Tuberculosis (TB) is a leading global cause of mortality owing to an infectious agent, accounting for almost one-third of antimicrobial resistance (AMR) deaths annually. We aimed to identify synergistic anti-TB drug combinations with the capacity to restore therapeutic efficacy against drug-resistant mutants of the causative agent, Mycobacterium tuberculosis.
KEYWORDS: Rv1258c, chlorpromazine, efflux, fusidic acid, potentiation, spectinomycin
ABSTRACT
Tuberculosis (TB) is a leading global cause of mortality owing to an infectious agent, accounting for almost one-third of antimicrobial resistance (AMR) deaths annually. We aimed to identify synergistic anti-TB drug combinations with the capacity to restore therapeutic efficacy against drug-resistant mutants of the causative agent, Mycobacterium tuberculosis. We investigated combinations containing the known translational inhibitors, spectinomycin (SPT) and fusidic acid (FA), or the phenothiazine, chlorpromazine (CPZ), which disrupts mycobacterial energy metabolism. Potentiation of whole-cell drug efficacy was observed in SPT-CPZ combinations. This effect was lost against an M. tuberculosis mutant lacking the major facilitator superfamily (MFS) efflux pump, Rv1258c. Notably, the SPT-CPZ combination partially restored SPT efficacy against an SPT-resistant mutant carrying a g1379t point mutation in rrs, encoding the mycobacterial 16S rRNA. Combinations of SPT with FA, which targets the mycobacterial elongation factor G, exhibited potentiating activity against wild-type M. tuberculosis. Moreover, this combination produced a modest potentiating effect against both FA-monoresistant and SPT-monoresistant mutants. Finally, combining SPT with the frontline anti-TB agents, rifampicin (RIF) and isoniazid, resulted in enhanced activity in vitro and ex vivo against both drug-susceptible M. tuberculosis and a RIF-monoresistant rpoB S531L mutant. These results support the utility of novel potentiating drug combinations in restoring antibiotic susceptibility of M. tuberculosis strains carrying genetic resistance to any one of the partner compounds.
INTRODUCTION
The increasing prevalence of multidrug-resistant tuberculosis (MDR-TB)—defined as resistance to the frontline anti-TB agents isoniazid (INH) and rifampicin (RIF)—necessitates the urgent development and implementation of new antimycobacterial drugs and therapeutic strategies (1, 2). A number of anti-TB compounds are currently in the drug discovery pipeline, with several others in advanced preclinical development (3). However, with the exception of bedaquiline (BDQ) and delamanid, no TB-specific drugs have been introduced into clinical use within the past 40 years (4). Therefore, new options need to be explored to address the problem of drug resistance.
Novel combination regimens comprising standard anti-TB agents and repurposed drugs represent a logical approach, especially where the new partner drug has already been approved for other clinical indications (5–7). Recent advances in understanding the physicochemical properties that determine drug distributions within complex tissue and cellular (micro)environments (8, 9), together with improved methods for rapid selection of multiple potentially synergistic drug partners in vitro and in vivo (8, 10–12), suggest the potential for rational development of novel combination approaches. This is important since it might address the long-held belief that developing combinations should be avoided owing to the complexities inherent in ensuring simultaneous and sustained delivery of the optimal partner compounds to the same target site (11). The impact of preexisting drug resistance on the utility of new drug combinations presents an additional challenge and is of particular concern when these combinations comprise current frontline anti-TB agents. To minimize the risks of exposing an individual to effective monotherapy, the likely preexistence of resistance to individual drugs must be recognized and informed combination approaches for drug therapies designed. These should incorporate multiple attributes beyond simply selecting individual molecules based on their biological activities as single agents (13).
In this study, we employed spectinomycin (SPT) as an anchor compound in combination with other experimental antibiotics and existing frontline anti-TB agents. SPT is an aminocyclitol antibiotic that inhibits protein synthesis by disrupting mRNA interactions with the 30S ribosome (14). Unlike other aminocyclitol antibiotics (including gentamicin and kanamycin [KAN]), SPT is not ototoxic (15) and has been used extensively in treating Neisseria gonorrhoeae infections in patients who cannot tolerate first-line treatments (16). From the perspective of new regimen design, SPT has been shown in combination screens against M. tuberculosis to synergize with several different classes of antimycobacterial compounds, both in vitro and in a macrophage model (10). Unfortunately, a key liability undermining its utility as a single agent is that SPT is subject to active efflux by M. tuberculosis—an observation that motivated an elegant medicinal chemistry solution in the development of the spectinamides (SPD) as derivative “efflux-resistant” anti-TB antibiotics (17–19). Spectinamides are also known to synergize with a variety of antibiotic classes (11), with lead spectinamide molecules, such as 1599, shown to be active against MDR M. tuberculosis strains and to synergize with existing and experimental anti-TB drugs in vivo (11, 20). However, developing suitable spectinamide formulations for therapeutic delivery remains an obstacle to the advancement of these compounds as novel anti-TB agents (21, 22).
We investigated the utility of potentiating combinations, anchored by SPT, to circumvent drug resistance and potentially restore (partial) susceptibility where genetically drug-resistant mutants preexist for one of the partner compounds. We applied combination screens utilizing (i) chlorpromazine (CPZ), a phenothiazine whose complex and unresolved mechanism of action involving disruption of the mycobacterial electron transport chain (23) has been implicated in efflux pump inhibition (24), and (ii) fusidic acid (FA), a translational inhibitor with demonstrated (albeit moderate) activity in vitro (25, 26). FA was selected owing to its potential for repositioning as anti-TB agent and because it possesses a unique mechanism of action, specifically, inhibition of bacterial protein synthesis by binding to elongation factor G (EF-G) (27). The antimicrobial-potentiating effect of FA with other antibiotics, including the frontline anti-TB drug ethambutol (EMB), as well as its lack of cross-resistance to other antimicrobial classes, provided additional motivation for our choice of FA (12, 28). By testing these combinations against both drug-susceptible M. tuberculosis H37Rv and selected drug-resistant mutants, we explored new potentiating combinations and demonstrated the utility of developing potent combinations against bacilli carrying preexisting genetic resistance to either of the partner drugs. In addition, this work revealed that the addition of SPT as third agent to the existing first-line anti-TB drug combination of RIF and INH restores activity in vitro against defined pre-MDR mutants of M. tuberculosis.
RESULTS
CPZ potentiates SPT activity by inhibiting Rv1258c-mediated efflux.
