Malassezia spp. are emerging fungal pathogens causing opportunistic skin and severe systemic infection. Nosocomial outbreaks are associated with azole resistance, and understanding of the underlying mechanisms is limited to knowledge of other fungal species.
KEYWORDS: intrinsic, acquired, resistance, azoles, Malassezia furfur, CYP51, PDR10, AMR, Malassezia, antifungal resistance, mechanisms of resistance
ABSTRACT
Malassezia spp. are emerging fungal pathogens causing opportunistic skin and severe systemic infection. Nosocomial outbreaks are associated with azole resistance, and understanding of the underlying mechanisms is limited to knowledge of other fungal species. Here, we identified distinct antifungal susceptibility patterns in 26 Malassezia furfur isolates derived from healthy and diseased individuals. A Y67F CYP51 mutation was identified in five isolates of M. furfur. However, this mutation alone was insufficient to induce reductions in azole susceptibility in the wild-type strain. RNA sequencing (RNA-seq) and differential gene analysis of strains derived from individuals with healthy or disease backgrounds exposed to clotrimazole in vitro identified several key metabolic pathways and transporter proteins which are involved in reducing azole susceptibility. The pleiotropic drug transporter gene PDR10 was the single most highly upregulated transporter gene in multiple strains of M. furfur after azole treatment, and increased expression of PDR10 is associated with reduced azole susceptibility in some systemic-disease isolates of M. furfur. Deletion of PDR10 in a pathogenic M. furfur strain with reduced susceptibility reduced MICs to those of susceptible isolates. In light of the current dearth of antifungal technologies, globally emerging multiazole resistance, and broad use of azoles in agriculture and consumer care, an improved understanding of the mechanisms underlying intrinsic and acquired azole resistance in Malassezia is crucial for development of antibiotic stewardship and antifungal treatment strategies.
INTRODUCTION
Malassezia, the lipid-dependent skin commensal, has also been shown to be an emerging pathogen not just in neonatal intensive care wards but also in Crohn’s disease and pancreatic cancers (1–4). Malassezia-associated superficial mycoses (dandruff, pityriasis versicolor, and folliculitis) affect up to 50% of the global population (5) and have the potential to cause severe systemic and invasive infection in immunocompromised individuals (6–9). Outbreaks in neonatal and intensive care wards have been increasingly reported (6). Azole antifungals are the primary antifungal treatment for Malassezia-associated diseases due to their intrinsic resistance to some antifungals, particularly echinocandins (10). Because reduced in vitro antifungal susceptibility may be an indicator of clinical failure (11), there is a need to profile the susceptibility of Malassezia isolates derived from individuals with healthy or disease backgrounds to elucidate and design prevention strategies for the underlying mechanisms contributing to current and emerging trends of antifungal resistance.
Azole resistance may be primary (intrinsic) or secondary (acquired) (12). The former is found naturally without prior (known) antifungal exposure. The latter is a result of a previously susceptible strain being exposed to antifungals or other selective pressure and may be a result of altered gene expression, point mutations, or allelic variations (13). Both may be attributed to an increase in (i) the prophylactic use of azole drugs, (ii) prolonged treatment regimens, (iii) agricultural use of azole fungicides for crop protection (14), or (iv) the broad-spectrum, long-term, and low-dose use of azoles in consumer care. Azole treatment efficacy is variable, depending on the involved species, and resistant strains are being documented with increasing frequency (10, 15). Among human commensal Malassezia species, Malassezia furfur and Malassezia pachydermatis are the species most commonly encountered in systemic infection of neonates and immunocompromised persons (15). An increasing number of ketoconazole-resistant M. pachydermatis strains have been reported to have been found on pet dogs in Korea and can be attributed to the use of antifungal shampoo (16). The prevalence of the normally zoophilic species M. pachydermatis in infection is worrying evidence of animal-to-human transmission (6).
Sterol 14α-demethylase (encoded by CYP51 [ERG11]) is a key cytochrome P450 enzyme involved in fungal cell wall ergosterol synthesis (17). Malassezia azole resistance is largely associated with mutations in the ERG11/CYP51 gene, identified from clinical isolates (15, 18, 19). In clinical isolates derived from subjects with severe dandruff, the CYP51 mutations Y127F, A169S, and K176 were identified in Malassezia globosa (15). Highly multiazole-resistant strains of M. pachydermatis have also been extensively documented and found to be associated with missense mutations in CYP51 (20, 21). A further genomic tandem quadruplication in the ERG5/ERG11 gene region has been described in a ketoconazole-resistant isolate of M. pachydermatis (18).
Fungal multidrug resistance is also associated with overexpression of transporters of the ATP-binding cassette (ABC) superfamily and major facilitator superfamily (MFS) (22–25). These transporters utilize ATP hydrolysis or electrochemical gradients to carry out efflux of antifungals, thus reducing susceptibility (22). Yeast ABC transporters are classified into six subfamilies of up to 30 unique proteins, such as CDR1 and MDR, which confer drug resistance in human pathogens, including Candida albicans. In M. furfur and M. pachydermatis, the role of efflux pumps has also been implicated through the synergistic use of efflux pump modulators together with azoles to inhibit growth (26). Pleiotropic drug resistance protein 10 (encoded by PDR10) is involved in fluconazole resistance and has been identified in M. furfur (27). Genomic multiplications in ATM1 and ERG11, encoding iron-sulfur transporters, have also been found to confer ketoconazole resistance in Malassezia restricta (16).
In this study, we analyzed antifungal susceptibility profiles of 26 strains of M. furfur derived from individuals with healthy or disease backgrounds and performed RNA sequencing (RNA-seq) to identify candidate genes driving intrinsic and acquired resistance. Although mutations in CYP51 were originally postulated to be the main driver of elevated MICs in M. furfur, differential gene analysis via RNA-seq and subsequent gene validation has identified genes from the ABC transporter protein family, particularly PDR10, to be key drivers of intrinsic and acquired resistance in the M. furfur strains CBS 7982 and CBS 14141. Knowledge of the functional mechanisms underlying antifungal susceptibility in Malassezia will be beneficial in identifying new therapeutic targets, understanding the emergence of new resistant strains, and developing global antifungal drug stewardship strategies.
RESULTS
Diseased-derived isolates of M. furfur have elevated terbinafine, clotrimazole, and miconazole MICs.
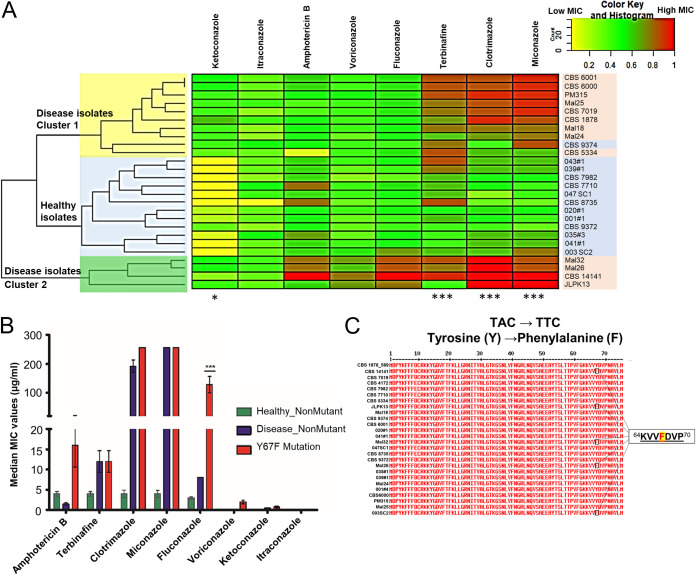
Antifungal susceptibility testing was performed on 26 strains of M. furfur comprising isolates from healthy human skin (referred to here as “healthy isolates”) and those from skin disease or systemic infection (“disease isolates”) (Table 1) using a panel of eight antifungals: amphotericin B (AMB), terbinafine (TRB), clotrimazole (CTZ), miconazole (MCZ), fluconazole (FLZ), voriconazole (VRZ), ketoconazole (KTZ) and itraconazole (ITZ) (Fig. 1). The MIC of each antifungal presented in Fig. 1 was normalized from 0 to 1, with 1 being the highest MIC (red) for each individual compound.
