoxic T lymphocytes (CTLs) should be considered. Here, we attempted to identify HLA-A*02:01-restricted CTL epitopes derived from the nonstructural polyprotein 1a of SARS-CoV-2. Eighty-two peptides were first predicted as epitope candidates based on bioinformatics. Fifty-four of the 82 peptides showed high or medium binding affinities to HLA-A*02:01. HLA-A*02:01 transgenic mice were then immunized with each of the 54 peptides encapsulated into liposomes. The intracellular cytokine staining assay revealed that 18 out of 54 peptides were active as CTL epitopes because of the induction of gamma interferon (IFN-γ)-producing CD8+ T cells. Of the 18 peptides, 10 peptides were chosen for the following analyses because of their high responses. To identify dominant CTL epitopes, mice were immunized with liposomes containing the mixture of the 10 peptides. Some peptides were shown to be statistically predominant over the other peptides. Surprisingly, all mice immunized with the liposomal 10-peptide mixture did not show the same reaction pattern to the 10 peptides. There were three response patterns, suggesting the existence of an immunodominance hierarchy following peptide vaccination, which may provide more variations in the epitope selection for designing CTL-based COVID-19 vaccines.
IMPORTANCE For the development of vaccines based on SARS-CoV-2-specific cytotoxic T lymphocytes (CTLs), we attempted to identify HLA-A*02:01-restricted CTL epitopes derived from the nonstructural polyprotein 1a of SARS-CoV-2. Out of 82 peptides predicted by bioinformatics, 54 peptides showed good binding affinities to HLA-A*02:01. Using HLA-A*02:01 transgenic mice, 18 in 54 peptides were found to be CTL epitopes in the intracellular cytokine staining assay. Out of 18 peptides, 10 peptides were chosen for the following analyses because of their high responses. To identify dominant epitopes, mice were immunized with liposomes containing the mixture of the 10 peptides. Some peptides were shown to be statistically predominant. Surprisingly, all immunized mice did not show the same reaction pattern to the 10 peptides. There were three reaction patterns, suggesting the existence of an immunodominance hierarchy following peptide vaccination, which may provide us more variations in the epitope selection for designing CTL-based COVID-19 vaccines.
INTRODUCTION
In December 2019, the coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first identified in Wuhan, Hubei province, China. Since then, the subsequent spread of global infection has continued to gain momentum. As of 16 September 2020, the COVID-19 pandemic has infected more than 29.4 million people around the world and caused more than 931,000 deaths. Although the clinical symptoms vary from asymptomatic or mild self-limited infection to severe life-threatening respiratory disease, the mechanism of disease outcome remains unclear. Many nations are struggling to find appropriate preventive and control strategies. However, there are as yet no vaccines or antiviral drugs available for the treatment of this infectious disease.
There are seven coronaviruses that infect humans. In addition to SARS-CoV-2, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) also cause severe pneumonia, whereas the other four human coronaviruses, HCoV-229E, -NL63, -OC43, and -HKU1, cause symptoms of the common cold (1). Like other coronaviruses, SARS-CoV-2 possesses a large single-stranded positive sense RNA genome (2). As shown in Fig. 1, the 5′-terminal two-thirds of the genome are composed of open reading frame 1a (ORF1a) and ORF1b. ORF1a encodes the polyprotein 1a (pp1a) containing nonstructural proteins (nsp1 to nsp11) (Fig. 1). The remaining one-third of the genome encodes the structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as accessory proteins (Fig. 1). Coronaviruses depend on the S protein for binding to host cells. The host cell receptor for SARS-CoV-2 is the angiotensin-converting enzyme 2 (ACE2), which is also the receptor for SARS-CoV (3, 4).
FIG 1.
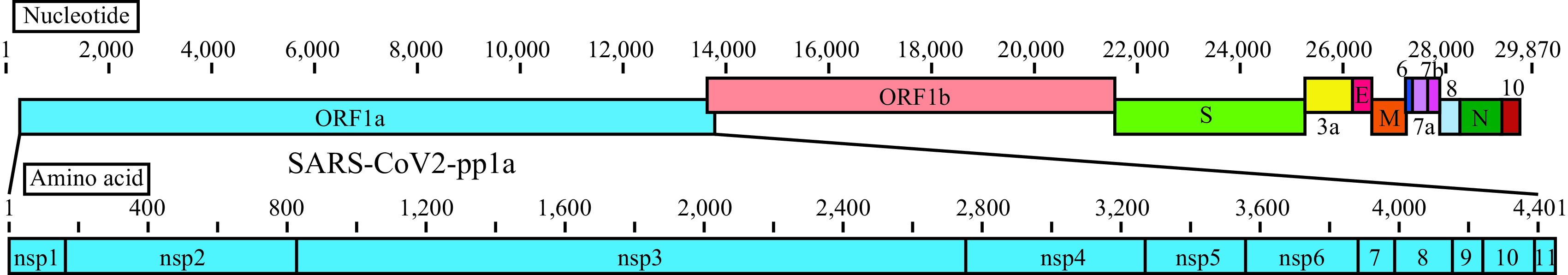
Linear diagrams of the SARS-CoV-2 genome and the protein subunits of ORF1a. The SARS-CoV-2 genome consists of ORF1a, ORF1b, spike (S), ORF3a, envelope (E), membrane (M), ORF6, ORF7a, ORF7b, ORF8, nucleocapsid (N), and ORF10. The ORF1a polyprotein is composed of 11 nonstructural proteins, nsp1 to nsp11.
It was shown that the increased cell numbers of antibody-secreting cells, follicular helper T cells, activated CD4+ T cells, and activated CD8+ T cells were observed in a non-severe COVID-19 patient (5), indicating that robust immune responses can be elicited to the newly emerged SARS-CoV-2. Therefore, the induction of protective immunity against SARS-CoV-2 is considered to help control COVID-19. Vaccines are being rapidly developed in the world, and human trials are under way for several vaccine candidates, ranging from traditional vaccines comprising inactivated SARS-CoV-2 preparations (6) to innovative vaccines such as subunit (7), RNA (8), DNA (9), and adenoviral (10) vaccines. Almost all of these vaccines have been designed to induce antibodies targeting the S protein because some antibodies specific for the receptor-binding domain (RBD) of S protein may have neutralizing activities and can interfere with the binding between the virus and ACE2 on host cells. In fact, it was reported that a DNA vaccine encoding the S protein induced neutralizing antibody in rhesus macaques and protected them from challenge with SARS-CoV-2 (9). However, there are two major issues concerning this vaccine. One issue is that nonneutralizing and subneutralizing antibodies to S protein induced by this vaccine are at risk of causing antibody-dependent enhancement (ADE) (11). ADE is the phenomenon by which binding of suboptimal antibodies to viruses enhances viral entry mediated by Fc receptors into immune cells, thereby promoting inflammation and tissue injury (12). ADE has been reported in the evaluation of vaccine candidates directed to S protein for SARS-CoV (13–16). Because of their similarities, these findings enable us to foresee the high risks of ADE in SARS-CoV-2 vaccines directed to S protein. It is worth noting that RBD-specific antibodies with potent neutralizing activity are extremely rare among S-specific antibodies in COVID-19-covalescent individuals (17, 18), suggesting that the development of effective vaccines might require novel strategies to selectively target the RBD of SARS-CoV-2. On the other hand, passive immunization of RBD-specific monoclonal antibodies obtained from convalescent individuals might be safe and effective for the elimination of SARS-CoV-2 (19–21), but much more expensive to produce for worldwide use than vaccines (22). A second issue is the short longevity of neutralizing antibodies to SARS-CoV-2. It was previously reported that the SARS-CoV-specific antibodies are short lived, persisting for at most about 2 to 3 years (23, 24), unlike SARS-CoV-specific memory T cells (25). The longevity of SARS-CoV-2-specific antibodies are likely to be even shorter, as indicated by antibody titers being undetectable or approaching baseline in the majority of SARS-CoV-2-infected individuals after 2 to 3 months post onset of symptoms (26). Taken together, these data strongly suggest that it does not seem wise to rely too heavily on the S-specific-antibody-inducing vaccine to control the COVID-19 pandemic.
In viral infections, CD8+ cytotoxic T lymphocytes (CTLs) play an important role for the clearance of virus, as do neutralizing antibodies. CTLs recognize virus-derived peptides in association with major histocompatibility complex class I (MHC-I) molecules on the surface of antigen-presenting cells and kill virus-infected target cells. It was reported that CD4+ T cells and CD8+ T cells are decreased in proportion to the disease severity and are exhausted in severe COVID-19 patients (27, 28), suggesting the significance of CD8+ CTLs in the clearance of SARS-CoV-2. Furthermore, SARS-CoV-specific memory T cells persisted up to 11 years (25), predicting the long life of SARS-CoV-2-specific memory T cells. Therefore, in addition to antibody-induced vaccines, novel vaccines based on SARS-CoV-2-specific CTLs should also be considered in future vaccine development for the prevention and disease control of COVID-19.
For the development of CTL-based COVID-19 vaccine, we here attempted to identify HLA-A*02:01-restricted, dominant CTL epitopes derived from pp1a of SARS-CoV-2. Pp1a is the largest protein (4,401 amino acids) among SARS-CoV-2 proteins and. therefore, it seems highly possible to find dominant epitopes in this protein. Furthermore, pp1a is produced first in SARS-CoV-2-infected cells, suggesting pp1a-specific CTLs could kill infected cells before the formation of mature virions. In addition, this protein is composed of 11 nonstructural regulatory proteins that are highly conserved among many different coronaviruses (29). To identify CTL epitopes, we used computational algorithms, HLA-A*02:01 transgenic mice, and peptide-encapsulated liposomes. In a similar way, we previously identified CTL epitopes of SARS-CoV pp1a (30).