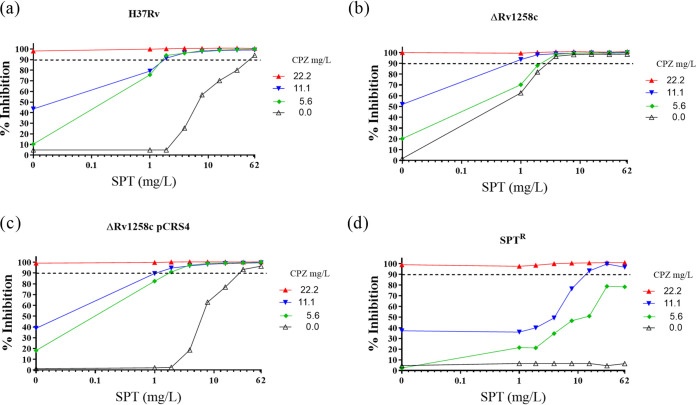
The combination of SPT and CPZ was previously reported as synergistic against Mycobacterium smegmatis (29). When tested against wild-type M. tuberculosis H37Rv (Fig. 1a), the same combination yielded a sum of fractional inhibitory concentration (ΣFIC) value of 0.09 (Table 1), confirming strong synergy (30). We investigated whether this effect resulted from CPZ-mediated disruption of the activity of the major facilitator superfamily (MFS) efflux pump, Rv1258c, which has been implicated in innate resistance to SPT (18). We performed checkerboard assays using the efflux-defective ΔRv1258c (“tap”) mutant, which had been used in the development of the spectinamides (SPD) (18), and its complemented derivative, ΔRv1258c pCRS4. Both the ΔRv1258c mutant and the complemented mutant exhibited the same MIC90 of 22 mg/liter for CPZ (Fig. 1 and Table 1), whereas the ΔRv1258c mutant was hypersusceptible to SPT, displaying an approximately 6-fold lower MIC90 of 3.9 mg/liter (Fig. 1b and Table 1; see also Table S1 in the supplemental material) as observed previously (31). Notably, the synergy detected on exposing wild-type M. tuberculosis to a combination of CPZ and SPT (Fig. 1a) was eliminated in the ΔRv1258c mutant (Fig. 1b)—which yielded a ΣFIC value of 0.75 (Table 1)—but was restored in the complemented ΔRv1258c pCRS4 strain (Fig. 1c), with a ΣFIC of 0.12 (Table 1). Previous studies have reported no significant alteration in Rv1258c transcription in response to CPZ treatment (32). Therefore, our observations suggested that CPZ treatment abrogated efflux-mediated intrinsic resistance to SPT in wild-type M. tuberculosis in a manner dependent on Rv1258c, perhaps as a result of CPZ-mediated inhibition of energy metabolism (23).
FIG 1.
Inhibition of Rv1258c-mediated efflux of SPT by addition of CPZ. Combinations of CPZ and SPT were applied in checkerboard assays against wild-type M. tuberculosis H37Rv (a), the ΔRv1258c mutant (b), the ΔRv1258c pCRS4 complemented mutant (c), and the SPTr strain (d). Bacterial growth inhibition was assessed in two independent experiments by fluorescence-based resazurin assay. The dashed horizontal line indicates 90% inhibition, and data are the means and standard deviations of two independent biological replicates.
TABLE 1.
Investigation of potential synergies between SPT and CPZ against different M. tuberculosis strains through the calculation of the FIC and sum of the FIC
| M. tuberculosis strain or mutant | Drug combination | MIC (mg/liter)a |
ΣFICb | Corresponding Fig. 1 panelc | |
|---|---|---|---|---|---|
| Alone | In combination | ||||
| H37Rv | SPT | 62 | 1.9 | 0.09 | a |
| CPZ | 22.2 | 1.4 | |||
| ΔRv1258c | SPT | 3.9 | 1.0 | 0.76 | b |
| CPZ | 22.2 | 11.1 | |||
| ΔRv1258c pCRS4 | SPT | 31.0 | 1.9 | 0.12 | c |
| CPZ | 22.2 | 1.4 | |||
| SPTr | SPT | >248 | 15.5 | 0.56 | d |
| CPZ | 22.2 | 11.1 | |||
The MIC was defined as the lowest drug concentration that inhibited growth by at least 90%.
The FIC of each drug was calculated as follows: (MIC of drug in combination)/(MIC of individual drug). The ΣFIC is the sum of the FICs of the two drugs where a ΣFIC of ≤0.5 is synergistic, ≥4.0 is antagonistic, and any value in the range 0.5 < x < 4.0 is considered additive or no interaction (64).
The respective figure panels show data of drug concentrations from which corresponding FIC values are calculated and the resulting ΣFIC computed.
CPZ synergizes with compounds other than SPT, but SPT potentiation arises solely from CPZ-mediated inhibition of Rv1258c.
To determine whether the synergy observed was specific for SPT or might apply to other antimycobacterial agents, CPZ was applied as the anchor compound in combination assays with a panel of anti-TB antibiotics of different classes and mechanisms of action (see Table S2 in the supplemental material). Of the eight compounds tested with CPZ, four exhibited clear synergy (ΣFIC ≤ 0.5) as follows: the frontline agents, RIF and INH, which returned ΣFICs of 0.37 and 0.5, respectively, and bedaquiline (BDQ) and nalidixic acid, both of which gave ΣFICs of ≤0.25. In contrast, no potentiation was observed with KAN or the fluoroquinolones, ciprofloxacin (CIP) and levofloxacin (LEV), all of which yielded ΣFICs of >0.5.
In a complementary approach, we also investigated if the potentiating effect observed with the SPT-CPZ combination was unique to CPZ. To this end, we assayed SPT in combination with an expanded panel of antimycobacterial agents (see Fig. S2 and Table S3 in the supplemental material). SPT was found to synergize with only 2 of the 11 compounds tested as follows: erythromycin (ERY), a macrolide targeting the ribosome, and verapamil (VER), a cationic amphiphile that was originally considered an M. tuberculosis efflux pump inhibitor but has been shown recently to disrupt membrane function (33). RIF, the mycobacterial RNA polymerase inhibitor, was just beyond the threshold determining synergistic activity.
To ascertain if inhibition of Rv1258c-mediated efflux resulted in the observed compound synergies, the ΔRv1258c mutant was tested for hypersensitivity to a corresponding panel of anti-TB agents (Fig. S3 and Table S1 in the supplemental material). The spectinamide 1599 had the same MIC90 for both wild-type M. tuberculosis H37Rv and the Rv1258c mutant, reflecting its successful modification to avoid Rv1258c-mediated efflux (18). Of the other 11 compounds tested, only SPT was associated with hypersensitivity in the Rv1258c-knockout mutant, returning an MIC90 value of 0.39 mg/liter compared to the MIC90 value of 62 to 125 mg/liter against the wild-type strain. In combination, these results strongly support the inference that the synergy detected with the CPZ-SPT combination arises from CPZ-mediated inhibition of Rv1258c.
The CPZ-SPT combination partially restores SPT sensitivity in an SPT-resistant mutant.
A spontaneous SPT-resistant mutant (SPTr) carrying a g1379t point mutation in the mycobacterial 16S rRNA, rrs, was associated with a >64-fold increase in the SPT MIC90 (see Table S1). In contrast, the activity of CPZ remained at the wild-type concentration for this strain, consistent with a mechanism of action of CPZ that was independent of rrs inhibition (34). While the SPTr mutant was resistant to SPT at concentrations of >248 mg/liter in the absence of CPZ, combinations utilizing CPZ at sub-MICs ([CPZ] ≤ 11.1 mg/liter) restored SPT sensitivity, at least partially (Fig. 1d). These results suggested the capacity for synergistic combinations to (partially) restore drug activity against mutant strains genetically resistant to either of the partner compounds.
Assessing synergy with SPT and fusidic acid, two antibiotics acting on the mycobacterial translational machinery.
