Recent outbreaks of cardiac surgery-associated Mycobacterium chimaera infections have highlighted the importance of species differentiation within the Mycobacterium avium complex and pointed to a lack of antibiotic susceptibility data for M. chimaera. Using the MGIT 960/EpiCenter TB eXiST platform, we have determined antibiotic susceptibility patterns of 48 clinical M. chimaera isolates and 139 other nontuberculous mycobacteria, including 119 members of the M. avium complex and 20 Mycobacterium kansasii isolates toward clofazimine and other drugs used to treat infections with slow-growing nontuberculous mycobacteria (NTM).
KEYWORDS: Mycobacterium chimaera, Mycobacterium avium complex, drug susceptibility testing, clofazimine, resistance, antibiotic resistance
ABSTRACT
Recent outbreaks of cardiac surgery-associated Mycobacterium chimaera infections have highlighted the importance of species differentiation within the Mycobacterium avium complex and pointed to a lack of antibiotic susceptibility data for M. chimaera. Using the MGIT 960/EpiCenter TB eXiST platform, we have determined antibiotic susceptibility patterns of 48 clinical M. chimaera isolates and 139 other nontuberculous mycobacteria, including 119 members of the M. avium complex and 20 Mycobacterium kansasii isolates toward clofazimine and other drugs used to treat infections with slow-growing nontuberculous mycobacteria (NTM). MIC50, MIC90, and tentative epidemiological cutoff (ECOFF) values for clofazimine were 0.5 mg/liter, 1 mg/liter, and 2 mg/liter, respectively, for M. chimaera. Comparable values were observed for other M. avium complex members, whereas lower MIC50 (≤0.25 mg/liter), MIC90 (0.5 mg/liter), and ECOFF (1 mg/liter) values were found for M. kansasii. Susceptibility to clarithromycin, ethambutol, rifampin, rifabutin, amikacin, moxifloxacin, and linezolid was in general similar for M. chimaera and other members of the M. avium complex, but increased for M. kansasii. The herein determined MIC distributions, MIC90, and ECOFF values of clofazimine for M. chimaera and other NTM provide the basis for the definition of clinical breakpoints. Further studies are needed to establish correlation of in vitro susceptibility and clinical outcome.
INTRODUCTION
Mycobacterium chimaera is a slow-growing nontuberculous mycobacterium (NTM) that was established in 2004 as a new species within the Mycobacterium avium complex (1). In the past, the number of infections with M. chimaera was underestimated, as commercial mycobacterial identification systems such as line probe assays failed to identify M. chimaera to species level and thus classified M. chimaera as M. avium complex, M. avium, or Mycobacterium intracellulare (1, 2). M. chimaera is differentiated from other members of the M. avium complex by a unique 16S rRNA gene sequence and internal transcribed spacer (ITS) region (1). Recently, a global outbreak of cardiac surgery-associated M. chimaera infections highlighted the importance of species identification within the M. avium complex (3, 4). The outbreak was linked to contaminated water reservoirs of heater-cooler devices that spread M. chimaera by aerosols during open chest surgery (5, 6). Severe, disseminated M. chimaera infections with a high case fatality rate were observed (7).
Due to the limited ability of commercial identification methods to adequately identify M. chimaera, few studies have reported drug susceptibility data on M. chimaera. Recent studies analyzed antimicrobial susceptibility of M. chimaera using a commercial microdilution system, the SLOWMYCO Sensititre panel from Trek Diagnostic Systems, and reported similar susceptibility patterns for M. chimaera as for other members of the M. avium complex (8–11). Recommended treatment options for disseminated M. chimaera infections include combination therapy with macrolides, rifamycins, ethambutol, amikacin, and clofazimine (7). Clofazimine is not yet included in the SLOWMYCO Sensititre antibiotic panel and consequently clofazimine MIC data for M. chimaera are scarce. We have previously established automated quantitative drug susceptibility testing (DST) for slow-growing NTM using the MGIT 960/EpiCenter TB eXiST platform (12, 13). We here report on MIC distributions of clofazimine and other drugs used to treat NTM infections for 48 clinical M. chimaera isolates and 139 other nontuberculous mycobacteria, including 119 members of the M. avium complex and 20 Mycobacterium kansasii isolates.
