The worldwide emergence of multidrug-resistant pathogenic fungi is a threat to human health. At this very moment, an emergence of Candida parapsilosis isolates harboring a resistance to fluconazole, one of the most popular antifungal drugs, is being described in several countries.
KEYWORDS: antifungal resistance, Candida, candidiasis, fungal outbreak, nosocomial infections, fluconazole, voriconazole
ABSTRACT
The worldwide emergence of multidrug-resistant pathogenic fungi is a threat to human health. At this very moment, an emergence of Candida parapsilosis isolates harboring a resistance to fluconazole, one of the most popular antifungal drugs, is being described in several countries. We seek to better understand the epidemiology, pathogenicity, and transmission of resistant Candida parapsilosis. Faced with an outbreak of invasive infections due to resistant isolates of C. parapsilosis, we performed a 7-year retrospective and prospective analysis of 283 C. parapsilosis isolates collected in 240 patients, among whom 111 had invasive candidiasis. The study included a review of hospital records, genotyping analysis, and susceptibility testing that allowed us to determine the type and outcome of infections, as well as the spatial and temporal spread of clusters. Overall, the incidence of azole resistance was 7.5%. Genotyping analysis unveiled several previously undetected outbreaks and clonal spread of susceptible and resistant isolates over a long period of time. In comparison with susceptible isolates, resistant ones have a more restricted genetic diversity and seem to be more likely to spread and more frequently associated with invasive infections. In intensive care units, patients with invasive infections due to resistant isolates had poorer outcomes (overall mortality at day 30 of 40%; 4/10) than susceptible ones (overall mortality at day 30 of 26.5%; 9/34). Our results suggest that the propensity of C. parapsilosis to spread in an epidemic fashion is underestimated, which warrants reinforced control and epidemiological survey of this species.
INTRODUCTION
Candida parapsilosis is one of the most common Candida species responsible for human infections, accounting for 15% to 30% of candidemia (1–4). It is generally susceptible to azole drugs, especially fluconazole and voriconazole (2, 5). Isolates of azole-resistant C. parapsilosis have occasionally been found in intensive care units (ICUs) (5) and resulted in high mortality rates in immunocompromised patients (6).
Outbreaks of fluconazole-resistant C. parapsilosis infections have been described in recent years (7–10). These isolates were responsible for infections occurring on an epidemic way with clonal spread and were associated with high morbidity and mortality. Our institution has recently faced several cases of invasive infections due to fluconazole-resistant isolates among patients in the same ICU. We have therefore initiated routine prospective screening of fluconazole resistance for all C. parapsilosis isolates and started a retrospective investigation of cases. Our study aimed at defining the antifungal susceptibility pattern of C. parapsilosis isolates as well as the incidence and evolution of azole resistance. We also performed a genotyping analysis using microsatellite markers to determine their genomic distribution and evolutionary dynamics.
RESULTS
Origin of the isolates.
The study involved 283 C. parapsilosis isolates obtained from 240 patients between March 2012 and October 2019 at the La Pitié-Salpêtrière Hospital, a 1,800-bed tertiary care center in Paris, France. It is a pavilion hospital that comprises 26 separate buildings destined for patient care. The origin of isolates and the clinical implications are presented in Table 1. A total of 131 isolates (46.3%) from 111 patients were responsible for invasive infections. Colonization involved 104 isolates in 93 patients and was predominantly superficial and cutaneous. Positive catheter cultures (29 isolates, 25 patients) that can be considered either colonization or infection related were accounted for separately.
TABLE 1.
Clinical origin and incidence of fluconazole resistance among 283 Candida parapsilosis isolates collected in 240 patients from a single hospital between 2012 and 2019
| Characteristic | No. (%) of patients (n = 240)a | No. (%) of fluconazole-susceptible isolates (n = 257) | No. (%) of fluconazole-resistant isolates (n = 26) | Total no. (%) of isolates (n = 283) |
|---|---|---|---|---|
| Infectionsb | 111 (46.3) | 115 (44.7) | 16 (61.5) | 131 (46.3) |
| Candidemias | 69 (62.1) | 70 (60.9) | 12 (75) | 82 (62.6) |
| Osteoarticular infections | 19 (17.1) | 19 (16.5) | 1 (6.3) | 20 (15.3) |
| Mediastinitis | 6 (5.4) | 7 (6) | 2 (12.5) | 9 (6.9) |
| Other deep infections | 19 (17.1) | 19 (16.5) | 1 (6.3) | 20 (15.3) |
| Colonizationc | 93 (38.8) | 98 (38.1) | 6 (23.1) | 104 (36.7) |
| Superficial/mucocutaneous | 57 (61.3) | 57 (58.2) | 3 (5) | 60 (57.7) |
| Respiratory tract | 23 (24.7) | 26 (26.5) | 1 (3.7) | 27 (26) |
| Urinary tract | 11 (11.8) | 10 (10.2) | 1 (9.1) | 11 (10.6) |
| Other | 6 (6.4) | 6 (6.1) | 0 (0) | 6 (5.8) |
| Catheter | 25 (10.4) | 25 (9.7) | 4 (15.4) | 29 (10.2) |
| Data not available | 18 (7.5) | 19 (7.4) | 0 (0) | 19 (6.7) |
Seven patients had isolates responsible for colonization and infection and/or were found in catheter culture.
