Abstract
Introduction
MiRNAs have been proven to modulate the progression of gastric cancer (GC). In this field, we evaluated the role and mechanism of miR-140-3p in GC.
Methods
Western blotting and qRT-PCR were used to detect the levels of miR-140-3p and BCL2. The interaction of miR-140-3p and BCL2 was confirmed by dual-luciferase reporter and miRNA pull-down assays. CCK-8, EdU, wound healing, and Transwell invasion assays were performed to evaluate cell proliferation, migration and invasion. Autophagy was analyzed using Western blot analysis of the LC3-II/I ratio and immunofluorescence staining. A xenograft model was established to reveal the role of miR-140-3p in tumorigenesis.
Results
In GC cell lines and tissues, miR-140-3p was highly expressed, and BCL2 was expressed at low levels. MiR-140-3p directly inhibited BCL2 expression and indirectly promoted BECN1 expression, and BCL2 inhibited BECN1 expression. MiR-140-3p overexpression or silencing restrained or facilitated migration, invasion and EMT in GC cells. Moreover, we noticed that overexpression or downregulation of miR-140-3p promoted or suppressed BECN1-dependent autophagy in GC cells. BCL2 introduction or BECN1 silencing in GC cells partially blocked the effects of miR-140-3p. In conclusion, miR-140-3p directly downregulated the expression of BCL2, BCL2 downregulation further activated BECN1-dependent autophagy, and autophagy activation further inhibited EMT.
Conclusion
miR-140-3p may act as a tumor suppressor by targeting BCL2 and regulating downstream BECN1-induced autophagy and metastasis in GC progression.
Keywords: miR-140-3p, gastric cancer, BCL2, BECN1, autophagy, EMT
Introduction
In 2018, there were 100,000 new cases of gastric cancer (GC) and 783,000 deaths worldwide.1 Genetics and diet are acknowledged risk factors for GC.2,3 Recurrence and metastasis occur in more than half of patients who undergo radical gastrectomy.4 A low BECN1 level is related to a poor prognosis for GC, which suggests autophagy as a potential antitumor target.5,6 Therefore, investigation of autophagy-related mechanisms is a new direction for the treatment of GC.
MicroRNAs (miRNAs) regulate the crosstalk between autophagy and metastasis in GC progression.7,8 MiR-140-3p, located on human chromosome 16q22.1, has been proven to inhibit the progression and metastasis of various cancers.9–13 A study reported that miR-140-3p inhibits inflammation and oxidative stress but enhances autophagy.14 However, the exact role of miR-140-3p in GC is still unknown.
The B-cell lymphoma 2 (BCL2) gene was originally described as an oncogene in follicular lymphoma and has been shown to inhibit cell apoptosis.15 A previous study observed that BCL2 inhibits BECN1-mediated autophagy and autophagic cell death.16 BCL2 is regulated by various microRNAs in many human cancers.17–19 MiR-140-3p directly targets BCL2 in aortic smooth muscle cells.20 However, whether miR-140-3p affects metastasis and autophagy in GC through the BCL2/BECN1 pathway has not yet been studied.
In this research, our team discovered that miR-140-3p was downregulated in GC cells and tissues. BCL2 was proven to be a target of miR-140-3p and involved in miR-140-3p-regulated cellular processes. In addition, the protein levels of BECN1 were negatively and positively regulated by BCL2 and miR-140-3p, respectively. Silencing BECN1 reversed the miR-140-3p-induced inhibition of cell proliferation, migration, invasion and EMT. In conclusion, the data suggested that miR-140-3p restrains GC progression and metastasis by inducing BCL2/BECN1-mediated autophagy.
Materials and Methods
Gastric Cancer Tissue Specimens
GC samples (n=55) and matched normal samples were taken from patients who agreed to be including in the experiments, and experiments were authorized by the Ethics Committee of the People’s Hospital of Jingjiang, and all procedures were performed in accordance with the Declaration of Helsinki. The samples were collected from January 2015 to the end of December 2016. All written informed consents were obtained from patients. At least two pathologists confirmed the pathological diagnosis of GC. The clinicopathological characteristics of the patients are shown in Table 1.
Table 1.
Correlations Between Target Gene Expression and Clinicopathological Features of GC Patients
| Characteristics | No. of Patients | miR-140-3p Level | P | BCL2 mRNA Level | P | ||
|---|---|---|---|---|---|---|---|
| n=55 | High (22) | Low (33) | High (25) | Low (30) | |||
| Sex | 0.913 | 0.101 | |||||
| Female | 22 | 9 | 13 | 7 | 15 | ||
| Male | 33 | 13 | 20 | 18 | 15 | ||
| Age (years) | 0.829 | 0.664 | |||||
| <60 | 29 | 12 | 17 | 14 | 15 | ||
| ≥60 | 26 | 10 | 16 | 11 | 15 | ||
| Differentiation | 0.277 | 0.157 | |||||
| Well, moderate | 30 | 14 | 16 | 11 | 19 | ||
| Poor | 25 | 8 | 17 | 14 | 11 | ||
| Tumor Size (cm) | 0.101 | 0.157 | |||||
| T<5 | 30 | 15 | 15 | 11 | 19 | ||
| T≥5 | 25 | 7 | 18 | 14 | 11 | ||
| TNM stage | 0.014 | 0.116 | |||||
| I, II | 24 | 14 | 10 | 8 | 16 | ||
| III, IV | 31 | 8 | 23 | 17 | 14 | ||
| Nerve involvement | 0.348 | 0.802 | |||||
| Negative | 50 | 19 | 31 | 23 | 27 | ||
| Positive | 5 | 3 | 2 | 2 | 3 | ||
| Vessel involvement | 0.151 | 0.672 | |||||
| Negative | 39 | 18 | 21 | 17 | 22 | ||
| Positive | 16 | 4 | 12 | 8 | 8 | ||
| Lymph node | 0.324 | 0.807 | |||||
| N0-1 | 23 | 11 | 12 | 10 | 13 | ||
| N2-3 | 32 | 11 | 21 | 15 | 17 | ||
| Tumor site | 0.711 | 0.825 | |||||
| Antrum | 41 | 17 | 24 | 19 | 22 | ||
| Cardia | 14 | 5 | 9 | 6 | 8 | ||
Note: p<0.05 is indicated in bold.
Abbreviations: TNM, tumor-node-metastasis; high, high expression; low, low expression.