TABLE 1.
Healthy and disease M. furfur isolates used in this study
| Isolate | Source |
|---|---|
| Healthy-skin isolates | |
| CBS 7982 | Ear |
| 020#01 | Nose |
| 041#01 | Nose |
| 039#01 | Nose |
| 035#03 | Nose |
| CBS 9372 | Back |
| CBS 9374 | Chest |
| CBS 8735 | Bronchial wash |
| 047 SC1 | Scalp |
| 003 SC2 | Scalp |
| CBS 7710 | Skin of man |
| 043#01 | Nose |
| 001#01 | Nose |
| Diseased-skin isolates | |
| CBS 5334 | Infected skin |
| CBS 6000 | Dandruff |
| CBS 6001 | Pityriasis versicolor |
| CBS 1878 | Dandruff |
| CBS 7019 | Pityriasis versicolor |
| Systemic-disease isolates | |
| CBS 14141 | Blood |
| JLPK13 | Urine |
| Mal18 | Blood |
| Mal24 | Blood |
| Mal25 | Skin catheter insertion |
| Mal26 | Blood |
| Mal32 | Skin catheter insertion |
| PM315 | Anal swab of neonate |
FIG 1.
The 26 strains of M. furfur used, including 13 healthy and 13 disease isolates. (A) Heat map of the MICs of 8 antifungals. MICs were normalized from 0 to 1, with 1 being the highest MIC (red) for each individual compound. Strains isolated from healthy individuals are highlighted in blue, and strains from individuals with disease are highlighted in red. A paired two-tailed t test (*, P < 0.05; ***, P < 0.005) was performed for median MICs between healthy and disease isolates. (B) Median MICs and standard deviations (SD) for healthy and disease wild-type and Y67F mutant strains. A one-way ANOVA was used with Dunnett’s test (***, P < 0.005). (C) CYP51 amino acid sequence showing the Y67F mutation in 5 isolates.
The isolates further clustered into 2 groups, with disease isolates in cluster 2 (Fig. 1A, green) having elevated MICs for almost all azole antifungals and disease isolates in cluster 1 (Fig. 1A, yellow) having high MICs specifically for TRB, CTZ, and MCZ (P < 0.001) (Fig. 1B; also, see Table S1 in the supplemental material). ITZ had the lowest MICs of all antifungals tested (Fig. 1B), although disease isolates of M. furfur still had significantly higher MICs for KTZ (P < 0.05) (Fig. 1B). FLZ and VRZ MICs were higher for disease isolates in cluster 2 (Fig. 1A, green), although these values were not significant.
CYP51A1 Y67F mutants have elevated fluconazole and voriconazole MICs.
The gene sequence of CYP51A1 in M. furfur was identified by a whole-genome-sequencing (WGS) BLAST search (BioProject ID 286710) of a known M. globosa CYP51A1 sequence (15) against M. furfur genomes and validated by the identification of the presence of the highly conserved heme-binding site (FGFGRHRCIG), EXXR motif (ERLR), and conserved threonine of the I helix involved in proton delivery (Fig. S1).
A tyrosine-to-phenylalanine mutation in residue 67 (Y67F) of CYP51A1 was identified in the blood isolate CBS 14141 (Fig. 1C; Fig. S2). The mutation was also identified in four other isolates, i.e., the systemic-disease isolates Mal24, Mal26, and JLPK13 and the healthy-skin isolate 003 SC2. The three systemic-disease isolates showed elevated MICs (Fig. 1B and C; Table S1) specifically for FLZ (P < 0.001), consistent with literature indicating the role of the tyrosine residue in drug binding (28). Healthy skin isolate 003 SC2 also showed slightly elevated MICs for MCZ and FLZ. This mutation is synonymous with the Y132F mutation in Candida albicans and is well known to affect binding to fluconazole due to the residue being 7 Å away from a key binding motif (29). It has been identified as a Y127F mutation in disease isolates of M. globosa (15).
Y67F knock-in and F67Y rescue mutants show no changes in azole susceptibility.
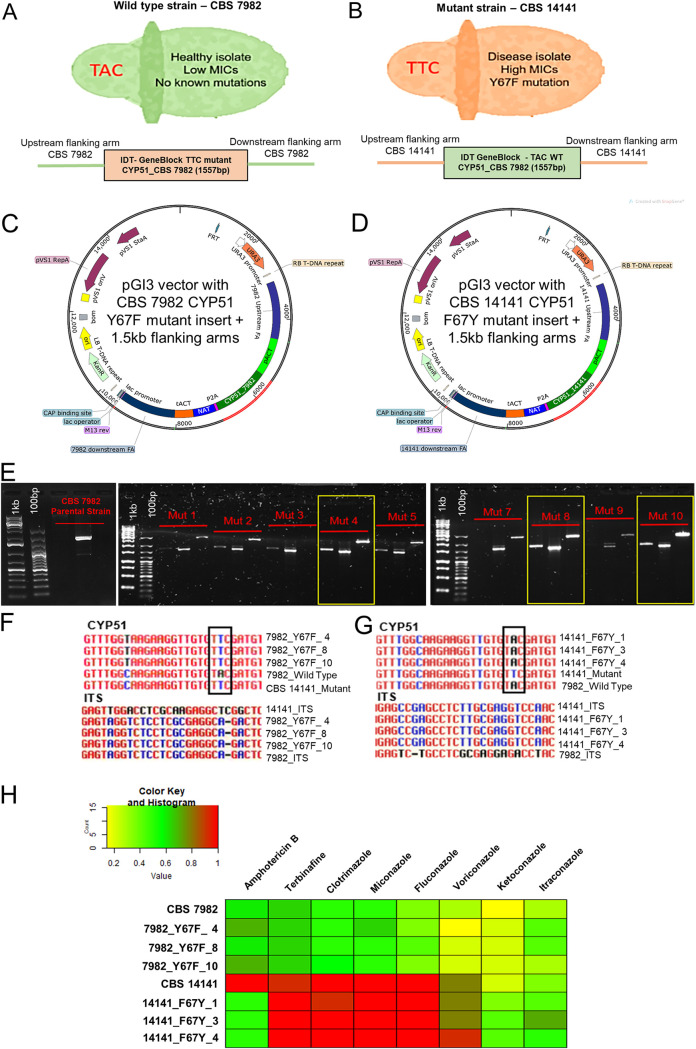
The single CYP51 nucleotide change coding for the Y67F mutation was inserted into the wild-type, low-MIC strain CBS 7982 via Agrobacterium tumefaciens-mediated transformation (ATMT) using a nourseothricin (NAT) expression vector (Fig. 2A to D). Similarly, a rescue F67Y CYP51 mutation was generated in the CYP51 mutant strain CBS 14141 (Fig. 2A to D). Colonies were screened for the presence of the correct 1.5-kb flanking arms and insert (Fig. 2E). Sanger sequencing of the CYP51 mutation regions and internal transcribed spacer regions (ITS) was performed to validate the presence of the correct single nucleotide mutation and strain background of the respective transformants (Fig. 2F and G).
FIG 2.
