Due to limited treatment options for carbapenem-resistant Acinetobacter baumannii (CR-AB) infections, antibiotic combinations are now considered potential treatments for CR-AB. This study aimed to explore the utility of fosfomycin-sulbactam combination (FOS/SUL) therapy against CR-AB isolates.
KEYWORDS: Acinetobacter baumannii, synergy, PK/PD model, Monte Carlo simulation, fosfomycin, sulbactam, in vitro
ABSTRACT
Due to limited treatment options for carbapenem-resistant Acinetobacter baumannii (CR-AB) infections, antibiotic combinations are now considered potential treatments for CR-AB. This study aimed to explore the utility of fosfomycin-sulbactam combination (FOS/SUL) therapy against CR-AB isolates. Synergism of FOS/SUL against 50 clinical CR-AB isolates was screened using the checkerboard method. Thereafter, time-kill studies against two CR-AB isolates were performed. The time-kill data were described using a semimechanistic pharmacokinetic/pharmacodynamic (PK/PD) model. Monte Carlo simulations were then performed to estimate the probability of stasis, 1-log kill, and 2-log kill after 24 h of combination therapy. The FOS/SUL combination demonstrated a synergistic effect against 74% of isolates. No antagonism was observed. The MIC50 and MIC90 of FOS/SUL were decreased 4- to 8-fold, compared to the monotherapy MIC50 and MIC90. In the time-kill studies, the combination displayed bactericidal activity against both isolates and synergistic activity against one isolate at the highest clinically achievable concentrations. Our PK/PD model was able to describe the interaction between fosfomycin and sulbactam in vitro. Bacterial kill was mainly driven by sulbactam, with fosfomycin augmentation. FOS/SUL regimens that included sulbactam at 4 g every 8 h demonstrated a probability of target attainment of 1-log10 kill at 24 h of ∼69 to 76%, compared to ∼15 to 30% with monotherapy regimens at the highest doses. The reduction in the MIC values and the achievement of a moderate PTA of a 2-log10 reduction in bacterial burden demonstrated that FOS/SUL may potentially be effective against some CR-AB infections.
TEXT
Acinetobacter baumannii possesses intrinsic resistance mechanisms and the ability to acquire new resistance genes proficiently (1). These abilities render it resistant to many antibiotics, thus restricting its treatment repertoire. Due to the limited treatment options for carbapenem-resistant A. baumannii (CR-AB) infections, antibiotic combinations have been used empirically. Antibiotics such as fosfomycin and sulbactam, in combination with other antibiotics, are now considered potential treatment options for CR-AB (2, 3). In vitro studies have reported synergy rates between 50 and 75% for fosfomycin in combination with colistin, imipenem, or sulbactam against multidrug-resistant (MDR) A. baumannii (4–6). Deveci et al. reported in vitro synergy rates ranging between 30 and 80% for sulbactam in combination with meropenem, gentamicin, colistin, or cefepime (7). Despite promising outcomes from in vitro studies, the role of combination therapy in clinical practice, particularly of fosfomycin-sulbactam combination, in the treatment CR-AB is still unclear.
Fosfomycin is a bactericidal broad-spectrum antibiotic that hinders bacterial cell wall synthesis via inhibition of enolpyruvyl transferase, an essential enzyme in peptidoglycan biosynthesis leading to bacterial cell lysis (8–10). On the other hand, sulbactam is a semisynthetic β-lactamase inhibitor which, when combined with certain β-lactams, extends their activity against bacteria that are usually resistant to the antibiotic due to the production of β-lactamases (11). Interestingly, sulbactam can bind to penicillin-binding proteins (PBP) of Acinetobacter spp. (12). It has a strong inclination to bind to PBP2 in A. baumannii (13). In vitro and in vivo studies have demonstrated clinically significant activity of sulbactam against Acinetobacter spp., making it unique from other β-lactamase inhibitors, such as tazobactam and clavulanic acid, which do not directly exhibit bacterial cell killing (14, 15).
Apart from the use of antibiotic combinations, optimization of dosing regimens of existing antibiotics has become increasingly important to respond to the threat of MDR bacterial infections. Optimal dosing strategies for antibiotics are essential to improve outcomes against multidrug-resistant infections (16). In this study, we endeavored to explore the utility of fosfomycin and sulbactam in combination against CR-AB isolates and describe the interaction between fosfomycin and sulbactam using a semimechanistic pharmacokinetic/pharmacodynamic (PK/PD) model based on time-kill experiments.
RESULTS
MICs and checkerboard and time-kill analyses.
All isolates were meropenem resistant, with a MIC ranging from 32 to 512 mg/liter. Table 1 summarizes the MICs, MIC50s, and MIC90s of fosfomycin and sulbactam, in monotherapy and in combination against each isolate, and the minimum (range) fractional inhibitory concentration index (FICI) for each isolate. The ranges of MICs were 16 to 256 mg/liter for sulbactam alone and 128 to 2,048 mg/liter for fosfomycin alone. The range of MICs of fosfomycin and sulbactam in combination were 8 to 256 mg/liter and 0.5 to 64 mg/liter, respectively. The MICs of the antibiotics in combination were lower than the MICs of the antibiotics in monotherapy for most isolates (49/50). The MIC50s of fosfomycin and sulbactam in combination were decreased 4- and 8-fold and the MIC90s were reduced 8- and 4-fold, respectively, compared to the MICs in the monotherapy setting. The fosfomycin-sulbactam combination was synergistic against 74% (37/50), additive against 24% (12/50), and indifferent against 2% (1/50) of the isolates. No antagonism was observed in the checkerboard study.
TABLE 1.