Cell Culture and RNAi Transfection
The human GC cell lines BGC-823, MKN45, MKN28, MGC-803, and SGC7901 and immortalized gastric mucosa GES-1 cells were purchased from the FuDan IBS Cell Center (Shanghai, China). All cells were cultured at 37°C in a humidified incubator with 5% CO2. The constructs for transfection were provided by GenePharma (Shanghai, China) and were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The sequences were as follows: si-BCL2 (5ʹ-UCUUUUCCUAAAAAGGAUGAC-3ʹ), si-BECN1 (5ʹ-ACUUUCUGUGGACAUCAUCCU-3ʹ), pc-BCL2 (forward: 5ʹ-CTCGGATCCGCGATGTCGACCTA-3ʹ; reverse: 5ʹ-CCGCCATGGTCTACTGTCTGGTCCA-3ʹ), pc-BECN1 (forward: 5ʹ-CCCTCGAGGGATGCTCGAATGACAT-3ʹ; reverse: 5ʹ-CCAAGCTTGGCTACGTGCCAGCCTGTT-3ʹ), miR-140-3p mimic (5ʹ-UACCACAGGGUAGAACCACGG-3ʹ), mimic-NC (5ʹ-GCAAGAGACAAGCGCUUAGCC-3ʹ), miR140-3p inhibitor (5ʹ-GCUAGCUUCGAUGCCAGUCG-3ʹ), and inhibitor-NC (5ʹ-AUGCCAGCUGGCCAUACGAU-3ʹ).
Real-Time PCR Analysis
Total RNA was isolated from GC tissues or normal stomach tissues and from BGC-823 and MKN45 cells using TRIzol (TaKaRa, Japan). Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect miRNA and mRNA levels. U6 and GAPDH were used as the internal controls, respectively. The primers were as follows: miR-140-3p, 5ʹ-CAGTGCTGTACCACAGGGTAGA-3ʹ (sense) and 5ʹ-TATCCTTGTTCACGACTCCTTCAC-3ʹ (antisense); BCL2, 5ʹ-CCTCGCTGCACAAATACTCC-3ʹ (sense) and 5ʹ-TGGAGAGAATGTTGGCGTCT-3ʹ (antisense); BECN1, 5ʹ-CCATGCAGGTGAGCTTCGT-3ʹ (sense) and 5ʹ-GAATCTGCGAGAGACACCATC-3ʹ (antisense); U6, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ (sense) and 5ʹ-GGAACGCTTCACGAATTTG-3ʹ (antisense); and GAPDH, 5ʹ-TGTTCGTCATGGGTGTGAAC-3ʹ (sense) and 5ʹ-ATGGCATGGACTGTGGTCAT-3ʹ (antisense).
Dual-Luciferase Reporter Assay
Possible targets of miR-140-3p were predicted by a bioinformatic prediction website. Wild-type and mutant BCL2 mRNA 3ʹ-UTR reporters (BCL2-wt and BCL2-mut) were constructed and cotransfected with the miR-140-3p mimic (or mimic control) into GC cells, and luciferase activity was determined. At 48 h post transfection, luciferase activities were analyzed in BGC-823 cells by a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
MiRNA Pull-Down Assay
For miRNA pull-down assays, 3′-end biotinylated miRNA Mimic (RiboBio, China) was transfected into GC cells at a final concentration of 20 nM by RNAiMax (Invitrogen) following the manufacturer’s protocol. After 24 h, the cells were lysed in lysis buffer as described above, and the same pull-down procedure was performed.
Western Blot Analysis
GC cells were collected and lysed in RIPA buffer (Servicebio Biotechnology, China). Equal amounts of proteins were separated by SDS-PAGE, transferred to PVDF membranes, blocked in 5% nonfat milk for 1 h, and then immunoblotted with primary antibodies at 4°C overnight. The primary antibodies were as follows: anti-BCL2, anti-BECN1, anti-LC3, anti-p62, anti-E-cadherin, anti-N-cadherin, anti-Vimentin, anti-β-actin (Abcam, USA) and anti-Bax (Cell Signaling Technology, USA). Following incubation with polyclonal HRP-conjugated secondary antibodies, the signals were detected by an ECL system (Biomiga, Agoura Hills, CA).
Immunofluorescence Staining
Cells were incubated with an anti-LC3 human polyclonal antibody at 4°C overnight. The cells were then incubated with a goat anti-human IgG secondary antibody (Abcam) at ambient temperature for 1 h. After washing with PBS, the samples were treated with an antifade reagent with DAPI (Thermo Fisher Scientific, P36931). Fluorescence images were acquired using an Olympus FV1200 confocal microscope.
Apoptosis Analysis
Apoptosis of GC cell lines was measured using an apoptosis detection kit (Beyotime, China). Briefly, 5×105 cells were washed with PBS and resuspended in binding buffer. After staining with Annexin V and propidium iodide for 15 min, the cells were analyzed by flow cytometry (BD Biosciences, China).
EdU Assay
For the EdU assay, 4 × 103 cells were seeded in a 96-well plate, and the Cell-Light EdU Apollo 567 Kit (RiboBio, China) was used to evaluate cell proliferation. After removing the medium, the cells were treated with a 50 μM EdU solution at 37°C. Then, the cells were fixed with 4% paraformaldehyde for 30 min and incubated with glycol for 5 min. After incubation with 100 μL 0.5% Triton X-100, the cells were washed with PBS, incubated with 100 μL Apollo staining reaction, and finally fixed and decolorized with methanol. Cells were visualized under a fluorescence microscope.
Migration and Invasion Assays
GC cells were cultured in six-well plates until confluent. The cells were vertically scraped with a micropipette tip (100 μL). Migration progression was measured at 24 h after wounding. The distance between the two edges of the wound was calculated at three different positions. A Transwell invasion assay was carried out in the manner described previously.21 Cells were seeded in the upper chamber with coated Matrigel, and the lower chamber comprised DMEM containing 10% FBS. After 24 h of cultivation, the cells that passed through the Matrigel-coated membrane were fixed, stained with crystal violet, and quantitated in 5 random microscopic fields.
Establishment of a GC Xenograft Mouse Model
A GC xenograft mouse model was established using BALB/C nude mice. Twenty nude mice were randomly divided into four groups. The xenograft in vivo assay was approved by the Institutional Animal Care and Use Committee of the People’s Hospital of Jingjiang. Groups received subcutaneous injections of anti-miR-control-, anti-miR-140-3p-, pre-miR-control-, or pre-miR-140-3p-transfected BGC-823 cells. Tumor volumes were calculated every week according to the formula: (length×width2)/2. The mice were sacrificed at 35 days after inoculation of tumor cells, and tumor weights were recorded.