Agrobacterium tumefaciens-mediated transformation (ATMT) of CYP51 mutation into wild-type CBS 7982 and mutant CBS 14141 M. furfur. (A and B) Phenotypes and genotypes of the M. furfur wild-type strain CBS 7982 (A) and the mutant strain CBS 14141 (B). (C and D) Vector maps for CBS 7982 (C) and CBS 14141 (D) ATMT. (E) Screening and selection of CBS 7982 transformant colonies with the correct CYP51 insert (∼700 bp) and 1.5-kb upstream/downstream flanking arms. (F and G) Sanger sequencing validation of the correct single nucleotide mutation and ITS sequences in CBS 7982 (F) and CBS 14141 (G). (H) Heat map of the MICs for CBS 7982 Y67F and CBS 14141 F67Y mutants. MICs were normalized from 0 to 1, with 1 being the highest MIC (red).
Insertion of the Y67F CYP51 mutation into the CBS 7982 strain did not yield any change in azole susceptibility (Fig. 2H; Table 2). Similarly, an F67Y rescue mutant generated from the CBS 14141 CYP51 mutant strain did not show any reduction in azole MICs when tested against our panel of antifungals. The F67Y rescue mutation restored amphotericin B MICs to wild-type levels in CBS 14141, although introduction into wild-type CBS 7982 increased AMB MICs only slightly.
TABLE 2.
MICs for CYP51 transformants
| Strain | MIC (μg/ml) ofa: |
|||||||
|---|---|---|---|---|---|---|---|---|
| AMB | TRB | CTZ | MCZ | FLZ | VRZ | KTZ | ITZ | |
| CBS 7982 | 4 | 2 | 4 | 4 | 1 | 0.03 | 0.03 | 0.03 |
| Y67F_4 | 8 | 2 | 4 | 16 | 1 | 0.008 | 0.06 | 0.125 |
| Y67F_8 | 4 | 2 | 4 | 4 | 2 | 0.015 | 0.06 | 0.125 |
| Y67F_10 | 8 | 2 | 8 | 4 | 1 | 0.015 | 0.06 | 0.03 |
| CBS 14141 | >32 | 32 | >256 | >256 | >256 | 4 | 0.06 | 0.06 |
| F67Y_1 | 4 | >32 | 256 | >256 | >256 | 4 | 0.25 | 0.125 |
| F67Y_3 | 4 | >32 | >256 | >256 | >256 | 4 | 0.5 | 2 |
| F67Y_4 | 4 | >32 | >256 | >256 | >256 | 16 | 0.5 | 0.25 |
AMB, amphotericin B; TRB, terbinafine; CTZ, clotrimazole; MCZ, miconazole; FLZ, fluconazole; VRZ, voriconazole; KTZ, ketoconazole; ITZ, itraconazole.
These observations suggest that reduced azole susceptibility may be mediated by factors other than CYP51 in M. furfur and that the Y67F mutation mainly impacts AMB susceptibility in our selected strains. This is supported by the observation that the healthy-skin isolate 003 SC2 contains the Y67F CYP51 mutation but does not show the level of reduced azole susceptibility observed in disease isolate cluster 2.
Genes involved in metabolism and secondary metabolite production are upregulated in disease strains of M. furfur.
To further elucidate the pathways potentially involved in M. furfur azole susceptibility, we performed RNA-seq in regular modified Dixon (mDixon) medium for representative candidate strains from each cluster (healthy isolates, disease isolate cluster 1, and disease isolate cluster 2) based on their antifungal susceptibility testing (AFST) profiles (Fig. 1). They were defined as the wild type, susceptible strain CBS 7982 (healthy isolate cluster), the disease CYP51 mutant strain CBS 14141 (disease isolate cluster 2), and the intermediate disease strain CBS 7019 (disease isolate cluster 1).
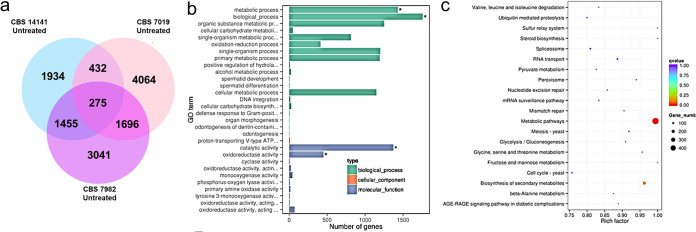
A list of the top 20 upregulated genes commonly expressed in disease isolates CBS 7019 and CBS 14141 is presented in Table S2. Genes involved in metabolism, biological process, catalytic activity, and extracellular activity were the key genes upregulated in the disease strains (Fig. 3b). Statistically enriched KEGG pathways include secondary metabolite production (Fig. 3c). Upregulation of some exons coding for ABC and MFS transporter proteins, such as YBT1, ITR1, and OPT1, was also observed.
FIG 3.
RNA-seq of CBS 7982, CBS 7019, and CBS 14141. (a) Venn diagram showing genes which are differentially expressed in CBS 7982, CBS 7019, and CBS 14141. (b) GO enrichment bar chart of differentially expressed terms. Asterisks indicate terms which are significantly enriched. (c) Scatterplot for KEGG enrichment results in CBS 7982 versus CBS 14141.
Healthy and disease isolates have different gene expression profiles following long-term exposure to clotrimazole in vitro.
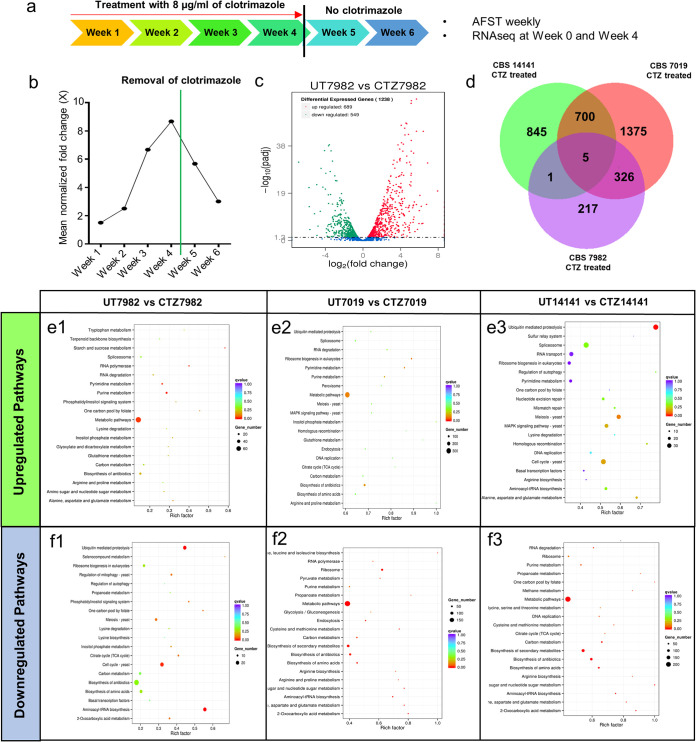
Long-term (up to 4 weeks) treatment of the wild-type healthy isolate CBS 7982 with clotrimazole (8 μg/ml) (Fig. 4a; Table S3) induced up to an 8-fold increase in clotrimazole MICs (Fig. 4b). After treatment removal at 4 weeks, MICs were observed to fall back to close to starting values. This suggests that elevated MICs may be induced in vitro via a transient mechanism. For the strains CBS 7019 and CBS 14141, long-term clotrimazole treatment in vitro did not produce any meaningful change in MIC readings, as their starting MICs were already high.
FIG 4.