MIC and FICIs of fosfomycin-sulbactam combination and monotherapy against 50 carbapenem-resistant A. baumannii strains
| Strain no. | MIC (mg/liter)a |
Minimal FICI (range) | Interpretation | |||
|---|---|---|---|---|---|---|
| FOS | SUL | FOScombination | SULcombination | |||
| 73 | 512 | 64 | 128 | 8 | 0.38 (0.38–1.01) | Synergistic |
| 74 | 128 | 64 | 32 | 16 | 0.50 (0.5–4.01) | Synergistic |
| 75 | 128 | 64 | ≤8 | 32 | 0.56 (0.75–4.01) | Additive |
| 76 | 128 | 64 | 32 | 16 | 0.50 (0.50–4.03) | Synergistic |
| 77 | 2,048 | 128 | 64 | 16 | 0.16 (0.16–0.50) | Synergistic |
| 78 | 128 | 128 | 32 | 16 | 0.38 (0.38–4.01) | Synergistic |
| 79 | 2,048 | 128 | 128 | 8 | 0.13 (0.13–0.31) | Synergistic |
| 80 | 256 | 256 | 64 | 16 | 0.31 (0.31–2.01) | Synergistic |
| 81 | 256 | 64 | 32 | 32 | 0.63 (0.63–2.12) | Additive |
| 82 | 512 | 32 | 128 | 8 | 0.50 (0.50–1.01) | Synergistic |
| 83 | 256 | 128 | 128 | 8 | 0.56 (0.56–2.00) | Additive |
| 84 | 1,024 | 128 | 256 | ≤0.5 | 0.25 (0.25–0.51) | Synergistic |
| 85 | 256 | 128 | 64 | 32 | 0.50 (0.50–2.00) | Synergistic |
| 86 | 1,024 | 128 | 128 | 16 | 0.25 (0.25–0.56) | Synergistic |
| 87 | 256 | 256 | 128 | 32 | 0.63 (0.63–2.00) | Additive |
| 88 | 256 | 128 | 64 | 32 | 0.50 (0.50–2.01) | Synergistic |
| 89 | 128 | 128 | ≤8 | 64 | 0.56 (0.56–4.00) | Additive |
| 90 | 256 | 128 | 64 | 32 | 0.50 (0.50–2.00) | Synergistic |
| 91 | 128 | 128 | ≤8 | 64 | 0.56 (0.56–4.01) | Additive |
| 92 | 512 | 128 | ≤8 | 32 | 0.27 (0.27–1.01) | Synergistic |
| 93 | 128 | 128 | ≤8 | 32 | 0.31 (0.31–4.01) | Synergistic |
| 94 | 256 | 128 | 32 | 32 | 0.38 (0.38–2.01) | Synergistic |
| 95 | 512 | 16 | 128 | 4 | 0.50 (0.50–1.03) | Synergistic |
| 96 | 1,024 | 64 | 64 | 16 | 0.31 (0.31–1.01) | Synergistic |
| 97 | 512 | 256 | 64 | 64 | 0.38 (0.38–1.25) | Synergistic |
| 98 | 256 | 64 | 64 | 16 | 0.50 (0.50–2.01) | Synergistic |
| 99 | 256 | 256 | ≤8 | 64 | 0.28 (0.28–2.13) | Synergistic |
| 100 | 256 | 32 | 64 | 8 | 0.50 (0.50–2.02) | Synergistic |
| 101 | 128 | 256 | 64 | 32 | 0.63 (0.63–4.00) | Additive |
| 102 | 128 | 64 | 32 | 16 | 0.50 (0.50–4.06) | Synergistic |
| 103 | 512 | 64 | 64 | 16 | 0.38 (0.38–1.25) | Synergistic |
| 104 | 128 | 64 | 32 | 8 | 0.38 (0.38–4.02) | Synergistic |
| 105 | 128 | 128 | ≤8 | 64 | 0.56 (0.56–4.00) | Additive |
| 106 | 256 | 128 | ≤8 | 64 | 0.53 (0.53–2.25) | Additive |
| 107 | 128 | 128 | ≤8 | 32 | 0.31 (0.31–4.01) | Synergistic |
| 108 | 256 | 32 | 32 | 16 | 0.63 (0.63–2.25) | Additive |
| 109 | 128 | 128 | 32 | 16 | 0.38 (0.38–4.02) | Synergistic |
| 110 | 512 | 256 | 64 | 32 | 0.25 (0.25–1.06) | Synergistic |
| 111 | 256 | 128 | 32 | 32 | 0.38 (0.38–2.02) | Synergistic |
| 112 | 128 | 256 | 128 | 16 | 1.06 (1.06–4.00) | Indifferent |
| 113 | 128 | 32 | 32 | 16 | 0.75 (0.75–4.02) | Additive |
| 114 | 256 | 256 | 64 | 32 | 0.38 (0.38–2.00) | Synergistic |
| 115 | 128 | 128 | ≤8 | 32 | 0.31 (0.31–4.03) | Synergistic |
| 116 | 256 | 128 | 32 | 16 | 0.25 (0.25–2.02) | Synergistic |
| 117 | 1,024 | 256 | 64 | 32 | 0.19 (0.19–0.56) | Synergistic |
| 118 | 128 | 256 | 32 | 8 | 0.28 (0.28–4.02) | Synergistic |
| 119 | 512 | 128 | 32 | 16 | 0.19 (0.19–1.06) | Synergistic |
| 120 | 1,024 | 256 | 64 | 16 | 0.13 (0.13–0.56) | Synergistic |
| 121 | 512 | 16 | 128 | 8 | 0.75 (0.75–2.02) | Additive |
| 122 | 512 | 128 | 128 | 32 | 0.50 (0.50–1.12) | Synergistic |
| MIC50 | 256 | 128 | 64 | 16 | ||
| MIC90 | 1,024 | 256 | 128 | 64 | ||
| MIC50 fold reduction | 4 | 8 | ||||
| MIC90 fold reduction | 8 | 4 | ||||
aFOS, fosfomycin; SUL, sulbactam; FOScombination, fosfomycin in combination with sulbactam; SULcombination, sulbactam in combination with fosfomycin.