RESULTS
Prediction of HLA-A*02:01-restricted CTL epitopes derived from SARS-CoV-2 pp1a.
We first attempted to predict HLA-A*02:01-restricted CTL epitopes derived from SARS-CoV-2 pp1a using four computer-based programs, SYFPEITHI (31), nHLAPred (32), ProPred-I (33), and IEDB (34). Scores of SARS-CoV-2 pp1a-derived peptides in the four programs were assessed by classifying into four ranks (A, excellent; B, very good; C, good; D, poor) with the following values: (i) SYFPEITHI: A ≥ 27, 24 ≤ B ≤ 26, 20 ≤ C ≤ 23, D < 20; (ii) nHLAPred: A = 1, 0.90 ≤ B <1, 0.5 ≤ C < 0.90, D <0.5; (iii) ProPred-I: A ≥ 1,000, B ≥ 500, C ≥ 100, D = ND (not detected); and (iv) IEDB: A > 0.1, 0.1 ≤ B < 0.5, 0.5 ≤ C < 1, D ≥ 1 (Table 1). All 28 peptides with rank A in the SYFPEITHI program were selected. Of the B- and C-ranked peptides in the SYFPEITHI program, 54 peptides were chosen because they were ranked from A to C in at least one of the remaining three programs (Table 1). A total of 82 peptides were synthesized into 9-mer peptides (Table 2). All of these peptides were then evaluated for their binding affinities to the HLA-A*02:01 molecule using TAP2-deficient RMA-S-HHD cells. As the half-maximal binding level (BL50) values of the influenza A virus matrix protein 1-derived peptide (FMP58-66) (35) as a high-binding control and the HIV reverse transcriptase-derived peptide (HIV-pol476-484) (36) as a medium-binding control were 2.3 µM and 80.6 µM, respectively, we defined a high binder as having a BL50 value below 10 µM, a medium binder as having a BL50 value ranging from 10 to 100 µM, and a low binder as having a BL50 value above 100 µM. As shown in Table 2, 20 peptides were high binders, whereas 34 peptides were medium binders, suggesting that the bioinformatics prediction was mostly successful. In contrast, the remaining 28 peptides displayed low affinities or no binding to HLA-A*02:01 molecules (Table 2). As shown in Table 3, none of the SYFPEITHI, nHLAPred, or ProPred-I algorithms successfully predicted the level of the peptide-binding affinity. However, the IEDB program is likely to have been able to predict it to some extent when we consider the binding affinity levels of A-ranked and D-ranked peptides using this program (Table 3). In the following experiments, 54 peptides shown to be high or medium binders were chosen to further investigate their abilities for peptide-specific CTL induction.
TABLE 1.
CTL candidate epitopes for the SARS-CoV-2 pp1a predicted by the 4 algorithms
| Peptide namea | Sequence | Rank by algorithmb |
Name | Sequence | Rank by algorithmb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | ||||
| pp1a-84 | VMVELVAEL | A | A | C | A | pp1a-2785 | AIFYLITPV | B | B | D | C |
| pp1a-106 | VLVPHVGEI | A | A | D | B | pp1a-3013 | SLPGVFCGV | B | B | D | B |
| pp1a-445 | GLNDNLLEI | A | A | D | A | pp1a-3083 | LLFLMSFTV | B | A | A | C |
| pp1a-468 | KLNEEIAII | A | B | D | A | pp1a-3115 | YLTNDVSFL | B | B | A | A |
| pp1a-600 | YITGGVVQL | A | C | D | A | pp1a-3183 | FLLNKEMYL | B | A | A | A |
| pp1a-692 | SIIIGGAKL | A | D | D | D | pp1a-3403 | FLNGSCGSV | B | A | D | C |
| pp1a-1161 | SLRVCVDTV | A | B | D | D | pp1a-3710 | TLMNVLTLV | B | A | B | A |
| pp1a-1312 | MLAKALRKV | A | A | D | B | pp1a-3753 | FLARGIVFM | B | A | C | A |
| pp1a-1433 | SLINTLNDL | A | C | D | B | pp1a-3886 | KLWAQCVQL | B | B | B | A |
| pp1a-1675 | YLATALLTL | A | A | C | A | pp1a-3917 | VLLSMQGAV | B | A | D | D |
| pp1a-1962 | DLNGDVVAI | A | D | D | D | pp1a-4094 | ALWEIQQVV | B | A | B | A |
| pp1a-2230 | IIWFLLLSV | A | B | C | C | pp1a-4283 | YLASGGQPI | B | C | D | B |
| pp1a-2235 | LLSVCLGSL | A | A | D | D | pp1a-38 | VLSEARQHL | C | A | D | B |
| pp1a-2242 | SLIYSTAAL | A | A | D | B | pp1a-52 | GLVEVEKGV | C | D | C | B |
| pp1a-2968 | YLEGSVRVV | A | B | D | B | pp1a-572 | GISQYSLRL | C | A | D | D |
| pp1a-3047 | IVAGGIVAI | A | D | D | B | pp1a-589 | DLATNNLVV | C | A | D | D |
| pp1a-3344 | SMQNCVLKL | A | B | D | B | pp1a-597 | VMAYITGGV | C | A | D | D |
| pp1a-3467 | VLAWLYAAV | A | A | D | C | pp1a-619 | TVYEKLKPV | C | D | C | B |
| pp1a-3482 | FLNRFTTTL | A | A | C | A | pp1a-685 | FLALCADSI | C | A | D | D |
| pp1a-3583 | LLLTILTSL | A | A | C | A | pp1a-1109 | NLAKHCLHV | C | A | D | B |
| pp1a-3587 | ILTSLLVLV | A | A | C | A | pp1a-1114 | CLHVVGPNV | C | A | D | D |
| pp1a-3639 | FLLPSLATV | A | A | A | A | pp1a-1148 | LLSAGIFGA | C | A | D | B |
| pp1a-3732 | SMWALIISV | A | A | B | A | pp1a-1214 | FITESKPSV | C | A | D | A |
| pp1a-3827 | GLLPPKNSI | A | A | D | B | pp1a-1367 | ILGTVSWNL | C | B | C | B |
| pp1a-3871 | VLLSVLQQL | A | A | C | A | pp1a-1387 | KLMPVCVET | C | B | D | B |
| pp1a-3839 | KLNIKLLGV | A | A | B | B | pp1a-1642 | FLGRYMSAL | C | A | C | B |
| pp1a-4032 | MLFTMLRKL | A | B | D | B | pp1a-2168 | YMPYFFTLL | C | A | B | B |
| pp1a-4183 | ALLSDLQDL | A | B | C | A | pp1a-2228 | NIIIWFLLL | C | A | D | D |
| pp1a-15 | QLSLPVLQV | B | A | D | B | pp1a-2249 | ALGVLMSNL | C | A | D | D |
| pp1a-103 | TLGVLVPHV | B | A | D | B | pp1a-2332 | ILFTRFFYV | C | A | A | A |
| pp1a-214 | TLSEQLDFI | B | A | C | B | pp1a-2348 | QLFFSYFAV | C | A | A | C |
| pp1a-641 | FLRDGWEIV | B | B | D | B | pp1a-2563 | QLMCQPILL | C | A | D | B |
| pp1a-702 | ALNLGETFV | B | A | C | B | pp1a-2884 | FLPRVFSAV | C | A | B | B |
| pp1a-881 | KTLQPVSEL | B | A | D | C | pp1a-2901 | KLIEYTDFA | C | A | C | B |
| pp1a-1143 | VLLAPLLSA | B | A | D | B | pp1a-3122 | FLAHIQWMV | C | A | A | A |
| pp1a-2076 | ILKPANNSL | B | A | D | B | pp1a-3128 | WMVMFTPLV | C | A | B | D |
| pp1a-2226 | LINIIIWFL | B | C | C | D | pp1a-3475 | VINGDRWFL | C | D | C | D |
| pp1a-2270 | YLNSTNVTI | B | A | D | A | pp1a-3662 | RIMTWLDMV | C | A | D | B |
| pp1a-2363 | WLMWLIINL | B | A | B | C | pp1a-3798 | CLLNRYFRL | C | A | B | D |
| pp1a-2364 | LMWLIINLV | B | A | A | D | pp1a-3807 | TLGVYDYLV | C | A | D | C |
| pp1a-2569 | ILLLDQALV | B | B | C | B | pp1a-4266 | VLSFCAFAV | C | A | B | C |
The number within each peptide name shows the first amino acid position in the SARS-CoV-2 pp1a.
Algorithm number 1, SYFPEITHI; number 2, nHLAPred; number 3, ProPred-I; number 4, IEDB. Scores of predicted peptides in the 4 algorithms were assessed by classifying into four ranks (A, excellent; B, very good; C, good; D, poor) as follows: SYFPEITHI, A ≥ 27, 24 ≤ B ≤ 26, 20 ≤ C ≤ 23, D < 20; nHLAPred, A = 1, 0.90 ≤ B <1, 0.5 ≤ C < 0.90, D < 0.5; ProPred-I, A ≥ 1000, B ≥ 500, C ≥ 100, D = ND (not detected); IEDB, A > 0.1, 0.1 ≤ B < 0.5, 0.5 ≤ C < 1, D ≥ 1).