Differentially expressed genes with long-term in vitro clotrimazole treatment. (a) Schematic of the 4-week treatment regimen with clotrimazole, followed by removal of clotrimazole after week 4 (black line). (b) Mean normalized fold change of CBS 7982 MICs during 4-week clotrimazole treatment and after clotrimazole removal (green line). (c) Volcano plot of the corresponding upregulated (red) and downregulated (green) genes between untreated CBS 7982 and clotrimazole-treated CBS 7982 at week 4. (d) Venn diagram showing numbers of differentially upregulated exons expressed in the clotrimazole treatment groups. (e and f) Scatterplots for KEGG enrichment showing upregulated (e) and downregulated (f) pathways in untreated (UT) versus clotrimazole treatment (CT) groups.
Differential gene expression analysis was performed on RNA-seq of isolates exposed to the above-mentioned MICs of clotrimazole for the three M. furfur strains (CBS 7982, CBS 7019 and CBS 14141). Of 1,238 differentially expressed genes between 4-week clotrimazole-treated and nontreated CBS 7982, 689 were upregulated (Fig. 4c and d). Examples of genes commonly upregulated by clotrimazole treatment include DNM1, encoding the dynamin protein, and RBP1P, encoding the yeast RNA-binding protein, (Table S4), which have been reported to affect antifungal susceptibility and calcineurin-dependent azole tolerance in C. albicans, respectively (30, 31). Metabolic pathways were commonly upregulated in 4-week clotrimazole-treated and nontreated groups for CBS 7982 and CBS 7019 (Fig. 4e1 and e2). KEGG pathways upregulated in clotrimazole-treated CBS 14141 were distinct from those in CBS 7982 and CBS 7019 and included the ubiquitin-mediated proteasome pathway and cell cycle (Fig. 4e3). A reverse trend was observed for KEGG pathways downregulated in the three strains, with metabolic pathways being downregulated in CBS7019 and CBS 14141 and the ubiquitin-mediated proteasome pathway being downregulated in CBS7982 (Fig. 4f1 to f3).
The ABC transporter encoded by PDR10 is upregulated after long-term exposure to clotrimazole in vitro.
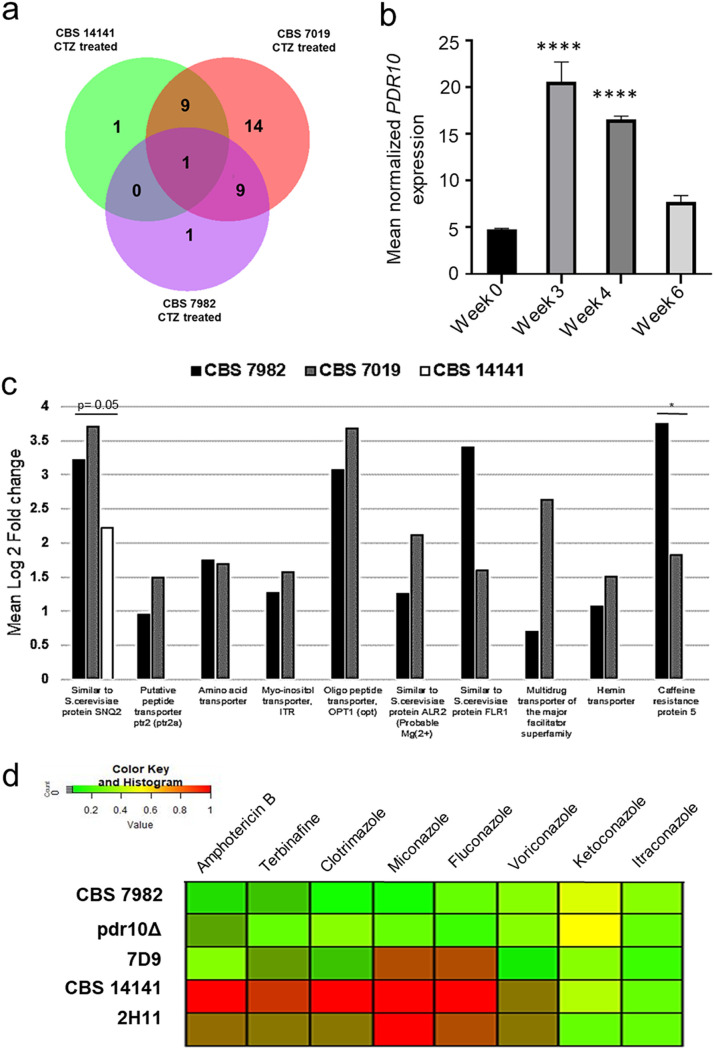
Narrowing RNA-seq gene identifications (IDs) to transporter pump proteins identified multiple reads matching exons mapping to “Similar to S. cerevisiae protein, SNQ2.” These reads were consistently the single most highly upregulated transporter gene in all three M. furfur strains (Fig. 5). Further, multiple-sequence alignment matched the exon sequence to that of the multidrug transporter encoded by PDR10, as described by Ianiri et al. (27). Further analysis of PDR10 gene expression via reverse transcription-quantitative PCR (RT-qPCR) at baseline and at 3-, 4-, and 6-week time points showed that PDR10 expression increased over the 4-week clotrimazole treatment period and dropped at 6 weeks, 2 weeks after treatment was withdrawn (Fig. 5b).
FIG 5.
Transporter pump gene expression. (a) Venn diagram showing transmembrane transporter genes which are differentially expressed in clotrimazole-treated CBS 7982, CBS 7019, and CBS 14141. (b) Mean normalized PDR10 gene expression of CBS 7982 during and after clotrimazole treatment. Values are means and SD. A one-way ANOVA was used to compare each time point against the no-treatment time point, followed by Dunnett’s test (****, P < 0.001). (c) Mean log2 fold change of key transporter pump genes upregulated after 4-week exposure to clotrimazole in CBS 7982, CBS 7019, and CBS 14141. (d) Heat map of MICs for CBS 7982, CBS 14141, the pdr10Δ mutant, and transformants 7D9 and 2H11. MICs were normalized from 0 to 1, with 1 being the highest MIC (red).
RNA-seq analysis also revealed that nine other transporter genes were upregulated in the clotrimazole-treated disease isolates CBS 7019 and CBS 14141 but not CBS 7982, including the previously reported mitochondrial ABC transporter, encoded by ATM1 (Fig. 5a; Table S5). Other highly upregulated transporter genes in clotrimazole-treated CBS 7982 and CBS 7019 include the oligopeptide protein transporter gene OPT1, the multidrug transporter gene FLR1, and the caffeine resistance protein gene CRP5 (Fig. 5c), which have not yet been sequence validated in all isolates of the divergent M. furfur strains. These findings suggest that differential expression of transporter genes between different isolates may also affect antifungal susceptibility.
Deletion of PDR10 abrogates elevated MICs in disease isolate CBS 14141.
The pleiotropic drug transporter gene PDR10 was identified to be significantly upregulated in clotrimazole-treated M. furfur strain CBS 7982 as described above. It was also found to play a role in Malassezia fluconazole resistance by Ianiri et al. (27), for which the CBS 14141 insertional mutants 7D9 and 2H11 and the CRISPR (clustered regularly interspaced palindromic repeats)/Cas9 pdr10Δ deletion mutant were constructed using the high-MIC mutant strain CBS 14141. 7D9 has a T-DNA insertion in the putative promoter region of PDR10, whereas 2H11 was previously described to have a chromosomal rearrangement mutant involving the gene ERG5 (27). We confirmed that the PDR10 gene was absent in the pdr10Δ mutant genome and that the pdr10Δ mutant showed null PDR10 expression (Fig. S3).