The time-kill curves for both isolates are shown in Fig. 1. The fosfomycin-sulbactam combination displayed bactericidal activity against both isolates at the highest clinically achievable concentrations (fosfomycin at 128 mg/liter and sulbactam at 128 mg/liter) (change in log CFU from 0 to 24 h [ΔlogCFU0 –24], −3.09 and −5.14 log10 CFU/ml for isolates 79 and 80, respectively). Synergism was observed only against isolate 80 at a concentration of 128 mg/liter for both fosfomycin and sulbactam (ΔlogCFU24 combination-monotherapy, −2.95 log10 CFU/ml). At concentrations equal to the fractional inhibitory concentration (FIC) (fosfomycin at 64 to 128 mg/liter and sulbactam at 8 to 16 mg/liter), there was a 2- to 3-log10 reduction in the bacterial density by 6 to 8 h, followed by regrowth, for both isolates 79 and 80. No bactericidal activity was observed against either isolate in the presence of fosfomycin monotherapy. When exposed to sulbactam monotherapy at a concentration of 128 mg/liter, there were 2.6- and 2.2-log10 reductions in bacterial burden at 24 h for isolates 79 and 80, respectively. No antagonism was observed in the time-kill study for either isolate.
FIG 1.
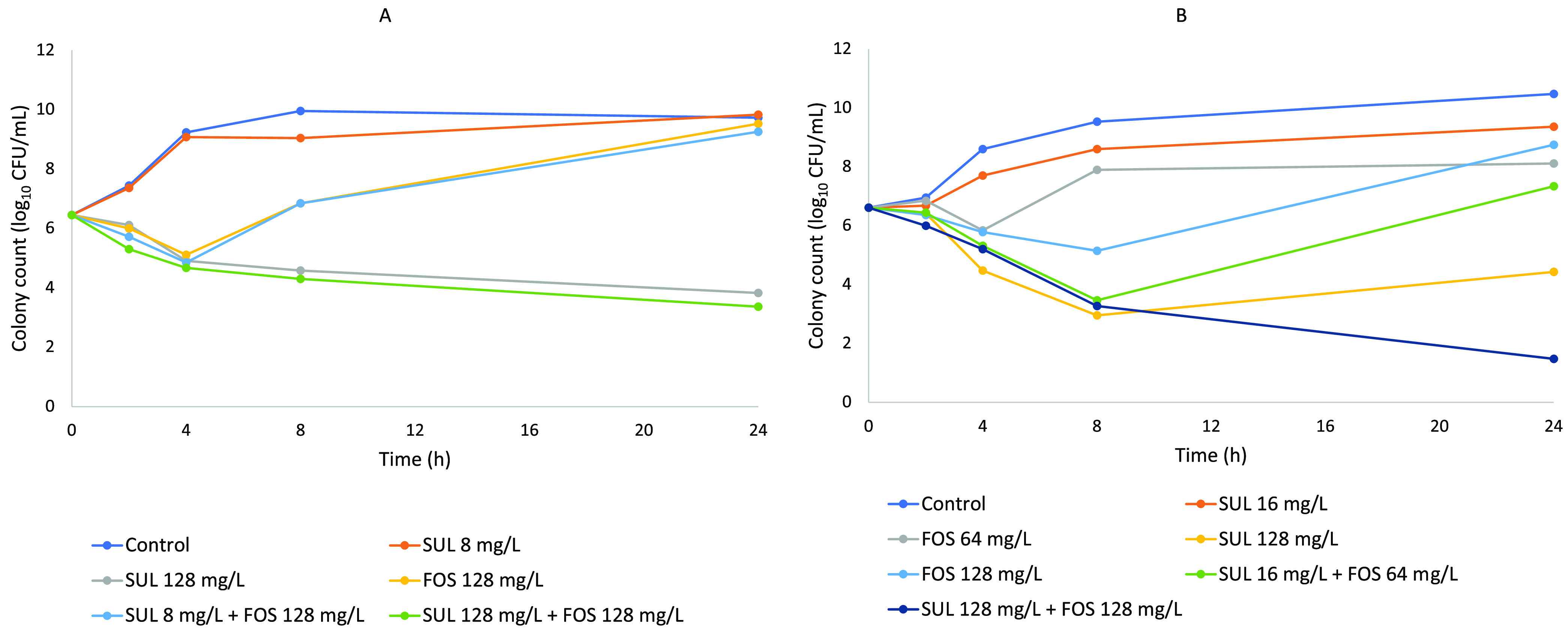
Time-kill curves of CR-AB isolates 79 (A) and 80 (B). FOS, fosfomycin; SUL, sulbactam.
Semimechanistic PK/PD model.
Interaction between fosfomycin and sulbactam and their effects on bacterial growth were best described using the general pharmacodynamic interaction (GPDI) model developed by Wicha et al. (17), with fosfomycin potentiating the effects of sulbactam and two bacterial subpopulations that were either sensitive or resistant to the combination therapy. The pharmacodynamic model is outlined by equations 1 and 2, which describe the bacterial growth and death, including the theoretical maximal bacterial density and drug-mediated killing, of both bacterial subpopulations. The parameter estimates for each isolate are detailed in Table S1 in the supplemental material. The observed versus individual and population predicted Bayesian posterior correlation coefficients (R2) are shown in Fig. 2.
| (1) |
| (2) |
FIG 2.