Statistical Analysis
Results are presented as the mean ± SEM. Analyses were carried out using GraphPad Prism 6.0 (GraphPad Software Inc., USA). Overall survival (OS) and disease-free survival (DFS) were evaluated by the Kaplan–Meier method. The Cox regression model was used for univariate and multivariate analyses. P < 0.05 was considered statistically significant.
Results
MiR-140-3p Was Downregulated in GC Tissues and Cells
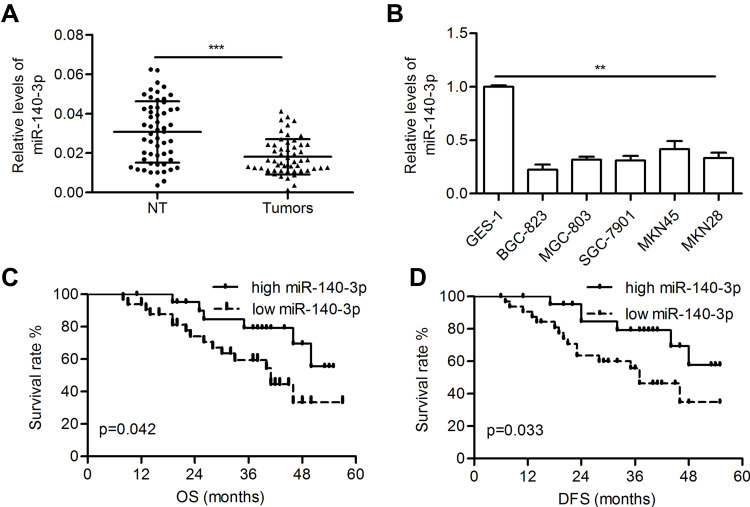
The miR-140-3p level was significantly decreased in GC tissues compared with paired normal tissues (0.018 ± 0.001 vs 0.031 ± 0.002; P<0.001; Figure 1A). Furthermore, reduced miR-140-3p expression was observed in five GC cell lines compared with GES-1 cells (P<0.01; Figure 1B). High levels of miR-140-3p (above the mean) indicated better patient OS than low miR-140-3p expression (below the mean) (P=0.042; Figure 1C). Furthermore, high expression of miR-140-3p indicated better disease-free survival in GC patients (P=0.033; Figure 1D).
Figure 1.
MiR-140-3p was downregulated in GC tissues and cell lines.
Notes: (A) The relative levels of miR-140-3p in tumors and corresponding nontumor gastric tissues were determined by qRT-PCR. (B) Relative levels of miR-140-3p in GC cell lines and a normal cell line. (C and D) Kaplan-Meier analysis of the overall survival and disease-free survival of GC patients stratified according to miR-140-3p expression. **P<0.01; ***P<0.001.
Abbreviations: NT, nontumor tissues; OS, overall survival; DFS, disease-free survival.
MiR-140-3p Directly Inhibited BCL2 Expression in GC Cell Lines
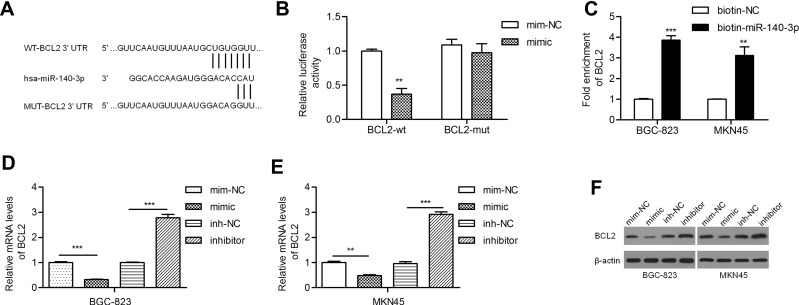
PicTar, miRanda and TargetScan predicted BCL2 as a target gene of miR-140-3p. Studies have revealed that BCL2 is relatively frequently expressed in GC patients with lymph node involvement.22,23 The potential binding sites of miR-140-3p in the BCL2 mRNA 3ʹ-untranslated region (3ʹ-UTR) are shown in Figure 2A. We found that miR-140-3p inhibited the luciferase activity of the wild-type BCL2 3ʹ-UTR but not that of mutant BCL2 3ʹ-UTRs (Figure 2B). A miRNA pull-down assay showed a nearly fourfold enrichment in BCL2 in the miR-140-3p group compared with the control group (Figure 2C). By qRT-PCR, we demonstrated that the BCL2 mRNA level was reduced by a miR-140-3p mimic and elevated by a miR-140-3p inhibitor (Figure 2D and E). Similarly, we found that the miR-140-3p mimic inhibited BCL2 protein expression and that the miR-140-3p inhibitor promoted BCL2 protein expression (Figure 2F).
Figure 2.
MiR-140-3p directly targeted BCL2 in GC cell lines.
Notes: (A) A putative miR-140-3p binding site in the 3ʹ-UTR of the BCL2 mRNA transcript. Mutations were generated in the complementary domain that directly binds to the seed region of miR-140-3p. (B) A luciferase reporter assay was performed with BGC-823 cells to detect the relative luciferase activities of WT and MUT BCL2 reporters. (C) Biotin-coupled miRNA pull-down assays determined that BCL2 was effectively enriched by miR-140-3p. (D and E) BCL2 mRNA levels were studied by qRT-PCR. (F) BCL2 was negatively regulated by miR-140-3p. **P<0.01; ***P<0.001.
Abbreviations: BCL2-wt, wild-type BCL2 reporter; BCL2-mut, mutant BCL2 reporter; NC, negative control.
Relative Expression of BCL2 in GC Tissues and Cell Lines
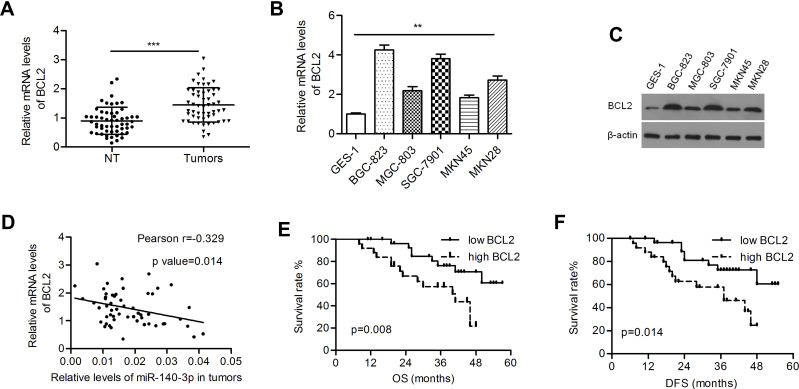
The BCL2 level was significantly higher in GC tissues than in paired normal tissues (1.44 ± 0.080 vs 0.896 ± 0.063; P<0.001; Figure 3A). Both the mRNA and protein levels of BCL2 were upregulated in GC cells (Figure 3B and C). Pearson’s correlation coefficient analysis demonstrated that BCL2 expression was inversely correlated with miR-140-3p expression in GC samples (r=−0.329, P=0.014; Figure 3D). High expression of BCL2 indicated poorer overall survival and disease-free survival in patients than low expression (P=0.008 and 0.014, respectively; Figure 3E and F).