One patient had mediastinitis, fungemia, and other deep infection.
Three patients had superficial and respiratory tract colonization, and one patient had superficial and urinary tract colonization.
Antifungal susceptibility testing.
Of the 283 isolates tested, 26 (9.2%) from 18 patients (7.5% of patients in whom C. parapsilosis was isolated) were classified as resistant to fluconazole (MIC > 4 mg/liter) by Etest and confirmed by EUCAST (Table S1 in the supplemental material). A panel of 24 isolates classified as susceptible to fluconazole by Etest (MIC ≤ 2 mg/liter) was taken as a control group and was found susceptible by the EUCAST method. In addition, these susceptible isolates did not show reduced susceptibility to any of the other antifungal drugs tested. The 26 fluconazole-resistant isolates showed different susceptibility profiles to other azole drugs (Table S1). Voriconazole-resistant isolates were those with the highest MICs for fluconazole (≥32 mg/liter). No azole-resistant isolates showed resistance to either echinocandins or amphotericin B.
erg11 sequencing.
The 26 fluconazole-resistant isolates and a random selection of 21 fluconazole-susceptible isolates were subjected to erg11 gene sequencing. Results highlighted the A395T mutation that confers the Y132F amino acid substitution in all fluconazole-resistant isolates except one (identification number PSL0172) for which no specific alteration was found. The latter showed a resistance profile to all azoles. The A395T mutation was absent in all 21 fluconazole-susceptible isolates tested as controls.
Microsatellite analysis.
The 26 fluconazole-resistant isolates and a random selection of 65 fluconazole-susceptible isolates were subjected to microsatellite genotyping. As shown by the unweighted pair group method with arithmetic mean (UPGMA) dendrogram in Fig. 1, azole-susceptible C. parapsilosis isolates were characterized by high genetic diversity. Of the 65 genotyped susceptible isolates, 58 isolates showed separate and unique genetic patterns, while 7 isolates were distributed into 3 clusters (referred to as S1 to S3). On the other hand, the 26 fluconazole-resistant isolates (18 patients) fell into only 6 clusters, 4 of which were restricted to one single isolate, while 2 clusters (referred to as R1 and R2) comprised 8 isolates (4 patients) and 14 isolates (10 patients), respectively. So, compared to fluconazole-susceptible isolates (55 colonized/infected patients, 61 clusters), fluconazole-resistant isolates (18 colonized/infected patients, 6 clusters) showed a reduced genetic diversity (P = 0.014 by chi-square test).
FIG 1.
UPGMA (unweighted pair group method with arithmetic mean) dendrogram of 93 Candida parapsilosis isolates typed by microsatellite approach. Fluconazole-resistant isolates (n = 26) are highlighted in yellow. Isolates from a single intensive care unit are in red. R1 and R2 represent 2 clusters of fluconazole-resistant isolates responsible for 2 outbreaks in 4 and 10 patients, respectively. S1 to S3 represent clusters of susceptible isolates.
Interestingly, antifungal susceptibility profiles were different between clones R1 and R2, the former having lower MICs to fluconazole (8 to 16 mg/liter) and being intermediate to voriconazole, the latter having higher MICs to fluconazole (≥32 mg/liter) and being resistant to voriconazole.
Spatial and temporal circulation of clustered isolates.
Most of the susceptible and resistant isolates from the clusters S1 to S3 (7 patients) and R1 to R2 (14 patients) came from patients related to the same ward (cardiac surgery ICU; 14/21, 66.7%) or the same building (16/21, 76.2%) that we will call for convenience “building A” (Fig. 2 and Table 2).