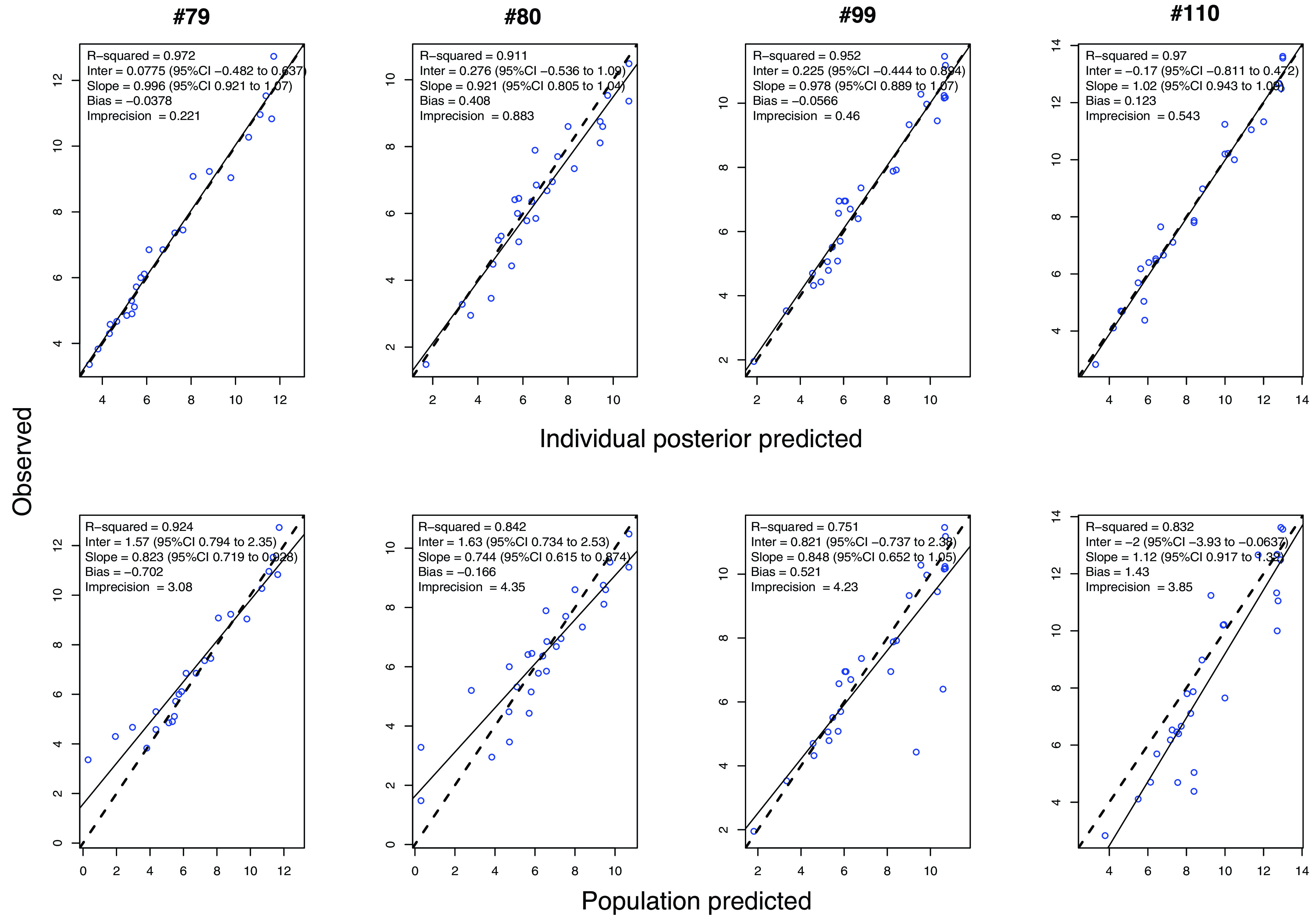
Observed versus individual and population-fitted viable counts for the fosfomycin-sulbactam combination against four CR-AB strains.
CFUs and CFUr represent the bacterial burdens for the sensitive and resistant bacterial subpopulations, respectively; Kgs and Kgr are the growth rate constants for the sensitive and resistant bacterial subpopulations, respectively; Bmax is the maximal bacterial burden; and represent the maximum rate of fosfomycin- and sulbactam-mediated bacterial killing, respectively (log10 CFU per milliliter per hour); CF and CS represent the concentration of fosfomycin and sulbactam, respectively; HF and HS are the power parameters (Hill coefficients) for fosfomycin and sulbactam effects on both subpopulations, respectively; and represent the fosfomycin and sulbactam concentrations for which the effect is 50% on the sensitive subpopulation, respectively; and represent the fosfomycin and sulbactam concentrations for which the effect is 50% on the resistant subpopulation, respectively; INTFS represents the maximum fractional change of the and caused by sulbactam; HIFS represents the power parameter (Hill coefficient) for sulbactam potentiation of the fosfomycin effect; and represents the sulbactam concentration needed to achieve 50% of INTFS.
The effect of both fosfomycin and sulbactam was best described by a sigmoidal Emax model, with the same Emax for both drugs for both subpopulations but different EC50s. The potentiation of the effect of sulbactam on both subpopulations by fosfomycin was best described by a modification of and by a sigmoidal Emax function of the fosfomycin concentration. The interaction parameters were set to the same values for both subpopulations.
Monte Carlo simulations.
The simulated time-concentration profiles of fosfomycin at 4 g, 6 g, and 8 g every 8 h given as a 1-h infusion and sulbactam at 2 g, 3 g, and 4 g every 8 h given as a 4-h infusion are shown in Fig. S1. The simulated bacterial killing of various fosfomycin-sulbactam monotherapy and combination dosing regimens are shown in Fig. S2 and S3. The probabilities of target attainment of 2-log10 kill, 1-log10 kill, and stasis at 24 h are summarized in Table 2.
TABLE 2.
Probability of target attainment of 2-log10 kill, 1-log10 kill, and stasis of various dosing regimens at 24 h against carbapenem-resistant A. baumannii isolate 79 at an initial inoculum of 107 CFU/ml
| Amt (g) of fosfomycin (1-h infusion) | Amt (g) of sulbactam (4-h infusion) | Probability of target attainment (%) |
||
|---|---|---|---|---|
| 2-log10 kill | 1-log10 kill | Stasis | ||
| 8 | 4 | 71.6 | 76.4 | 81.6 |
| 6 | 4 | 68.5 | 73.6 | 78.5 |
| 4 | 4 | 61.4 | 69.1 | 74.8 |
| 8 | 3 | 59.5 | 66.7 | 71.4 |
| 6 | 3 | 53.2 | 61 | 68.4 |
| 4 | 3 | 46.5 | 54.5 | 61.2 |
| 8 | 2 | 45.5 | 50.1 | 56.7 |
| 6 | 2 | 39.8 | 45.7 | 51.3 |
| 4 | 2 | 31 | 38.3 | 44.1 |
| 8 | 15.5 | 19.8 | 23.3 | |
| 4 | 32.5 | 46.5 | 53.5 | |
Against an initial inoculum of 107 CFU/ml, both fosfomycin and sulbactam monotherapies resulted in no or limited bacterial killing at 24 h (50th percentile) (Fig. S2A and B). At the 50th percentile, there was a >3.5-log10 reduction in bacterial burden by 24 h in combination regimens including sulbactam at 4 g every 8 h (Fig. S3D to F), with the combination of fosfomycin at 8 g every 8 h and sulbactam at 4 g every 8 h displaying the most extensive bacterial killing by 24 h (∼5.5-log10 reduction in colony count) (Fig. S3F). For combination regimens comprising sulbactam at 3 g every 8 h with 4 g, 6 g, or 8 g of fosfomycin every 8 h, bacterial killing ranged between ∼1 and 3.5 log10 (Fig. S3A to C). Limited or no bacterial killing was observed for combination regimens containing sulbactam at 2 g every 8 h (Fig. S2C to E).