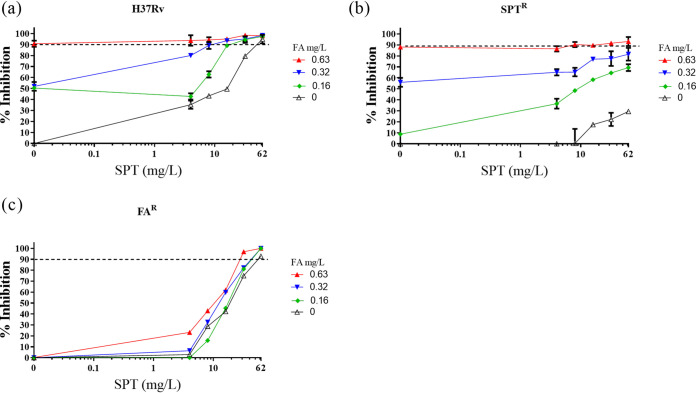
The systematic application of drug combinations can reveal synergistic interactions. One form of synergy occurs when drug(s) that perturb normal cell physiology trigger (compensatory) cellular responses that can, in turn, affect (potentiate) the activities of other drugs (35). Nichols et al. demonstrated that the synergistic interaction between sulfamethoxazole and trimethoprim was a result of the two drugs targeting tetrahydrofolate biosynthesis (36). With this in mind, a combination comprising SPT and FA—translation inhibitors, which act at discrete steps of the elongation process—was tested against wild-type M. tuberculosis and two resistant strains, FAr and SPTr mutants. Isolation of spontaneous FAr mutants in vitro yielded a single strain on 25× MIC FA at a frequency of ∼10−8. Whole-genome sequencing identified a c1384t (H462Y) substitution in fusA1 (Rv0684), encoding the essential mycobacterial elongation factor G (EF-G) (37). The histidine residue is highly conserved across multiple bacterial species; therefore, using the Thermus thermophilus structure as template (38), it can be inferred that M. tuberculosis H462 corresponds to T. thermophilus H458 (39), mutations of which are likely to alter the FA-binding pocket (40). In MIC assays, the H462Y mutant consistently yielded an MIC90 of ≥25 mg/liter, confirming heritable FAr (Table 2).
TABLE 2.
Investigation of potential synergies between SPT and FA against different M. tuberculosis strains through the calculation of the FIC and sum of the FIC
| M. tuberculosis strain | Drug combination | MIC (mg/liter)a |
ΣFICb | Corresponding Fig. 2 panelc | |
|---|---|---|---|---|---|
| Alone | In combination | ||||
| H37Rv | SPT | 62 | 16.3 | 0.5 | a |
| FA | 0.63 | 0.16 | |||
| SPTr | SPT | 62 | 62 | 1.5 | b |
| FA | 0.63 | 0.32 | |||
| FAr | SPT | 62 | 31.5 | 1.5 | c |
| FA | 25 | 25 | |||
The MIC was defined as the lowest drug concentration that inhibited growth by at least 90%.
The FIC of each drug was calculated as follows: (MIC of drug in combination)/(MIC of individual drug). The ΣFIC is the sum of the FICs of the two drugs where a ΣFIC of ≤0.5 is synergistic, ≥4.0 is antagonistic, and any value in the range 0.5 < x < 4.0 is considered additive or no interaction (64).
The respective figure panels show data of drug concentrations from which corresponding FIC values are calculated and the resulting ΣFIC computed.
With these strains in hand, we evaluated the interaction between SPT and FA and, furthermore, assessed whether this combination—which is synergistic against the parental, drug-susceptible M. tuberculosis H37Rv—might counter preexisting genetic resistance to either compound. The combination of SPT and FA exhibited a ΣFIC value of 0.50 (Table 2) against wild-type H37Rv; upon addition of FA at sub-MIC ([FA] = 0.16 mg/liter), the MIC90 of SPT exhibited an ∼4-fold decrease from 62 mg/liter to 16.3 mg/liter (Fig. 2a), P < 0.001. FA at subinhibitory concentration ([FA] = 0.32 and [FA] = 0.16 mg/liter) enhanced the activity of SPT ([SPT] = 62 mg/liter) against an SPTr mutant. The inhibitory effect was significantly different from the results observed with similar concentrations of SPT in the absence of FA ([FA] = 0 mg/liter), P < 0.001 (Fig. 2b). Although the calculated sum FIC did not satisfy the criterion for “synergistic” (ΣFIC ≤ 0.5) (Table 2), the effect was marked and reproducible in two independent biological replicates (Fig. 2b). Notably, the same combination did not return enhanced activity against the FAr mutant (Fig. 2c), strongly suggesting that FA was the major contributor to the SPT-FA combination. A summary of the inhibitory effects of the CPZ-SPT and FA-SPT combinations is presented in Table 3.
FIG 2.
In vitro interaction between SPT and FA. Combinations of SPT and FA were applied in checkerboard assays against wild-type M. tuberculosis H37Rv (a), the SPTr mutant (b), and the FAr mutant (c). Bacterial viability was assessed by fluorescence-based resazurin assay. Dashed horizontal lines indicate 90% inhibition, and data are the means and standard deviations of two independent biological replicates.
TABLE 3.
Summary of in vitro drug activities against M. tuberculosis H37Rv strains
Confirmation that fluorescence intensities correlate with cell density readings.
The centrality of the resazurin microtiter assay (REMA) in determining the synergies reported in this study demanded orthogonal evidence supporting the claimed results. To this end, a 96-well-based antimycobacterial assay was performed with a selected panel of drugs having different mechanisms of action (see Fig. S4 in the supplemental material). The experiments were conducted using the H37Rv::GFP reporter mutant in which expression of the fluorophore is constitutive (41). After 8 days of incubation in the presence of 2-fold dilutions of the antimicrobial agents, green fluorescent protein (GFP) and resazurin fluorescence intensities were determined, and the corresponding optical density readouts recorded in parallel. The values of the respective fluorescence intensities and that of optical density at 600 nm (OD600) were converted to percent inhibition. Strong agreement was generally discerned when comparing GFP and OD600 methods with the standard resazurin readout (Fig. S4), supporting the use of the resazurin assay for both MIC and FIC determinations.
A three-drug combination comprising SPT, RIF, and INH enhances in vitro activity against M. tuberculosis.
The premise that synergistic combinations might be usefully applied to overcome existing drug resistance was further explored using SPT in combination with the frontline anti-TB agents RIF and INH. In a two-dimensional (2D) pairwise screening assay, we evaluated two-drug permutations of RIF, INH, and SPT. The RIF-INH combination showed no interaction (see Fig. S5a in the supplemental material), while sub-MICs of INH (0.125 mg/liter) and RIF (0.003 mg/liter) reproducibly resulted in a modest (2-fold) reduction in the effective SPT concentration from 125 mg/liter to 62 mg/liter (Fig. S5b and c). To leverage the potential effect of SPT, a three-dimensional (3D) combination assay was performed in which RIF and INH were titrated against decreasing sub-MIC90 concentrations of SPT (1/2×, 1/4×, 1/8×, 1/16×, and 1/32× MIC90) using the format illustrated in Fig. S1b in the supplemental material. When tested against drug-susceptible M. tuberculosis H37Rv, RIF at sub-MIC ([RIF] = 0.003 mg/liter) plus SPT at both 1/4× and 1/2× MIC resulted in an 8-fold decrease in the effective concentration of INH from 0.25 mg/liter to 0.03 mg/liter (see Fig. S6 in the supplemental material). A kill kinetics assay showed that the addition of SPT to the RIF-INH combination elicited ∼1 log10 unit reduction (P < 0.001) in the viable bacillary population following 8-day exposure to the three-drug combination (see Fig. S7a in the supplemental material). In contrast, when the ΔRv1258c “tap” knockout mutant was tested, RIF at sub-MIC ([RIF] = 0.003 mg/liter) plus 1/2× MIC SPT resulted in only an ∼3-fold decrease in the effective concentration of INH, from 0.25 mg/liter to 0.09 mg/liter (Fig. S7b), again implicating Rv1258c in intrinsic antibiotic resistance in M. tuberculosis.