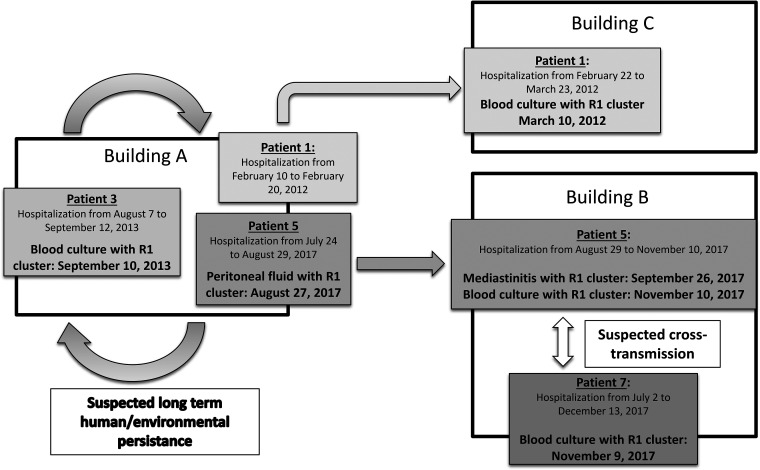
FIG 2.
Spatial and temporal circulation of Candida parapsilosis fluconazole-resistant isolates belonging to a single cluster (R1) among patients with invasive candidiasis hosted in different buildings of a single hospital between 2012 and 2017.
TABLE 2.
Information for 26 fluconazole-resistant Candida parapsilosis isolates from 18 patients
| Patient | Isolate identification no. | Sample type | Date of sample | Genotype | Unit | Building | Previous stay in building A | Underlying condition | Day 30 status |
|---|---|---|---|---|---|---|---|---|---|
| 1 | PSL0010 | Blood culture | 10 March 2012 | R1 | Surgical critical care 1 | C | Yes (18 days before) | Cardiothoracic surgery | Alive |
| 2 | PSL0020 | Blood culture | 5 August 2013 | Other | Neurological critical care | D | No | Guillain-Barré syndrome | Alive |
| 3 | PSL0014 | Central catheter | 5 September 2013 | R1 | Cardiac surgery intensive care unit | A | NAa | Cardiothoracic surgery | Death |
| PSL0011 | Blood culture | 10 September 2013 | R1 | Cardiac surgery intensive care unit | A | ||||
| 4 | PSL1009 | Blood culture | 4 April 2014 | Other | Cardiac surgery intensive care unit | A | NA | Cardiothoracic surgery | Death |
| 5 | PSL0007 | Peritoneal fluid | 27 August 2017 | R1 | Cardiac surgery intensive care unit | A | NA | Cardiothoracic surgery | Death |
| PSL0015 | Mediastinitis | 26 September 2017 | R1 | Surgical critical care 2 | B | ||||
| PSL0001 | Pelvic urine | 31 October 2017 | R1 | Vascular surgery | B | ||||
| PSL0016 | Blood culture | 10 November 2017 | R1 | Surgical critical care 2 | B | ||||
| 6 | PSL0004 | Central catheter | 6 November 2017 | R2 | Cardiac surgery intensive care unit | A | NA | Cardiothoracic surgery | Death |
| PSL0018 | Mediastinitis | 20 November 2017 | R2 | Cardiac surgery intensive care unit | A | ||||
| 7 | PSL0003 | Blood culture | 9 November 2017 | R1 | Surgical critical care 2 | B | No | Abdominal surgery | Alive |
| 8 | PSL0008 | Blood culture | 13 January 2018 | R2 | Surgical critical care 2 | B | Yes (14 days before) | Cardiothoracic surgery | Alive |
| PSL0005 | Urine | 27 January 2018 | R2 | Surgical critical care 2 | B | ||||
| 9 | PSL0013 | Pacemaker pocket | 16 August 2018 | R2 | Cardiac surgery intensive care unit | A | NA | Heart transplantation | Alive |
| 10 | PSL0019 | Blood culture | 29 September 2018 | R2 | Medical critical care | A | NA | Heart transplantation | Alive |
| PSL0106 | Blood culture | 18 October 2018 | R2 | Medical critical care | A | ||||
| 11 | PSL0102 | Bronchoalveolar lavage | 15 October 2018 | R2 | Cardiac surgery intensive care unit | A | NA | Heart transplantation | Death |
| PSL0126 | Blood culture | 18 December 2018 | R2 | Cardiac surgery intensive care unit | A | ||||
| 12 | PSL0107 | Anal swab | 28 October 2018 | R2 | Cardiac surgery intensive care unit | A | NA | Cardiothoracic surgery | Alive |
| 13 | PSL0119 | Blood culture | 29 November 2018 | Other | Medical critical care | A | NA | Cardiogenic shock/ECMO | Death |
| 14 | PSL0151 | Catheter | 25 March 2019 | R2 | Cardiac surgery intensive care unit | A | NA | Cardiogenic shock/ECMO | Alive |
| 15 | PSL0172 | Diabetic foot | 1 July 2019 | Other | Diabetology | E | No | Diabetes mellitus | Alive |
| 16 | PSL0201 | Nasal swab | 31 July 2019 | R2 | Surgical critical care 2 | B | No | Bariatric surgery | Alive |
| 17 | PSL0211 | Nasal swab | 3 September 2019 | R2 | Cardiac surgery intensive care unit | A | NA | Cardiothoracic surgery | Alive |
| 18 | PSL0225 | Catheter | 15 October 2019 | R2 | Surgical critical care 2 | B | Yes (7 months before) | Abdominal surgery | Alive |
NA, not available.