DISCUSSION
To the best of our knowledge, this is the first study to describe the interaction between fosfomycin and sulbactam in combination against CR-AB isolates using semimechanistic pharmacodynamic modeling with Monte Carlo simulations. As mentioned above, we found the GPDI model (17) to best describe the interaction between the two antibiotics. In general, this model allows for measurement of the interaction between two drugs by measuring the change in Emax or EC50, which provides a quantitative and statistically interpretable (point) estimate of a pharmacodynamic interaction (17). The GPDI model is also able to identify the perpetrator and victim of a pharmacodynamic interaction, whereby a perpetrator alters the Emax or EC50 of the victim drug, leading to either synergism or antagonism (17).
Our pharmacodynamic model indicated that fosfomycin (perpetrator) enhanced the bacterial killing of sulbactam despite the lack of susceptibility to fosfomycin in these isolates. This observation is illustrated in Fig. S3, which shows that the increment of sulbactam from 3 g to 4 g improved bacterial killing by 2 log10. This observation also suggests that when sulbactam is used at a sufficiently high dose, the fosfomycin dose could be reduced to achieve a similar decrease in the colony count. As shown in Fig. S3C and D, the combination regimen of sulbactam at 4 g every 8 h with fosfomycin at 4 g every 8 h was able to achieve a bacterial kill of ∼3.5 log10, similar to that of the regimen containing sulbactam at 3 g every 8 h and fosfomycin at 8 g every 8 h.
The potentiation of one antibiotic by another is not uncommon. Fosfomycin has been previously described to enhance the uptake of tobramycin in Pseudomonas aeruginosa, leading to greater repression of bacterial protein synthesis (18). Unfortunately, the mechanism underlying the enhancement of the in vitro activity of sulbactam by fosfomycin has not been previously described. Furthermore, we still do not fully understand the mechanisms by which sulbactam acts against A. baumannii (19). At this juncture, we know that both sulbactam and fosfomycin act by disrupting the biosynthesis of bacterial cell wall via two distinct pathways (10, 19). Fosfomycin acts by targeting mucopeptide synthesis, thereby inhibiting phosphoenolpyruvate transferase, which is the first enzyme involved in peptidoglycan synthesis (10). It therefore acts on the first stage of peptidoglycan synthesis, inhibiting bacterial cell wall production at an earlier stage than most antibiotic classes (10), which could potentially lead to an increased uptake of sulbactam by fosfomycin.
In addition, our simulation illustrated that clinically achievable plasma fosfomycin and fosfomycin concentrations may be able to attain synergistic killing (≥2-log10 kill) when used in combination in ∼60% to 70% of the simulated patients, compared to ∼15% to 30% with fosfomycin or sulbactam monotherapies at the highest doses (Table 2), against CR-AB isolates. Our simulations suggest that a combination of fosfomycin at 4 g every 8 h (1-h infusion) with sulbactam at 4 g every 8 h (4-h infusion), as a minimum, may provide a favorable effect in lowering the bacterial load to a level where the immune system in immunocompetent patients can eradicate remaining bacteria, as exhibited in a murine pneumonia model by Louie et al. (20).
Through the use of mathematical modeling and Monte Carlo simulations, we were able to appraise a range of fosfomycin-sulbactam dosing regimens, also incorporating between-patient pharmacokinetic variability that is often seen in critically ill patients (21). With the absence of an immune response in vitro, our pharmacodynamic model and simulations may represent an immunocompromised patient population. Moreover, to emulate a worst-case scenario, we simulated a high inoculum (107 CFU/ml) of an extremely difficult-to-treat isolate (fosfomycin MIC of 2,048 mg/liter, sulbactam MIC of 128 mg/liter, and meropenem MIC of 128 mg/liter).
Nevertheless, this study has some limitations. To avoid over- or underestimation of the drugs’ plasma concentrations and for ease of comparison, we fixed the simulated patient weight to 70 kg and creatinine clearance (CLCR) to 100 ml/min. Hence, weight and renal clearance should be taken into account when extrapolating the results of the simulation, as they were essential covariates in the pharmacokinetic model (22, 23). Nonetheless, these parameters can be easily modified in the simulation, allowing for the possibility of various weight and renal function combinations, depending on the patient of interest in the clinical setting. Furthermore, the pharmacodynamic model utilized data from static time-kill studies, which do not mirror the changing drug concentrations in vivo.
Also, in this model, a fixed, non-concentration-dependent interaction term, INTFS, was used, whereby the mean INTFS estimate was derived from the best-case scenario and applied for subsequent simulations. We acknowledge that the time-kill data used for the pharmacodynamic simulation can restrict the application unless it is inclusive of the best- and worst-case scenarios. We have used time-kill data from four isolates with various responses to the combination in the model building and used the time-kill data of isolate 79 to derive the final model parameter estimates used for the simulation, as this isolate had the highest fosfomycin, sulbactam, and meropenem MICs (and therefore was the most resistant isolate) and synergism was not observed when the isolate was exposed to the combination therapy, to mirror a worst-case-scenario. Further evaluation of this promising combination in across various CR-AB isolates, and testing using dynamic infection models, should be pursued to provide a clearer picture of the potential utility of this antibiotic combination in the treatment of CR-AB.