RESULTS
M. chimaera clofazimine MIC distribution.
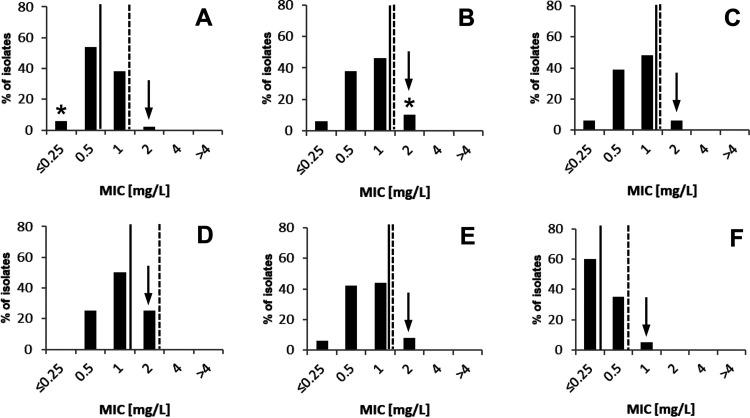
MICs of clofazimine were determined for 48 clinical, nonduplicate M. chimaera isolates using the MGIT 960/EpiCenter TB eXiST system (Becton Dickinson, Sparks, MD). A clofazimine concentration range of 0.25 mg/liter to 4 mg/liter was tested in 2-fold serial dilutions. Out of 48 M. chimaera isolates, 34 (71%) were of respiratory origin and 13 (27%) isolates were of nonrespiratory origin (Table 1). For one isolate, the source was unknown. Clofazimine MIC values for M. chimaera ranged from ≤0.25 mg/liter to 2 mg/liter (Fig. 1A, Table 2). MIC50 and MIC90 values of 0.5 mg/liter and 1 mg/liter were determined. A tentative epidemiological cutoff (ECOFF) was set at 2 mg/liter by visual inspection of the MIC distribution (Fig. 1). The clofazimine MIC distribution of M. chimaera was compared to MIC distributions of 119 M. avium complex isolates, including M. avium (n = 80), M. intracellulare (n = 31), Mycobacterium yongonense (n = 3), Mycobacterium timonense (n = 2), Mycobacterium bouchedurhonense (n = 1), Mycobacterium colombiense (n = 1), and Mycobacterium vulneris (n = 1) (Fig. 1B to E, Table 2). The MIC range, MIC50, MIC90, and tentative ECOFF values of M. chimaera and other M. avium complex isolates were comparable. The clofazimine MIC distribution of M. kansasii, a slow-growing nontuberculous mycobacterium not related to the M. avium complex, showed lower MIC50 (≤0.25 mg/liter), MIC90 (0.5 mg/liter), and tentative ECOFF value (1 mg/liter) compared to M. chimaera and other M. avium complex species (Fig. 1F).
TABLE 1.
Number and origin of NTM isolates included in this study
| Speciesa | No. (%) of respiratory isolates | No. (%) of nonrespiratory isolates | No. (%) of isolates with unknown origin | Total |
|---|---|---|---|---|
| M. avium (MAC) | 55 (69) | 8 (10) | 17 (21) | 80 |
| M. intracellulare (MAC) | 30 (97) | 0 (0) | 1 (3) | 31 |
| M. chimaera (MAC) | 34 (71) | 13 (27) | 1 (2) | 48 |
| Other MAC | 6 (75) | 1 (12.5) | 1 (12.5) | 8 |
| M. kansasii | 12 (60) | 5 (25) | 3 (15) | 20 |
MAC, M. avium complex.
FIG 1.
MIC distributions of clofazimine for M. chimaera (n = 48) (A), M. avium (n = 80) (B), M. intracellulare (n = 31) (C), other MAC (n = 8) (D), M. avium complex overall (n = 167) (E), and M. kansasii (n = 20) (F). Tentative ECOFF (arrow), MIC50 (solid line), and MIC90 (dashed line) are indicated. The clofazimine MIC values of the type strains M. avium ATCC 19421 and M. chimaera DSM 44623 are indicated (*).
TABLE 2.
Assignment of NTM isolates to susceptibility categories in the MGIT 960 system
| Drug/speciesa | MIC (mg/liter) |
In vitro DST category of isolates (n)b |
No. of isolates | ||
|---|---|---|---|---|---|
| S | I | R | |||
| Clarithromycin | |||||
| M. avium | 4 | 67 | 11 | 2 | 80 |
| 16 | 78 | 0 | 2 | ||
| 32 | 78 | 0 | 2 | ||
| 64 | 78 | 0 | 2 | ||
| M. chimaera | 4 | 47 | 0 | 1 | 48 |
| 16 | 47 | 0 | 1 | ||
| 32 | 47 | 1 | 0 | ||
| 64 | 48 | 0 | 0 | ||
| M. intracellulare | 4 | 29 | 1 | 1 | 31 |
| 16 | 30 | 0 | 1 | ||
| 32 | 30 | 0 | 1 | ||
| 64 | 31 | 0 | 0 | ||
| Other MAC | 4 | 8 | 0 | 0 | 8 |
| 16 | 8 | 0 | 0 | ||
| 32 | 8 | 0 | 0 | ||
| 64 | 8 | 0 | 0 | ||
| M. kansasii | 4 | 20 | 0 | 0 | 20 |
| 16 | 20 | 0 | 0 | ||
| 32 | 20 | 0 | 0 | ||
| 64 | 20 | 0 | 0 | ||
| Rifampin | |||||