R1 cluster isolates (R1 clone) were detected in 4 patients between 10 March 2012 and 10 November 2017. While the initial detection of an R1 clone occurred in another building (building C), this patient (patient 1) was hospitalized in building A a few weeks earlier (Fig. 2). Three of the 4 patients carrying the R1 clone were linked to the same ICU. Furthermore, a review of hospital records showed that 1 of these 3 patients (patient 5) was transferred from the cardiac surgery ICU to another unit (surgical critical care unit 2) where the fourth patient (patient 7) was cared for only a few days before he was detected with an R1 clone.
R2 cluster isolates (R2 clone) were detected in 10 patients between 6 November 2017 and 15 October 2019. They were found in 7 patients hospitalized in building A and in 2 patients hospitalized in another building but who previously stayed in building A.
Comparison of patient profiles between susceptible and resistant isolates.
We then analyzed whether there were differences between patients who developed invasive infection due to a fluconazole-susceptible isolate and those who developed invasive infection due to a fluconazole-resistant isolate. Relevant data were available for 78 patients (Table 3). No differences were observed with respect to the clinical form (candidemia, mediastinitis, or others) or the presence or absence of risk factors usually associated with invasive candidiasis (surgical procedure, immunosuppression, broad-spectrum antibiotic therapy, and presence of external devices). Importantly, preexposure to an azole antifungal drug was also not a related factor. On the other hand, ICU stay was statistically associated with infection due to a fluconazole-resistant Candida parapsilosis isolate.
TABLE 3.
Data for 78 patients who developed an invasive infection due to Candida parapsilosisb
| Characteristic | Invasive infections due to fluconazole-susceptible isolates | Invasive infections due to fluconazole-resistant isolates | P value |
|---|---|---|---|
| No. of patients | 67 | 11 | |
| Mean age (median [IQR]) (yrs) | 59.4 (62 [51–72]) | 59.6 (65 [56–65.5]) | 0.78 |
| Sex (no. male/no. female) | 52/15 | 9/2 | 1 |
| Clinical presentation [no. (%)] | |||
| Candidemia | 55 (82.1) | 9/11 (81.8) | 1 |
| Mediastinitis | 3 (4.5) | 2/11 (18.2) | 0.14 |
| Others | 9 (13.4) | 0 | 0.34 |
| Hospital ward [no. (%)] | |||
| ICU | 34 (50.7) | 10 (90.9) | 0.019 |
| Non-ICU | 22 (32.8) | 0 | 0.028 |
| Chirurgical unit | 11 (16.4) | 1 (9.1) | 1 |
| Immunosuppression [no. (%)]a | 25/60 (41.7) | 3/11 (27.3) | 0.5 |
| Presence of catheter and/or external devices [no. (%)] | 46/53 (86.8) | 11/11 (100) | 0.34 |
| Recent chirurgical intervention (last 30 days) [no. (%)] | 42/57 (73.7) | 10/11 (90.9) | 0.44 |
| Broad-spectrum antibiotic therapy [no. (%)] | 40/50 (80) | 10/10 (100) | 0.19 |
| Preexposition to antifungal drugs (last 3 mo) [no. (%)] | 8/47 (17) | 2/10 (20) | 1 |
| Targeted antifungal therapy [no. (%)] | 36/39 (92.3 | 8/10 (80) | 0.27 |
| Echinocandin based | 12/36 (33.3) | 5/8 (62.5) | 0.22 |
| Fluconazole based | 21/36 (58.3) | 1/8 (12.5) | 0.046 |
| Other | 3/36 (8.3) | 2/8 (25) | 0.22 |
| All-cause mortality [no. (%)] | |||
| All patients | |||
| Day 30 | 15/67 (22.4) | 5/11 (45.5) | 0.082 |
| Day 90 | 19/64 (29.7) | 6/11 (54.5) | 0.12 |
| ICU patients | |||
| Day 30 | 9/34 (26.5) | 4/10 (40) | 0.40 |
| Day 90 | 11/31 (35.5) | 5/10 (50) | 0.46 |
Immunocompromised conditions include solid organ transplantation, HIV infection, neutropenia (<500 cells/μl), long-term corticosteroid therapy (>3 weeks), malignancies, and allogeneic hematopoietic stem cell transplantation.