The RIF-INH-SPT combination is active in M. tuberculosis-infected macrophages and against monoresistant pre-MDR strains.
Since M. tuberculosis survives and replicates in macrophages (42), the synergy of RIF-INH plus SPT was evaluated against intracellular bacilli in M. tuberculosis-infected THP-1 cells. This three-drug combination showed inhibitory activity at 1× MIC90 of the combined drugs (see Fig. S8 in the supplemental material). In contrast, the inhibitory effect was reduced when similar concentrations of each drug were applied individually or when the standard RIF-INH combination was used without SPT. The intracellular activity of this triple combination was further confirmed by CFU enumeration (see Fig. S9 in the supplemental material), which revealed a 2-log10 decrease in CFU/ml when 1× MIC90 RIF-INH-SPT was applied compared to the untreated control.
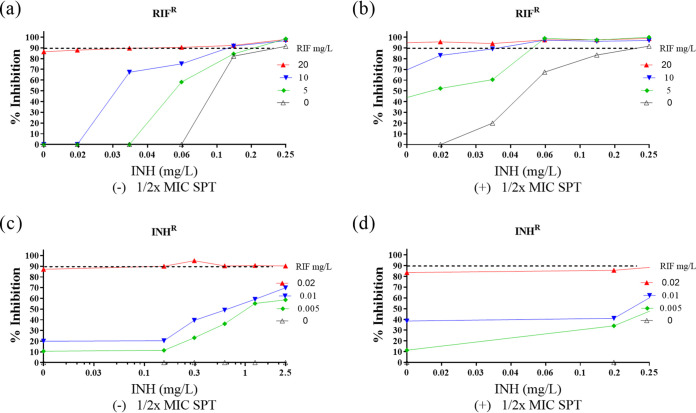
To evaluate the efficacy of RIF-INH-SPT against known drug-resistant strains, the combination was tested against two pre-MDR M. tuberculosis mutants as follows: an RIF-monoresistant mutant carrying the common rpoB S531L allele and an INH-monoresistant strain harboring a −c15t mutation in the inhA promoter region that confers low-level INH resistance (Fig. 3). Duplicate checkerboard experiments showed that, for the rpoBS531L (the S-to-L change at position 531 of RpoB) mutant, addition of 1/2× MIC SPT to the RIF-INH plate resulted in a decrease in the effective concentrations of both RIF (20 to 10 mg/liter) and INH (0.25 to 0.03 mg/liter) (Fig. 3a and b), indicating partial restoration of drug susceptibility in the presence of SPT. Enhanced susceptibility was also observed for the INHr mutant, albeit to a lesser extent: a sub-MIC RIF concentration of 0.01 mg/liter and INH at 2.5 mg/liter achieved ∼80% bacterial inhibition (Fig. 3c and d).
FIG 3.
Activity against pre-MDR M. tuberculosis strains. In vitro activity of RIF-INH against RIFr M. tuberculosis rpoBS531L mutant in the absence (a) or presence (b) of 1/2× MIC SPT and against the INHr M. tuberculosis inhA mutant in the absence (c) or presence (d) of 1/2× MIC SPT. The dashed horizontal line indicates 90% inhibition, and data are the means and standard deviations of two independent biological replicates.
DISCUSSION
Notwithstanding recent promising claims (43), the bacterial capacity for acquisition of resistance by multiple mechanisms means that it is difficult, perhaps even conceivably impossible, to overcome antibiotic resistance sustainably (44). Different approaches can be used to circumvent resistance transiently, ensuring antibiotic efficacy despite the preexistence of resistant organisms in an infecting population. Combination therapy represents one such approach.
In a previous study, Chen et al. demonstrated a synergistic interaction between SQ109 and RIF when tested against RIFr isolates: at 0.5× MIC, SQ109 was able to increase RIF’s activity against de facto resistant organisms in a dose-dependent manner (45). Recently, Yang et al. reported the enhanced efficacy of the imipenem-colistin combination against multiple drug-resistant Enterobacter cloacae bacteria in vitro and in an infection model (46). Relatively few studies have been undertaken to illustrate the association between potentiating drug interactions and the ability of the particular drug combination to overcome preexisting genetic resistance. In a clinical study, Ankomah et al. suggested that drugs acting synergistically can prevent treatment failure even when bacteria resistant to one of these drugs are present at the beginning of therapy (47). Our interaction studies between SPT and CPZ reaffirmed the susceptibility of SPT to Rv1258c-mediated efflux, an observation which suggests that efforts to modify SPT—including through novel chemical modifications that engineer resistance to efflux (18)—should be pursued.
We tested the susceptibility of the ΔRv1258c mutant to a small panel of antimycobacterial compounds with different mechanisms of action and observed that SPT alone was associated with hypersusceptibility. A similar hypersusceptibility phenotype was achieved against wild-type M. tuberculosis H37Rv via chemical potentiation of SPT using CPZ as the combination agent. However, the potentiating effect of CPZ was not specific to SPT and was instead observed for a handful of other agents. This suggests the likelihood that CPZ might disable more than one intrinsic resistance mechanism in M. tuberculosis. Further work is required to ascertain the precise mechanism for each compound, with evidence to date implicating multiple potential efflux systems in intrinsic resistance to INH, RIF, and the fluoroquinolones (48). For BDQ, the multisubstrate RND family transporter MmpS5-MmpL5 appears to be a strong candidate based on previous reports (49).
Our results revealed synergy between FA and SPT against drug-susceptible bacteria via a mechanism independent of the efflux inhibition seen with SPT-CPZ. Notably, the same FA-SPT combination exerted an enhanced inhibitory effect against a genotypically confirmed SPTr mutant compared to an FAr mutant. Although there is no definitive explanation for this finding, we postulate that the relative potency of FA (∼0.63 mg/liter) against the drug-susceptible H37Rv parent compared to that of SPT (∼50 mg/liter) could impact the FA-SPT combination against the SPTr mutant, restoring susceptibility. A similar effect was not evident in an SPT-FA combination against the FAr mutant, owing to the diminished activity of FA and high MIC90 of SPT. Of interest is the impact of individual active drugs in driving synergy.
Other explanations for potentiation include the sustained drug pressure emanating from the drug interactions, which leads to an increased effective dose of the drug combination. Moreover, some studies have demonstrated that the drug susceptibility of pathogens can be significantly enhanced as a result of a reduced efflux pump efficiency, either by genetic manipulation (50) or addition of efflux pump inhibitors (51, 52). The clinical relevance of this finding is that, despite the existence of bacterial resistance against a combination partner, it would still be possible to achieve optimal therapeutic outcomes via the use of appropriate potent drug combinations.