TABLE 2.
Binding affinities of predicted SARS-CoV-2 pp1a peptides to HLA-A*02:01
| Peptide namea | Sequence | BL50b | Namea | Sequence | BL50b |
|---|---|---|---|---|---|
| High binders | |||||
| pp1a-84 | VMVELVAEL | 6.8 ± 0.3 | pp1a-103 | TLGVLVPHV | 1.3 ± 0.2 |
| pp1a-214 | TLSEQLDFI | 2.0 ± 0.2 | pp1a-445 | GLNDNLLEI | 6.0 ± 0.2 |
| pp1a-597 | VMAYITGGV | 9.5 ± 0.9 | pp1a-619 | TVYEKLKPV | 5.6 ± 0.7 |
| pp1a-641 | FLRDGWEIV | 0.9 ± 0.1 | pp1a-1675 | YLATALLTL | 8.4 ± 0.2 |
| pp1a-2270 | YLNSTNVTI | 8.9 ± 0.3 | pp1a-2569 | ILLLDQALV | 9.6 ± 1.5 |
| pp1a-2785 | AIFYLITPV | 5.1 ± 1.6 | pp1a-2968 | YLEGSVRVV | 7.5 ± 0.1 |
| pp1a-3013 | SLPGVFCGV | 3.1 ± 0.2 | pp1a-3115 | YLTNDVSFL | 4.8 ± 0.3 |
| pp1a-3122 | FLAHIQWMV | 6.9 ± 0.4 | pp1a-3128 | WMVMFTPLV | 9.8 ± 1.1 |
| pp1a-3587 | ILTSLLVLV | 8.3 ± 0.9 | pp1a-3710 | TLMNVLTLV | 2.0 ± 0.4 |
| pp1a-3732 | SMWALIISV | 8.9 ± 0.4 | pp1a-4094 | ALWEIQQVV | 6.4 ± 0.4 |
| Medium binders | |||||
| pp1a-15 | QLSLPVLQV | 79.4 ± 4.4 | pp1a-38 | VLSEARQHL | 76.8 ± 6.8 |
| pp1a-52 | GLVEVEKGV | 14.5 ± 4.9 | pp1a-106 | VLVPHVGEI | 76.0 ± 2.0 |
| pp1a-468 | KLNEEIAII | 56.2 ± 4.8 | pp1a-600 | YITGGVVQL | 48.5 ± 7.9 |
| pp1a-685 | FLALCADSI | 10.2 ± 0.5 | pp1a-702 | ALNLGETFV | 40.7 ± 2.8 |
| pp1a-1109 | NLAKHCLHV | 38.5 ± 8.2 | pp1a-1114 | CLHVVGPNV | 62.9 ± 2.3 |
| pp1a-1161 | SLRVCVDTV | 85.7 ± 1.9 | pp1a-1312 | MLAKALRKV | 95.2 ± 0.7 |
| pp1a-2168 | YMPYFFTLL | 99.0 ± 2.4 | pp1a-2332 | ILFTRFFYV | 96.5 ± 14.0 |
| pp1a-2563 | QLMCQPILL | 11.1 ± 1.8 | pp1a-2884 | FLPRVFSAV | 29.3 ± 6.5 |
| pp1a-3047 | IVAGGIVAI | 50.5 ± 2.5 | pp1a-3083 | LLFLMSFTV | 39.4 ± 0.9 |
| pp1a-3183 | FLLNKEMYL | 12.9 ± 6.8 | pp1a-3403 | FLNGSCGSV | 18.7 ± 3.2 |
| pp1a-3467 | VLAWLYAAV | 45.0 ± 10.8 | pp1a-3475 | VINGDRWFL | 85.0 ± 5.4 |
| pp1a-3482 | FLNRFTTTL | 79.0 ± 9.6 | pp1a-3583 | LLLTILTSL | 63.8 ± 3.1 |
| pp1a-3639 | FLLPSLATV | 15.0 ± 4.4 | pp1a-3662 | RIMTWLDMV | 76.5 ± 7.1 |
| pp1a-3753 | FLARGIVFM | 71.2 ± 12.9 | pp1a-3798 | CLLNRYFRL | 81.6 ± 20.2 |
| pp1a-3807 | TLGVYDYLV | 61.7 ± 8.7 | pp1a-3871 | VLLSVLQQL | 44.9 ± 6.0 |
| pp1a-3886 | KLWAQCVQL | 27.2 ± 9.4 | pp1a-4183 | ALLSDLQDL | 42.4 ± 6.1 |
| pp1a-4266 | VLSFCAFAV | 20.1 ± 6.1 | pp1a-4283 | YLASGGQPI | 39.8 ± 6.2 |
| Low binders | |||||
| pp1a-572 | GISQYSLRL | ND | pp1a-589 | DLATNNLVV | ND |
| pp1a-692 | SIIIGGAKL | ND | pp1a-881 | KTLQPVSEL | 161.7 ± 28.3 |
| pp1a-1143 | VLLAPLLSA | ND | pp1a-1148 | LLSAGIFGA | ND |
| pp1a-1214 | FITESKPSV | 122.1 ± 31.7 | pp1a-1367 | ILGTVSWNL | 123.2 ± 11.3 |
| pp1a-1387 | KLMPVCVET | 174.2 ± 33.5 | pp1a-1433 | SLINTLNDL | 123.4 ± 11.0 |
| pp1a-1642 | FLGRYMSAL | 124.1 ± 1.9 | pp1a-1962 | DLNGDVVAI | ND |
| pp1a-2076 | ILKPANNSL | ND | pp1a-2226 | LINIIIWFL | ND |
| pp1a-2228 | NIIIWFLLL | ND | pp1a-2230 | IIWFLLLSV | 176.7 ± 6.3 |
| pp1a-2235 | LLSVCLGSL | ND | pp1a-2242 | SLIYSTAAL | 134.0 ± 7.6 |
| pp1a-2249 | ALGVLMSNL | ND | pp1a-2348 | QLFFSYFAV | 146.4 ± 19.7 |
| pp1a-2363 | WLMWLIINL | 102.7 ± 6.4 | pp1a-2364 | LMWLIINLV | 159.0 ± 42.8 |
| pp1a-2901 | KLIEYTDFA | 126.0 ± 2.5 | pp1a-3344 | SMQNCVLKL | 104.5 ± 2.2 |
| pp1a-3827 | GLLPPKNSI | ND | pp1a-3839 | KLNIKLLGV | 213.5 ± 85.7 |
| pp1a-3917 | VLLSMQGAV | 112.3 ± 7.2 | pp1a-4032 | MLFTMLRKL | 165.8 ± 21.7 |
The number within each peptide name shows the first amino acid position in the SARS-CoV-2 pp1a.
Data of peptide binding assays are shown as BL50, indicating a concentration (µM) of each peptide that yields the 50% relative binding, as described in the Materials and Methods. Experiments were performed in triplicate and repeated twice with similar results. Data are given as mean values ± SD. High binders, BL50 < 10 µM; medium binders, 10 µM ≤ BL50 < 100 µM; low binders, BL50 ≥ 100 µM or ND (not detected).
TABLE 3.
Comparison between the peptide binding affinity and the rank of peptides in the 4 algorithms
| Algorithm | Ranka | No. high binder BL50 < 10 µM (%) | No. medium binder 10 µM ≤ BL50 < 50 µM (%) | No. medium binder 50 µM ≤ BL50 < 100 µM (%) | No. low binder BL50 ≥ 100 µM (%) |
|---|---|---|---|---|---|
| SYFPEITHI | A | 6/82 (7.3) | 5/82 (6.1) | 7/82 (8.5) | 10/82 (12.2) |
| B | 10/82 (12.2) | 6/82 (7.3) | 2/82 (2.4) | 7/82 (8.5) | |
| C | 4/82 (4.9) | 6/82 (7.3) | 8/82 (9.8) | 11/82 13.4) | |
| D | 0/82 (0) | 0/82 (0) | 0/82 (0) | 0/82 (0) | |
| nHLAPred | A | 13/82 (15.9) | 12/82 (14.6) | 13/82 (15.9) | 19/82 (23.2) |
| B | 6/82 (7.3) | 2/82 (2.4) | 2/82 (2.4) | 5/82 (6.1) | |
| C | 0/82 (0) | 2/82 (2.4) | 0/82 (0) | 2/82 (2.4) | |
| D | 1/82 (1.2) | 1/82 (1.2) | 2/82 (2.4) | 2/82 (2.4) | |
| ProPred-I | A | 2/82 (2.4) | 3/82 (3.7) | 1/82 (1.2) | 2/82 (2.4) |
| B | 4/82 (4.9) | 3/82 (3.7) | 2/82 (2.4) | 2/82 (2.4) | |
| C | 6/82 (7.3) | 4/82 (4.9) | 4/82 (4.9) | 5/82 (6.1) | |
| D | 8/82 (9.8) | 7/82 (8.5) | 10/82 (12.2) | 19/82 (23.2) | |
| IEDB | A | 10/82 (12.2) | 6/82 (7.3) | 5/82 (6.1) | 1/82 (1.2) |
| B | 7/82 (8.5) | 6/82 (7.3) | 7/82 (8.5) | 13/82 (15.9) | |
| C | 1/82 (1.2) | 4/82 (4.9) | 1/82 (1.2) | 4/82 (4.9) | |
| D | 2/82 (2.4) | 2/82 (2.4) | 4/82 (4.9) | 10/82 (12.2) |
Peptides were classified into four ranks (A, excellent; B, very good; C, good; D, poor) in each of the four algorithms (SYFPEITHI, nHLAPred, ProPred-I, and IEDB).