AFST was performed on the insertional mutants 7D9, 2H11, and the CRISPR/Cas9 pdr10Δ mutant alongside wild-type CBS 7982 and mutant CBS 14141 M. furfur strains (Fig. 5d; Table 3). Deletion of PDR10 in CBS 14141 completely reversed the elevated MIC phenotype. The pdr10Δ strain showed an AFST profile close to that of the wild type (i.e., CBS 7982) (Fig. 5d). The 7D9 mutant exhibited intermediate susceptibility to the tested azoles, with amphotericin B, terbinafine, and clotrimazole having lower MICs, whereas 2H11 had elevated MICs comparable to those of CBS 14141. Treatment with ABC transport inhibitors (verapamil, carbamazepine, and trifluoperazine) did not reduce antifungal susceptibility in M. furfur (data not shown).
TABLE 3.
MICs for PDR10 transformants
| Strain | MIC (μg/ml) ofa: |
|||||||
|---|---|---|---|---|---|---|---|---|
| AMB | TRB | CTZ | MCZ | FLZ | VRZ | KTZ | ITZ | |
| CBS 7982 | 4 | 2 | 4 | 4 | 1 | 0.03 | 0.03 | 0.03 |
| PDR10Δ | 2 | 0.25 | 1 | 2 | 2 | 0.03 | 0.125 | 0.125 |
| 7D9 | 1 | 2 | 8 | 32 | 4 | 0.5 | 0.25 | 0.06 |
| CBS 14141 | >32 | 32 | >256 | >256 | >256 | 4 | 0.06 | 0.06 |
| 2H11 | 4 | 1 | 8 | >256 | 128 | 8 | 0.25 | 0.125 |
AMB, amphotericin B; TRB, terbinafine; CTZ, clotrimazole; MCZ, miconazole; FLZ, fluconazole; VRZ, voriconazole; KTZ, ketoconazole; ITZ, itraconazole.
These observations are primarily relevant to M. furfur strains from the CBS 14141 genomic background (the pdr10Δ mutant, 7D9, and 2H11). Analysis of PDR10 gene expression in other healthy and disease isolates did not show correlation between PDR10 expression and MIC phenotype (Fig. S3B), suggesting that baseline PDR10 gene expression may not be an indicator of reduced azole susceptibility in all M. furfur strains.
Wild-type and mutant strains show differences in rhodamine 6G efflux.
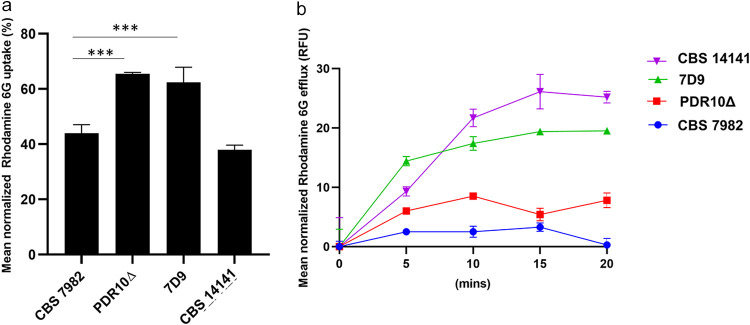
The fluorescent small-molecule dye rhodamine 6G is commonly used to assess the efflux activity of ABC transporters (32). The pdr10Δ mutant and 7D9 were observed to have a higher uptake of rhodamine 6G (Fig. 6a) than CBS 7982 and CBS 14141 (Fig. 6a). This is consistent with literature which has reported that rhodamine 6G efflux is correlated with CDR1 expression (33). CBS 14141 was observed to have the highest rhodamine 6G efflux of the four strains, and CBS 7982 had the lowest (Fig. 6b). Surprisingly, the pdr10Δ mutant had rhodamine 6G efflux levels close to those of CBS 7982, suggesting that PDR10 is a key effector of rhodamine 6G efflux. The insertional mutant 7D9 showed intermediate rhodamine 6G efflux levels between those of CBS 7982 and CBS 14141.
FIG 6.
Rhodamine 6G efflux in PDR10 mutant strains. (a) Percent normalized rhodamine uptake (mean and SD). A one-way ANOVA was used to compare each time point against the no-treatment time point, followed by Dunnett’s test (***, P < 0.005). (b) Mean normalized rhodamine 6G efflux (RFU, relative fluorescence units) of wild-type (CBS 7982), mutant (CBS 14141), and transformant (pdr10Δ and 7D9) strains over 20 min.
DISCUSSION
In this study, we profiled the antifungal susceptibility of multiple M. furfur isolates collected from healthy and disease states and elucidated mechanisms underlying their antifungal susceptibility. Strains isolated from individuals with skin or systemic disease had elevated antifungal MICs relative to strains isolated from healthy individuals. Isolates of M. furfur from healthy and disease individuals cluster into groups which reflect their health/disease origin based on their antifungal susceptibility profiles. Isolates from individuals with systemic disease (e.g., blood, urine, and catheter isolates) were the most likely to have elevated MICs. However, exceptions include CBS 9374, which is from a healthy individual but still shows elevated MICs of terbinafine and miconazole. It is likely that the patterns of antifungal susceptibility observed in our disease isolates are stable, given that many of the disease strains have been maintained in culture and passaged repeatedly in fungal banks without antifungal exposure. However, a lack of information on the clinical background of many disease isolates (i.e., antifungal treatment prior to culture isolation and treatment dosage/duration) hampers further interpretation regarding how reduced azole susceptibility could have arisen and whether it is intrinsic or acquired.
Although mutations in CYP51 are well documented to result in reduced azole susceptibility in fungi (17), this has not been functionally established for Malassezia. For Malassezia, only M. globosa (MGL_2415), Malassezia sympodialis (MSYG_3973), and M. pachydermatis (Malapachy_4010) have annotated CYP51 genes available, with limited literature on the functional role of CYP51 mutations. Kim et al. described three CYP51 mutations in an azole-resistant isolate of M. globosa, Y127F, A169, and K176N (15), although the extent to which they confer azole resistance was not reported. The large-scale construction of transformants from different strain backgrounds is currently limited by the need for strain-specific sequence data (preferably whole-genome sequencing data) for the design of 1.5-kb flanking arms for homologous recombination required in Agrobacterium tumefaciens-mediated transformation.
The presence of the Y67F mutation in the healthy-scalp isolate 003 SC2 suggests that the mutation alone is insufficient to confer reduced azole susceptibility. A synonymous Y134F mutation in the plant rust fungus Puccinia triticina was documented to have limited impact on its susceptibility to epoxiconazole (34). Our F67Y mutation in CBS 14141 was observed to result in increased amphotericin B susceptibility. This is consistent with observations in Leishmania mexicana, in which a single nucleotide mutation in CYP51 was also reported to induce amphotericin B resistance (35), and suggests a role for this gene in amphotericin B susceptibility.
Rapid transient increases in the mRNA expression of CDR efflux pump genes in C. albicans after exposure to fluconazole have been reported (36, 37). Transporter pump genes such as PDR10 were observed to be upregulated after exposure to clotrimazole based on RNA-seq data. These genes were validated based on known homology to similar proteins in Saccharomyces cerevisiae or based on existing literature. While the transporter genes PDR10 (27), FLR1 (38), and ITR1 (39) have been reported to be associated with azole resistance, there is little information on the role of other transporter genes, such as OPT1 and CRP5, in multidrug resistance. A putative paralog of PDR10 was identified immediately adjacent and 3′ to PDR10 and was identified as PDR10_2 by Ianiri et al. (27). This warranted further gene validation to ensure that the pdr10Δ mutant did not include a knockout of PDR10_2 (Fig. S3). However, we confirmed that a single knockout of the PDR10 gene (i.e., pdr10Δ) was sufficient to reduce MICs to wild-type levels.
While we demonstrated an increase in clotrimazole MICs after 4 weeks of successive treatment, the effect was dose dependent, and the phenomenon is best observed at antifungal concentrations two to four steps higher than the known MIC for the specific strain. While higher treatment doses may promote higher fold changes in MICs, they are less well tolerated, and the subsequent low viable inoculum impedes susceptibility testing.