Previous work has demonstrated the potential of having three-drug combinations when compared to individual or two-drug regimens (53, 54). Recently, Tekin et al. reported that combinations of three different antibiotics can often overcome antimicrobial resistance to antibiotics, even when none of the three antibiotics on their own—or even two of the three together—is effective (55). In addition, based on drug interaction studies, Ramon-Garcia et al. hypothesized that the synergistic activity of the triplet combination might have multiplicative effects (10). Here, SPT was deployed as part of a three-drug regimen, which also included RIF and INH, the two drugs that form the cornerstone of TB treatment. Other studies have shown the interaction between RIF and INH against M. tuberculosis to have no interaction or to be mildly antagonistic (8, 56). The inclusion of SPT in this drug regimen was underpinned by reports that 24 out of 70 random combinations tested were synergistically active in M. smegmatis (10). This suggests a large unexplored pool of synergistic combinations. Notably, SPT exhibited synergy with several compounds both in vitro and ex vivo (10), even though the compound has a high MIC90 against M. tuberculosis when administered on its own (18).
In the three-drug combination assay, synergy resulted when subinhibitory concentrations (1/2× and 1/4× MICs) of SPT were titrated into media containing RIF and INH. This finding correlated well with the results of time-kill kinetics. However, the time-kill assay suggested that the inhibitory effect of the three-drug interaction was bacteriostatic (≤3 log10 CFU reduction) and not bactericidal. This observation reveals that the combination of RIF and INH—two bactericidal drugs that are most potent against actively dividing cells—shows bacteriostatic effects. Furthermore, the inhibition of growth induced by a bacteriostatic drug, SPT, results in an overall static effect when the drug is used in combination with a bactericidal drug. Other studies have shown that, in similar interactions, the resulting effect achieves a more efficient clearance at lower concentrations (31).
In attempting to exploit synergy for potential optimal treatment outcomes, an investigation of the RIF-INH plus SPT interaction was performed in rpoB and inhA mutants. The RIFr rpoB mutant had an MIC value of >2,000 times the MIC90 for drug-susceptible M. tuberculosis. Notably, addition of 1/2× MIC of SPT restored partial drug efficacy against this resistant mutant. As with the drug-susceptible H37Rv strain, a mechanistic explanation for the synergy observed using the RIF-INH plus SPT combination against the rpoB mutant is presently lacking. INH targets mycobacterial cell envelope biosynthesis, possibly enhancing permeation of SPT into the bacilli. However, access alone may not necessarily contribute to the synergistic interaction. Chen et al. reported synergy between SQ109, a presumed cell envelope inhibitor, and RIF (45). Conversely, EMB, which also affects mycobacterial cell envelope synthesis, did not exhibit synergy with RIF (57).
Prior reports have shown RIF to be an efficient inducer of cytochrome P450 (CYP 450), a superfamily of heme-containing enzymes involved in the biosynthesis of compounds, such as sterols, steroids, and fatty acids, as well as detoxification of xenobiotics and chemicals (58). RIF has been linked with the induction of CYP both in humans and in M. tuberculosis (59). The elevated levels of CYP have been associated with drug resistance due to the enhanced rate of elimination of the drugs by metabolism and detoxification pathways. INH, conversely, inhibits CYP in M. tuberculosis (59). This ability of INH to inhibit CYP may contribute to synergy in the RIF-INH plus SPT combination when the active form of INH is not rapidly eliminated inside M. tuberculosis and when SPT acts by further reducing the activity of CYPs.
There are prospects to combine SPT with RIF-INH in treatment regimens. SPT is given by intramuscular injection to achieve therapeutic concentrations in serum of about 100 mg/liter 1 h after a single 2-g dose. An over 4-fold increase in its effectiveness within a triple SPT-RIF-INH combination, as indicated by these data, would potentially allow for oral formulation, a critical delivery format when administering treatment to TB outpatients. In summary, these in vitro and ex vivo results suggest that the RIF-INH plus SPT triple-combination may be an effective therapeutic option for the treatment of both drug-susceptible and -resistant M. tuberculosis infections. They also reinforce a growing body of evidence supporting the utility of drug potentiation strategies in improving treatment outcomes.
MATERIALS AND METHODS
Chemicals and reagents.
All chemicals and solvents were purchased from Sigma-Aldrich. Working solutions of all antimicrobial agents were prepared in dimethyl sulfoxide (DMSO).
Bacterial strains and culture conditions.
The laboratory strain, M. tuberculosis H37Rv, its derivative mutants, and a reporter strain that has been used previously in high-throughput antimicrobial drug screening and constitutively expresses green fluorescent protein (GFP), H37Rv pMSP::eGFP (41), were maintained as freezer stocks. Strains were inoculated in standard Middlebrook 7H9 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) (Difco) and incubated as stationary cultures at 37°C for approximately 3 days, subcultured, and incubated until culture density was an OD600 of ∼0.5. A second reporter mutant, M. tuberculosis H37Rv::(pSMYC::mCherry) (60), which constitutively expresses the mCherry fluorophore, was grown in medium containing 50 mg/liter hygromycin. Cell suspensions were diluted to give an expected final concentration of 105 cells/ml at the time of inoculation into the microplate for the MIC assays.
Drug susceptibility assays.
The resazurin microtiter assay (REMA) was used to determine the susceptibility of drugs against M. tuberculosis strains as described (61). Briefly, 2-fold serial dilutions of compounds were performed on clear, round-bottom 96-well plates using 7H9-OADC medium. M. tuberculosis cultures, grown to an OD600 of 0.5 (∼108 cells/ml) and diluted 1,000-fold, were added at equal volume for a total volume of 100 μl per well. The plates were sealed in zip-lock bags and incubated at 37°C for 7 days, consistent with EUCAST guidelines (62) and published literature (63) recommending that MIC plates should be read after 7 and 14 days postinoculation. Resazurin dye was added and the plates incubated for a further 24 h. Fluorescence readings, at excitation and emission wavelengths of 540 and 590 nm, respectively, were recorded using a BMG Labtech POLARstar Omega plate reader (BMG Labtech, Offenburg, Germany) or a SpectraMax i3x plate reader (Molecular Devices). The lowest drug concentration that inhibited growth by at least 90% was determined from the dose-response curve; this concentration was defined as the MIC90 value.
Checkerboard assays. (i) 2D checkerboard.
Standard “two-dimensional” (2D) drug-drug interactions were determined by checkerboard titration in a 96-well plate (see Fig. S1a in the supplemental material). The 2D microdilution was carried out as described (45) with slight modification. Briefly, the first drug (A) to be serially diluted was dispensed (2 μl) along the x axis (columns 3 to 11; row B) at a starting concentration 100 times higher than the final concentration in the well, and 2 μl per well of the second drug (B) was serially dispensed along the y axis (from row B to H) at a starting concentration 100 times higher than the final concentration in the 96-well microtiter plate. The first column (column 1) and last column (column 12) contained drug-free controls (with 1% DMSO as a diluent) and a control drug concentration giving maximum inhibition, respectively. The second column from B2 to H2 and first row from A3 to A11 contained individual drugs, thus providing the MIC for each drug alone in each assay (each plate). The plates were placed in zip-lock bags and incubated at 37°C for 7 days. Resazurin dye was then added and the plates incubated for a further 24 h. A visible color change from blue to pink indicated growth of bacteria, and the visualized MIC was defined as the lowest concentration of drug that prevented growth (at which the color change did not occur) (61). Fluorescence readings (excitation, 544 nm; emission, 590 nm) were obtained using a BMG Labtech POLARstar Omega plate reader (BMG Labtech, Offenburg, Germany) or a SpectraMax i3x plate reader (Molecular Devices). The mean fluorescence value for the “maximum inhibition” wells (column 12) was subtracted from all other wells to control for background fluorescence. Percent inhibition was defined as 1 − (test well fluorescence units/mean fluorescence units of maximum inhibition wells) × 100 on day 8 after incubation. The lowest drug concentration effecting inhibition of 90% was considered the MIC90. In addition, synergy was interpreted according to the sum of fractional inhibitory concentration (ΣFIC). The fractional inhibitory concentration for each compound was calculated as follows (64): FICA − (MIC of compound A in the presence of compound B)/(MIC of compound A alone), where FICA is the fractional inhibitory concentration of compound A. Similarly, the FIC for compound B was calculated. The ΣFIC was calculated as FICA + FICB. Synergy was defined by values of ΣFIC of ≤0.5, antagonism by ΣFIC of >4.0, and no interaction by ΣFIC values from 0.5 to 4.0 (30).