Detection of SARS-CoV-2 pp1a-specific CD8+ T cell responses in mice immunized with peptide-encapsulated liposomes.
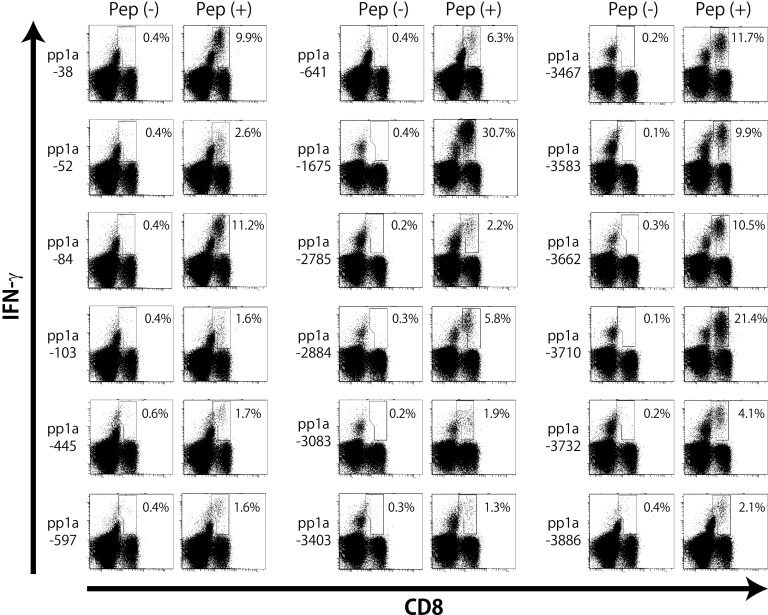
Each of the 54 peptides selected were encapsulated into liposomes as described in the Materials and Methods. HLA-A*02:01 transgenic (HHD) mice (37) were then subcutaneously (s.c.) immunized twice at a 1-week interval with each of peptide-encapsulated liposomes together with CpG adjuvant. One week later, spleen cells of immunized mice were prepared, stimulated in vitro with a relevant peptide for 5 h, and stained for the expression of cell-surface CD8 and intracellular gamma interferon (IFN-γ). As shown in Fig. 2, the intracellular cytokine staining (ICS) assay showed that significant numbers of IFN-γ-producing CD8+ T cells were elicited in mice immunized with 18 liposomal peptides, including pp1a-38, -52, -84, -103, -445, -597, -641, -1675, -2785, -2884, -3083, -3403, -3467, -3583, -3662, -3710, -3732, and -3886, revealing that these 18 peptides are HLA-A*02:01-restricted CTL epitopes derived from SARS-CoV-2 pp1a. As shown in Table 4, high-binding peptides did not always elicit IFN-γ-producing CD8+ T cells (>1% of IFN-γ+ cells in CD8+ T cells). However, the proportion of high-binding peptides that induced IFN-γ-producing CD8+ T cells was higher than that of medium-binding peptides (Table 4). As indicated in Table 5, multiple epitopes were located in small proteins such as nsp1 (180 amino acids [aa]) and nsp6 (290 aa), whereas only one epitope was seen in the large nsp3 composed of 1,945 amino acids. On the other hand, the remaining 36 out of 54 peptides in liposomes were not able to stimulate peptide-specific CTLs in mice (data not shown), demonstrating the necessity to generate data through wet-lab experiments. Interestingly, four epitopes, including pp1a-103, -2884, -3403, and -3467, are also located in the amino acid sequence of SARS-CoV pp1a (Table 5), and pp1a-3467 was previously reported by us in the identification of SARS-CoV-derived CTL epitopes (30). However, none of the18 epitopes are found in the amino acid sequence of either MERS-CoV or the four human common cold coronaviruses, HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1.
FIG 2.
Intracellular IFN-γ staining of CD8+ T cells specific for peptides derived from SARS-CoV-2 pp1a. HHD mice were immunized twice with each of the predicted peptides of SARS-CoV-2 pp1a in liposomes together with CpG. After 1 week, spleen cells were prepared and stimulated with (+) or without (−) a relevant peptide for 5 h. Cells were then stained for their surface expression of CD8 (x axis) and their intracellular expression of IFN-γ (y axis). The numbers shown indicate the percentages of intracellular IFN-γ+ cells within CD8+ T cells. The data shown are representative of three independent experiments. Three to five mice per group were used in each experiment, and the spleen cells of the mice per group were pooled.
TABLE 4.
Correlation between the peptide binding affinity and the peptide immunogenicity
| ICSa | High binder peptides (BL50 < 10 µM) (%) | Medium binder peptides 10 µM ≤ BL50 < 50 µM (%) | Medium binder peptides 50 µM ≤ BL50 < 100 µM (%) |
|---|---|---|---|
| 10% ≤ | 3 (15.0) | 1 (5.9) | 1 (5.9) |
| 1–10% | 6 (30.0) | 5 (29.4) | 2 (11.8) |
| 1% > | 11 (55.0) | 11 (64.7) | 14 (82.4) |
| Total No. | 20 | 17 | 17 |
ICS, intracellular cytokine staining indicates the % of intracellular IFN-γ+ cells in CD8+ T cells.
TABLE 5.
Comparison of amino acid sequences of SARS-CoV-2 pp1a CTL epitopes with the amino acid sequence of SARS-CoV
| SARS-CoV-2 |
SARS-CoV |
||||
|---|---|---|---|---|---|
| Peptide name | Start–End | Sequence | Protein | Sequence | Identity (%) |
| pp1a-38 | 38–46 | VLSEARQHL | nsp1 | ALSEAREHL | 89 |
| pp1a-52 | 52–60 | GLVEVEKGV | nsp1 | GLVELEKGV | 89 |
| pp1a-84 | 84–92 | VMVELVAEL | nsp1 | KVVELVAEM | 67 |
| pp1a-103 | 103–11 | TLGVLVPHV | nsp1 | TLGVLVPHV | 100 |
| pp1a-445 | 445–53 | GLNDNLLEI | nsp2 | TLNEDLLEI | 67 |
| pp1a-597 | 597–605 | VMAYITGGV | nsp2 | IMAYVTGGL | 67 |
| pp1a-641 | 641–9 | FLRDGWEIV | nsp2 | FLKDAWEIL | 67 |
| pp1a-1675 | 1675–83 | YLATALLTL | nsp3 | ||
| pp1a-2785 | 2785–93 | AIFYLITPV | nsp4 | ||
| pp1a-2884 | 2884–92 | FLPRVFSAV | nsp4 | FLPRVFSAV | 100 |
| pp1a-3083 | 3083–91 | LLFLMSFTV | nsp4 | LLFLMSFTI | 89 |
| pp1a-3403 | 3403–11 | FLNGSCGSV | nsp5 | FLNGSCGSV | 100 |
| pp1a-3467 | 3467–75 | VLAWLYAAV | nsp5 | VLAWLYAAV | 100 |
| pp1a-3583 | 3583–91 | LLLTILTSL | nsp6 | ||
| pp1a-3662 | 3662–70 | RIMTWLDMV | nsp6 | RIMTWLELA | 67 |
| pp1a-3710 | 3710–8 | TLMNVLTLV | nsp6 | TLMNVITLV | 89 |
| pp1a-3732 | 3732–40 | SMWALIISV | nsp6 | SMWALVISV | 89 |
| pp1a-3886 | 3886–94 | KLWAQCVQL | nsp7 | ||
Of the 18 positive peptides, 10 peptides, including pp1a-38, -84, -641, -1675, -2884, -3467, -3583, -3662, -3710, and -3732, were selected for the following analyses because of the high ratios of IFN-γ+ cells in CD8+ T cells (Fig. 2).
Identification of dominant CTL epitopes.
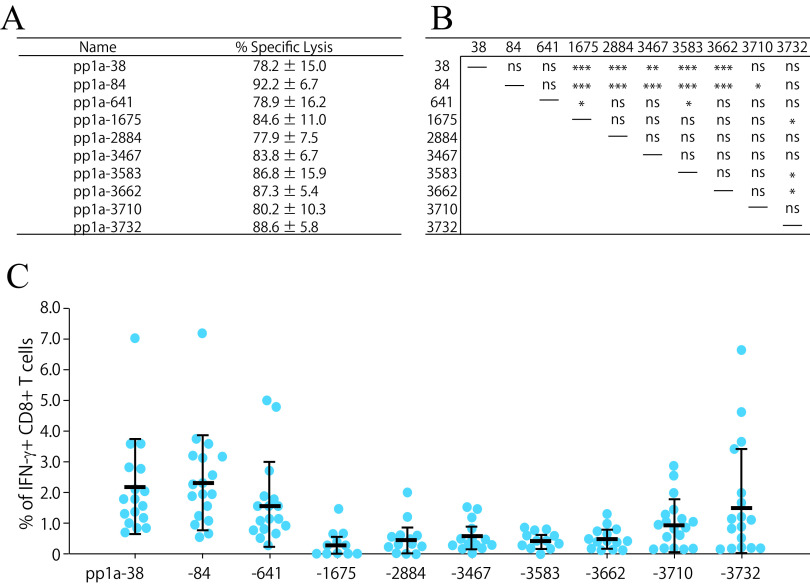
To confirm that the 10 peptides are effective epitopes for peptide-specific CTL responses, we examined whether peptide-specific killing activities were elicited in mice with each of 10 peptides in liposomes. HHD mice were immunized s.c. twice with each of peptide-encapsulated liposomes and CpG adjuvant. One week later, equal numbers of peptide-pulsed and -unpulsed target cells were transferred into immunized mice via intravenous (i.v.) injection, and peptide-specific lysis was analyzed by flow cytometry. In support of the data seen with ICS (Fig. 2), peptide-specific killing was observed in mice immunized with any of 10 liposomal peptides (Fig. 3A).