Screening of synergistic activity for the fosfomycin-sulbactam combination was done using the broth microdilution instead of the agar dilution method. Nonetheless, the broth microdilution and agar dilution methods have been shown to correlate reasonably well (24, 25). One study demonstrated that MIC values were within one 2-fold dilution when tested in broth microdilution compared to those tested in agar dilution for 86 of 106 isolates (81.1%) (24). Additionally, there are no studies which investigated the PK/PD target attainment of fosfomycin and sulbactam in combination. The pharmacodynamic targets (stasis and 1-log10 and 2-log10 reductions) used in this study were based on the European Medicines Agency’s guideline on the use of pharmacokinetics and pharmacodynamics in the development of antimicrobial medicinal products (26).
As mentioned above, simulation of a different patient weight and renal function combination, according to the patient of interest, is possible, by adjusting these parameters during the simulation. Hence, we propose that the model may have potential use in the clinical setting, particularly when dealing with critically ill patients with multidrug-resistant A. baumannii infections, as these patients are at heightened risk of suboptimal drug dosing due to altered pharmacokinetics (27). The use of the PK/PD model and simulation in the clinical setting would allow clinicians to predict the probability of attaining a particular pharmacodynamic target (stasis and 1-log10 and 2-log10 reductions) for a patient of interest, based on the individual’s weight and renal function. However, as this combination is synergistic in only ∼70% of isolates, as observed in our checkerboard study, we are unable to extrapolate the use of this model to all patients with CR-AB infection, particularly those with CR-AB isolates against which the fosfomycin and sulbactam combination lacks synergism. Therefore, we suggest the application of this PK/PD model in combination with in vitro antibiotic combination testing, e.g., the checkerboard assay, to screen for synergism of the fosfomycin and sulbactam combination. The model would provide better quantification of the effect of the combination compared to the checkerboard testing alone. In vitro antibiotic combination testing in the clinical setting has been previously reported and appears to hold a promising role in improving patient outcome and judicious antibiotic use (28).
Conclusion.
Favorable results for the fosfomycin and sulbactam combination were observed through synergism analysis and PK/PD evaluation. Our study demonstrated that the fosfomycin and sulbactam combination could be a plausible option to treat CR-AB infections for a number of clinically relevant isolates. Further dynamic infection models and in vivo preclinical or clinical studies are needed to validate these in vitro and in silico results.
MATERIALS AND METHODS
Bacterial isolates, antimicrobial agents, susceptibility testing, checkerboard assay, and static time-kill assay.
The methodology of the pharmacodynamic component of this study has been published elsewhere (29). Briefly, the CR-AB isolates were obtained from The University of Queensland Centre of Clinical Research (UQCCR). Fifty isolates were chosen from samples previously analyzed by Zowawi et al. (30). The study by Zowawi et al. suggested that the 107 isolates were from diverse clonal lineages. The 50 isolates were chosen from the 107 randomly to avoid any systematic selection bias and to ensure that the high genetic diversity of the baseline population of isolates was represented in our study isolates.
Carbapenem resistance was determined in their study by the disk diffusion susceptibility testing for imipenem (10 μg) and meropenem (10 μg) following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) methodology and in reference to the updated breakpoint defined by EUCAST (31). To confirm carbapenem resistance, the MICs of meropenem against the isolates were retested in this study using the broth microdilution method (32).
Sulbactam (Toronto Research Chemicals) and fosfomycin (Sigma-Aldrich) were obtained from their respective manufacturers. Stock solutions of sulbactam and fosfomycin were prepared in sterile Milli-Q water, filter sterilized with a 0.22-μm polyvinylidene difluoride (PVDF) syringe filter, aliquoted, and stored at −80°C until required. Broth or agar containing fosfomycin was supplemented with 25 mg/liter of glucose-6-phosphate (Sigma-Aldrich).
The MICs of sulbactam against the 50 A. baumannii isolates were determined by the broth microdilution method, in quadruplicate, following the recommendations of the Clinical and Laboratory Standards Institute (CLSI) as described in the CLSI M100 approved standard (32). The mode MIC was reported for each isolate. Fosfomycin susceptibility testing was performed by agar dilution (32). Klebsiella pneumoniae (ATCC 700603) and Pseudomonas aeruginosa (ATCC 27853) strains were used as quality control strains for sulbactam and fosfomycin, respectively.
The synergistic activities of fosfomycin and sulbactam in combination against the 50 CR-AB isolates were then screened using the checkerboard assay, which was done twice for each isolate. The ranges of concentrations of the antibiotics used were as follows: fosfomycin, 8 to 512 mg/liter, and sulbactam, 0.5 to 64 mg/liter, with the highest concentrations being those that are clinically achievable in critically ill patients, based on previously published data (22, 23). The FICI was calculated by the summation of MICdrug A in combination/MICdrug A alone and MICdrug B in combination/MICdrug B alone (33). The FICIs were interpreted as follows: ≤0.5, synergistic; 0.5 < FICI ≤ 1, additive; 1 < FICI ≤ 4, indifferent; and >4, antagonistic (34). The minimum FICI was reported for each isolate. The mode MICs were reported.
Subsequently, static time-kill studies were conducted against two A. baumannii isolates (79 and 80), which demonstrated synergistic activity in the checkerboard assay (fosfomycin MIC, 256 to 2,048 mg/liter, and sulbactam MIC, 128 to 256 mg/liter). The concentrations of the antibiotics equal to the FIC exhibiting synergy by the checkerboard and the highest clinically achievable antibiotic concentrations, alone and in combination, were tested against an inoculum of approximately 106 CFU/ml. A decrease of ≥3 log10 at 24 h compared to the number of viable cells at the initial time point (ΔlogCFU0 –24) was indicative of a bactericidal effect (33). A synergistic effect was determined by a decrease of ≥2 log10 in CFU per milliliter at 24 h when comparing the antibiotics in combination to the most active drug alone at that time point (ΔlogCFU24 combination-monotherapy), while an increase of >2 log10 was considered antagonism (33). Indifference was interpreted as any other outcome that did not meet the criteria for either synergy or antagonism (35).
Semimechanistic PK/PD modeling.