(ii) 3D checkerboard.
In the three-drug (“three-dimensional” [3D]) combinations (Fig. S1b), microdilutions for the first two drugs were initially set up principally following the standard 2D checkerboard assay protocol described above. The third drug (2 μl) was then added at a starting concentration 100 times higher than the final concentration in the well as an overlay at five subinhibitory concentrations ranging from 1/32 to 1/2 of the single-drug MIC. Well A2 on all plates contained the third drug only, providing the single-drug MIC for the third drug in each assay (set of 5 plates). After inoculation with a log-phase culture, an OD600 of 0.5 (∼108 cells/ml) of M. tuberculosis, 50 μl to each well, the plates were placed in zip-lock bags and incubated for 7 days at 37°C before addition of resazurin. The plates were further incubated for 24 h, and the results were read in the BMG Labtech plate reader (excitation, 544 nm; emission, 590 nm) (63). The percent inhibition was calculated as described above.
Macrophage assays. (i) Cell culture and maintenance.
Human promonocytic THP-1 cells were maintained in RPMI 1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) at an initial density of 8 × 105 cells/ml at 37°C in a humidified, 5% CO2 atmosphere. Prior to plating of the cells, viability was assessed by trypan blue exclusion method (65). Maturation of THP-1 cells into macrophages was induced by adding 200 nM PMA (phorbol 12-myristate 13-acetate; Sigma) in cell culture medium for 24 h. Differentiated macrophages were then washed three times with prewarmed phosphate-buffered saline (PBS) to remove the PMA and replenished with cell culture medium.
(ii) Infection of macrophages and drug treatment.
To check the efficacy of drugs in macrophages, 5 × 104 THP-1 cells/well (100-μl final volume) in 96-well flat-bottomed tissue culture plates were differentiated into macrophages. To infect macrophages, an exponentially growing M. tuberculosis H37Rv::(pSMYC::mCherry) culture was harvested by centrifugation and washed twice with PBS. The pellet was resuspended in PBS and passed through a 5-μm filter to generate a suspension of single-cell bacilli. The bacterial suspension density was estimated by measuring OD at 600 nm, corresponding an OD600 of ∼0.5 to 1 × 108 CFU/ml. Infection medium comprised cell culture medium containing a number of bacteria required to achieve a multiplicity of infection (MOI) of 5:1 (5 bacilli for every THP-1 cell) (66). The macrophage cells were overlaid with infection medium and incubated at 37°C in 5% CO2 for the phagocytic period of 3 h. Cells were then washed gently and thoroughly with prewarmed PBS to remove extracellular bacteria. The cells from three wells were lysed by adding Triton X-100 (0.05% in PBS), and the lysate was plated onto 7H10 to score CFU for untreated day zero. The cells in remaining wells were treated with the indicated antibiotic either alone or in combination at 1× MIC90 or 5× MIC90 as determined in liquid culture. Hygromycin at 50 mg/liter was added into the cell culture medium for all wells to maintain the plasmid expressing mCherry.
(iii) Fluorescence measurement.
The fluorescence of the mCherry reporter (excitation, 590 nm; emission, 610 nm) was measured at different time points on a BMG Labtech plate reader.
(iv) CFU enumeration.
To estimate the numbers of live bacilli after drug treatment, untreated and drug-treated M. tuberculosis-infected cells were lysed in Triton X-100 (0.05% in PBS) on days 2, 4, and 6, and serial dilutions of the cell lysate were plated onto 7H10 agar. Colonies were counted after 3 to 4 weeks of incubation at 37°C.
Statistical analyses.
Statistical analyses were performed using Prism 9.0.0.121 (GraphPad). Means were compared via analysis of variance (ANOVA) with posttest evaluation using Dunnett's or Bonferroni's test. P values are abbreviated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council (SAMRC) with funds received from the South African Department of Science Innovation (to K.C. and D.F.W.). We acknowledge the support of the University of Cape Town, the SAMRC (to K.C. and V.M.), the South African Research Chairs Initiative (to K.C.) and Department of Science Innovation administered through the National Research Foundation, the Schlumberger Foundation Faculty for the Future scheme (to E.K. and A.W.), and the Research Council of Norway (to D.F.W.). Funding was provided by the following: The University of Cape Town, Novartis Research Foundation, South African Medical Research Council, Strategic Health Innovations Partnerships Unit of the South African Medical Research Council, South African Research Chairs Initiative of the Department of Science and Innovation administered through the South African National Research Foundation, Carnegie Mellon Scholarship Awards, and the Research Council of Norway.