FIG 3.
Killing activities and IFN-γ+ CD8+ T cell induction by the 10 most active peptides. (A) In vivo killing activities specific for the 10 peptides. HHD mice were immunized twice with either one of the 10 liposomal peptides (pp1a-38, -84, -641, -1675, -2884, -3467, -3583, -3662, 3710, or -3732) or else liposomes alone, together with CpG. One week later, in vivo peptide-specific killing activities were measured. Three to five mice per group were used, and the data of % specific lysis are shown as the mean ± standard deviation (SD). (B and C) Comparison of the 10 peptides in the induction of IFN-γ+ CD8+ T cells. Seventeen HHD mice were immunized once with the mixture of 10 peptides (pp1a-38, -84, -641, -1675, -2884, -3467, -3583, -3662, -3710, and -3732) in liposomes with CpG. After 1 week, spleen cells were stimulated with or without each of the 10 peptides, and intracellular IFN-γ in CD8+ T cells was stained. (C) The y axis indicates the relative percentages of IFN-γ+ cells in CD8+ T cells, which were calculated by subtracting the % of IFN-γ+ cells in CD8+ T cells without a peptide from the % of IFN-γ+ cells in CD8+ T cells with a relevant peptide. Each blue circle represents an individual mouse. Data are shown as the mean (horizontal bars) ± SD. (B) Statistical comparisons of the relative % values of IFN-γ+ CD8+ T cells among the 10 peptides in Fig. 3C were made by one-way ANOVA followed by post hoc tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
We next attempted to identify dominant CTL epitopes among the 10 CTL epitopes. The same amounts of the 10 peptide solutions at an equal concentration were mixed together and encapsulated into liposomes. Seventeen mice were immunized once with the liposomes containing the mixture of 10 peptides. One week later, spleen cells were incubated with each of the 10 peptides for 5 h, and the ICS assay was performed. As shown in Fig. 3B and C, pp1a-38 and -84 were statistically predominant over almost all other peptides in the induction of peptide-specific IFN-γ+ CD8+ T cells. Furthermore, pp1a-641 and pp1a-3732 were significantly superior to pp1a-1675/-3583 and pp1a-1675/-3583/-3662 in the stimulation of IFN-γ-producing CD8+ T cells, respectively (Fig. 3B). One month after vaccination, the same experiments were performed and similar results were obtained (data not shown), suggesting that CD8+ T cells recognizing the different epitopes establish memory to a similar extent.
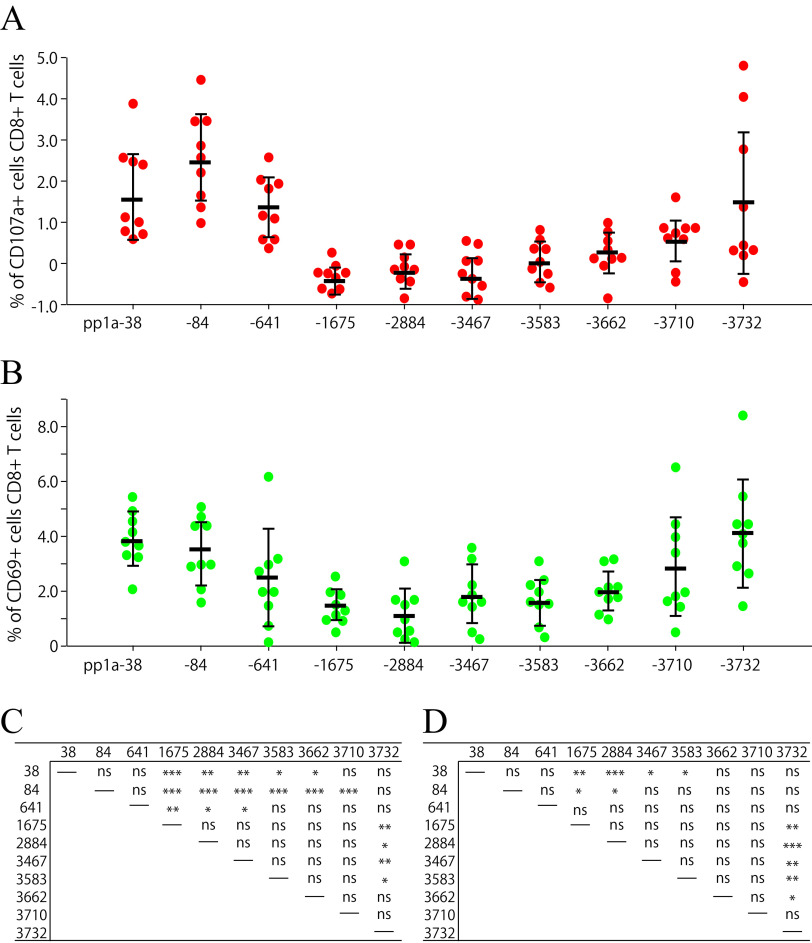
We also examined the peptide-specific induction of CD107a+ CD8+ T cells and CD69+ CD8+ T cells. CD107a and CD69 are markers of degranulation and early activation on CD8+ T lymphocytes, respectively. Nine mice were immunized once with the liposomes encapsulating the mixture of the 10 peptides. After 1 week, spleen cells were stimulated with each peptide and stained for their expression of CD107a or CD69 of CD8+ T cells. At first glance, the graphs of CD107a (Fig. 4A) and CD69 (Fig. 4B) expression were similar to that of IFN-γ expression of CD8+ T cells (Fig. 3C). As shown in Fig. 4A and C, both pp1a-38 and pp1a-84 were superior to almost all other peptides for the CD107a expression of CD8+ T cells. Moreover, pp1a-641 and pp1a-3732 elicited CD107a-positive T cells better than pp1a-1675/-2884/-3467 and pp1a-1675/-2884/-3467/-3583, respectively. In the case of CD69 expression (Fig. 4B and D), pp1a-38 and pp1a-3732 were more dominant than pp1a-1675/-2884/-3467/-3583 and pp1a-1675/-2884/-3467/-3583/-3662, respectively. Further, pp1a-84 was superior to pp1a-1675/-2884.
FIG 4.
Comparison of the 10 peptides in the induction of CD107a+ CD8+ T cells (A) and CD69+ CD8+ T cells (B). Nine HHD mice were immunized once with the mixture of 10 peptides (pp1a-38, -84, -641, -1675, -2884, -3467, -3583, -3662, -3710, and -3732) in liposomes with CpG. After 1 week, spleen cells were stimulated with or without each of the 10 peptides, and the expression of CD107a (A) or CD69 (B) on CD8+ T cells was analyzed. Data indicate the relative percentages of CD107a+ (A) and CD69+ (B) cells in CD8+ T cells, which were obtained by subtracting the % of CD107a+ and CD69+ cells in CD8+ T cells without a peptide from the % of CD107a+ and CD69+ cells in CD8+ T cells with a peptide, respectively. Each red (A) or green (B) circle represents an individual mouse. Data are shown as the mean (horizontal bars) ± SD. (C and D) Statistical analyses of the data among the 10 peptides in A and B were performed by one-way ANOVA followed by post hoc tests for CD107a (C) and CD69 (D) data: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Taken together, the 10 peptides differed significantly in their ability to induce SARS-CoV-2 pp1a-specific CTLs when mice were immunized with the mixture of 10 peptides in liposomes. Thus, some peptides were found to be dominant CTL epitopes despite that each peptide alone had the capability to efficiently activate peptide-specific CTL (Fig. 2, Fig. 3A).
Existence of an immunodominance hierarchy.
The data in Fig. 5 indicate reactivity to the 10 peptides in each of 15 mice immunized with the mixture of the 10 peptides in liposomes. Each graph represents reactivity of an individual mouse (Fig. 5). Unexpectedly, all mice did not show the same reaction pattern against the 10 peptides. It appears there were roughly three types of reaction patterns to the 10 peptides. Type A is the group of mice in which pp1a-38, -84, -641-specific IFN-γ+ CD8+ T cells were predominantly elicited, whereas the remaining 7 peptides were not able to activate peptide-specific IFN-γ+ CD8+ T cells. In the case of type B, pp1a-3732 stimulated peptide-specific IFN-γ+ CD8+ T cells as well as pp1a-38, and -84. In addition to these three peptides, several other peptides also induced IFN-γ+ CD8+ T cells in type C mice. These data suggest there seems to be three patterns of immunodominance hierarchy in CD8+ T cell responses following peptide vaccination. The immunodominance hierarchy may provide us more variations for designing CTL-based COVID-19 vaccines.
FIG 5.