At present, PDR10 expression in CBS 7982 appears to be strongly inducible by the presence of clotrimazole and likely azoles and other stress factors such as chemicals, UV exposure, elevated temperature and nutrient-limiting conditions (27). As described previously, a deletion in PDR10 for CBS 14141 resulted in increased sensitivity to fluconazole and benomyl, which are antifungal drugs that have different mechanisms of action (27). This suggests a critical role of PDR10 in nonspecific (or pleiotropic) cellular detoxification in M. furfur CBS 14141, most likely through active efflux of xenobiotics, including antifungal drugs, in line with the known function of ABC transporters. Further transcriptional analysis and deletion models in more strains and species of Malassezia are required to validate the role of ABC transporters in Malassezia azole resistance. This current progress is limited by the poor practical efficiency of generating ATMT plasmid constructs and the need for strain-specific knowledge of genome regions upstream and downstream of the gene of interest. While this has been improved by the development of a CRISPR/Cas9 system (27), challenges remain in the targeting of specific genes (versus unknown homologs) and the role of random insertions as confounding factors.
While rhodamine 6G assays have been useful in giving us an approximation of the relative ABC transporter efflux activity in strains from the same background (i.e., mainly CBS 14141), many transporter pumps are pleiotropic, and more specific binding assays are likely required to narrow down the role of specific pumps across multiple M. furfur strains. For the same reasons, the use of generic ABC transport inhibitors, such as verapamil and carbamazepine, was not useful for comparison across different strains.
In summary, we identified differences in azole antifungal susceptibility patterns of 26 isolates of M. furfur (13 from healthy subjects and 13 from those with disease). While a Y67F mutation was identified as a contributor to intrinsic resistance in some systemic-disease isolates, its impact on antifungal susceptibility must be interpreted on a strain-by-strain basis in context with other physiological factors. In CBS 7982 and CBS 14141, acquired resistance to clotrimazole and likely other azoles appear to be largely associated with differential activity of ABC transporters, particularly that encoded by PDR10. This has been validated by gene expression and functional deletion studies. There are likely additional factors contributing to interstrain differences, which are a result of multiple environmental stresses and other selection pressures. Understanding the functional mechanisms underlying azole resistance in Malassezia across a spectrum of strains, sources, and genetic backgrounds will be useful for the identification of new therapeutic targets to prepare for the emergence of new resistant strains.
MATERIALS AND METHODS
Strains and culture conditions.
No active primary culture isolation was performed in this study. All 26 isolates used in this study were either obtained from the Westerdijik Fungal Diversity Institute or obtained from previously published studies (40–42). Based on data available in fungal bank records and from previous publications, (13 of the strains were isolated from healthy individuals and 13 from patients with preexisting skin or medical conditions) (Table 1). Strains CBS 14141, JLPK13, Mal18, Mal24, Mal25, Mal26, Mal32, and PM315 were a kind contribution from Bart Theelen and Claudia Cafarchia. All Malassezia furfur strains were maintained on modified Dixon agar or broth at 32°C as described previously (10, 40).
Antifungal susceptibility testing.
Antifungal susceptibility testing was performed using a broth microdilution method as described by Leong et al. (10). Briefly, 200× drug stock dilutions were prepared at a 2× concentration in fresh OptiMAL medium. Amphotericin B, terbinafine, clotrimazole, miconazole, itraconazole, fluconazole, voriconazole, and ketoconazole were purchased from Sigma-Aldrich, Singapore. Stock and drug plate dilutions were prepared in accordance with CLSI and EUCAST guidelines. Yeast inocula were obtained from 4- to 7-day-old strains of Malassezia spp. A 50-μl yeast inoculum was added to 50 μl of 2× concentrated antifungals to achieve a final cell density of 5 × 103 to 5 × 104 CFU/ml. A 10-μl portion of 2× yeast inoculum that had been diluted 10 times was also plated onto a modified Leeming-Notman agar plate and incubated for 4 to 7 days at 35°C for postverification of the CFU inoculum (10 to 100 colonies per 10 μl). Each assay was performed in triplicate plates for a single culture at every individual time point or reading.
Long-term in vitro treatment with clotrimazole.
CBS 7982 was maintained in triplicate cultures of mDixon broth containing 8 μg/ml of clotrimazole for 4 weeks at 32°C, with fresh medium and antifungal supplied every alternate day. After 4 weeks, medium was replaced with fresh medium only (i.e., no antifungal) every alternate day for another 2 weeks. A 100-μl portion of culture was removed every 7 days for antifungal susceptibility testing as described above. For CBS 7019 and CBS 14141, clotrimazole concentrations were both 256 μg/ml. RNA extraction was performed on aliquots of each triplicate samples at log phase in week 0 (before addition of clotrimazole) and week 4 (after 4-week clotrimazole treatment) for RNA-seq analysis.
Gene analysis.
All primer sequences used in this study are listed in Table S5. Unannotated gene sequences for CYP51 and PDR10 were derived from a nucleotide BLAST of the whole-genome sequencing (WGS) data (NCBI BioProject database no. PRJNA286710 [43]) or by sequencing the specific gene using the self-designed primers (Table S5). PCR was performed using Platinum Taq DNA polymerase (Thermo Fisher, Singapore), TaKaRa HS DNA polymerase (TaKaRa Bio USA Inc.) for 1.5-kb flanking arms, and LongAmp (New England Biolabs [NEB], Singapore). Gel extraction of the PCR product was performed with a QIAquick gel extraction kit per the manufacturer’s instructions (Qiagen, Singapore). Sanger sequencing was performed on the purified PCR product using BigDye (Thermo Fisher, Singapore) per the manufacturer’s instructions. RT-qPCR was performed using the GoTaq one-step RT-qPCR system (Promega, Singapore) or a Superscript transcriptase III kit (Thermo Fisher, Singapore) with the respective gene-specific primers using the Malassezia actin gene ACT1 as the housekeeping gene. PCR with reverse-transcribed cDNA for actin was used to detect genomic-DNA contamination. Relative gene expression was analyzed using the Q-Gene module as described by Muller et al. (44).
Gene sequences derived from Sanger sequencing were translated into protein sequences using the ExPASy translate tool (https://web.expasy.org/translate/) and analyzed using multiple-sequence alignment (http://multalin.toulouse.inra.fr/multalin/multalin.html) (45).
Agrobacterium tumefaciens-mediated transformation.
For ATMT, 1.5-kb upstream and downstream flanking arms of CYP51 were derived from PCR (TaKaRa Bio USA Inc.) of CBS 7982 or CBS 14141 genomic DNA and assembled with CBS 7982 CYP51 gene blocks (TTC/TAC) (Integrated DNA Technologies, Singapore) in the binary vector, pGI3, and the nourseothricin (NAT) and neomycin (NEO) plasmid cassettes pAIM2 and pAIM6 under the control of the M. sympodialis ACT1 promoter and terminator. Insertional mutagenesis was performed using the A. tumefaciens strain EHA105 as described previously (46).
RNA-seq.
Total RNA was extracted and purified from log-phase cultures of CBS 7982, CBS 7019, and CBS 14141. Three biological replicates of each sample were analyzed. Briefly, cell pellets were washed three times in sterile phosphate-buffered saline (PBS) before resuspension in TRIzol and frozen at −80°C overnight. On thawing, samples were subjected to bead beating followed by use of the Direct-zol RNA miniprep kit (Zymo Research, USA), following the manufacturer's instructions. An additional DNase step was performed with the Turbo DNase kit (Thermo Fisher, Singapore) per the manufacturer’s instructions. RNA sequencing and differential analysis services were provided by Novogene AIT, Singapore. One microgram of RNA was used for library preparation using a NEBNext Ultra RNA library prep kit for Illumina (NEB, USA) per the manufacturer’s instructions. Next, 125-bp/150-bp paired-end reads were generated from an Illumina-based sequencing platform and processed with the appropriate quality controls. Differential expression analysis (untreated versus clotrimazole treated, healthy versus disease isolate) was performed using the DESeq R package (1.18.0).
