The relatedness of the equine-associated Escherichia coli strain ST1250 and its single- and double-locus variants (ST1250-SLV/DLV), obtained from horses in Europe, was studied by comparative genome analysis. A total of 54 isolates of E. coli ST1250 and ST1250-SLV/DLV from healthy and hospitalized horses across Europe (Czech Republic [n = 23], The Netherlands [n = 18], Germany [n = 9], Denmark [n = 3], and France [n = 1]) from 2008 to 2017 were subjected to whole-genome sequencing.
KEYWORDS: Escherichia coli, IncHI1, ST1250, horses, multidrug resistance, plasmids
ABSTRACT
The relatedness of the equine-associated Escherichia coli strain ST1250 and its single- and double-locus variants (ST1250-SLV/DLV), obtained from horses in Europe, was studied by comparative genome analysis. A total of 54 isolates of E. coli ST1250 and ST1250-SLV/DLV from healthy and hospitalized horses across Europe (Czech Republic [n = 23], The Netherlands [n = 18], Germany [n = 9], Denmark [n = 3], and France [n = 1]) from 2008 to 2017 were subjected to whole-genome sequencing. An additional 25 draft genome assemblies of E. coli ST1250 and ST1250-SLV/DLV were obtained from the public databases. The isolates were compared for genomic features, virulence genes, clade structure, and plasmid content. The complete nucleotide sequences of eight IncHI1/ST9 plasmids and one IncHI1/ST2 plasmid were obtained using long-read sequencing by PacBio or MinION. In the collection of 79 isolates, only 10 were phylogenetically close (<8 single nucleotide polymorphisms [SNP]). The majority of isolates belonged to phylogroup B1 (73/79 [92.4%]) and carried blaCTX-M-1 (58/79 [73.4%]). The plasmid content of the isolates was dominated by IncHI1 of ST9 (56/62 [90.3%]) and ST2 (6/62 [9.7%]), while 84.5% (49/58) of the blaCTX-M-1 genes were associated with the presence of the IncHI1 replicon of ST9 and 6.9% (4/58) with the IncHI1 replicon of ST2 within the corresponding isolates. The operon for the utilization of short-chain fructooligosaccharides (the fos operon) was present in 55 of 79 (69.6%) isolates, and all of these carried IncHI1/ST9 plasmids. The eight complete IncHI1/ST9 plasmid sequences showed the presence of blaCTX-M-1 and the fos operon within the same molecule. Sequences of IncHI1/ST9 plasmids were highly conserved (>98% similarity) regardless of country of origin and differed only in the structure and integration site of the multidrug resistance (MDR) region. E. coli ST1250 and ST1250-SLV/DLV are phylogenetically diverse strains associated with horses. A strong linkage of E. coli ST1250 with the epidemic multidrug resistance plasmid lineage IncHI1/ST9 carrying blaCTX-M-1 and the fos operon was identified.
INTRODUCTION
Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, such as ESBL-producing Escherichia coli (ESBL-PEc), are widespread in humans, food-producing and companion animals, wildlife, and the environment (1). ESBLs can be encoded by a variety of genes, among which the blaCTX-M gene family dominates. The spread of blaCTX-M in ESBL-PEc is facilitated by mobile genetic elements, including multidrug resistance (MDR) plasmids of various incompatibility (Inc) groups (2, 3).
Antimicrobial resistance is of concern for a wide range of equine pathogens. ESBL-producing Enterobacteriaceae have been increasingly isolated from wounds, as well as from individuals with respiratory and urinary tract infections (4). Several studies have reported the occurrence of CTX-M-1-producing E. coli in hospitalized but also nonhospitalized horses in many European countries, such as the Czech Republic (5, 6), Denmark (7, 8), The Netherlands (9, 10), the United Kingdom (11), Germany (12, 13), Sweden (14), Switzerland, and France (14), including Guadeloupe (15).