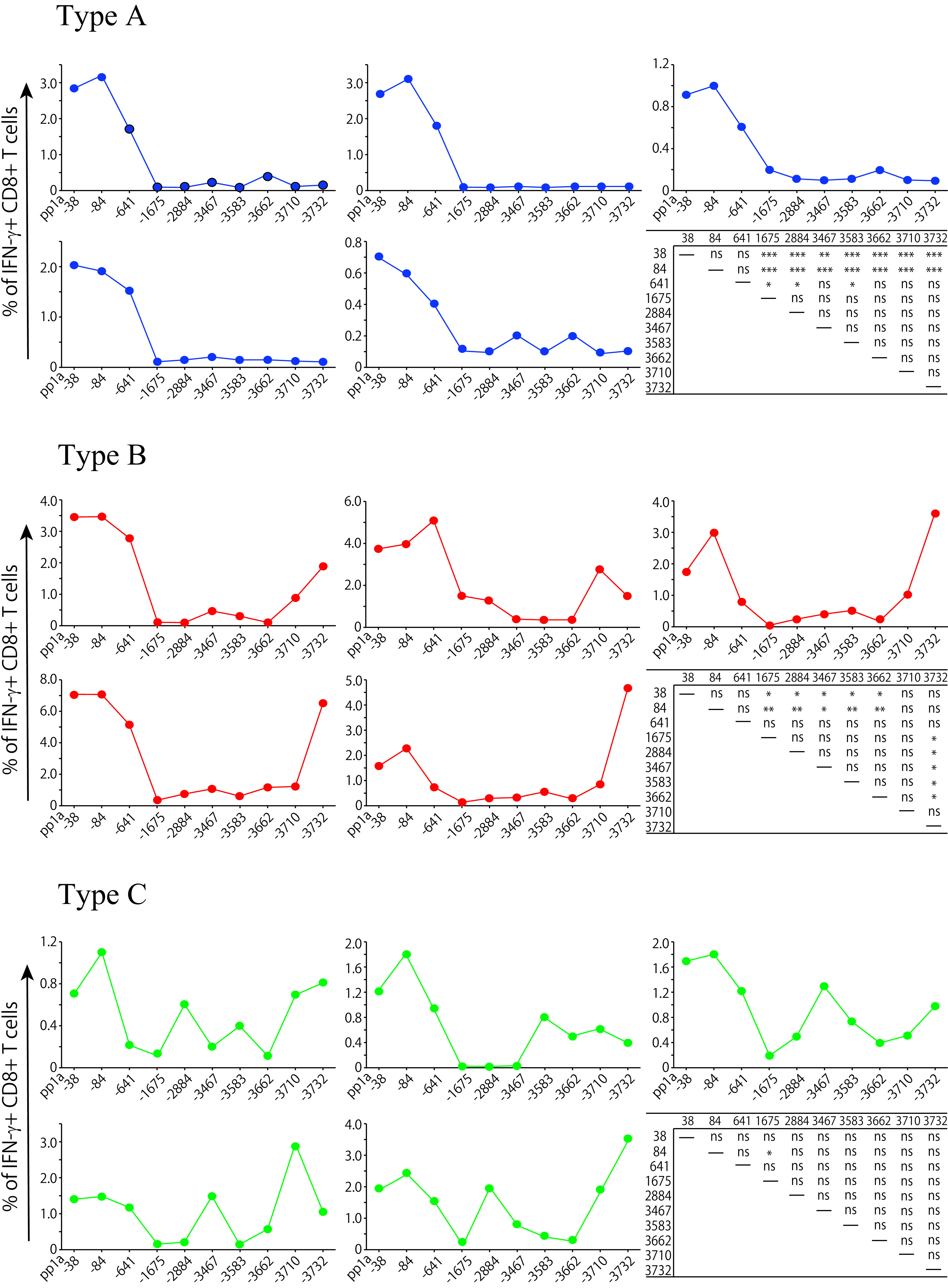
Three types of reactivity in mice immunized with the mixture of 10 peptides. Fifteen mice were immunized once with the mixture of 10 peptides, including pp1a-38, -84, -641, -1675, -2884, -3467, -3583, -3662, -3710, and -3732, in liposomes with CpG. After 1 week, spleen cells were stimulated with or without each of the 10 peptides, and intracellular IFN-γ in CD8+ T cells was stained. Based on the reactivity pattern to the 10 peptides, 15 mice were divided into three types, A, B, and C. Each graph represents reactivity of an individual mouse. The y axis indicates the relative percentage of IFN-γ+ cells in CD8+ T cells, which was calculated by subtracting the % of IFN-γ+ cells in CD8+ T cells without a peptide from the % of IFN-γ+ cells in CD8+ T cells with a relevant peptide. Statistical analyses of the relative % values to 10 peptides in each type were performed by one-way ANOVA followed by post hoc tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
DISCUSSION
After the epidemics of SARS and MERS, scientists have not yet succeeded in developing preventive or therapeutic vaccines available to avoid reemergence of them. In the SARS and MERS vaccine development, the full-length S protein or its S1 subunit have frequently been used as an antigen to produce anti-RBD neutralizing antibodies. However, these vaccine candidates provided partial protection against virus challenge in animal models, accompanied by safety concerns such as ADE (1). Furthermore, antibody responses to coronaviruses rapidly wane following infection or immunization (23, 24, 26). Considering the above, it should be necessary to consider CTL-based vaccines against SARS-CoV-2 to provide robust, long-lived T cell memory, although neutralizing antibody responses are a primary vaccine target.
In the current study, we aimed to find HLA-A*02:01-resctricted CTL dominant epitopes derived from SARS-CoV-2. Dominant epitopes induce a strong immune response to eliminate a certain pathogen quickly and effectively, and also contribute to the memory T cell pool. We focused on pp1a of SARS-CoV-2 to find CTL epitopes because pp1a is the largest and best-conserved polyprotein among the constituent proteins. Further, pp1a is produced earlier than structural proteins, suggesting that pp1a-specific CTLs can eliminate infected cells before the formation of mature virions. To predict CTL epitopes, we utilized bioinformatics to select 82 peptides with high scores in four kinds of computer-based programs (Table 2). The value of each peptide in the four programs was assessed by classifying into four ranks. However, evaluations of each peptide did not always match in the four algorithms (Table 1), suggesting that multiple programs are needed for predicting epitopes. In the evaluation of peptide binding, 54 peptides showed high or medium binding affinities to HLA-A*02:01 molecules, whereas the remaining 28 peptides displayed low binding affinities or no binding (Table 2). Compared with the other three programs, the IEDB program seems to be most useful for the selection of peptides with high binding affinity (Table 3). Out of the 54 peptides, only 18 peptides were found to be CTL epitopes. Hence, we have to keep in mind that currently available algorithms have a limited ability to accurately predict CTL epitopes, although the bioinformatics approach is useful for quickly predicting a number of epitopes in a large protein (38, 39).
Among the 18 epitopes that we have identified in the current study, 4 epitopes, including pp1a-103, -2884, -3403, and -3467 are present in the amino acid sequence of SARS-CoV pp1a (100% identity) (Table 5). Therefore, CTLs induced by these four epitopes could work fine for the clearance of SARS-CoV as well as SARS-CoV-2. In support of these data, Le Bert et al. reported that long-lasting memory T cells in SARS-recovered individuals cross-reacted to the N protein of SARS-CoV-2 (40). Recently, several studies found that SARS-CoV-2-reactive CD4+ and CD8+ T cells were detected in a substantial proportion of healthy donors who have never been infected with SARS-CoV-2 or SARS-CoV (40–44). It is most likely that these individuals were previously infected with one of the four human coronaviruses (HCoV-229E, -NL63, -OC43, and -HKU1) that cause seasonal common cold. Nelde et al. demonstrated that the amino acid sequences of several SARS-CoV-2 T cell epitopes recognized by unexposed individuals are similar to some amino acid sequences in the four seasonal common cold human coronaviruses, with identities ranging from 10% to 89% (not 100% identity) (44). However, no one has shown evidence that people with this cross-reactivity are less susceptible to COVID-19. It may also be possible to assume that preexisting T cell immunity might be detrimental through mechanisms such as original antigenic sin or ADE (45). In the current data, none of the 18 epitopes was found in the amino acid sequences of the four human common cold coronaviruses, suggesting that effective common CTL epitopes derived from pp1a, if any, might be very few.
Here, we focused on CTL epitopes restricted by HLA-A*02:01, which is the most common HLA class I allele in the world, and used highly reactive HLA-A*02:01 transgenic mice, termed HHD mice (37). Although we can use lymphocytes of SARS-CoV-2-infected individuals to identify CTL epitopes, there are two main advantages to using HHD mice. First, a large amount of blood of patients is required for examining many candidates of CTL epitopes, but any number of mice can be prepared for this purpose. Second, when using patients’ lymphocytes, we are only testing whether the peptide candidates are recognized by memory CTLs. When using naive mice, however, we can find whether the epitope candidates are able to prime peptide-specific CTLs, which may be a better criterion to judge them as vaccine antigens. It is supposed that the efficient epitope for CTL recognition is not always efficient for CTL priming. Using HLA-A*02:01 transgenic mice, we have identified 18 CTL candidate epitopes derived from SARS-CoV-2 pp1a (Table 5). However, we should take into account that the immunogenic variation in HLA class I transgenic mice may not be identical to that in humans because the antigen processing and presentation differ between them. In addition, there is no data showing that a viral infection in a mouse model induces T cells targeting these epitopes because liposomal peptides were used as an immunogen. Therefore, there is no validation that the candidate epitopes identified here are actually presented during live infection of human cells or that T cells from COVID-19 patients recognize these epitopes. Recently, four epitopes with the same amino acid sequences as pp1a-641, -3403, -3467, and -3886 (Table 5) have been submitted to the Virus Pathogen Database and Analysis Resource (ViPR) by the Adaptive Biotechnologies Corporation (ImmuneCODE-Release001.1). Because these epitopes were tested in an ELISPOT-like assay using human CD8+ T cells and lymphocytes of COVID-19 patients, these four epitopes should be real CTL epitopes that are presented by human cells and recognized by human CD8+ T cells during the infection with SARS-CoV-2. In other words, the remaining 14 epitopes in Table 5 represent new candidate epitopes that have not been previously identified.