Data and statistical analysis.
All assays were performed as three independent experiments, each with triplicate readings unless otherwise stated. For pairwise comparisons, a paired two-tailed Student's t test was performed with Microsoft Excel. For grouped comparisons, a one-way analysis of variance (ANOVA) with Dunnett’s test was performed with GraphPad Prism 8 (GraphPad Software, CA, USA). For heat map plotting of MICs, all MICs were normalized across the different antifungal concentrations such that the highest values were set at 1 and the lowest value were calculated as 1/total number of concentrations. Heat maps and Venn diagrams were plotted using the gplots package in R (version 3.03).
Rhodamine 6G assay.
The rhodamine 6G efflux assay was performed as described previously (30). Briefly, equal volumes of cells were normalized to an optical density at 600 nm (OD600) of 0.2 and incubated with 10 μM rhodamine 6G (Tee Hai Chem, Singapore) in PBS for 30 min at 32°C. Next, cells were transferred to 4°C and spun down to collect the supernatant. Pellets were washed twice with PBS and resuspended in equal volumes of PBS containing 2% glucose and incubated at 32°C. An aliquot of solution was removed at 0-, 5-, 10-, 15-, and 20-min intervals and spun down to collect the supernatant. Each sample was plated in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bart Theelen and Claudia Cafarchia for the use of the Mal24, Mal25, Mal26, Mal32, PM315, CBS 14141, and JLPK13 disease isolates.
This work was supported by an Asian Skin Microbiome Program IAF-PP grant (H1801a016).
C.L., J.W.K.C., S.M.L., G.I., and T.L.D. were involved in the conception and design of the study. Data collection was performed by C.L., J.W.K.C., and S.M.L. C.L., J.W.K.C., S.M.L., Y.I.L., J.P.Z.G., and T.L.D. were involved in the data interpretation and analysis. C.L., G.I., and T.L.D. were involved in the writing and critical revision of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hyde KD, Al-Hatmi AMS, Andersen B, Boekhout T, Buzina W, Dawson TL, Eastwood DC, Jones EBG, de Hoog S, Kang Y, Longcore JE, McKenzie EHC, Meis JF, Pinson-Gadais L, Rathnayaka AR, Richard-Forget F, Stadler M, Theelen B, Thongbai B, Tsui CKM. 2018. The world’s ten most feared fungi. Fungal Divers 93:161–194. 10.1007/s13225-018-0413-9. [DOI] [Google Scholar]
- 2.Theelen B, Cafarchia C, Gaitanis G, Bassukas ID, Boekhout T, Dawson TL. 2018. Malassezia ecology, pathophysiology, and treatment. Med Mycol 56:S10–S25. 10.1093/mmy/myx134. [DOI] [PubMed] [Google Scholar]
- 3.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM. 2019. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25:377–388.E6. 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, Guo Y, Saxena A, Vardhan M, Diskin B, Wang W, Leinwand J, Kurz E, Kochen Rossi JA, Hundeyin M, Zambrinis C, Li X, Saxena D, Miller G. 2019. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574:264–267. 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havlickova B, Czaika VA, Friedrich M. 2008. Epidemiological trends in skin mycoses worldwide. Mycoses 51:2–15. 10.1111/j.1439-0507.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- 6.Chang HJ, Miller HL, Watkins N, Arduino MJ, Ashford DA, Midgley G, Aguero SM, Pinto-Powell R, von Reyn CF, Edwards W, McNeil MM, Jarvis WR, Pruitt R. 1998. An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonization of health care workers’ pet dogs. N Engl J Med 338:706–711. 10.1056/NEJM199803123381102. [DOI] [PubMed] [Google Scholar]
- 7.Chinn R, Pong A, Schultz K, Kim J, Kaegi D, Rasmussen M, Woerle C, Malagon-Maldonado G, Neder C, Beer K, Chow N, Glowicz J, Lockhart S, Jackson B, Litvintseva A. 2017. A cluster of fluconazole-resistant Malassezia pachydermatis in a neonatal intensive care unit—California, 2015–2016. Open Forum Infect Dis 4:S176–S177. 10.1093/ofid/ofx163.321. [DOI] [Google Scholar]
- 8.Ilahi A, Hadrich I, Goudjil S, Kongolo G, Chazal C, Léké A, Ayadi A, Chouaki T, Ranque S. 2018. Molecular epidemiology of a Malassezia pachydermatis neonatal unit outbreak. Med Mycol 56:69–77. 10.1093/mmy/myx022. [DOI] [PubMed] [Google Scholar]
- 9.Archer-Dubon C, Icaza-Chivez ME, Orozco-Topete R, Reyes E, Baez-Martinez R, De León SP. 1999. An epidemic outbreak of Malassezia folliculitis in three adult patients in an intensive care unit: a previously unrecognized nosocomial infection. Int J Dermatol 38:453–456. 10.1046/j.1365-4362.1999.00718.x. [DOI] [PubMed] [Google Scholar]
- 10.Leong C, Buttafuoco A, Glatz M, Bosshard PP. 2017. Antifungal susceptibility testing of Malassezia spp. with an optimized colorimetric broth microdilution method. J Clin Microbiol 55:1883–1893. 10.1128/JCM.00338-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beardsley J, Halliday CL, Chen SCA, Sorrell TC. 2018. Responding to the emergence of antifungal drug resistance: perspectives from the bench and the bedside. Future Microbiol 13:1175–1191. 10.2217/fmb-2018-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeina A, Kanafani JRP. 2008. Resistance to antifungal sgents: mechanisms and clinical impact. Clin Infect Dis 46:120–128. 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 13.Rogers PD, Barker KS. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 47:1220–1227. 10.1128/aac.47.4.1220-1227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman M, Cloud GA, Smedema M, Lemonte A, Connolly P, Mckinsey DS, Kauffman CA, Moskovitz B, Wheat LJ, Flanigan C, Gutsch H, Weissinger B, Baruch A, Dine A, Smith D, Lee B, Nixon H, Bamberger D, Simpson D, Black J, Norris S, Slama T, Ryan S, Richardson J, McKinsey J, Lancaster D, Ray D, Threlkeld M. 2000. Does long-term itraconazole prophylaxis result in in vitro azole resistance in mucosal Candida albicans isolates from persons with advanced human immunodeficiency virus infection? Antimicrob Agents Chemother 44:1585–1587. 10.1128/aac.44.6.1585-1587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D, Lim YR, Ohk SO, Kim BJ, Chun YJ. 2011. Functional expression and characterization of CYP51 from dandruff-causing Malassezia globosa. FEMS Yeast Res 11:80–87. 10.1111/j.1567-1364.2010.00692.x. [DOI] [PubMed] [Google Scholar]
- 16.Park M, Cho YJ, Lee YW, Jung WH. 2020. Genomic multiplication and drug efflux influence ketoconazole resistance in Malassezia restricta. Front Cell Infect Microbiol 10:191. 10.3389/fcimb.2020.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker JE, Warrilow AGS, Price CL, Mullins JGL, Kelly DE, Kelly SL. 2014. Resistance to antifungals that target CYP51. J Chem Biol 7:143–161. 10.1007/s12154-014-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M, Cho YJ, Park M, Choi Y, Hwang SY, Jung WH. 2018. Genomic tandem quadruplication is associated with ketoconazole resistance in malassezia pachydermatis. J Microbiol Biotechnol 28:1937–1945. 10.4014/jmb.1810.10019. [DOI] [PubMed] [Google Scholar]