| M. avium | 1 | 2 | 16 | 62 | 80 |
| 4 | 25 | 32 | 23 | ||
| 20 | 60 | 18 | 2 | ||
| M. chimaera | 1 | 9 | 14 | 25 | 48 |
| 4 | 28 | 19 | 1 | ||
| 20 | 48 | 0 | 0 | ||
| M. intracellulare | 1 | 1 | 2 | 28 | 31 |
| 4 | 3 | 26 | 2 | ||
| 20 | 30 | 1 | 0 | ||
| Other MAC | 1 | 4 | 1 | 3 | 8 |
| 4 | 5 | 3 | 0 | ||
| 20 | 8 | 0 | 0 | ||
| M. kansasii | 1 | 19 | 0 | 1 | 20 |
| 4 | 20 | 0 | 0 | ||
| 20 | 20 | 0 | 0 | ||
| Rifabutin | |||||
| M. avium | 0.1 | 11 | 15 | 54 | 80 |
| 0.4 | 39 | 34 | 7 | ||
| 2 | 78 | 0 | 2 | ||
| M. chimaera | 0.1 | 9 | 7 | 32 | 48 |
| 0.4 | 36 | 10 | 2 | ||
| 2 | 47 | 1 | 0 | ||
| M. intracellulare | 0.1 | 0 | 1 | 30 | 31 |
| 0.4 | 10 | 19 | 2 | ||
| 2 | 31 | 0 | 0 | ||
| Other MAC | 0.1 | 3 | 2 | 3 | 8 |
| 0.4 | 5 | 3 | 0 | ||
| 2 | 8 | 0 | 0 | ||
| M. kansasii | 0.1 | 20 | 0 | 0 | 20 |
| 0.4 | 20 | 0 | 0 | ||
| 2 | 20 | 0 | 0 | ||
| Ethambutol | |||||
| M. avium | 5 | 46 | 19 | 15 | 80 |
| 12.5 | 71 | 4 | 5 | ||
| 50 | 78 | 0 | 2 | ||
| M. chimaera | 5 | 19 | 4 | 25 | 48 |
| 12.5 | 47 | 1 | 0 | ||
| 50 | 48 | 0 | 0 | ||
| M. intracellulare | 5 | 27 | 2 | 2 | 31 |
| 12.5 | 30 | 0 | 1 | ||
| 50 | 31 | 0 | 0 | ||
| Other MAC | 5 | 7 | 1 | 0 | 8 |
| 12.5 | 8 | 0 | 0 | ||
| 50 | 8 | 0 | 0 | ||
| M. kansasii | 5 | 19 | 0 | 1 | 20 |
| 12.5 | 20 | 0 | 0 | ||
| 50 | 20 | 0 | 0 | ||
| Amikacin | |||||
| M. avium | 1 | 0 | 0 | 80 | 80 |
| 4 | 5 | 27 | 48 | ||
| 20 | 72 | 6 | 2 | ||
| M. chimaera | 1 | 0 | 1 | 47 | 48 |
| 4 | 18 | 14 | 16 | ||
| 20 | 48 | 0 | 0 | ||
| M. intracellulare | 1 | 1 | 0 | 30 | 31 |
| 4 | 3 | 11 | 17 | ||
| 20 | 28 | 1 | 2 | ||
| Other MAC | 1 | 3 | 0 | 5 | 8 |
| 4 | 4 | 2 | 2 | ||
| 20 | 8 | 0 | 0 | ||
| M. kansasii | 1 | 3 | 0 | 17 | 20 |
| 4 | 20 | 0 | 0 | ||
| 20 | 20 | 0 | 0 | ||
| Moxifloxacin | |||||
| M. avium | 0.5 | 29 | 26 | 25 | 80 |
| 2.5 | 75 | 3 | 2 | ||
| 10 | 79 | 1 | 0 | ||
| M. chimaera | 0.5 | 11 | 24 | 13 | 48 |
| 2.5 | 48 | 0 | 0 | ||
| 10 | 48 | 0 | 0 | ||
| M. intracellulare | 0.5 | 3 | 8 | 20 | 31 |
| 2.5 | 30 | 1 | 0 | ||
| 10 | 31 | 0 | 0 | ||
| Other MAC | 0.5 | 3 | 1 | 4 | 8 |
| 2.5 | 7 | 0 | 1 | ||
| 10 | 8 | 0 | 0 | ||
| M. kansasii | 0.5 | 20 | 0 | 0 | 20 |
| 2.5 | 20 | 0 | 0 | ||
| 10 | 20 | 0 | 0 | ||
| Linezolid | |||||
| M. avium | 1 | 1 | 1 | 78 | 80 |
| 4 | 8 | 8 | 64 | ||
| 16 | 27 | 34 | 19 | ||
| M. chimaera | 1 | 0 | 2 | 46 | 48 |
| 4 | 3 | 13 | 32 | ||
| 16 | 40 | 8 | 0 | ||
| M. intracellulare | 1 | 0 | 1 | 30 | 31 |
| 4 | 2 | 5 | 24 | ||
| 16 | 12 | 14 | 5 | ||
| Other MAC | 1 | 0 | 0 | 8 | 8 |
| 4 | 1 | 3 | 4 | ||
| 16 | 5 | 2 | 1 | ||
| M. kansasii | 1 | 15 | 4 | 1 | 20 |
| 4 | 20 | 0 | 0 | ||
| 16 | 20 | 0 | 0 | ||
| Clofazimine | |||||
| M. avium | 0.25 | 1 | 4 | 75 | 80 |
| 0.5 | 9 | 26 | 45 | ||
| 1 | 44 | 28 | 8 | ||
| 2 | 80 | 0 | 0 | ||
| 4 | 80 | 0 | 0 | ||
| M. chimaera | 0.25 | 3 | 0 | 45 | 48 |
| 0.5 | 24 | 5 | 19 | ||
| 1 | 39 | 8 | 1 | ||
| 2 | 48 | 0 | 0 | ||
| 4 | 48 | 0 | 0 | ||
| M. intracellulare | 0.25 | 0 | 2 | 29 | 31 |
| 0.5 | 2 | 12 | 17 | ||
| 1 | 17 | 12 | 2 | ||
| 2 | 30 | 1 | 0 | ||
| 4 | 31 | 0 | 0 | ||
| Other MAC | 0.25 | 0 | 0 | 8 | 8 |
| 0.5 | 1 | 1 | 6 | ||
| 1 | 4 | 2 | 2 | ||
| 2 | 8 | 0 | 0 | ||
| 4 | 8 | 0 | 0 | ||
| M. kansasii | 0.25 | 9 | 3 | 8 | 20 |
| 0.5 | 18 | 1 | 1 | ||
| 1 | 20 | 0 | 0 | ||
| 2 | 20 | 0 | 0 | ||
| 4 | 20 | 0 | 0 | ||
MAC, M. avium complex.