In previous studies, we used peptide-linked liposomes as an immunogen (30). The surface-linked liposomal peptides were effective for peptide-specific CTL induction in mice. However, attaching peptides to the surface of liposomes followed by purifying them through the column is a fairly complicated process and time consuming. In the current study, peptide-encapsulated liposomes were used as an immunogen. In contrast to the peptide-linked liposomes, the peptide-encapsulated liposomes are prepared by just mixing liposomes and the peptide. In addition, the peptide-encapsulated liposomes are able to prime peptide-specific CTLs in mice as efficiently as the peptide-linked liposomes. Liposome itself, consisting of lipid bilayers, is a very safe material for humans. Therefore, the peptide-encapsulated liposomes are considered to be promising as a CTL-based vaccine candidate.
Understanding the mechanism of immunodominance is obviously important for the development of effective vaccines. When mice were immunized with liposomes containing the mixture of 10 peptides, it was found that some peptides induced peptide-specific CTLs stronger than other peptides (Fig. 3, Fig. 4). In this case, these peptides were not subjected to proteolysis, or editing by chaperones such as Tapasin in antigen-presenting cells, allowing for true selection, and, therefore, it should be noted that the current data do not show the true immunodominance. As shown in Fig. 3 and Fig. 4, pp1a-38 and -84 are considered to be relatively dominant in comparison with other peptides. In general, dominant epitope-specific CTLs are activated sooner and proliferate faster than subdominant epitope-specific CTLs. This immunodominance may be associated with the affinity of peptide to MHC-I molecules and the affinity of T-cell receptor (TCR) to the peptide-MHC-I complex. Table 4 may provide evidence that the peptide-binding affinity to MHC-I is related to the immunodominance of CTL epitopes. As shown in Table 2, the peptide affinity of pp1a-84 to HLA-A*02:01 is very high (BL50 = 6.8 µM), while pp1a-38 is a medium binder (BL50 = 76.8 µM). Interestingly, the peptide affinity of pp1a-38 is lowest among the 10 peptides selected (Table 2). These data suggest that the affinity of TCR to the peptide-MHC-I complex is critical for CTL immunodominance. In the selection of antigenic epitopes for the CTL-based vaccine against SARS-CoV-2, dominant epitopes such as pp1a-38 and -84 should be chosen because they produce strong CTL response to eliminate virus-infected cells effectively. However, the immunological pressure exerted by dominant epitopes may allow the epitope sequences of SARS-CoV-2 to be mutated and, therefore, a vaccine containing multiple antigenic epitopes should be recommended for a successful COVID-19 vaccine.
It was surprising that all of the genetically identical mice did not show the same reactive pattern when they were immunized with liposomes containing the mixture of 10 peptides (Fig. 5). There were roughly three patterns in CD8+ T cell responses following peptide vaccination with the 10 peptides (Fig. 5). The differences among the three types might be explained by the timing of CTL expansion. In the type A, dominant peptides pp1a-38, -84, and -641 presumably activated T cells more efficiently than the other peptides and, hence, dominant peptide-specific CTLs proliferated faster and curtailed the expansion of CTLs specific for the other peptides. In the type B, it is likely that the expansion of dominant CTLs specific for pp1a-38 and -84 was delayed for some reason compared to that in type A, and thereby subdominant CTLs specific for pp1a-3732 could afford to expand. It is also thought that even nondominant CTLs proliferated because the expansion of both dominant CTLs and subdominant CTLs in the type C was later than that in the type B. Although it is difficult to explain what caused differences in the timing of CTL expansion among the three reaction types, it should be important to find out what causes these differences for the development of CTL-based peptide vaccines.
In summary, we have identified 18 kinds of HLA-A*02:01-restricted CTL epitopes derived from pp1a of SARS-CoV-2 using computational algorithms, HLA-A*02:01 transgenic mice, and peptide-encapsulated liposomes. Out of 18 epitopes, we have also found some dominant CTL epitopes such as pp1a-38 and -84. In the process of finding dominant epitopes, we showed the existence of an immunodominance hierarchy in CD8+ T cell responses to these epitopes. These data may provide important information for further studies of T cell immunity in COVID-19 and the development of preventive and/or therapeutic CTL-based vaccines against SARS-CoV-2.
MATERIALS AND METHODS
Prediction of CTL epitopes.
Four computer-based programs, including SYFPEITHI (31), nHLAPred (32), ProPred-I (33), and IEDB (34) were used to predict HLA-A*02:01-restricted CTL epitopes derived from pp1a of SARS-CoV-2 (GenBank accession numbers LC528232.1 and LC528233.1). As shown in Table 1, 82 peptides with superior scores were selected and were synthesized by Eurofins Genomics (Tokyo, Japan). Two control peptides, FMP58-66 (sequence: GILFGVFTL) (35) and HIV-pol476-484 (sequence: ILKEPVHGV) (36), were synthesized as well.
Mice.
We used the HLA-A*02:01 transgenic and H-2Db−/− β2m−/− double knockout mice (37) that express transgenic HLA-A*02:01 monochains, designated HHD, in which human β2m is covalently linked to a chimeric heavy chain composed of HLA-A*02:01 (α1 and α2 domains) and H-2Db (α3, transmembrane, and cytoplasmic domains). Six- to ten-week-old mice were used for all experiments. Mice were housed in appropriate animal care facilities at Saitama Medical University and were handled according to the international guidelines for experiments with animals. This study was approved by the Animal Research Committee of Saitama Medical University.
Cell lines.
RMA-S-HHD is a TAP2-deficient mouse lymphoma cell line, RMA-S (H-2b), transfected with the HHD gene (37). RMA-S-HHD was cultured in RPMI 1640 medium (Nacalai Tesque Inc., Kyoto, Japan) with 10% fetal calf serum (FCS) (Biowest, Nuaille, France) and 500 µg/ml G418 (Nacalai Tesque Inc.).
Peptide binding assay.
Binding of peptides to the HLA-A*02:01 molecule was measured using RMA-S-HHD cells, as previously described (46). Briefly, RMA-S-HHD cells were precultured overnight at 26°C in a CO2 incubator and then pulsed with each peptide at various concentrations ranging from 0.01 µM to 100 µM for 1 h at 26°C. After 3 h of incubation at 37°C, peptide-pulsed cells were stained with anti-HLA-A2 monoclonal antibody, BB7.2 (47), followed by fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG antibody (Sigma-Aldrich, St. Louis, MO). Mean fluorescence intensity (MFI) of HLA-A2 expression on the surface of RMA-S-HHD cells was measured by flow cytometry (FACSCanto II, BD Biosciences, Franklin Lakes, NJ), and standardized as the percent cell surface expression by the following formula: % relative binding = ({[MFI of cells pulsed with each peptide] − [MFI of cells incubated at 37°C without a peptide]}/{[MFI of cells incubated at 26°C without a peptide] − [MFI of cells incubated at 37°C without a peptide]}) × 100. The concentration of each peptide that yields the 50% relative binding was calculated as the half-maximal binding level (BL50). FMP58-66 and HIV-pol476-484 were used as control peptides.
Peptide-encapsulated liposomes.
Peptide-encapsulated liposomes were prepared using Lipocapsulater FD-U PL (Hygieia BioScience, Osaka, Japan) according to the manufacturer’s instructions with a slight modification. In brief, each of 54 synthetic peptides was dissolved in dimethyl sulfoxide (DMSO) at a final concentration of 10 mM. For the first screening, the same amounts of 4 to 5 kinds of 10 mM peptides were mixed to make a total 100 µl, which was then diluted with 1.9 ml of H2O. For the second screening, 20 µl of each peptide at 10 mM was diluted to 2 ml with H2O. For the identification of dominant epitopes among the 10 peptides selected, 20 µl of each of the 10 peptide solutions at a concentration of 10 mM was mixed together, and the total volume was increased to 2 ml by adding 1.8 ml of H2O. The peptide solution was added into a vial of Lipocapsulater containing 10 mg of dried liposomes and incubated for 15 min at room temperature. The resultant solution contained peptide-encapsulated liposomes.
Immunizations.
Mice were immunized s.c. once or twice at a 1-week interval with peptide-encapsulated liposomes (100 µl/mouse) together with CpG-ODN (5002: 5′-TCCATGACGTTCTTGATGTT-3′; Hokkaido System Science, Sapporo, Japan) (5 µg/mouse) in the footpad.
Intracellular cytokine staining.
Intracellular cytokine staining (ICS) was performed as previously described (30). In brief, after 1 wk or 1 month following immunization, spleen cells were incubated with 50 µM of each peptide for 5 h at 37°C in the presence of brefeldin A (GolgiPlug, BD Biosciences) and were stained with FITC-conjugated anti-mouse CD8 monoclonal antibody (MAb) (BioLegend, San Diego, CA). Cells were then fixed, permeabilized, and stained with phycoerythrin (PE)-conjugated rat anti-mouse IFN-γ MAb (BD Biosciences). After washing the cells, flow cytometric analyses were performed using flow cytometry (FACSCanto II, BD Biosciences).
Detection of CD107a and CD69 molecules on CD8+ T cells.