Figure 3.
BCL2 was negatively regulated by miR-140-3p in GC tissues and cell lines.
Notes: (A) Relative mRNA levels of BCL2 in tumors and nontumor gastric tissues. (B and C) Relative BCL2 mRNA and protein expression in GC cell lines and a normal cell line. (D) The mRNA levels of BCL2 were inversely correlated with the levels of miR-140-3p in tumor tissues. (E and F) Kaplan-Meier analysis of overall survival and disease-free survival in GC patients. **P<0.01; ***P<0.001.
Abbreviations: NT, nontumor tissues; OS, overall survival; DFS, disease-free survival.
Studies on the relationships between clinicopathological factors and miR-140-3p expression showed that only tumor-node-metastasis (TNM) stage was correlated with low miR-140-3p expression (P = 0.014, Table 1). No correlation was found between miR-140-3p expression and age, sex or tumor size (Table 1). As shown in Table 2, high expression of BCL2 (HR=3.159; 95% CI, 1.271–7.852; p=0.013), tumor size (HR=2.350; 95% CI, 1.008–5.480; p=0.048), and TNM stage (HR=5.280; 95% CI, 1.770–15.748; p=0.003) were significant prognostic factors for predicting the OS of GC patients using univariate analysis. However, multivariate Cox regression analysis showed that only TNM stage (HR=4.434; 95% CI, 1.463–13.435; p=0.008) was an independent prognostic factor. To evaluate the impact of miR-140-3p and BCL2 on DFS, Cox regression analysis was performed. We found that TNM stage, miR-140-3p levels, and BCL2 mRNA levels were significantly correlated with DFS probability by univariate analysis, while only TNM stage was an independent factor verified by multivariate analysis (Table 3).
Table 2.
Cox Univariate and Multivariate Analyses of Overall Survival
| Characteristics | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | p value | 95% CI | HR | p value | 95% CI | |
| Sex (female vs male) | 1.216 | 0.660 | 0.508–2.910 | |||
| Age (<60 vs ≥60) | 1.831 | 0.159 | 0.789–4.251 | |||
| Differentiation (well+moderate vs poor) | 1.633 | 0.253 | 0.705–3.785 | |||
| Tumor Size (<5 vs ≥5 cm) | 2.350 | 0.048 | 1.008–5.480 | 2.068 | 0.138 | 0.792–5.400 |
| TNM stage (I, II vs III, IV) | 5.280 | 0.003 | 1.770–15.748 | 4.434 | 0.008 | 1.463–13.435 |
| Nerves (negative vs positive) | 2.223 | 0.152 | 0.745–6.629 | |||
| Vessels (negative vs positive) | 0.975 | 0.956 | 0.396–2.400 | |||
| Lymph node (N0-1 vs N2-3) | 1.559 | 0.334 | 0.633–3.840 | |||
| Tumor site (Antrum vs Cardia) | 1.127 | 0.686 | 0.632–2.010 | |||
| miR-140-3p level (high vs low) | 2.571 | 0.052 | 0.992–6.664 | |||
| BCL2 level (low vs high) | 3.159 | 0.013 | 1.271–7.852 | 2.402 | 0.062 | 0.956–6.037 |
Note: p<0.05 is indicated in bold.
Abbreviations: TNM, tumor-node-metastasis; HR, hazard ratio; CI, confidence interval; high, high expression; low, low expression.
Table 3.
Cox Univariate and Multivariate Analyses of Disease-Free Survival
| Characteristics | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | p value | 95% CI | HR | p value | 95% CI | |
| Sex (female vs male) | 1.264 | 0.597 | 0.529–3.019 | |||
| Age (<60 vs ≥60) | 1.705 | 0.214 | 0.735–3.953 | |||
| Differentiation (well+moderate vs poor) | 1.651 | 0.242 | 0.713–3.827 | |||
| Tumor Size (<5 vs ≥5) | 2.284 | 0.056 | 0.980–5.326 | |||
| TNM stage (I, II vs III, IV) | 5.322 | 0.003 | 1.780–15.913 | 4.480 | 0.008 | 1.473–13.625 |
| Nerves (negative vs positive) | 2.291 | 0.137 | 0.768–6.829 | |||
| Vessels (negative vs positive) | 1.067 | 0.888 | 0.433–2.628 | |||
| Lymph node (N0-1 vs N2-3) | 1.562 | 0.333 | 0.633–3.853 | |||
| Tumor site (Antrum vs Cardia) | 1.079 | 0.874 | 0.420–2.773 | |||
| miR-140-3p level (high vs low) | 2.700 | 0.041 | 1.043–6.990 | 1.294 | 0.636 | 0.446–3.755 |
| BCL2 level (low vs high) | 2.836 | 0.019 | 1.183–6.799 | 2.112 | 0.098 | 0.870–5.126 |
Note: p<0.05 is indicated present in bold.
Abbreviations: TNM, tumor-node-metastasis; HR, hazard ratio; CI, confidence interval; high, high expression; low, low expression.