Semimechanistic PK/PD modeling was performed to quantify the exposure-effect relationships of both drugs provided alone or in combination, based on the static time-kill data. As mentioned above, we have performed static time-kill studies on two isolates (unpublished data) and further enriched the simulation with data from two additional isolates from a previous publication (29), for a total of four isolates. Parameter estimation was performed using the Pmetrics package (v1.52; Laboratory of Applied Pharmacokinetics and Bioinformatics, Los Angeles, CA) (36) for R (v3.6.2; R Core Team, Vienna, Austria). Model diagnostics, including Akaike information criteria, log likelihood, coefficient of determination (R2) from the observed-versus-expected plots, plausibility of the parameter estimates, and visual predictive checks, were used to evaluate and compare models. Various structural models were tested to find the best-fitting model.
Simulation of bacterial kill at clinically achievable concentrations.
To better understand fosfomycin and sulbactam pharmacodynamic interactions in a dynamic drug concentration as in the clinical setting, Monte Carlo simulations of various fosfomycin and sulbactam clinical dosing regimens were performed for 1,000 virtual patients. The final pharmacodynamic model was combined with previously reported human population pharmacokinetic models for fosfomycin (22) and sulbactam (23) in critically ill patients. These Monte Carlo simulations accounted for between-patient variability in the pharmacokinetics of each antibiotic. The mean parameter estimates and the coefficient of variation of the parameters were also included in the simulation to account for between-patient variability of the parameter estimates.
The simulated patients were assumed to have bacteremia caused by a difficult-to-treat CR-AB isolate, isolate 79, against which the combination therapy did not exhibit synergism (fosfomycin MIC of 2,048 mg/liter, sulbactam MIC of 128 mg/liter, and meropenem MIC of 128 mg/liter) and lacked any aspect of the immune system, to paint a worst-case scenario. The weight of the simulated patient was fixed at 70 kg to represent the weight of a typical adult (37, 38), and the creatinine clearance (CLCR) was fixed at 100 ml/min/1.73 m2 to represent patient with normal renal clearance. The simulated regimens included fosfomycin given as a 1-h infusion of 4 g, 6 g, or 8 g every 8 h and sulbactam given as a 4-h infusion of 2 g, 3 g, or 4 g every 8 h. The probabilities of target attainment (PTA) of stasis, 1-log10 kill, and 2-log10 kill at 24 h for the simulated exposures of the 1,000 virtual patients were calculated.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Australian National Health and Medical Research Council (NHMRC) for a Centre of Research Excellence fund (APP1099452). J.A.R. is funded in part by a Practitioner Fellowship (APP1117065) from the NHMRC. S.M.S.L. acknowledges funding from the University of Queensland Research Training Scholarship. A.J.H. acknowledges funding from a Griffith School of Medicine Research Higher degree scholarship. F.B.S. is funded in part by an NHMRC Investigator Grant (APP1197866).
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, Lee SH. 2017. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. 2009. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents 34:111–120. 10.1016/j.ijantimicag.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Liu Q, Chen Z, Li C. 2017. Efficacy of sulbactam for the treatment of Acinetobacter baumannii complex infection: a systematic review and meta-analysis. J Infect Chemother 23:278–285. 10.1016/j.jiac.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Santimaleeworagun W, Wongpoowarak P, Chayakul P, Pattharachayakul S, Tansakul P, Garey KW. 2011. In vitro activity of colistin or sulbactam in combination with fosfomycin or imipenem against clinical isolates of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemases. Southeast Asian J Trop Med Public Health 42:890. [PubMed] [Google Scholar]
- 5.Zhu W, Wang Y, Cao W, Cao S, Zhang J. 2018. In vitro evaluation of antimicrobial combinations against imipenem-resistant Acinetobacter baumannii of different MICs. J Infect Public Health 11:856–860. 10.1016/j.jiph.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Wei W, Yang H, Liu Y, Ye Y, Li J. 2016. In vitro synergy of colistin combinations against extensively drug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase. J Chemother 28:159–163. 10.1179/1973947815Y.0000000030. [DOI] [PubMed] [Google Scholar]
- 7.Deveci A, Coban AY, Acicbe O, Tanyel E, Yaman G, Durupinar B. 2012. In vitro effects of sulbactam combinations with different antibiotic groups against clinical Acinetobacter baumannii isolates. J Chemother 24:247–252. 10.1179/1973947812Y.0000000029. [DOI] [PubMed] [Google Scholar]
- 8.Hendlin D, Stapley EO, Jackson M, Wallick H, Miller AK, Wolf FJ, Miller TW, Chaiet L, Kahan FM, Foltz EL, Woodruff HB, Mata JM, Hernandez S, Mochales S. 1969. Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science 166:122–123. 10.1126/science.166.3901.122. [DOI] [PubMed] [Google Scholar]
- 9.Brown ED, Vivas EI, Walsh CT, Kolter R. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol 177:4194–4197. 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci 235:364–386. 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 11.Campoli-Richards DM, Brogden RN. 1987. Sulbactam/ampicillin. Drugs 33:577–609. 10.2165/00003495-198733060-00003. [DOI] [PubMed] [Google Scholar]
- 12.Williams JD. 1997. β-Lactamase inhibition and in vitro activity of sulbactam and sulbactam/cefoperazone. Clin Infect Dis 24:494–497. 10.1093/clinids/24.3.494. [DOI] [PubMed] [Google Scholar]
- 13.Yokota T, Sekiguchi R, Azuma E, Suzuki E. 1984. Sulbactam: permanent inactivation of various types of betalactamases and the affinity to penicillin-binding proteins in bacteria. Chemotherapy (Tokyo) 32:11–19. [Google Scholar]