The categories susceptible (S), intermediate (I), and resistant (R) are used in this study to describe presence or absence of in vitro growth at a defined drug concentration and neither represent clinical breakpoints nor predicted clinical outcome. Intermediate growth inhibition represents significant (>99%) but not complete inhibition and was categorized susceptible (S) for calculating MIC values and depicting distributions at the population level.
Susceptibility distributions of additional drugs used for the treatment of M. chimaera infections.
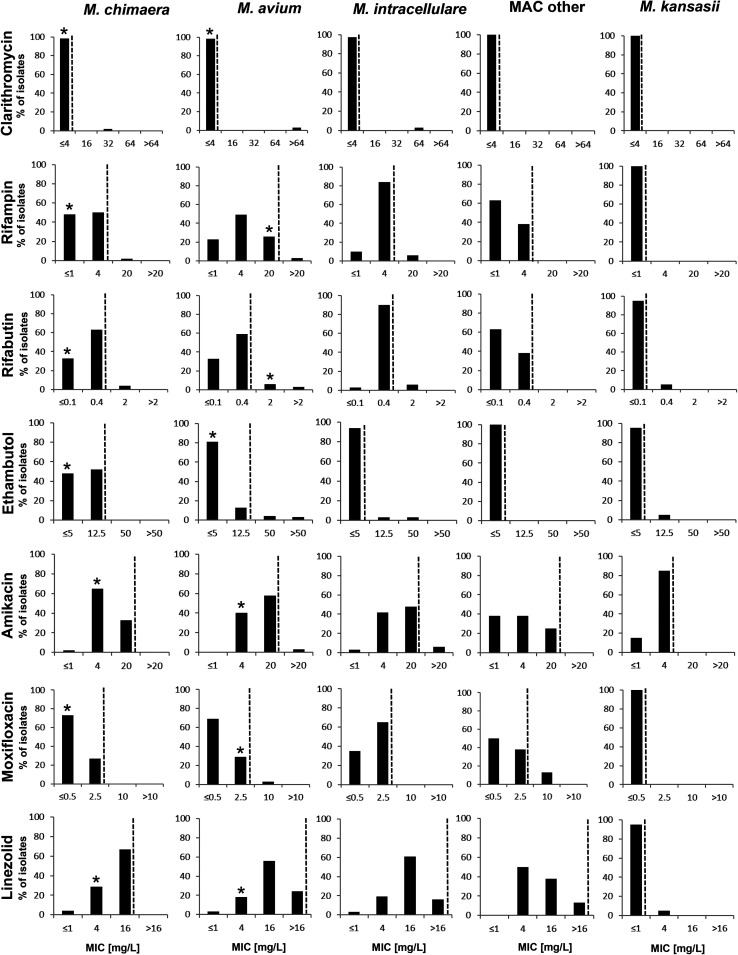
Susceptibility patterns of additional drugs used for the treatment of M. chimaera infections, such as clarithromycin, ethambutol, rifampin, rifabutin, amikacin, moxifloxacin, and linezolid, are shown in Fig. 2 and Table 2 for M. chimaera, M. avium complex species, and M. kansasii. Susceptibility to these drugs was in general comparable for M. chimaera and other members of the M. avium complex. Lower MIC values were observed for M. kansasii toward amikacin, linezolid, moxifloxacin, rifampin, and rifabutin compared to M. chimaera and the M. avium complex.
FIG 2.
Susceptibility distributions for different drugs and NTM species based on quantitative drug susceptibility testing data using MGIT TB eXiST. Approximated MIC90 values are indicated (dashed line). MIC values of the type strains M. avium ATCC 19421 and M. chimaera DSM 44623 are indicated (*).