We thank J. A. Aínsa for providing the M. tuberculosis ΔRv1258c (“tap”) knockout mutant and its complemented derivative, R. E. Lee for providing stocks of the spectinamide 1599, Krupa Naran for advice with checkerboard assays, and Mandy Mason for critical review of the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sullivan T, Ben Amor Y. 2016. Global introduction of new multidrug-resistant tuberculosis drugs-balancing regulation with urgent patient needs. Emerg Infect Dis 22:e151228. 10.3201/eid2203.151228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoagland DT, Liu J, Lee RB, Lee RE. 2016. New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv Drug Deliv Rev 102:55–72. 10.1016/j.addr.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans JC, Mizrahi V. 2018. Priming the tuberculosis drug pipeline: new antimycobacterial targets and agents. Curr Opin Microbiol 45:39–46. 10.1016/j.mib.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 12:388–404. 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 5.Nzila A, Ma Z, Chibale K. 2011. Drug repositioning in the treatment of malaria and TB. Future Med Chem 3:1413–1426. 10.4155/fmc.11.95. [DOI] [PubMed] [Google Scholar]
- 6.Oprea TI, Bauman JE, Bologa CG, Buranda T, Chigaev A, Edwards BS, Jarvik JW, Gresham HD, Haynes MK, Hjelle B, Hromas R, Hudson L, Mackenzie DA, Muller CY, Reed JC, Simons PC, Smagley Y, Strouse J, Surviladze Z, Thompson T, Ursu O, Waller A, Wandinger-Ness A, Winter SS, Wu Y, Young SM, Larson RS, Willman C, Sklar LA. 2011. Drug repurposing from an academic perspective. Drug Discov Today Ther Strateg 8:61–69. 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumla AI, Schito M, Maeurer M. 2014. Advancing the portfolio of tuberculosis diagnostics, drugs, biomarkers, and vaccines. Lancet Infect Dis 14:267–269. 10.1016/S1473-3099(14)70028-3. [DOI] [PubMed] [Google Scholar]
- 8.Cokol M, Kuru N, Bicak E, Larkins-Ford J, Aldridge BB. 2017. Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci Adv 3:e1701881. 10.1126/sciadv.1701881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenwood DJ, Dos Santos MS, Huang S, Russell MRG, Collinson LM, MacRae JI, West A, Jiang H, Gutierrez MG. 2019. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science 364:1279–1282. 10.1126/science.aat9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramon-Garcia S, Ng C, Anderson H, Chao JD, Zheng X, Pfeifer T, Av-Gay Y, Roberge M, Thompson CJ. 2011. Synergistic drug combinations for tuberculosis therapy identified by a novel high-throughput screen. Antimicrob Agents Chemother 55:3861–3869. 10.1128/AAC.00474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruhn DF, Scherman MS, Liu J, Scherbakov D, Meibohm B, Bottger EC, Lenaerts AJ, Lee RE. 2015. In vitro and in vivo evaluation of synergism between anti-tubercular spectinamides and non-classical tuberculosis antibiotics. Sci Rep 5:13985. 10.1038/srep13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yilancioglu K, Cokol M. 2019. Design of high-order antibiotic combinations against M. tuberculosis by ranking and exclusion. Sci Rep 9:11876. 10.1038/s41598-019-48410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks BD, Brooks AE. 2014. Therapeutic strategies to combat antibiotic resistance. Adv Drug Deliv Rev 78:14–27. 10.1016/j.addr.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 14.Hoeksema H, Knight JC. 1975. The production of dihydrospectinomycin by Streptomyces spectabilis. J Antibiot (Tokyo) 28:240–241. 10.7164/antibiotics.28.240. [DOI] [PubMed] [Google Scholar]
- 15.Novak E, Gray JE, Pfeifer RT. 1974. Animal and human tolerance of high-dose intramuscular therapy with spectinomycin. J Infect Dis 130:50–55. 10.1093/infdis/130.1.50. [DOI] [PubMed] [Google Scholar]
- 16.Boslego JW, Tramont EC, Takafuji ET, Diniega BM, Mitchell BS, Small JW, Khan WN, Stein DC. 1987. Effect of spectinomycin use on the prevalence of spectinomycin-resistant and of penicillinase-producing Neisseria gonorrhoeae. N Engl J Med 317:272–278. 10.1056/NEJM198707303170504. [DOI] [PubMed] [Google Scholar]
- 17.Cully M. 2014. Antibacterial drugs: redesigned antibiotic combats drug-resistant tuberculosis. Nat Rev Drug Discov 13:257. 10.1038/nrd4287. [DOI] [PubMed] [Google Scholar]
- 18.Lee RE, Hurdle JG, Liu J, Bruhn DF, Matt T, Scherman MS, Vaddady PK, Zheng Z, Qi J, Akbergenov R, Das S, Madhura DB, Rathi C, Trivedi A, Villellas C, Lee RB, Rakesh, Waidyarachchi SL, Sun D, McNeil MR, Ainsa JA, Boshoff HI, Gonzalez-Juarrero M, Meibohm B, Bottger EC, Lenaerts AJ. 2014. Spectinamides: a new class of semisynthetic antituberculosis agents that overcome native drug efflux. Nat Med 20:152–158. 10.1038/nm.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Bruhn DF, Lee RB, Zheng Z, Janusic T, Scherbakov D, Scherman MS, Boshoff HI, Das S, Rakesh, Waidyarachchi SL, Brewer TA, Gracia B, Yang L, Bollinger J, Robertson GT, Meibohm B, Lenaerts AJ, Ainsa J, Bottger EC, Lee RE. 2017. Structure-activity relationships of spectinamide antituberculosis agents: a dissection of ribosomal inhibition and native efflux avoidance contributions. ACS Infect Dis 3:72–88. 10.1021/acsinfecdis.6b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson GT, Scherman MS, Bruhn DF, Liu J, Hastings C, McNeil MR, Butler MM, Bowlin TL, Lee RB, Lee RE, Lenaerts AJ. 2017. Spectinamides are effective partner agents for the treatment of tuberculosis in multiple mouse infection models. J Antimicrob Chemother 72:770–777. 10.1093/jac/dkw467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathi C, Lukka PB, Wagh S, Lee RE, Lenaerts AJ, Braunstein M, Hickey A, Gonzalez-Juarrero M, Meibohm B. 2019. Comparative pharmacokinetics of spectinamide 1599 after subcutaneous and intrapulmonary aerosol administration in mice. Tuberculosis (Edinb) 114:119–122. 10.1016/j.tube.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart IE, Lukka PB, Liu J, Meibohm B, Gonzalez-Juarrero M, Braunstein MS, Lee RE, Hickey AJ. 2019. Development and characterization of a dry powder formulation for anti-tuberculosis drug spectinamide 1599. Pharm Res 36:136. 10.1007/s11095-019-2666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci U S A 102:4548–4553. 10.1073/pnas.0500469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal I, Jain A, Verma SK, Singh P, Kant S, Singh M. 2017. Effect of efflux pump inhibitors on the susceptibility of Mycobacterium tuberculosis to isoniazid. Lung India 34:499–505. 10.4103/0970-2113.217567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castanheira M, Watters AA, Bell JM, Turnidge JD, Jones RN. 2010. Fusidic acid resistance rates and prevalence of resistance mechanisms among Staphylococcus spp. isolated in North America and Australia, 2007–2008. Antimicrob Agents Chemother 54:3614–3617. 10.1128/AAC.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collignon P, Turnidge J. 1999. Fusidic acid in vitro activity. Int J Antimicrob Agents 12(Suppl):S45–S58. 10.1016/s0924-8579(98)00073-9. [DOI] [PubMed] [Google Scholar]
- 27.Turnidge J. 1999. Fusidic acid pharmacology, pharmacokinetics and pharmacodynamics. Int J Antimicrob Agents 12(Suppl):S23–S34. 10.1016/s0924-8579(98)00071-5. [DOI] [PubMed] [Google Scholar]
- 28.Fuursted K, Askgaard D, Faber V. 1992. Susceptibility of strains of the Mycobacterium tuberculosis complex to fusidic acid. APMIS 100:663–667. 10.1111/j.1699-0463.1992.tb03983.x. [DOI] [PubMed] [Google Scholar]
- 29.Kigondu EM, Njoroge M, Singh K, Njuguna N, Warner DF, Chibale K. 2014. Synthesis and synergistic antimycobacterial screening of chlorpromazine and its metabolites. MedChemComm 5:502–506. 10.1039/C3MD00387F. [DOI] [Google Scholar]