An epidemic IncHI1/ST9 plasmid lineage conferring multidrug resistance and harboring blaCTX-M-1 has been identified in E. coli isolates from hospitalized horses in the Czech Republic (5) and later in isolates from healthy and diseased horses in France (16), Sweden (14), and The Netherlands (10). Moreover, the fos operon, consisting of seven genes (fosK, fosY, fosGH2, fosX, fosGH1, fosT, fosR) involved in the metabolism of short-chain fructooligosaccharides (scFOS), has been detected on pEQ1, a conjugative IncHI1/ST9 plasmid obtained from E. coli T23, originating from the feces of a hospitalized horse in the Czech Republic (6). The importance of the presence of the fos operon arises from the fact that until Schouler et al. (17) detected the fos operon in extraintestinal pathogenic E. coli, the fos operon occurred only in probiotic, not pathogenic, bacteria and only as part of a chromosomally encoded genomic island. However, none of the studies mentioned above, except for that of Dolejska et al. (6), characterize the complete sequence of an epidemic IncHI1/ST9 plasmid. The presence of the fos operon might play a role in the coselection of multiresistance plasmids, since it provides bacteria with a metabolic advantage and enables them to grow in environments where pathogenic bacteria would normally not survive. The association of the blaCTX-M-1 gene and the fos operon on plasmids is particularly worrying because of the specific role of fructooligosaccharides in the horse diet and the ability of plasmids to be transferred horizontally. scFOS are commonly used prebiotics, and they are often part of a supplementary horse diet which should support gut health and beneficial microflora in horses (18). However, our results indicate that using scFOS in a horse diet may contribute to the spread of antibiotic resistance genes, so it is doubtful whether scFOS are indeed beneficial. Moreover, the use of scFOS as prebiotics is not limited to horses; they are part of many human probiotics and nutritional supplements for newborns. Therefore, more attention should be paid to the relation between the ability to utilize scFOS and antibiotic resistance.
In the current study and publicly available sequence data, E. coli ST1250 is found to be a rare sequence type (ST) in hosts other than horses. Moreover, the close association of E. coli ST1250 with IncHI1/ST9 has been observed (10, 14, 16). Clinical and nonclinical isolates of E. coli ST1250 and its single- and double-locus variants (ST1250-SLV/DLV) collected from horses in the Czech Republic, The Netherlands, Denmark, France, and Germany were subjected to short-read DNA sequencing. E. coli ST1250 was selected due to its frequent occurrence in horse samples. Other STs included are single- or double-locus variants of ST1250 and were selected both for their close relationship (with regard to the multilocus sequence type [MLST]) to ST1250 and for their frequent occurrence in horse samples. For the selected isolates, long-read sequencing was applied to obtain complete nucleotide sequences of chromosomes and IncHI1 plasmids. Additional sequences of E. coli ST1250 and ST1250-SLV/DLV were recovered from EnteroBase before genomic features, clade structure, and plasmid content were examined by detailed phylogenetics. Our hypothesis is that E. coli ST1250 is adapted to horses and has low pathogenic potential itself, while this study shows frequent association of E. coli ST1250 with HI1/ST9 multiresistance plasmids encoding blaCTX-M-1 and the fos operon.
(This work was partially presented at the One Health European Joint Program Annual Meeting, Prague, Czech Republic, 27 to 29 May 2020 [online], in the poster section [P 153].)
RESULTS AND DISCUSSION
Genomic characterization of the isolates.
A total of 79 draft genomes of E. coli ST1250 or ST1250-SLV/DLV, consisting of 54 sequences obtained in this study and 25 retrieved from EnteroBase or GenBank, were analyzed. The majority of isolates belonged to phylogroup B1 (73/79 [92.4%]), followed by phylogroups A (5/79 [6.3%]) and E (1/79 [1.2%]). Since the collection studied was aimed at E. coli ST1250 and ST1250-SLV/DLV, the majority of the isolates in the collection indeed belonged to ST1250 (60/79 [75.9%]), followed by ST1250-SLV/DLV: DLV-ST4164 (4/79 [5.0%]), SLV-ST826 (3/79 [3.3%]), SLV-ST4527 (2/79 [2.5%]), SLV-ST1686 (2/79 [2.5%]), SLV-ST11123 (2/79 [2.5%]), and SLV-ST2336, SLV-ST4552, SLV-ST7434, DLV-ST11033, SLV-ST11121, and SLV-ST11122 (1/79 [1.2%] each). The common detection of E. coli ST1250, along with ST1250-SLV/DLV, in horses as opposed to other sources suggests that this ST is horse associated. This assumption is supported by our unpublished study of hospitalized and healthy horses across the Czech Republic, where 31.7% (20/63) of E. coli isolates belonged to ST1250 (6/63 [9.5%]) or ST1250-SLV/DLV (14/63 [22.2%]). Similarly, in a nationwide study conducted on nonhospitalized horses from The Netherlands (10), the most prevalent group of isolates whose whole genomes had been sequenced belonged to ST1250 (8/48 [16.7%]) or ST1250-SLV/DLV (9/48 [18.8%]). Isolates from both the Czech Republic (unpublished data) and The Netherlands are included in this study. Despite the limited number of publicly available draft genome assemblies of E. coli ST1250, the association of E. coli ST1250 with horses (7/10 [70%]) was observed in EnteroBase as well, where only 1 of 10 ST1250 isolates was of human origin, while 2 were of undefined origin.