Macrolide and amikacin resistance.
For two M. avium isolates and one isolate each of M. chimaera and M. intracellulare, MIC values of ≥32 mg/liter were observed for clarithromycin, which indicates macrolide resistance according to CLSI guidelines (14). Sequence analysis of the 23S rRNA gene of both M. avium isolates and M. intracellulare revealed mutations at nucleotide position A2059G (E. coli numbering), thereby providing a genotypic confirmation of the high-level macrolide-resistance phenotype. However, for the M. chimaera isolate, no mutation could be detected at nucleotide positions A2058/A2059. Repeated clarithromycin testing confirmed the decreased in vitro macrolide susceptibility of this isolate that was observed after prolonged macrolide treatment. Two M. avium and two M. intracellulare isolates exhibited MIC values of ≥20 mg/liter for amikacin. One M. intracellulare isolate exhibited an A1408G mutation in the 16S rRNA gene (E. coli numbering), which is known to confer high-level aminoglycoside resistance (15, 16). In contrast, the second M. intracellulare isolate and the two M. avium isolates carried a wild-type 16S rRNA allele. Therefore, the molecular mechanisms underlying decreased susceptibility in these strains remain elusive.
DISCUSSION
Treatment of M. chimaera and M. avium complex infections is complicated and requires multidrug regimens. Treatment options are limited, especially for macrolide-resistant isolates (7). Clofazimine, a drug traditionally used in leprosy therapy and recently recommended by the World Health Organization (WHO) for the treatment of multidrug-resistant tuberculosis (MDR-TB), is also increasingly used to treat severe M. avium complex infections (17, 18). Elevated MICs for clofazimine have been reported for M. avium and M. intracellulare and suggest the occurrence of resistant isolates (19). Whereas for Mycobacterium tuberculosis complex the WHO has released guidelines on clofazimine susceptibility testing and defined clinical breakpoints, i.e., critical concentrations, to separate resistant from susceptible isolates, such guidelines are lacking for NTM (20). Determination of MIC distributions and ECOFFs is a prerequisite for the assignment of clinical breakpoints.
Clofazimine MIC distribution data have to our knowledge not yet been reported for M. chimaera. Pang et al. reported a MIC of 0.5 mg/liter for clofazimine for the type strain M. chimaera DSM 44623 (21). We determined the MIC50, MIC90, and ECOFF values to be 0.5 mg/liter, 1 mg/liter, and 2 mg/liter, respectively, based on the MIC distribution of 48 clinical isolates of M. chimaera using the MGIT 960/EpiCenter TB eXiST platform and showed that these values are comparable to clofazimine MIC50, MIC90, and ECOFF values of other members of the M. avium complex, including M. avium sensu stricto and M. intracellulare. Our data are in agreement with different reports of clofazimine susceptibility data for M. avium complex (19, 22–24). A MIC50 of 1 mg/liter for M. avium complex was found by van Ingen et al. (24), and MIC90 values of 4 mg/liter and 1 mg/liter were described by Huang et al. for M. avium and M. intracellulare, respectively (23). Luo et al. determined a clofazimine ECOFF of 2 mg/liter for M. avium and M. intracellulare (19). The clofazimine MIC distribution of M. kansasii was shifted toward lower MICs compared to members of the M. avium complex in our study. This is in line with findings that M. kansasii is in general more susceptible to NTM drugs than the M. avium complex and reports of a clofazimine ECOFF of 0.5 mg/liter for M. kansasii by Luo et al. (19).
Clofazimine resistance in NTM has been associated with mutations in the TetR family of regulators of adjacent MmpS5-MmpL5 efflux pumps: mmpT5 in M. intracellulare (25) and MAB_2299 in Mycobacterium abscessus (26). The NTM isolates characterized in this study were therapy naive regarding clofazimine, and no elevated MICs were observed. Exploratory investigations into 10 randomly selected M. chimaera isolates did not reveal genetic diversity within the putative homologs RS13290 (mmpt5; 100% amino acid [aa] sequence identity) and RS24730 (MAB_2299; 70% aa sequence identity) of M. chimaera strain DSM 44623T (CP015278.1) (data not shown). In M. tuberculosis, mutations in the Rv0678 (mmpR5) locus are associated with clofazimine and bedaquiline resistance (27, 28). The closest homologs of Rv0678 were RS18640 (35% aa identity), RS15530 (35% aa identity), and RS06670 (24% aa identity) of M. chimaera DSM 44623T (data not shown). These findings confirm reports of others that there is no ortholog of Rv0678 (MmpR5) in the M. avium complex (25). Furthermore, unlike RS13290 and RS24730, the latter three genes are not located in the proximity of mmpL genes.