- 30.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 31.Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. 2000. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 182:3142–3150. 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Gu Y, Li J, Xie L, Li X, Xie J. 2017. Mycobacterium tuberculosis major facilitator superfamily transporters. J Membr Biol 250:573–585. 10.1007/s00232-017-9982-x. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Gardete S, Jansen RS, Shetty A, Dick T, Rhee KY, Dartois V. 2018. Verapamil targets membrane energetics in Mycobacterium tuberculosis. Antimicrob Agents Chemother 62:e02107-17. 10.1128/AAC.02107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubin H. 2007. The respiratory chain of M. tuberculosis. FASEB J 21:A207. 10.1096/fasebj.21.5.A207-b. [DOI] [Google Scholar]
- 35.Bollenbach T. 2015. Antimicrobial interactions: mechanisms and implications for drug discovery and resistance evolution. Curr Opin Microbiol 27:1–9. 10.1016/j.mib.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326:694–699. 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramakrishnan G, Chandra NR, Srinivasan N. 2015. Recognizing drug targets using evolutionary information: implications for repurposing FDA-approved drugs against Mycobacterium tuberculosis H37Rv. Mol Biosyst 11:3316–3331. 10.1039/c5mb00476d. [DOI] [PubMed] [Google Scholar]
- 40.Besier S, Ludwig A, Brade V, Wichelhaus TA. 2003. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol Microbiol 47:463–469. 10.1046/j.1365-2958.2003.03307.x. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava V, Rouanet C, Srivastava R, Ramalingam B, Locht C, Srivastava BS. 2007. Macrophage-specific Mycobacterium tuberculosis genes: identification by green fluorescent protein and kanamycin resistance selection. Microbiology 153:659–666. 10.1099/mic.0.2006/000547-0. [DOI] [PubMed] [Google Scholar]
- 42.Takaki K, Davis JM, Winglee K, Ramakrishnan L. 2013. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat Protoc 8:1114–1124. 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance (Trends in Microbiology 24, 862–871; October 17, 2016). Trends Microbiol 24:928. 10.1016/j.tim.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Chen P, Gearhart J, Protopopova M, Einck L, Nacy CA. 2006. Synergistic interactions of SQ109, a new ethylene diamine, with front-line antitubercular drugs in vitro. J Antimicrob Chemother 58:332–337. 10.1093/jac/dkl227. [DOI] [PubMed] [Google Scholar]
- 46.Yang H, Chen G, Hu L, Liu Y, Cheng J, Ye Y, Li J. 2018. Enhanced efficacy of imipenem-colistin combination therapy against multiple-drug-resistant Enterobacter cloacae: in vitro activity and a Galleria mellonella model. J Microbiol Immunol Infect 51:70–75. 10.1016/j.jmii.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Ankomah P, Johnson PJ, Levin BR. 2013. The pharmaco-, population and evolutionary dynamics of multi-drug therapy: experiments with S. aureus and E. coli and computer simulations. PLoS Pathog 9:e1003300. 10.1371/journal.ppat.1003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodrigues L, Cravo P, Viveiros M. 2020. Efflux pump inhibitors as a promising adjunct therapy against drug resistant tuberculosis: a new strategy to revisit mycobacterial targets and repurpose old drugs. Expert Rev Anti Infect Ther 18:741–757. 10.1080/14787210.2020.1760845. [DOI] [PubMed] [Google Scholar]
- 49.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, Boyer E, Chamberland S, Lee VJ. 1999. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:1340–1346. 10.1128/AAC.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markham PN, Neyfakh AA. 1996. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 40:2673–2674. 10.1128/AAC.40.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markham PN. 1999. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pneumoniae by the multidrug efflux inhibitor reserpine. Antimicrob Agents Chemother 43:988–989. 10.1128/AAC.43.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cokol-Cakmak M, Bakan F, Cetiner S, Cokol M. 2018. Diagonal method to measure synergy among any number of drugs. J Vis Exp 136:57713. 10.3791/57713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cokol M, Weinstein ZB, Yilancioglu K, Tasan M, Doak A, Cansever D, Mutlu B, Li S, Rodriguez-Esteban R, Akhmedov M, Guvenek A, Cokol M, Cetiner S, Giaever G, Iossifov I, Nislow C, Shoichet B, Roth FP. 2014. Large-scale identification and analysis of suppressive drug interactions. Chem Biol 21:541–551. 10.1016/j.chembiol.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tekin E, Beppler C, White C, Mao Z, Savage VM, Yeh PJ. 2016. Enhanced identification of synergistic and antagonistic emergent interactions among three or more drugs. J R Soc Interface 13:20160332. 10.1098/rsif.2016.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhusal Y, Shiohira CM, Yamane N. 2005. Determination of in vitro synergy when three antimicrobial agents are combined against Mycobacterium tuberculosis. Int J Antimicrob Agents 26:292–297. 10.1016/j.ijantimicag.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Torella JP, Chait R, Kishony R. 2010. Optimal drug synergy in antimicrobial treatments. PLoS Comput Biol 6:e1000796. 10.1371/journal.pcbi.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarathy JP, Zuccotto F, Hsinpin H, Sandberg L, Via LE, Marriner GA, Masquelin T, Wyatt P, Ray P, Dartois V. 2016. Prediction of drug penetration in tuberculosis lesions. ACS Infect Dis 2:552–563. 10.1021/acsinfecdis.6b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Yan K, Zhang Y, Huang R, Bian J, Zheng C, Sun H, Chen Z, Sun N, An R, Min F, Zhao W, Zhuo Y, You J, Song Y, Yu Z, Liu Z, Yang K, Gao H, Dai H, Zhang X, Wang J, Fu C, Pei G, Liu J, Zhang S, Goodfellow M, Jiang Y, Kuai J, Zhou G, Chen X. 2007. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc Natl Acad Sci U S A 104:4606–4611. 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carroll P, Schreuder LJ, Muwanguzi-Karugaba J, Wiles S, Robertson BD, Ripoll J, Ward TH, Bancroft GJ, Schaible UE, Parish T. 2010. Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS One 5:e9823. 10.1371/journal.pone.0009823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montoro E, Lemus D, Echemendia M, Martin A, Portaels F, Palomino JC. 2005. Comparative evaluation of the nitrate reduction assay, the MTT test, and the resazurin microtitre assay for drug susceptibility testing of clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother 55:500–505. 10.1093/jac/dki023. [DOI] [PubMed] [Google Scholar]
- 62.Schon T, Werngren J, Machado D, Borroni E, Wijkander M, Lina G, Mouton J, Matuschek E, Kahlmeter G, Giske C, Santin M, Cirillo DM, Viveiros M, Cambau E. 2020. Antimicrobial susceptibility testing of Mycobacterium tuberculosis complex isolates - the EUCAST broth microdilution reference method for MIC determination. Clin Microbiol Infect 26:1488–1492. 10.1016/j.cmi.2020.07.036. [DOI] [PubMed] [Google Scholar]
- 63.Martin A, Camacho M, Portaels F, Palomino JC. 2003. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother 47:3616–3619. 10.1128/aac.47.11.3616-3619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramon-Garcia S, Martin C, Ainsa JA, De Rossi E. 2006. Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J Antimicrob Chemother 57:252–259. 10.1093/jac/dki436. [DOI] [PubMed] [Google Scholar]
- 65.Strober W. 2001. Trypan blue exclusion test of cell viability. Curr Protoc Immunol Appendix 3:Appendix 3B. 10.1002/0471142735.ima03bs21. [DOI] [PubMed] [Google Scholar]
- 66.Stokes RW, Doxsee D. 1999. The receptor-mediated uptake, survival, replication, and drug sensitivity of Mycobacterium tuberculosis within the macrophage-like cell line THP-1: a comparison with human monocyte-derived macrophages. Cell Immunol 197:1–9. 10.1006/cimm.1999.1554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.