Comparison between susceptible and fluconazole-resistant isolates. Statistically significant values are in bold. ICU, intensive care unit; IQR, interquartile range.
We then focused on patients in ICUs, as these are the main source of invasive infections related to fluconazole-resistant isolates. During the prospective part of the study (where the analysis of C. parapsilosis isolates is exhaustive) and focusing on 36 ICU patients, we observed that 5 out of 9 patients (55.5%) with a resistant isolate developed invasive candidiasis, while only 6 out of 27 patients (22.2%) with a susceptible isolate developed invasive candidiasis (P = 0.09; Fisher’s exact test). Considering the number of Candida isolates rather than the number of patients, 6 out of 10 resistant isolates were responsible for invasive infection, while 8 out of 35 susceptible isolates were responsible for invasive infection (P = 0.049; Fisher's exact test).
Ecological investigations.
The clustering of resistant isolates and their predominant presence in a single unit, along with some susceptible clonal isolates, raised the question of an existing environmental and/or human reservoir and the possibility of patient-to-patient transmission. Following discussion with the hospital's hygiene team, the local nosocomial infection control committee, and the unit's medical head, we conducted an epidemiological investigation in the mentioned ICU in search of an environmental reservoir. We took 100 swab samples. Different rooms and corridors were examined. The floor, bed rails, ultrasound scanners, washbasins, care trolleys, and ECMO (extracorporeal membrane oxygenation) devices were swabbed. Only two colonies of C. parapsilosis were found in one sample (washbasin). They were susceptible to azole drugs, had the same microsatellite profile, and were not linked to any of the clinical isolates (Fig. 1).
DISCUSSION
Historically, the first described C. parapsilosis outbreaks had been related to environmental reservoirs and medical devices, with direct contamination of the patients. In 1977, Plouffe et al. reported an outbreak of C. parapsilosis fungemia related to the contamination of a vacuum system in the preparation room for intravenous infusions (11). Years later, outbreaks due to contaminated hospital environmental reservoirs (12), as well as epidemics related to hand contamination by health care workers, have also been reported (13). Thus, the epidemic transmission of C. parapsilosis seems to be related both to human and/or environmental reservoirs with direct or indirect contamination of the patients. But in most cases, it is very difficult or even impossible to detect an irrefutable unique source of contamination (14). Moreover, several works report outbreaks, but few provide genetic analysis of the isolates to confirm their clonal nature.
Recently, outbreaks of C. parapsilosis infection due to fluconazole-resistant isolates have been described in several countries (7–10) with no explanation for this emergence. In our study, resistance to fluconazole (and voriconazole) revealed two major C. parapsilosis clones involved in two separate outbreaks. The A395T mutation that confers the Y132F amino acid substitution was found in 96.1% (25/26) of the fluconazole-resistant isolates and is presumably the main mechanism that confers azole resistance. This mutation, which affects an amino acid located close to the drug-target interaction area (15), is now widely reported among resistant C. parapsilosis (9, 16, 17) and has also been shown to confer fluconazole resistance to Candida albicans (18) or Candida tropicalis (19). However, the existence of different susceptibility profiles between the different clusters raises the question of other acquired resistance mechanisms, such as those involving CDR1, MDR1, MRR1, TAC1, or UPC2 (20). This aspect is worth consideration and requires investigation in further study. Of note, apart from two patients, none had received or been preexposed to azole therapy when the resistant isolates were identified, and preexposure to an azole antifungal drug was not related to the occurrence of invasive infection due to a resistant isolate.
The results of our study call for several comments. First and importantly, genotyping of C. parapsilosis isolates may reveal undetected epidemic transmission, even over a very large time scale. Indeed, the possibility of transmission leading to colonization without further infection, the low number of cases and their sporadic nature, and the significant delay that may exist between two cases all make it very difficult to detect epidemic transmission.