The MGIT 960/EpiCenter TB eXiST platform (Becton Dickinson) is recommended by the WHO for drug susceptibility testing of M. tuberculosis, including the testing of clofazimine, and therefore available in many mycobacteria laboratories worldwide (20). We have previously adapted MGIT 960 testing for automated quantitative drug susceptibility testing of slow-growing NTM and expanded this method for the testing of clofazimine within this study (12, 13). Commercial microdilution systems that lack clofazimine testing, e.g., the SLOWMYCO Sensititre panel from Trek Diagnostic Systems, are broadly used for drug susceptibility testing of slow-growing NTM (8–11). MGIT 960 testing of clofazimine, a method established in many laboratories worldwide for M. tuberculosis complex, could complement commercial microdilution testing for slow-growing NTM in these laboratories. Our data support the addition of clofazimine to future commercial microdilution panels for NTM.
MIC90 values of M. chimaera for drugs other than clofazimine, such as amikacin, clarithromycin, ethambutol, moxifloxacin, linezolid, and rifampin, are in agreement with the findings of previous studies for M. chimaera and comparable to values reported for the M. avium complex (1, 8–11, 22).
In conclusion, we provide MIC distribution, MIC90, and ECOFF values of clofazimine for M. chimaera and demonstrate comparable values for other members of the M. avium complex. Further studies are needed to correlate in vitro susceptibility with clinical outcome.
MATERIALS AND METHODS
Mycobacterial strains and culture conditions.
Drug susceptibility was measured for 48 nonduplicate clinical isolates of M. chimaera and 139 additional slow-growing NTM from respiratory and nonrespiratory origin, including the M. avium complex isolates M. avium (n = 80), M. intracellulare (n = 31), M. yongonense (n = 3), M. timonense (n = 2), M. bouchedurhonense (n = 1), M. colombiense (n = 1), and M. vulneris (n = 1), together with M. kansasii (n = 20) isolates, that were submitted to or isolated at our mycobacteriological laboratory from 2014 to 2018 (Table 1). In addition, the type strains M. avium ATCC 19421 and M. chimaera DSM 44623 were analyzed. The isolates were identified by partial 16S rRNA gene sequence analysis as described previously (29). M. kansasii was differentiated by sequence analysis of the hsp65 gene (30). Mycobacteria were grown in mycobacterium growth indicator tube (MGIT) medium supplemented with oleic acid albumin dextrose catalase (OADC) (Becton Dickinson, Sparks, MD) at 37°C.
Drug susceptibility testing.
Drug susceptibility distributions of NTM were determined by automated, quantitative DST using the MGIT 960 system and the Epicenter TB eXIST system (Becton Dickinson) as previously described (12, 13). The antibiotics amikacin (1, 4, and 20 mg/liter), clarithromycin (4, 16, 32, and 64 mg/liter), clofazimine (0.25, 0.5, 1, 2, and 4 mg/liter), ethambutol (5, 12.5, and 50 mg/liter), linezolid (1, 4, and 16 mg/liter), moxiflocaxin (0.5, 2.5, and 10 mg/liter), rifabutin (0.1, 0.4, and 2 mg/liter), and rifampin (1, 4, and 20 mg/liter) were analyzed. Clofazimine was purchased from Sigma-Aldrich (Buchs, Switzerland) and dissolved in 100% dimethyl sulfoxide (DMSO). The terms susceptible (S), intermediate (I), and resistant (R) are used in this study to describe presence or absence of in vitro growth at a defined drug concentration and neither represent clinical breakpoints nor predicted clinical outcome. Intermediate growth inhibition represents significant (>99%) but not complete inhibition and was categorized susceptible (S) for calculating MIC values and depicting distributions at the population level.
Clarithromycin and amikacin resistance analysis.
Phenotypic clarithromycin and amikacin resistance was confirmed by sequence analysis of the 23S rRNA gene and 16S rRNA gene, respectively, as described elsewhere (31, 32). Mutations at nucleotide positions A2058 and A2059 (E. coli equivalent) of the 23S rRNA gene were considered resistance markers for macrolides, and mutations at nucleotide position A1408 and C1409 (E. coli equivalent) of the 16S rRNA gene were considered amikacin resistance markers.
Determination of ECOFF, MIC50, and MIC90 values.
MIC distributions were generated from the quantitative DST results. ECOFF values were determined by visual inspection of the MIC distributions (33). MIC50 and MIC90 were defined as drug concentrations that inhibit growth of 50% and 90%, respectively, of the population of a given species.
ACKNOWLEDGMENTS
We thank the laboratory technicians of the Institute of Medical Microbiology for assistance in performing the drug susceptibility testing, and we thank E. C. Böttger and R. Hömke for critical reading of the manuscript.
This study was supported by the University of Zurich.