- 19.Warrilow AGS, Price CL, Parker JE, Rolley NJ, Smyrniotis CJ, Hughes DD, Thoss V, Nes WD, Kelly DE, Holman TR, Kelly SL. 2016. Azole antifungal sensitivity of sterol 14α-demethylase (CYP51) and CYP5218 from malassezia globosa. Sci Rep 6:27690. 10.1038/srep27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angileri M, Pasquetti M, De Lucia M, Peano A. 2019. Azole resistance of Malassezia pachydermatis causing treatment failure in a dog. Med Mycol Case Rep 23:23–58. 10.1016/j.mmcr.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kano R, Yokoi S, Kariya N, Oshimo K, Kamata H. 2019. Multi-azole-resistant strain of Malassezia pachydermatis isolated from a canine Malassezia dermatitis. Med Mycol 57:346–350. 10.1093/mmy/myy035. [DOI] [PubMed] [Google Scholar]
- 22.Prasad R, Rawal MK. 2014. Efflux pump proteins in antifungal resistance. Front Pharmacol 5:202. 10.3389/fphar.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasi M, Kumar Khandelwal N, Moorhouse AJ, Nair R, Vishwakarma P, Bravo Ruiz G, Ross ZK, Lorenz A, Rudramurthy SM, Chakrabarti A, Lynn AM, Mondal AK, Gow NAR, Prasad R. 2019. ABC transporter genes show upregulated expression in drug-resistant clinical isolates of candida auris: a genome-wide characterization of atp-binding cassette (abc) transporter genes. Front Microbiol 10:1445. 10.3389/fmicb.2019.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa C, Dias PJ, Sá-Correia I, Teixeira MC. 2014. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front Physiol 5:197. 10.3389/fphys.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi MJ, Won EJ, Shin JH, Kim SH, Lee WG, Kim MN, Lee K, Shin MG, Suh SP, Ryang DW, Im YJ. 2016. Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with fluconazole-less-susceptible isolates. Antimicrob Agents Chemother 60:3653–3661. 10.1128/AAC.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cafarchia C, Iatta R, Immediato D, Puttilli MR, Otranto D. 2015. Azole susceptibility of Malassezia pachydermatis and Malassezia furfur and tentative epidemiological cut-off values. Med Mycol 53:743–748. 10.1093/mmy/myv049. [DOI] [PubMed] [Google Scholar]
- 27.Ianiri G, Dagotto G, Sun S, Heitman J. 2019. Advancing functional genetics through Agrobacterium-mediated insertional mutagenesis and CRISPR/Cas9 in the commensal and pathogenic yeast Malassezia. Genetics 212:1163–1179. 10.1534/genetics.119.302329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CK, Leung SSF, Guilbert C, Jacobson MP, Mckerrow JH, Podust LM. 2010. Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole. PLoS Negl Trop Dis 4:e651. 10.1371/journal.pntd.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warrilow AG, Nishimoto AT, Parker JE, Price CL, Flowers SA, Kelly DE, David Rogers P, Kelly SL. 2019. The evolution of Azole resistance in Candida albicans Sterol 14-demethylase (CYP51) through incremental amino acid substitutions. Antimicrob Agents Chemother 63:e02586-18. 10.1128/AAC.02586-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truong T, Zeng G, Lim TK, Cao T, Pang LM, Lee YM, Lin Q, Wang Y, Seneviratne CJ. 2020. Proteomics analysis of Candida albicans dnm1 haploid mutant unraveled the association between mitochondrial fission and antifungal susceptibility. Proteomics 20:1900240. 10.1002/pmic.201900240. [DOI] [PubMed] [Google Scholar]
- 31.Cannon RD, Lamping E, Holmes AR, Niimi K, Tanabe K, Niimi M, Monk BC. 2007. Candida albicans drug resistance—another way to cope with stress. Microbiology (Reading) 153:3211–3217. 10.1099/mic.0.2007/010405-0. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharya S, Sobel JD, White TC. 2016. A combination fluorescence assay demonstrates increased efflux pump activity as a resistance mechanism in azole-resistant vaginal Candida albicans isolates. Antimicrob Agents Chemother 60:5858–5866. 10.1128/AAC.01252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi CH. 2005. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer Cell Int 5:30. 10.1186/1475-2867-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stammler G, Cordero J, Koch A, Semar M, Schlehuber S. 2009. Role of the Y134F mutation in cyp51 and overexpression of cyp51 in the sensitivity response of Puccinia triticina to epoxiconazole. Crop Prot 28:891–897. 10.1016/j.cropro.2009.05.007. [DOI] [Google Scholar]
- 35.Mwenechanya R, Kovářová J, Dickens NJ, Manikhandan M, Herzyk P, Vincent IM, Weidt SK, Burgess KE, Burchmore RJS, Pountain AW, Smith TK, Creek DJ, Kim DH, Lepesheva GI, Barrett MP. 2017. Sterol 14α-demethylase mutation leads to amphotericin B resistance in Leishmania mexicana. PLoS Negl Trop Dis 11:e0005649. 10.1371/journal.pntd.0005649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marr KA, Lyons CN, Rustad T, Bowden RA, White TC. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob Agents Chemother 42:2584–2589. 10.1128/AAC.42.10.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marr KA, Lyons CN, Ha K, Rustad TR, White TC. 2001. Inducible azole resistance associated with a heterogeneous phenotype in Candida albicans. Antimicrob Agents Chemother 45:52–59. 10.1128/AAC.45.1.52-59.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brôco N, Tenreiro S, Viegas CA, Sá-Correia I. 1999. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance inSaccharomyces cerevisiae and its benomyl-induced expression is dependent on Pdr3 transcriptional regulator. Yeast 15:1595–1608. . [DOI] [PubMed] [Google Scholar]
- 39.Claus S, Jezierska S, Van Bogaert INA. 2019. Protein-facilitated transport of hydrophobic molecules across the yeast plasma membrane. FEBS Lett 593:1508–1527. 10.1002/1873-3468.13469. [DOI] [PubMed] [Google Scholar]
- 40.Leong C, Schmid B, Toi MJ, Wang J, Irudayaswamy AS, Goh JPZ, Bosshard PP, Glatz M, Dawson TL. 2019. Geographical and ethnic differences influence culturable commensal yeast diversity on healthy skin. Front Microbiol 10:1891. 10.3389/fmicb.2019.01891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boekhout T, Kamp M, Guého E. 1998. Molecular typing of Malassezia species with PFGE and RAPD. Med Mycol 36:365–372. 10.1080/02681219880000581. [DOI] [PubMed] [Google Scholar]
- 42.Iatta R, Figueredo LA, Montagna MT, Otranto D, Cafarchia C. 2014. In vitro antifungal susceptibility of Malassezia furfur from bloodstream infections. J Med Microbiol 63:1467–1473. 10.1099/jmm.0.078709-0. [DOI] [PubMed] [Google Scholar]
- 43.Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, Sun S, Billmyre BR, Li W, Averette AF, Mieczkowski P, Heitman J, Theelen B, Schröder MS, De Sessions PF, Butler G, Maurer-Stroh S, Boekhout T, Nagarajan N, Dawson TL. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet 11:e1005614. 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller PY, Janovjak H, Miserez AR, Dobbie Z. 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32:1372–1374, 1376, 1378–1379. [PubMed] [Google Scholar]
- 45.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ianiri G, Averette AF, Kingsbury JM, Heitman J, Idnurm A. 2016. Gene function analysis in the ubiquitous human commensal and pathogen Malassezia genus. mBio 7:e01853-16. 10.1128/mBio.01853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.