Analysis of the R1 clone is particularly informative at this level. It was first detected in a patient in March 2012 (patient 1) and again a year and a half later (patient 3), which suggests the possibility of a persistent environmental or human reservoir among health care personnel. Surprisingly, it was found again more than 4 years later (patient 5). It should be noted that some isolates were responsible for colonization only and therefore not tested for MICs or kept frozen, and may have been missed during this period. One of the patients infected by an R1 clone (patient 5) and hospitalized in the cardiac surgery ICU was transferred to another ICU (surgical critical care 2) where one more patient (patient 7) was hospitalized just a few days before he was detected positive with the R1 clone. This strongly suggests a transmission from patient to patient mediated by health care workers or medical devices. Also surprisingly, the R1 clone disappeared and has not been detected since.
The second point deals with genotyping analysis as well. It shows that genetic diversity is reduced for resistant isolates compared to susceptible isolates and that resistant isolates have an increased ability to spread in a clonal mode. Moreover, they were more frequently related to invasive infections than the azole-susceptible isolates. Whether the Y132F substitution that confers azole resistance might also be linked to a particular fitness of C. parapsilosis, which would favor either its human/environmental persistence and/or its pathogenicity, requires further investigation. Moreover, patients diagnosed with an invasive infection due to resistant isolates had a poorer outcome than the patients infected by susceptible ones, but this result has to be confirmed on larger series, as it did not reach statistical significance. Thus, the 30-day mortality rate of invasive infections due to susceptible isolates was comparable to those reported in other studies (1, 21). Intriguingly, Grossman et al. also reported that C. parapsilosis isolates harboring the Y132F alteration tended to be closely related genetically and limited to a small number of hospitals, whereas other resistant isolates had no hospital specificity (16). More recently, Choi et al. have also provided arguments for a greater propensity of Y132F isolates to cause clonal spread and to persist in hospitals (9). Our results are in accordance with these observations and suggest that resistant C. parapsilosis isolates with Y132F modification have a capacity to persist over a long period of time. As they are thought to support important mechanisms of pathogen spreading and virulence, it would be interesting to evaluate the isolates harboring the Y132F modification for their fitness, ability of adherence, biofilm formations (22), or their survival after exposure to surface disinfection agents such as quaternary ammonium.
Finally, it should be noted that the genotyping also revealed 3 grouped cases due to wild-type azole-susceptible isolates. It indicates that susceptible isolates might also be related to clonal spreading, although their susceptible phenotype makes them less intriguing and possibly more difficult to detect.
As a consequence of our study, hygiene recommendations have been taken, notably strengthening hand disinfection with hydroalcoholic solutions and the cleaning of rooms. As C. parapsilosis has been shown to accommodate to various environments (23), alive (human skin, body surface of insects, fruits, or pine trees) or inert (tap water, surfaces in habitations), we also collected hundreds of environmental swab samples but failed to find any source for the C. parapsilosis azoles-resistant clones. Investigations should be extended to health care workers.
In conclusion, our study shed light on the propensity of C. parapsilosis to lead to an epidemic over a long period of time. The particular fluconazole-resistant phenotype linked to the Y132F substitution seems related to clonal spreading, invasive infection, and high mortality. Our results also underline the importance of genotyping C. parapsilosis isolates, even for susceptible isolates, to unveil unsuspected clustered cases and thus apply hygiene measures to limit nosocomial transmission.
MATERIALS AND METHODS
Study design.
The study covers the period from March 2012 to October 2019 (92 months) and includes a prospective part and a retrospective part. From March 2012 to September 2018, C. parapsilosis isolates responsible for infections and/or for which MICs of antifungal drugs had been determined were kept and therefore available for the study; others isolates, e.g., responsible for colonization without MIC determinations, were not kept. From October 2018 to October 2019, any C. parapsilosis isolate found in a hospitalized patient was considered and prospectively included in the study. All isolates were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry using the MSI database.
MICs of antifungal drugs.
Fluconazole MIC was determined for all isolates included in the study by a gradient concentration strip method (Etest; bioMérieux). For all fluconazole-resistant isolates (MICs above 4 mg/liter by Etest) and a random selection of susceptible isolates, fluconazole susceptibility was also assessed by broth microdilution (EUCAST method). Antifungal susceptibility testing was extended to other antifungal drugs. C. parapsilosis ATCC 22019 or Candida krusei ATCC 6258 strains were used as quality control. Isolates were classified as susceptible, intermediate, or resistant according to the EUCAST clinical breakpoints available at http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/.
Genotyping and erg11 sequencing.