REFERENCES
- 1.Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int J Syst Evol Microbiol 54:1277–1285. doi: 10.1099/ijs.0.02777-0. [DOI] [PubMed] [Google Scholar]
- 2.Lecorche E, Haenn S, Mougari F, Kumanski S, Veziris N, Benmansour H, Raskine L, Moulin L, Cambau E, MyRMA CNR, CNR-MyRMA . 2018. Comparison of methods available for identification of Mycobacterium chimaera. Clin Microbiol Infect 24:409–413. doi: 10.1016/j.cmi.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 3.Kohler P, Kuster SP, Bloemberg G, Schulthess B, Frank M, Tanner FC, Rossle M, Boni C, Falk V, Wilhelm MJ, Sommerstein R, Achermann Y, Ten Oever J, Debast SB, Wolfhagen MJ, Brandon Bravo Bruinsma GJ, Vos MC, Bogers A, Serr A, Beyersdorf F, Sax H, Böttger EC, Weber R, van Ingen J, Wagner D, Hasse B. 2015. Healthcare-associated prosthetic heart valve, aortic vascular graft, and disseminated Mycobacterium chimaera infections subsequent to open heart surgery. Eur Heart J 36:2745–2753. doi: 10.1093/eurheartj/ehv342. [DOI] [PubMed] [Google Scholar]
- 4.van Ingen J, Kohl TA, Kranzer K, Hasse B, Keller PM, Katarzyna Szafrańska A, Hillemann D, Chand M, Schreiber PW, Sommerstein R, Berger C, Genoni M, Rüegg C, Troillet N, Widmer AF, Becker SL, Herrmann M, Eckmanns T, Haller S, Höller C, Debast SB, Wolfhagen MJ, Hopman J, Kluytmans J, Langelaar M, Notermans DW, Ten Oever J, van den Barselaar P, Vonk ABA, Vos MC, Ahmed N, Brown T, Crook D, Lamagni T, Phin N, Smith EG, Zambon M, Serr A, Götting T, Ebner W, Thürmer A, Utpatel C, Spröer C, Bunk B, Nübel U, Bloemberg GV, Böttger EC, Niemann S, Wagner D, Sax H. 2017. Global outbreak of severe Mycobacterium chimaera disease after cardiac surgery: a molecular epidemiological study. Lancet Infect Dis 17:1033–1041. doi: 10.1016/S1473-3099(17)30324-9. [DOI] [PubMed] [Google Scholar]
- 5.Sax H, Bloemberg G, Hasse B, Sommerstein R, Kohler P, Achermann Y, Rossle M, Falk V, Kuster SP, Böttger EC, Weber R. 2015. Prolonged outbreak of Mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis 61:67–75. doi: 10.1093/cid/civ198. [DOI] [PubMed] [Google Scholar]
- 6.Sommerstein R, Rüegg C, Kohler P, Bloemberg G, Kuster SP, Sax H. 2016. Transmission of Mycobacterium chimaera from heater-cooler units during cardiac surgery despite an ultraclean air ventilation system. Emerg Infect Dis 22:1008–1013. doi: 10.3201/eid2206.160045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasse B, Hannan MM, Keller PM, Maurer FP, Sommerstein R, Mertz D, Wagner D, Fernández-Hidalgo N, Nomura J, Manfrin V, Bettex D, Hernandez Conte A, Durante-Mangoni E, Tang TH-C, Stuart RL, Lundgren J, Gordon S, Jarashow MC, Schreiber PW, Niemann S, Kohl TA, Daley CL, Stewardson AJ, Whitener CJ, Perkins K, Plachouras D, Lamagni T, Chand M, Freiberger T, Zweifel S, Sander P, Schulthess B, Scriven JE, Sax H, van Ingen J, Mestres CA, Diekema D, Brown-Elliott BA, Wallace RJ, Baddour LM, Miro JM, Hoen B, Athan E, Bayer A, Barsic B, Corey GR, Chu VH, Durack DT, Fortes CQ, Fowler B, Hoen V, Krachmer AW, Durante-Magnoni E, Miro JM, Wilson WR, Infectious Diseases Specialists, Hospital Epidemiologists, Microbiologists and Molecular Typing Specialists, Cardiac Surgeons/Perfusionists/Cardiologists, Ophthalmology, Anaesthesiologists, Public Health . 2020. International Society of Cardiovascular Infectious Diseases guidelines for the diagnosis, treatment and prevention of disseminated Mycobacterium chimaera infection following cardiac surgery with cardiopulmonary bypass. J Hosp Infect 104:214–235. doi: 10.1016/j.jhin.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Maurer FP, Pohle P, Kernbach M, Sievert D, Hillemann D, Rupp J, Hombach M, Kranzer K. 2019. Differential drug susceptibility patterns of Mycobacterium chimaera and other members of the Mycobacterium avium-intracellulare complex. Clin Microbiol Infect 25:371–379. doi: 10.1016/j.cmi.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Chen LC, Huang HN, Yu CJ, Chien JY, Hsueh PR. 2020. Clinical features and treatment outcomes of Mycobacterium chimaera lung disease and antimicrobial susceptibility of the mycobacterial isolates. J Infect 80:437–443. doi: 10.1016/j.jinf.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Truden S, Zolnir-Dovc M, Sodja E, Starcic Erjavec M. 2020. Nationwide analysis of Mycobacterium chimaera and Mycobacterium intracellulare isolates: frequency, clinical importance, and molecular and phenotypic resistance profiles. Infect Genet Evol 82:104311. doi: 10.1016/j.meegid.2020.104311. [DOI] [PubMed] [Google Scholar]