MiR-140-3p/BCL2 Regulated Cell Proliferation and Apoptosis
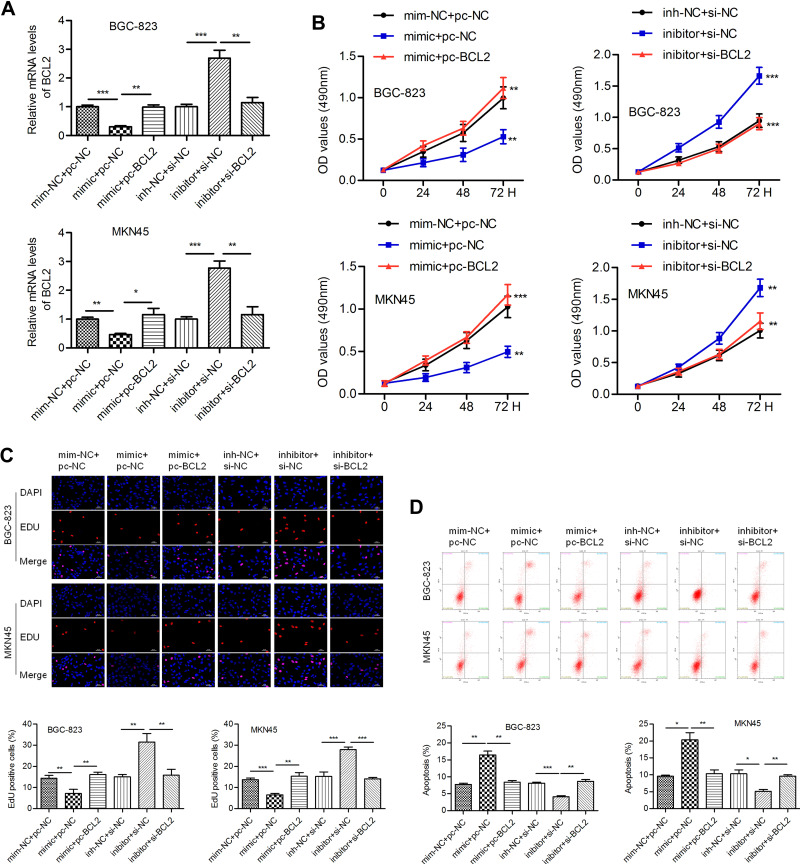
We intended to verify whether miR-140-3p regulates cell behavior by targeting BCL2. The miR-140-3p mimic (or inhibitor) and BCL2-expressing vectors (or si-BCL2-expressing vectors) were cotransfected into BGC-823 and MKN45 cells. QPCR was conducted to determine the transfection efficiency (Figure 4A). We noticed that miR-140-3p significantly inhibited cell proliferation, and this phenomenon could be reversed by BCL2. Knockdown of BCL2 expression impaired the miR-140-3p-induced increase in cell proliferation (Figure 4B). The results of an EdU assay also showed that proliferation was inhibited by miR-140-3p but promoted by BCL2 in GC cells (Figure 4C). MiR-140-3p promoted apoptosis in BGC-823 and MKN45 cells, whereas BCL2 reversed these effects. Similarly, silencing BCL2 attenuated the miR-140-3p inhibitor-induced decrease in cell apoptosis (Figure 4D).
Figure 4.
MiR-140-3p inhibits proliferation and promotes apoptosis by targeting BCL2.
Notes: (A) Transfection efficiency was determined by a qRT-PCR assay. (B) The viability of GC cells was detected by a CCK-8 assay. (C) An EdU assay was conducted to evaluate the proliferation of GC cells; overexpression of BCL2 reversed miR-140-3p-induced cell growth inhibition; scale bar = 100 μm. (D) Flow cytometric analysis was used to detect cell apoptosis. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: pc-NC, empty control vector; pc-BCL2-, BCL2-expressing vectors; min-NC, mimic negative control; inh-NC, inhibitor negative control.
MiR-140-3p/BCL2 Regulated Cell Migration, Invasion and Autophagy
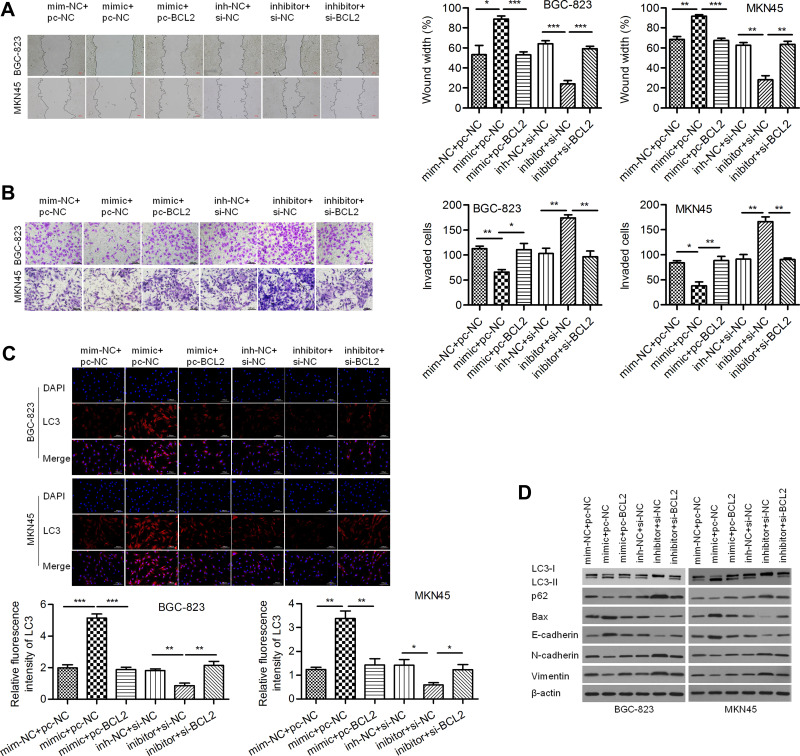
A wound healing assay showed that the relative migration distance in the miR-140-3p mimic group was lower than that in the negative control group and that BCL2 reversed this effect to increase the migration distance. Similarly, the relative migration distance in the miR-140-3p inhibitor group was higher than that in the negative control group, and si-BCL2 reversed this effect to decrease the migration distance (Figure 5A). A Transwell invasion assay showed that miR-140-3p decreased the invasive of GC cells and that reintroduction of BCL2 reversed this effect (Figure 5B). Moreover, we found that the formation of LC3-labeled vacuoles in the cytoplasm was markedly increased after transfection of miR-140-3p and could be reversed by BCL2 (Figure 5C). In addition, we observed increases in E-cadherin and Bax and decreases in N-cadherin, Vimentin and p62 proteins in the miR-140-3p mimic group, and BCL2 reversed these effects. MiR-140-3p markedly upregulated the LC3-II/I ratio, which could be reversed by BCL2 (Figure 5D). Our study suggested that miR-140-3p promotes cellular autophagic death and suppresses cell motility and EMT, probably by targeting BCL2.
Figure 5.
MiR-140-3p inhibited migration and invasion and enhanced autophagy by targeting BCL2.
Notes: (A) A wound healing assay was used to detect the migration of BGC-823 and MKN45 cells; scale bar = 100 μm. (B) A Transwell assay was used to detect the invasion of GC cells; scale bar = 100 μm. (C) Immunofluorescence assay for LC3; scale bar = 100 μm. (D) The expression of Vimentin, N-cadherin, E-cadherin, Bax, p62 and LC3 II/I was determined using Western blot analysis. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: NC, negative control; si-BCL2, specific siRNA targeting BCL2.