Microsatellite genotyping was performed as previously described (24). Briefly, a panel of 6 short tandem repeats was used, resulting in a unique 12-marker microsatellite profile for each isolate. The resulting microsatellite profiles were then exported and submitted to an unweighted pair group method with arithmetic mean (UPGMA) cluster analysis (Dendro-UPGMA, available at http://genomes.urv.es/UPGMA/) to generate a dendrogram, considering data as categorical values. Isolates with 100% identical genotypes by microsatellite typing were considered clonal. The erg11 gene sequencing was performed using primers and conditions previously described by Grossman et al. (16).
Statistical analysis.
Statistical analyses were performed using Prism 5 (GraphPad Software). Continuous and categorical variables are presented as mean (median [interquartile range {IQR}]) and number (percentage), respectively. Categorical variables were compared using Fisher’s exact test, and the Mann-Whitney U test was used for continuous variables. Survival distributions were compared using the log-rank (Mantel-Cox) test.
Supplementary Material
ACKNOWLEDGMENTS
Part of the data was presented (oral communication) during the 8th Trends in Medical Mycology Congress (11 to 14 October 2019, Nice, France).
Internal funding supported this work.
We declare we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wu YM, Huang PY, Lu JJ, Shie SS, Ye JJ, Wu TS, Huang CT. 2018. Risk factors and outcomes of candidemia caused by Candida parapsilosis complex in a medical center in northern Taiwan. Diagn Microbiol Infect Dis 90:44–49. 10.1016/j.diagmicrobio.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Prigitano A, Cavanna C, Passera M, Gelmi M, Sala E, Ossi C, Grancini A, Calabro M, Bramati S, Tejada M, Lallitto F, Farina C, Rognoni V, Fasano MA, Pini B, Romano L, Cogliati M, Esposto MC, Tortorano AM. 2019. Evolution of fungemia in an Italian region. J Mycol Med 30:100906. 10.1016/j.mycmed.2019.100906. [DOI] [PubMed] [Google Scholar]
- 3.Puig-Asensio M, Padilla B, Garnacho-Montero J, Zaragoza O, Aguado JM, Zaragoza R, Montejo M, Munoz P, Ruiz-Camps I, Cuenca-Estrella M, Almirante B, CANDIPOP Project, GEIH-GEMICOMED (SEIMC), REIPI. 2014. Epidemiology and predictive factors for early and late mortality in Candida bloodstream infections: a population-based surveillance in Spain. Clin Microbiol Infect 20:O245–O254. 10.1111/1469-0691.12380. [DOI] [PubMed] [Google Scholar]
- 4.Santolaya ME, Thompson L, Benadof D, Tapia C, Legarraga P, Cortes C, Rabello M, Valenzuela R, Rojas P, Rabagliati R, Chilean Invasive Mycosis Network. 2019. A prospective, multi-center study of Candida bloodstream infections in Chile. PLoS One 14:e0212924. 10.1371/journal.pone.0212924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ng KP, Colombo A, Finquelievich J, Barnes R, Wadula J, Global Antifungal Surveillance Group. 2008. Geographic and temporal trends in isolation and antifungal susceptibility of Candida parapsilosis: a global assessment from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J Clin Microbiol 46:842–849. 10.1128/JCM.02122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghuram A, Restrepo A, Safadjou S, Cooley J, Orloff M, Hardy D, Butler S, Koval CE. 2012. Invasive fungal infections following liver transplantation: incidence, risk factors, survival, and impact of fluconazole-resistant Candida parapsilosis (2003–2007). Liver Transpl 18:1100–1109. 10.1002/lt.23467. [DOI] [PubMed] [Google Scholar]
- 7.Govender NP, Patel J, Magobo RE, Naicker S, Wadula J, Whitelaw A, Coovadia Y, Kularatne R, Govind C, Lockhart SR, Zietsman IL, TRAC-South Africa group. 2016. Emergence of azole-resistant Candida parapsilosis causing bloodstream infection: results from laboratory-based sentinel surveillance in South Africa. J Antimicrob Chemother 71:1994–2004. 10.1093/jac/dkw091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinhati HM, Casulari LA, Souza AC, Siqueira RA, Damasceno CM, Colombo AL. 2016. Outbreak of candidemia caused by fluconazole resistant Candida parapsilosis strains in an intensive care unit. BMC Infect Dis 16:433. 10.1186/s12879-016-1767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi YJ, Kim YJ, Yong D, Byun JH, Kim TS, Chang YS, Choi MJ, Byeon SA, Won EJ, Kim SH, Shin MG, Shin JH. 2018. Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg Infect Dis 24:1768–1770. 10.3201/eid2409.180625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomaz DY, de Almeida JN, Jr., Lima GME, Nunes MO, Camargo CH, Grenfell RC, Benard G, Del Negro GMB. 2018. An azole-resistant Candida parapsilosis outbreak: clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front Microbiol 9:2997. 10.3389/fmicb.2018.02997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plouffe JF, Brown DG, Silva J, Jr., Eck T, Stricof RL, Fekety FR, Jr.. 1977. Nosocomial outbreak of Candida parapsilosis fungemia related to intravenous infusions. Arch Intern Med 137:1686–1689. 10.1001/archinte.1977.03630240022010. [DOI] [PubMed] [Google Scholar]
- 12.Qi L, Fan W, Xia X, Yao L, Liu L, Zhao H, Kong X, Liu J. 2018. Nosocomial outbreak of Candida parapsilosis sensu stricto fungaemia in a neonatal intensive care unit in China. J Hosp Infect 100:e246–e252. 10.1016/j.jhin.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez-Castro R, Arroyo-Escalante S, Carrillo-Casas EM, Moncada-Barron D, Alvarez-Verona E, Hernandez-Delgado L, Torres-Narvaez P, Lavalle-Villalobos A. 2010. Outbreak of Candida parapsilosis in a neonatal intensive care unit: a health care workers source. Eur J Pediatr 169:783–787. 10.1007/s00431-009-1109-7. [DOI] [PubMed] [Google Scholar]
- 14.Johnston BL, Schlech WF, III, Marrie TJ. 1994. An outbreak of Candida parapsilosis prosthetic valve endocarditis following cardiac surgery. J Hosp Infect 28:103–112. 10.1016/0195-6701(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 15.Mane A, Vidhate P, Kusro C, Waman V, Saxena V, Kulkarni-Kale U, Risbud A. 2016. Molecular mechanisms associated with fluconazole resistance in clinical Candida albicans isolates from India. Mycoses 59:93–100. 10.1111/myc.12439. [DOI] [PubMed] [Google Scholar]
- 16.Grossman NT, Pham CD, Cleveland AA, Lockhart SR. 2015. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob Agents Chemother 59:1030–1037. 10.1128/AAC.04613-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castanheira M, Deshpande LM, Messer SA, Rhomberg PR, Pfaller MA. 2019. Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int J Antimicrob Agents 55:105799. 10.1016/j.ijantimicag.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan X, Xiao M, Zhang D, Huang JJ, Wang H, Hou X, Zhang L, Kong F, Chen SC, Tong ZH, Xu YC. 2019. Molecular mechanisms of azole resistance in Candida tropicalis isolates causing invasive candidiasis in China. Clin Microbiol Infect 25:885–891. 10.1016/j.cmi.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Neji S, Hadrich I, Trabelsi H, Abbes S, Cheikhrouhou F, Sellami H, Makni F, Ayadi A. 2017. Virulence factors, antifungal susceptibility and molecular mechanisms of azole resistance among Candida parapsilosis complex isolates recovered from clinical specimens. J Biomed Sci 24:67. 10.1186/s12929-017-0376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H, Yoshimura Y, Suido Y, Shimizu H, Ide K, Sugiyama Y, Matsuno K, Nakajima H. 2019. Mortality and risk factor analysis for Candida blood stream infection: a multicenter study. J Infect Chemother 25:341–345. 10.1016/j.jiac.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Soldini S, Posteraro B, Vella A, De Carolis E, Borghi E, Falleni M, Losito AR, Maiuro G, Trecarichi EM, Sanguinetti M, Tumbarello M. 2018. Microbiologic and clinical characteristics of biofilm-forming Candida parapsilosis isolates associated with fungaemia and their impact on mortality. Clin Microbiol Infect 24:771–777. 10.1016/j.cmi.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Dogen A, Sav H, Gonca S, Kaplan E, Ilkit M, Novak Babic M, Gunde-Cimerman N, de Hoog GS. 2017. Candida parapsilosis in domestic laundry machines. Med Mycol 55:813–819. 10.1093/mmy/myx008. [DOI] [PubMed] [Google Scholar]
- 24.Diab-Elschahawi M, Forstner C, Hagen F, Meis JF, Lassnig AM, Presterl E, Klaassen CH. 2012. Microsatellite genotyping clarified conspicuous accumulation of Candida parapsilosis at a cardiothoracic surgery intensive care unit. J Clin Microbiol 50:3422–3426. 10.1128/JCM.01179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.