- 11.Mok S, Hannan MM, Nölke L, Stapleton P, O’Sullivan N, Murphy P, McLaughlin AM, McNamara E, Fitzgibbon MM, Rogers TR. 2019. Antimicrobial susceptibility of clinical and environmental Mycobacterium chimaera isolates. Antimicrob Agents Chemother 63:e00755-19. doi: 10.1128/AAC.00755-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hombach M, Somoskövi A, Homke R, Ritter C, Böttger EC. 2013. Drug susceptibility distributions in slowly growing non-tuberculous mycobacteria using MGIT 960 TB eXiST. Int J Med Microbiol 303:270–276. doi: 10.1016/j.ijmm.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Lucke K, Hombach M, Friedel U, Ritter C, Böttger EC. 2012. Automated quantitative drug susceptibility testing of non-tuberculous mycobacteria using MGIT 960/EpiCenter TB eXiST. J Antimicrob Chemother 67:154–158. doi: 10.1093/jac/dkr399. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2018. Performance standards for susceptibility testing of Mycobacteria, Nocardia spp., and other aerobic Actinomyces, 1st ed CLSI supplement M62. Clinical and Laboratory Standards Institute, Wayne. PA. [Google Scholar]
- 15.Hobbie SN, Pfister P, Brüll C, Westhof E, Böttger EC. 2005. Analysis of the contribution of individual substituents in 4,6-aminoglycoside-ribosome interaction. Antimicrob Agents Chemother 49:5112–5118. doi: 10.1128/AAC.49.12.5112-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, Zhang Y, Böttger EC, Wallace RJ Jr.. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis 177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 17.Haworth CS, Banks J, Capstick T, Fisher AJ, Gorsuch T, Laurenson IF, Leitch A, Loebinger MR, Milburn HJ, Nightingale M, Ormerod P, Shingadia D, Smith D, Whitehead N, Wilson R, Floto RA. 2017. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax 72:ii1–ii64. doi: 10.1136/thoraxjnl-2017-210927. [DOI] [PubMed] [Google Scholar]
- 18.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. 2020. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis 71:905–913. doi: 10.1093/cid/ciaa1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J, Yu X, Jiang G, Fu Y, Huo F, Ma Y, Wang F, Shang Y, Liang Q, Xue Y, Huang H. 2018. In vitro activity of clofazimine against nontuberculous Mycobacteria isolated in Beijing, China. Antimicrob Agents Chemother 62:e00072-18. doi: 10.1128/AAC.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. 2018. Technical manual for drug susceptibility testing of medicines used in the treatment of tuberculosis. World Health Organization, Geneva, Switzerland. Licence: CC BY-NC-SA 3.0. [Google Scholar]
- 21.Pang H, Jiang Y, Wan K. 2017. Drug susceptibility of 33 reference strains of slowly growing Mycobacteria to 19 antimicrobial agents. Biomed Res Int 2017:1584658. doi: 10.1155/2017/1584658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowman S, Burns K, Benson S, Wilson R, Loebinger MR. 2016. The antimicrobial susceptibility of non-tuberculous mycobacteria. J Infect 72:324–331. doi: 10.1016/j.jinf.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Huang CC, Wu MF, Chen HC, Huang WC. 2018. In vitro activity of aminoglycosides, clofazimine, d-cycloserine and dapsone against 83 Mycobacterium avium complex clinical isolates. J Microbiol Immunol Infect 51:636–643. doi: 10.1016/j.jmii.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 24.van Ingen J, van der Laan T, Dekhuijzen R, Boeree M, van Soolingen D. 2010. In vitro drug susceptibility of 2275 clinical non-tuberculous Mycobacterium isolates of 49 species in The Netherlands. Int J Antimicrob Agents 35:169–173. doi: 10.1016/j.ijantimicag.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Alexander DC, Vasireddy R, Vasireddy S, Philley JV, Brown-Elliott BA, Perry BJ, Griffith DE, Benwill JL, Cameron AD, Wallace RJ Jr.. 2017. Emergence of mmpT5 variants during bedaquiline treatment of Mycobacterium intracellulare lung disease. J Clin Microbiol 55:574–584. doi: 10.1128/JCM.02087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez AV, Richard M, Roquet-Baneres F, Viljoen A, Kremer L. 2019. The TetR family transcription factor MAB_2299c regulates the expression of two distinct MmpS-MmpL efflux pumps involved in cross-resistance to clofazimine and bedaquiline in Mycobacterium abscessus. Antimicrob Agents Chemother 63:e01000-19. doi: 10.1128/AAC.01000-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, de Jong BC, Koul A. 2014. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogall T, Flohr T, Böttger EC. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol 136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 30.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 31:175–178. doi: 10.1128/JCM.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier A, Kirschner P, Springer B, Steingrube VA, Brown BA, Wallace RJ, Jr., Böttger EC. 1994. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother 38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfister P, Hobbie S, Brüll C, Corti N, Vasella A, Westhof E, Böttger EC. 2005. Mutagenesis of 16S rRNA C1409-G1491 base-pair differentiates between 6'OH and 6'NH3+ aminoglycosides. J Mol Biol 346:467–475. doi: 10.1016/j.jmb.2004.11.073. [DOI] [PubMed] [Google Scholar]
- 33.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]