BECN1 Interferes with miR-140-3p-Regulated Autophagy
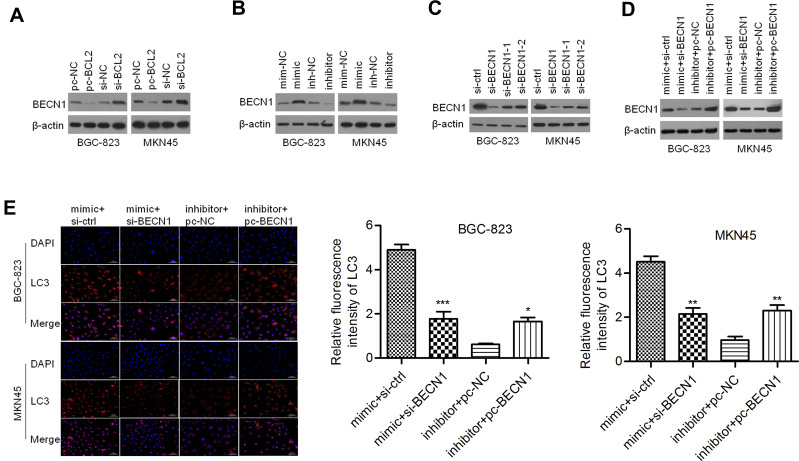
Considering that BCL2 may be a regulator of BECN1, we evaluated the effect of BCL2 on BECN1 expression. As shown in Figure 6A, ectopic expression of BCL2 reduced the protein expression of BECN1, whereas silencing BCL2 elevated BECN1 protein expression in BGC-823 and MKN45 cells. Thus, we further validated whether miR-140-3p regulates BECN1. We discovered that the protein levels of BECN1 were positively regulated by miR-140-3p in GC cells (Figure 6B). These data indicated that miR-140-3p might regulate BECN1 protein expression by targeting BCL2. To investigate whether BECN1-related autophagy is involved in miR-140-3p-regulated cell migration and invasion, GC cells were cotransfected with the miR-140-3p mimic/inhibitor and si-BECN1/pc-BECN1. Three siRNAs for knocking down of BECN1 were designed and the most effective siRNA was used to knockdown BECN1 in GC cells (Figure 6C). The results showed that BECN1 knockdown could reverse the promotion of BECN1 protein expression by miR-140-3p (Figure 6D). In BGC-823 and MKN45 cells, the formation of LC3-labeled vacuoles was significantly increased in the cytoplasm after transfection of miR-140-3p and could be prevented with si-BECN1. The formation of LC3-labeled vacuoles was significantly decreased after transfection of the miR-140-3p inhibitor and could be promoted with pc-BECN1 (Figure 6E). Our study indicated that knockdown of BECN1 could reverse the promotion of GC cellular autophagic processes by miR-140-3p.
Figure 6.
BECN1 interfered with autophagy regulated by miR-140-3p in GC cell lines.
Notes: (A) BECN1 was negatively regulated by BCL2. (B) miR-140-3p affected the expression of BCL2. (C) Three siRNAs for knocking down BECN1 were designed and Western blot was used to detect BECN1 expression. (D) Transfection efficiency was determined by Western blot analysis. (E) Immunofluorescence staining for LC3 was performed with GC cells; scale bar = 100 μm. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: si-BCL2, siRNA targeting BCL2; pc-BECN1, BECN1-expressing vectors; NC, negative control.
BECN1 Interferes with Cell Motility Regulated by miR-140-3p in GC Cells
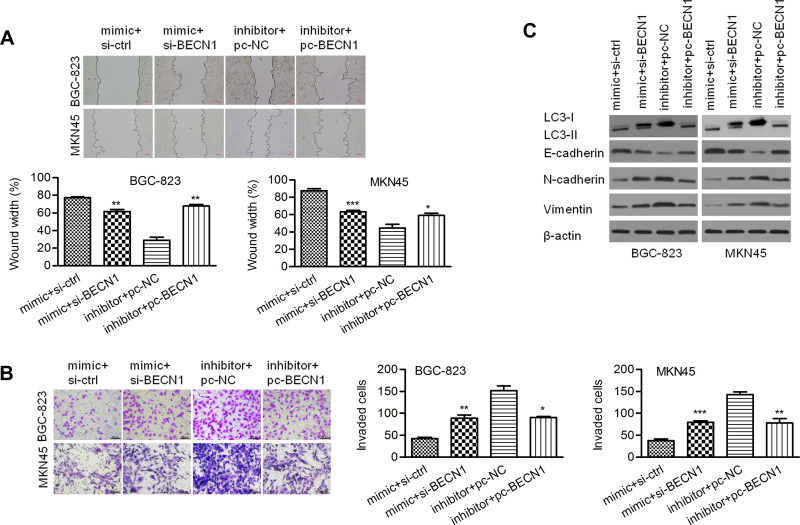
As shown in Figure 7A, a wound healing assay showed that knockdown of BECN1 could reverse the negative effect of the miR-140-3p mimic on migration distance. A Transwell invasion assay showed that knockdown of BECN1 could reverse the negative effect on GC cell invasion (Figure 7B). MiR-140-3p markedly upregulated the LC3-II/I ratio in GC cells, and knockdown of BECN1 reversed this effect to decrease the LC3-II/I ratio (Figure 7C). In addition, knockdown of BECN1 decreased the E-cadherin protein levels that were increased by the miR-140-3p mimic. In contrast, knockdown of BECN1 increased the N-cadherin and Vimentin protein levels that were decreased by the miR-140-3p mimic (Figure 7C).
Figure 7.
BECN1 interfered with migration, invasion and EMT regulated by miR-140-3p. (A) A wound healing assay was used to detect the migration of BGC-823 and MKN45 cells; scale bar = 100 μm. (B) A Transwell assay was used to detect cell invasion; scale bar = 100 μm. (C) The protein expression of Vimentin, N-cadherin, E-cadherin, and LC3 II/I was determined using Western blot analysis. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: si-BCL2, siRNA targeting BCL2; pc-BECN1, BECN1-expressing vectors; NC, negative control.