For the detection of CD107a, spleen cells of immunized mice were incubated with 50 µM of each peptide for 6 h at 37°C in the presence of monensin (GolgiStop, BD Biosciences) and 0.8 µg of FITC-conjugated anti-mouse CD107a MAb (BioLegend). Cells were then stained with PE-Cy5-conjugated anti-mouse CD8 MAb (BioLegend). For examination of the CD69 marker, spleen cells of immunized mice were stimulated with 50 µM of each peptide for 4 h at 37°C. Cells were then stained with PE-conjugated anti-mouse CD69 MAb (BioLegend) and FITC-conjugated anti-mouse CD8 MAb. Stained cells were analyzed by flow cytometry (FACSCanto II, BD Biosciences).
In vivo CTL assay.
The in vivo CTL assay was carried out as previously described (46). In brief, spleen cells from naive HHD mice were equally split into two populations. One population was pulsed with 50 µM of a relevant peptide and labeled with a high concentration (2.5 µM) of carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes, Eugene, OR). The other population was unpulsed and labeled with a lower concentration (0.25 µM) of CFSE. An equal number (1 × 107) of cells from each population was mixed together and adoptively transferred i.v. into mice that had been immunized once with a liposomal peptide. Sixteen hours later, spleen cells were prepared and analyzed by flow cytometry. To calculate specific lysis, the following formula was used: % specific lysis = (1 − {[number of CFSElow cells in normal mice]/[number of CFSEhigh cells in normal mice]}/{[number of CFSElow cells in immunized mice]/[number of CFSEhigh cells in immunized mice]}) × 100.
Statistical analyses.
One-way ANOVA followed by post hoc tests were performed for statistical analyses among multiple groups using GraphPad Prism 5 software (GraphPad software, San Diego, CA). A value of P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
The authors are grateful to François A. Lemonnier (Pasteur Institute, Paris, France) for providing HHD mice and RMA-S-HHD cells. This work was supported by a Grant-in-Aid for Scientific Research (C) (JSPS KAKENHI grant number JP18K06631) to M.M., a Grant-in-Aid for Early-Career Scientists (JSPS KAKENHI grant number JP18K15430) to A.T. the from Japan Society for the Promotion of Science, and a Grant from the Ochiai Memorial Award 2018 to A.T.
The authors have no conflicting financial interests.
REFERENCES
- 1.Sariol A, Perlman S. 2020. Lessons for COVID-19 immunity from other coronavirus infections. Immunity 53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srinivasan S, Cui H, Gao Z, Liu M, Lu S, Mkandawire W, Narykov O, Sun M, Korkin D. 2020. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses 12:360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, Jia X, Nicholson S, Catton M, Cowie B, Tong SYC, Lewin SR, Kedzierska K. 2020. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med 26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, Li J, Gong X, Lou X, Shi W, Wu D, Zhang H, Zhu L, Deng W, Li Y, Lu J, Li C, Wang X, Yin W, Zhang Y, Qin C. 2020. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guebre-Xabier M, Patel N, Tian J-H, Zhou B, Maciejewski S, Lam K, Portnoff AD, Massare MJ, Frieman MB, Piedra PA, Ellingsworth L, Glenn G, Smith G. 2020. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine 38:7892–7896. doi: 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh EE, Frenck R, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, TüReci Z, Tompkins KR, Lyke KE, Raabe V, Dormitzer PR, Jansen KU, Şahin U, Gruber WC. 2020. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Ry AV, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, Alter G, Andersen H, Lewis MG, Barouch DH. 2020. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, Chen RE, Winkler ES, Wessel AW, Case JB, Kashentseva E, McCune BT, Bailey AL, Zhao H, VanBlargan LA, Dai Y-N, Ma M, Adams LJ, Shrihari S, Danis JE, Gralinski LE, Hou YJ, Schäfer A, Kim AS, Keeler SP, Weiskopf D, Baric RS, Holtzman MJ, Fremont DH, Curiel DT, Diamond MS. 2020. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell 183:169–184. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki A, Yang Y. 2020. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, Lanzavecchia A, Corti D, Virgin HW. 2020. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 13.Weingart H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, Smith G, Jones S, Proulx R, Deschambault Y, Grudeski E, Andonov A, He R, Li Y, Copps J, Grolla A, Dick D, Berry J, Ganske S, Manning L, Cao J. 2004. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol 78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, Peters CJ, Couch RB. 2012. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, Chen KH, Liu FT, Liu WT, Chen YM, Huang JC. 2014. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun 451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, Wu T, Cheung K-W, Chan K-H, Alvarez X, Qin C, Lackner A, Perlman S, Yuen K-Y, Chen Z. 2019. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hägglöf T, Oliveira TY, Viant C, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP, Jr, Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC. 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juno JA, Tan H-X, Lee WS, Reynaldi A, Kelly HG, Wragg K, Esterbauer R, Kent HE, Batten J, Mordant FL, Gherardin NA, Pymm P, Dietrich MH, Scott NE, Tham W-H, Godfrey DI, Subbarao K, Davenport MP, Kent SJ, Wheatley AK. 2020. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med 26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 19.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici A, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. 2020. Cross-neutralization of SARS-COV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JF-W, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen K-W, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD. 2020. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 21.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo Y-M, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe JE, Jr. 2020. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ledford H. 2020. Antibody therapies could be a bridge to a coronavirus vaccine—but will the world benefit? Nature 584:333–334. doi: 10.1038/d41586-020-02360-y. [DOI] [PubMed] [Google Scholar]
- 23.Cao W-C, Liu W, Zhang P-H, Zhang F, Richardus JH. 2007. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med 357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 24.Mo H, Zeng G, Ren X, Li H, Ke C, Tan Y, Cai C, Lai K, Chen R, Chan-Yeung M, Zhong N. 2006. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 11:49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, Tan YJ. 2016. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seow J, Graham C, Merrick B, Acors S, Steel KJA, Hemmings O, O’Bryne A, Kouphou N, Pickering S, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Temperton N, Snell L, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, O’Connell L, O’Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Edgeworth JD, Neil SJD, Malim MH, Doores KJ. 2020. Logitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Liu Y, Wang G, Yuan Z, Feng Z, Zhang Y, Wu Y, Chen Y. 2020. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A, Yale IMPACT Team. 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohyama S, Ohno S, Suda T, Taneichi M, Yokoyama S, Mori M, Kobayashi A, Hayashi H, Uchida T, Matsui M. 2009. Efficient induction of cytotoxic T lymphocytes specific for severe acute respiratory syndrome (SARS)-associated coronavirus by immunization with surface-linked liposomal peptides derived from a non-structural polyprotein 1a. Antiviral Res 84:168–177. doi: 10.1016/j.antiviral.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 32.Bhasin M, Raghava GPS. 2007. A hybrid approach for predicting promiscuous MHC class I restricted T cell epitopes. J Biosci 32:31–42. doi: 10.1007/s12038-007-0004-5. [DOI] [PubMed] [Google Scholar]
- 33.Singh H, Raghava GPS. 2003. ProPred1: prediction of promiscuous MHC class-I binding sites. Bioinformatics 19:1009–1014. doi: 10.1093/bioinformatics/btg108. [DOI] [PubMed] [Google Scholar]
- 34.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B. 2019. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res 47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. 1987. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature 326:881–882. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 36.Tsomides TJ, Walker BD, Eisen HN. 1991. An optimal viral peptide recognized by CD8+ T cells binds very tightly to the restricting class I major histocompatibility complex protein on intact cells but not to the purified class I protein. Proc Natl Acad Sci U S A 88:11276–11280. doi: 10.1073/pnas.88.24.11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Pérarnau B. 1997. HLA-A2.1-restricted education and cytolytic activity of CD8+ T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med 185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. 2020. A sequence homology and bioinformatics approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 27:671–680. 2020. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ong E, Wong MU, Huffman A, He Y. 2020. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol 11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI, Wang LF, Ooi EE, Kalimuddin S, Tambyah PA, Low JG, Tan YJ, Bertoletti A. 2020. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 41.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, Antunes RS, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D. 2020. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, Baysal E, Mangold M, Henze L, Lauster R, Mall MA, Beyer K, Röhmel J, Voigt S, Schmitz J, Miltenyi S, Demuth I, Müller MA, Hocke A, Witzenrath M, Suttorp N, Kern F, Reimer U, Wenschuh H, Drosten C, Corman VM, Giesecke-Thiel C, Sander LE, Thiel A. 2020. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 44.Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lübke M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Traenkle B, Kaiser PD, Othbauer U, Becker M, Junker D, Krause G, Strengert M, Schneiderhan-Marra N, Templin MF, Joos TO, Kowalewski DJ, Stos-Zweifel V, Fehr M, Graf M, Gruber L-C, Rachfalski D, Preuß B, Hagelstein I, Märklin M, Bakchoul T, Gouttefangeas C, Kohlbacher O, Klein R, Stevanović S, Rammensee H-G, Walz JS. 2020. SARS-CoV-2-epitopes define heterologous and COVID-19-induced T-cell recognition. Nat Immunol doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 45.Sette A, Crotty S. 2020. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol 20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsui M, Kawano M, Matsushita S, Akatsuka T. 2014. Introduction of a point mutation into an HLA class I single-chain trimer induces enhancement of CTL priming and antitumor immunity. Mol Ther Methods Clin Dev 1:14027. doi: 10.1038/mtm.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parham P, Brodsky FM. 1981. Partial purification and some properties of BB7.2 a cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum Immunol 3:277–299. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]