Antibiotic susceptibility testing of the isolates that were physically available (n = 54) showed that all exhibited a multidrug (≥3 antibiotics) resistance profile corresponding to a resistance genotype, except for one (d16-1) that was resistant to ciprofloxacin but susceptible to nalidixic acid, yet no qnr gene was detected. Among the 79 sequences analyzed for antibiotic resistance gene (ARG) content, the most prevalent ESBL genes included blaCTX-M-1 (58/79 [73.4%]), blaCTX-M-2 (6/79 [7.6%]), and blaSHV-12 (7/79 [8.9%]). While 4 isolates carried both blaCTX-M-1 and blaSHV-12, 12 isolates did not encode any ESBL. The whole-genome sequence (WGS) data from 79 isolates showed the presence of genes conferring resistance to tetracycline, trimethoprim, sulfonamides, gentamicin, streptomycin, and chloramphenicol (see Fig. S1 in the supplemental material). Neither the resistance profile nor the plasmid content was influenced by the source of isolation (feces or clinical material). A complete profile of ARGs, along with the results of antibiotic susceptibility testing, can be found in Fig. S1.
Analysis of virulence genes showed the presence of the siderophore synthase-encoding operon ent and the enterobactin export-encoding gene fes in all 79 isolates and the enterobactin uptake-encoding operon fep in 75 (94.9%) isolates. These genes are common for the majority of Gram-negative bacteria (19) and may be considered beneficial for the animal hosting the bacteria carrying these genes. This has been observed in Caenorhabditis elegans, where the bacterial enterobactin promoted the growth of C. elegans via mitochondrial iron uptake (20). More than half of the isolates (56/79 [70.9%]) harbored the fli operon, which is involved in the biosynthesis and functioning of the flagellar organelle (21). Twelve of the 79 isolates (15.2%) carried a fim operon, encoding type I fimbriae. The results of in silico screening for virulence genes suggest that the isolates did not carry any virulence factors enabling their classification into E. coli pathotypes. These findings support the observation that E. coli ST1250 and related STs are part of the commensal equine microbiota (10).
Regarding the plasmid content, the most common plasmid replicons among the collection of 79 isolates were three IncHI1-related replicons (62/79 [78.5%]) of ST9 (56/62 [90.3%]) and ST2 (6/62 [9.7%]). The high prevalence of conjugative IncHI1/ST9 and its association with blaCTX-M-1 have been reported recently (16). IncHI1/ST9 appears to be very well adapted to E. coli in horses, especially E. coli ST1250.
Comparison of E. coli ST1250 and E. coli K-12 chromosomes.
The results revealed that unlike E. coli K-12, the majority of E. coli ST1250 and ST1250-SLV/DLV strains did not carry genes encoding products facilitating adhesion and colonization (yadN, ygiL, paa, yde) (22, 23), a type II secretion pathway (gspCO) (24, 25), the stress response (ygfK, yqeK, ydfO) (26–28), short-chain fatty acid and xanthosine metabolism (ato, xapA) (29, 30), or a toxin-antitoxin system (rnlAB) (31). This finding suggests that E. coli ST1250 does not contain many virulence genes and represents a lineage with low pathogenicity that is adapted to the horse gut. Generally, in terms of genetic features that are conserved and specific for E. coli ST1250, no per se pathogenic genes were detected in the genome.
High diversity of ST1250 and ST1250-SLV/DLV.