MiR-140-3p regulated tumorigenesis in a nude mouse model
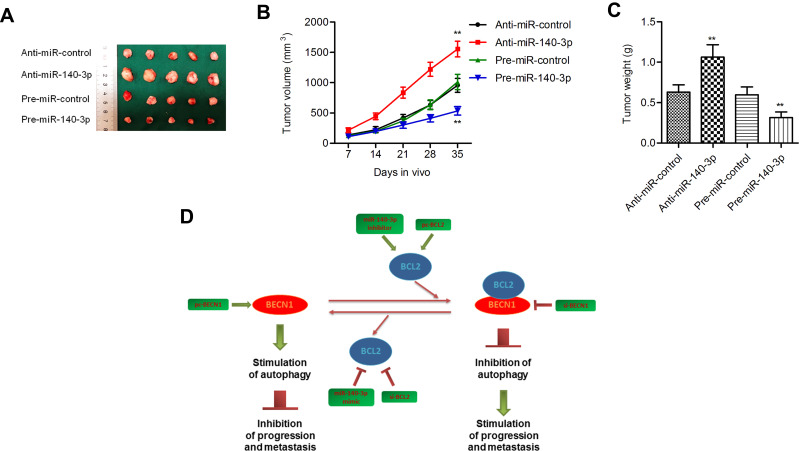
Pre-miR-140-3p-, anti-miR-140-3p- or negative control-transfected cell lines were injected into nude mice. The volumes and weights of tumors in the miR-140-3p overexpression group were smaller than those in the corresponding control group, and the tumor volumes and weights of mouse tumors in the miR-140-3p-silenced group were higher than those in the corresponding control group (P < 0.01; Figure 8A–C). These data suggested that miR-140-3p inhibited tumor growth in vivo.
Figure 8.
MiR-140-3p regulated tumorigenesis in a nude mouse model.
Notes: (A) Representative mouse tumors from the four groups. (B and C) Both tumor volumes and weights were increased in the miR-140-3p-silenced group but decreased in the miR-140-3p-overexpressing group. (D) Schematic summary of the antitumor effect of miR-140-3p in GC cells. miR-140-3p impedes GC progression and metastasis by directly downregulating the expression of BCL2. BCL2 downregulation further activates BECN1-dependent autophagy. **P<0.01.
Discussion
GC metastasis greatly limits treatment success.24 The occurrence and progression of GC are closely correlated with abnormal expression of miRNAs.25,26 In the present study, we discovered that miR-140-3p was underexpressed in GC tissues and cells. MiR-140-3p has been reported to impede proliferation and migration in breast cancer.12 In addition, miR-140-3p is known to suppress cell growth in colorectal cancer.9 In lung cancer, miR-140-3p inhibits cell metastasis by targeting ATP6AP213 and ATP8A1.27 A study found that miR-140-3p inhibits EMT and invasion in hepatocellular carcinoma by targeting granulin.10 All these studies suggest that miR-140-3p may have the ability to inhibit progression in various solid tumors. Our data expanded the knowledge on miR-140-3p as a tumor suppressor in GC development. In clinical samples, lower expression of miR-140-3p was related to an advanced pathological stage and shorter OS time. We also identified that in BGC-823 and MKN45 cells, migration, invasion and EMT were inhibited and apoptosis and autophagy were enhanced after transfection with miR-140-3p. MiR-140-3p downregulation promoted GC migration, invasion and EMT and inhibited GC autophagy and apoptosis. MiR-140-3p inhibited tumor growth, and silencing miR-140-3p promoted tumor growth in vivo. Our results imply that miR-140-3p may act as a promising prognostic factor and suppressor of GC by decreasing cell proliferation and enhancing cell autophagy.
MiR-140-3p targeting BCL2 was predicted by combined results from PicTar, miRanda, and TargetScan. Zhu et al found that BCL2 is a direct target of miR-140-3p in airway smooth muscle cells.20 We identified BCL2 as a downstream target of miR-140-3p in BGC-823 cells. BCL2 has been shown to inhibit cell apoptosis.28 BCL2 is an oncogene that increases cellular proliferation and inhibits programmed cell death.29,30 Contrary to the expression of miR-140-3p, BCL2 expression was upregulated in GC tissues and cells, and GC patients with higher expression of BCL2 had shorter OS times than those with lower expression. In addition, the expression of BCL2 was a significant prognostic factor for predicting the OS of GC patients. An inverse correlation was found between miR-140-3p and BCL2 in GC tissues, suggesting that dysregulation of miR-140-3p/BCL2 may have roles in GC carcinogenesis. Therefore, we speculated that miR-140-3p may participate in metastasis and autophagy in GC by targeting BCL2. We found that miR-140-3p upregulation significantly decreased BCL2 mRNA and protein levels in GC cells. In further experiments on cellular function, we found that BCL2 inhibited the effect of miR-140-3p overexpression, suggesting that BCL2 promotes GC cell migration, invasion and EMT and inhibits GC cell autophagic death. Our study showed that BCL2 might be a functional target of miR-140-3p in GC cells.
BECN1 was identified as a new binding partner of BCL2 in 1998.31 BECN1 was the first tumor suppressor gene found to play a part in the lysosomal degradation pathway of autophagy.32 A study noted that BCL2 inhibits BECN1-dependent autophagy.16 Yun Du et al indicated that BCL2 downregulation induced BECN1-dependent autophagy in human SGC-7901 cells.33 Thus, we intended to study whether BCL2 regulates BECN1 in our study. We showed that BCL2 inhibited BECN1 protein expression in GC cells, whereas miR-140-3p promoted BECN1 protein expression. In further experiments on cellular function, we found that silencing BECN1 could reverse the effect of miR-140-3p overexpression, suggesting that BECN1 promotes GC cell autophagy and inhibits GC cell migration, invasion and EMT. These results suggested that miR-140-3p indirectly promotes BECN1 expression by directly inhibiting the expression of BCL2. Furthermore, BECN1-dependent autophagy activation inhibits GC progression and metastasis. Activation of EMT is generally seen in a variety of malignancies, including GC.34 Autophagy can inhibit cancer metastasis by blocking EMT.35 Our study indicated that miR-140-3p might inhibit EMT by inducing BECN1-dependent autophagy and thus impeding GC metastasis. Overall, miR-140-3p may impede GC progression and metastasis by regulating the BCL2-BECN1-autophagy axis.
In summary, we discovered that miR-140-3p was downregulated in GC and associated with an advanced tumor stage and a poor prognosis. MiR-140-3p directly targets BCL2, which reverses the inhibitory effects of miR-140-3p on cell proliferation, migration, and invasion and the promotive effects of miR-140-3p on cell apoptosis and autophagy. In addition, we found that BECN1 is regulated by miR-140-3p/BCL2. MiR-140-3p was found to inhibit cell migration, invasion, and EMT by regulating BECN1-related autophagy (Figure 8D). Our data suggest that miR-140-3p acts as a promising tumor suppressor in GC by targeting BCL2, thereby regulating BECN1-regulated autophagy, providing a new therapeutic strategy for the treatment of GC.
Disclosure
The authors declare no competing interests in this work.