- 14.Appleman MD, Belzberg H, Citron DM, Heseltine PNR, Yellin AE, Murray J, Berne TV. 2000. In vitro activities of nontraditional antimicrobials against multiresistant Acinetobacter baumannii strains isolated in an intensive care unit outbreak. Antimicrob Agents Chemother 44:1035–1040. 10.1128/aac.44.4.1035-1040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoyama Y, Matsumoto K, Ikawa K, Watanabe E, Shigemi A, Umezaki Y, Nakamura K, Ueno K, Morikawa N, Takeda Y. 2014. Pharmacokinetic/pharmacodynamic evaluation of sulbactam against Acinetobacter baumannii in in vitro and murine thigh and lung infection models. Int J Antimicrob Agents 43:547–552. 10.1016/j.ijantimicag.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Kollef MH, Golan Y, Micek ST, Shorr AF, Restrepo MI. 2011. Appraising contemporary strategies to combat multidrug resistant gram-negative bacterial infections—proceedings and data from the Gram-Negative Resistance Summit. Clin Infect Dis 53:S33–S55. 10.1093/cid/cir475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicha S, Chen C, Clewe O, Simonsson U. 2017. A general pharmacodynamic interaction model identifies perpetrators and victims in drug interactions. Nat Commun 8:2129. 10.1038/s41467-017-01929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLeod DL, Velayudhan J, Kenney TF, Therrien JH, Sutherland JL, Barker LM, Baker WR. 2012. Fosfomycin enhances the active transport of tobramycin in Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:1529–1538. 10.1128/AAC.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penwell WF, Shapiro AB, Giacobbe RA, Gu RF, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BL, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louie A, Brown D, Baluya D, Rodriquez J, Robbins N, Kurhanewicz S, Fikes S, Liu W, Drusano GL, Cirz R. 2014. Interaction of drug- and granulocyte-mediated killing of Pseudomonas aeruginosa in a murine pneumonia model. J Infect Dis 210:1319–1324. 10.1093/infdis/jiu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts JA, Lipman J. 2009. Pharmacokinetic issues for antibiotics in the critically ill patient. Crit Care Med 37:840–851; quiz, 859. 10.1097/CCM.0b013e3181961bff. [DOI] [PubMed] [Google Scholar]
- 22.Parker SL, Frantzeskaki F, Wallis SC, Diakaki C, Giamarellou H, Koulenti D, Karaiskos I, Lipman J, Dimopoulos G, Roberts JA. 2015. Population pharmacokinetics of fosfomycin in critically ill patients. Antimicrob Agents Chemother 59:6471–6476. 10.1128/AAC.01321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaruratanasirikul S, Wongpoowarak W, Wattanavijitkul T, Sukarnjanaset W, Samaeng M, Nawakitrangsan M, Ingviya N. 2016. Population pharmacokinetics and pharmacodynamics modeling to optimize dosage regimens of sulbactam in critically ill patients with severe sepsis caused by Acinetobacter baumannii. Antimicrob Agents Chemother 60:7236–7244. 10.1128/AAC.01669-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flamm RK, Rhomberg PR, Huynh HK, Sader HS, Ellis-Grosse E. 2016. In vitro activity of fosfomycin (ZTI-01, fosfomycin for injection) against contemporary Gram-negative and Gram-positive isolates: a comparison of intermethod testing. Open Forum Infect Dis 3(Suppl 1):1833. 10.1093/ofid/ofw172.1381. [DOI] [Google Scholar]
- 25.Ballestero-Téllez M, Docobo-Pérez F, Rodríguez-Martínez J-M, Conejo MC, Ramos-Guelfo M, Blázquez J, Rodríguez-Baño J, Pascual A. 2017. Role of inoculum and mutant frequency on fosfomycin MIC discrepancies by agar dilution and broth microdilution methods in Enterobacteriaceae. Clin Microbiol Infect 23:325–331. 10.1016/j.cmi.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 26.European Medicines Agency. 2016. Guideline on the use of pharmacokinetics and pharmacodynamics in the development of antimicrobial medicinal products. European Medicines Agency, London, United Kingdom. [Google Scholar]
- 27.Udy AA, Roberts JA, Boots RJ, Paterson DL, Lipman J. 2010. Augmented renal clearance: implications for antibacterial dosing in the critically ill. Clin Pharmacokinet 49:1–16. 10.2165/11318140-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 28.Cai Y, Chua NG, Lim T-P, Teo JQ-M, Lee W, Kurup A, Koh T-H, Tan T-T, Kwa AL. 2016. From bench-top to bedside: a prospective in vitro antibiotic combination testing (iACT) service to guide the selection of rationally optimized antimicrobial combinations against extensively drug resistant (XDR) Gram negative bacteria (GNB). PLoS One 11:e0158740. 10.1371/journal.pone.0158740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohd Sazlly Lim S, Naicker S, Ayfan AK, Zowawi H, Roberts JA, Sime FB. 2020. Non-polymyxin-based combinations as potential alternatives in treatment against carbapenem-resistant Acinetobacter baumannii infections. Int J Antimicrob Agents 56:106115. 10.1016/j.ijantimicag.2020.106115. [DOI] [PubMed] [Google Scholar]
- 30.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, Al Salman J, Dashti AA, Johani K, Paterson DL. 2015. Molecular epidemiology of carbapenem resistant Acinetobacter baumannii in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J Clin Microbiol 53:896–903. 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters, on version. https://eucast.org/clinical_breakpoints/. Accessed 08/06/2020.
- 32.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. Approved standard M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Doern CD. 2014. When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J Clin Microbiol 52:4124–4128. 10.1128/JCM.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiman L. 2007. Clinical utility of synergy testing for multidrug-resistant Pseudomonas aeruginosa isolated from patients with cystic fibrosis: ‘the motion for.’ Paediatr Respir Rev 8:249–255. 10.1016/j.prrv.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Petersen PJ, Labthavikul P, Jones CH, Bradford PA. 2006. In vitro antibacterial activities of tigecycline in combination with other antimicrobial agents determined by chequerboard and time-kill kinetic analysis. J Antimicrob Chemother 57:573–576. 10.1093/jac/dki477. [DOI] [PubMed] [Google Scholar]
- 36.Neely M, van Guilder M, Yamada W, Schumitzky A, Jelliffe R. 2012. Accurate detection of outliers and subpopulations with Pmetrics, a non-parametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 34:467–476. 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mould DR, Upton RN. 2013. Basic concepts in population modeling, simulation, and model‐based drug development—part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacometrics Syst Pharmacol 2:e38. 10.1038/psp.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goulooze SC, Völler S, Välitalo PA, Calvier EA, Aarons L, Krekels EH, Knibbe CA. 2019. The influence of normalization weight in population pharmacokinetic covariate models. Clin Pharmacokinet 58:131–138. 10.1007/s40262-018-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