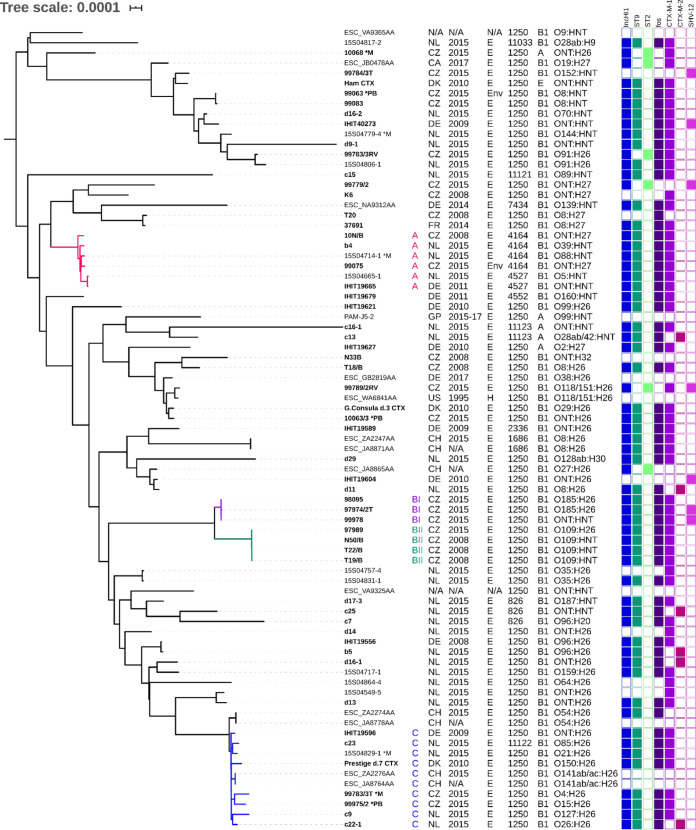
Phylogenetic analysis based on the core genome from whole-genome sequences of 79 isolates showed high diversity of E. coli ST1250 and ST1250-SLV/DLV (Fig. 1; also Fig. S1). The core genome alignment was created from 2,556 core genes and had a length of 3,382,416 bp. The isolates differed in 0 to 10,490 single nucleotide polymorphisms (SNP), with 10 phylogenetically close isolates differing in only 0 to 8 SNP.
FIG 1.
Phylogenetic tree depicting the diversity of E. coli ST1250 and ST1250-SLV/DLV isolates with highlighted clusters A, BI, BII, and C. Isolates marked *M or *PB were sequenced on MinION or PacBio, respectively. Metadata for country of origin, year of isolation, source, ST, phylogroup, and serotype are included. Colored boxes indicate the presence of the IncHI1 plasmid and its ST, the fos operon, and ESBL genes. The designations of physically obtained isolates are set in boldface, and data sets from EnteroBase start with “ESC.” N/A, not available; E, equine; Env, equine clinic environment; H, human; NT, not typeable; NL, The Netherlands; DK, Denmark; CZ, Czech Republic; DE, Germany; CH, Switzerland; US, United States; GP, Guadeloupe; FR, France; CA, Canada.
The isolates in the minimum spanning tree were clustered regardless of their geographical origin and the carriage of IncHI1 plasmids (Fig. S2). Despite the high variability, three noteworthy clusters with one subcluster, marked A, BI, BII, and C, emerged (Fig. 1). Cluster A consisted of six ST1250-SLV/DLV isolates, including ST4164 (four isolates) and ST4527 (two isolates), from The Netherlands, the Czech Republic, and Germany. Their SNP differences ranged from 29 to 321, while the lowest SNP difference (29 SNP) was between a Dutch and a German isolate (15S04665-1 and IHIT_19665). Cluster B was made of subclusters BI and BII, consisting of three and four Czech ST1250 isolates with SNP differences of 1 to 16 and 0 to 8, respectively. Cluster C consisted of nine ST1250 isolates and one novel SLV. The isolates in this cluster differed in 0 to 530 SNP and originated from Switzerland, The Netherlands, the Czech Republic, Denmark, and Germany. However, presumed clonal spread of some isolates occurred only sporadically, usually within the same country. For example, no SNP differences in cluster C were observed between two couples of Swiss isolates (ESC_JA8778AA with ESC_ZA2274AA and ESC_JA8764AA with ESC_ZA2276AA).
Our findings suggest that even though there are clusters (A and C) of closely related international isolates, the isolates in these clusters belonged to several different serotypes (Fig. 1), and no clonal spread (<10 SNP) of E. coli ST1250 was demonstrated. Closely related isolates were observed only within subclusters BI and BII, which consist largely of isolates from the same country, sampling site, and year. Subcluster BI consisted of three isolates obtained within the same horseback riding center in 2015. Subcluster BII consisted of four isolates, of which three were obtained from a horse clinic in 2008 and one (isolate 97989) from a horseback riding center in 2015. Isolate 97989 was closely related (8 SNP) to the three isolates obtained from the horseback riding center. However, it is not clear whether this relatedness is caused by the persistence of the clone within the sampling sites over several years or by repeated introduction of the clone to the horse outside the hospital.