References
- 1.Bray F, J-Ferlay I, Soerjomataram RL, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–628. doi: 10.11622/smedj.2014174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sitarz R, Skierucha M, Mielko J, et al. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrelli DA, De Stefano G, de Manzoni P, et al. Prediction of recurrence after radical surgery for gastric cancer: a scoring system obtained from a prospective multicenter study. Ann Surg. 2005;241:247–255. doi: 10.1097/01.sla.0000152019.14741.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu S, Li G, Wang Z, et al. Low expression of MAP1LC3B, associated with low Beclin-1, predicts lymph node metastasis and poor prognosis of gastric cancer. Tumour Biol. 2016;37:15007–15017. doi: 10.1007/s13277-016-5383-5 [DOI] [PubMed] [Google Scholar]
- 6.Cao YY, Luo J, Zou J, et al. Autophagy and its role in gastric cancer. Clin Chim Acta. 2019;489:10–20. doi: 10.1016/j.cca.2018.11.028 [DOI] [PubMed] [Google Scholar]
- 7.Li H, He C, Wang X, et al. MicroRNA-183 affects the development of gastric cancer by regulating autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR signals. Artif Cells Nanomed Biotechnol. 2019;47:3163–3171. doi: 10.1080/21691401.2019.1642903 [DOI] [PubMed] [Google Scholar]
- 8.Guo W, Chen Z, Chen Z, et al. Promotion of cell proliferation through inhibition of cell autophagy signalling pathway by Rab3IP is restrained by MicroRNA-532-3p in Gastric Cancer. J Cancer. 2018;9:4363–4373. doi: 10.7150/jca.27533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Li T, Wang J, et al. miR-140-3p suppresses cell growth and induces apoptosis in colorectal cancer by targeting PD-L1. Onco Targets Ther. 2019;12:10275–10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang QY, Men CJ, Ding XW. Upregulation of microRNA-140-3p inhibits epithelial-mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN. J Cell Biochem. 2019;120:14885–14898. doi: 10.1002/jcb.28750 [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Zhang F, Sun P. miR-140-3p impedes the proliferation of human cervical cancer cells by targeting RRM2 to induce cell-cycle arrest and early apoptosis. Bioorg Med Chem. 2020;28:115283. doi: 10.1016/j.bmc.2019.115283 [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Wang B, Wang Y, et al. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett. 2019;17:3835–3841. doi: 10.3892/ol.2019.10038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong XM, Zhang GH, Huo YK, et al. MicroRNA-140-3p inhibits proliferation, migration and invasion of lung cancer cells by targeting ATP6AP2. Int J Clin Exp Pathol. 2015;8:12845–12852. [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Modawi RN, Brinchmann JE, Karlsen TA. Multi-pathway protective effects of microRNAs on human chondrocytes in an in vitro model of osteoarthritis. Mol Ther Nucleic Acids. 2019;17:776–790. doi: 10.1016/j.omtn.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korsmeyer SJ. Bcl-2 initiates a new category of oncogenes: regulators of cell death. Blood. 1992;80:879–886. doi: 10.1182/blood.V80.4.879.879 [DOI] [PubMed] [Google Scholar]
- 16.Pattingre S, Levine B. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 2006;66:2885–2888. doi: 10.1158/0008-5472.CAN-05-4412 [DOI] [PubMed] [Google Scholar]
- 17.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin AR, Bao M, Roth L, et al. microRNA-23a contributes to asthma by targeting BCL2 in airway epithelial cells and CXCL12 in fibroblasts. J Cell Physiol. 2019;234:21153–21165. doi: 10.1002/jcp.28718 [DOI] [PubMed] [Google Scholar]
- 19.Sims EK, Lakhter AJ, Anderson-Baucum E, Al ET. MicroRNA 21 targets BCL2 mRNA to increase apoptosis in rat and human beta cells. Diabetologia. 2017;60:1057–1065. doi: 10.1007/s00125-017-4237-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu ZR, He Q, Wu WB, et al. MiR-140-3p is involved in in-stent restenosis by targeting c-myb and bcl-2 in peripheral artery disease. J Atheroscler Thromb. 2018;25:1168–1181. doi: 10.5551/jat.44024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Zhang QY, Zou JL, et al. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. 2016;7:4817–4828. doi: 10.18632/oncotarget.6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gryko M, Pryczynicz A, Zareba K, et al. The expression of Bcl-2 and BID in gastric cancer cells. J Immunol Res. 2014;2014:953203. doi: 10.1155/2014/953203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan W, Ishii H, Ebihara Y, et al. Prognostic use of growth characteristics of early gastric cancer and expression patterns of apoptotic, cell proliferation, and cell adhesion proteins. J Surg Oncol. 2003;82:104–110. doi: 10.1002/jso.10204 [DOI] [PubMed] [Google Scholar]
- 24.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 25.Hwang J, Min BH, Jang J, et al. MicroRNA expression profiles in gastric carcinogenesis. Sci Rep. 2018;8:14393. doi: 10.1038/s41598-018-32782-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan HW, Li SC, Tsai KW. MicroRNA dysregulation in gastric cancer. Curr Pharm Des. 2013;19:1273–1284. doi: 10.2174/138161213804805621 [DOI] [PubMed] [Google Scholar]
- 27.Dong W, Yao C, Teng X, et al. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016;37:2973–2985. doi: 10.1007/s13277-015-3452-9 [DOI] [PubMed] [Google Scholar]
- 28.Geng YJ. Molecular signal transduction in vascular cell apoptosis. Cell Res. 2001;11:253–264. doi: 10.1038/sj.cr.7290094 [DOI] [PubMed] [Google Scholar]
- 29.McDonnell TJ, Deane N, Platt FM, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8 [DOI] [PubMed] [Google Scholar]
- 30.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0 [DOI] [PubMed] [Google Scholar]
- 31.Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/JVI.72.11.8586-8596.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. [DOI] [PubMed] [Google Scholar]
- 33.Du Y, Ji X. Bcl-2 down-regulation by small interfering RNA induces Beclin1-dependent autophagy in human SGC-7901 cells. Cell Biol Int. 2014;38:1155–1162. doi: 10.1002/cbin.10333 [DOI] [PubMed] [Google Scholar]
- 34.Wang HC, Wang W, Tian Y, et al. The crucial role of SRPK1 in IGF-1-induced EMT of human gastric cancer. Oncotarget. 2017;8:72157–72166. doi: 10.18632/oncotarget.20048 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Gugnoni MV, Sancisi G, Gandolfi G, et al. Cadherin-6 promotes EMT and cancer metastasis by restraining autophagy. Oncogene. 2017;36:667–677. doi: 10.1038/onc.2016.237 [DOI] [PubMed] [Google Scholar]