Phylogenetic analysis of our international collection of E. coli ST1250 isolates, including ST1250-SLV/DLV, indicates that E. coli ST1250 is genetically highly variable yet apparently associated with horses, since to our knowledge, this ST was not commonly detected in nonequine studies. However, there is no evidence of transmission of ESBL-PEc ST1250 to humans, except for one record of E. coli ST1250 of human origin in EnteroBase, with no further information. Moreover, E. coli ST1250-SLV/DLV are closely related to E. coli ST1250 clonal lineages, highlighting the importance of whole-genome sequencing and analyses based on the core genome rather than conventional 7-allele MLST profiling.
Comparative genomics of IncHI1 plasmids.
IncHI1 plasmids were identified in the genomes of 62 E. coli strains in this study. Comparison of draft genomes and complete IncHI1 plasmids revealed two lineages of IncHI1: ST2 (6/62 [9.7%]) and ST9 (56/62 [90.3%]). These lineages are distinguished by one SNP in repHIA (allele HCM1_064), plasmid nucleotide identity (between p10068 [IncHI1/ST2] and p99063 [IncHI1/ST9]) of >99.9%, and >96% query coverage. They differ in the structure of the multidrug resistance (MDR) region and by the presence of the fos operon in IncHI1/ST9 plasmids. Once these regions were removed, comparison of the plasmid backbones of p10068 and p99063 showed nucleotide identity and query coverage of 99.98% and 99%, respectively.
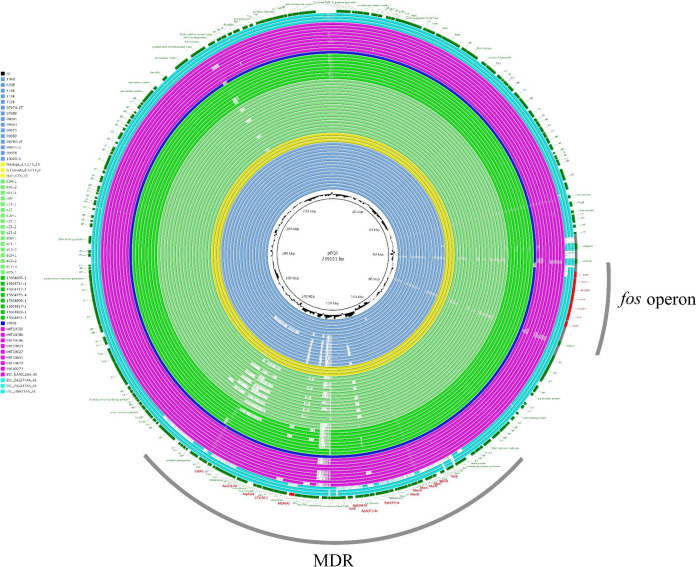
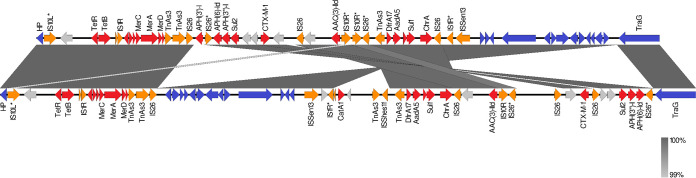
The majority (45/56 [80.4%]) of isolates with IncHI1/ST9 plasmids harbored a class 1 integron with a dfrA17-aadA5 gene cassette. Isolates carrying IncHI1/ST2 plasmids (5/6 [83.3%]) were associated with a class 1 integron with a dfrA12-aadA2 cassette array. The other genetic features of the isolates harboring IncHI1/ST2 and IncHI1/ST9 plasmids are described in the figures presented below and in the supplemental materials. Eight complete nucleotide sequences of IncHI1/ST9 were obtained using PacBio (n = 4) and MinION (n = 4) long-read sequencing. Overall, they exhibited high nucleotide identities (>98%) to pEQ1 and to each other. Figure 2 shows a comparison of complete and draft genome assemblies of IncHI1/ST9 plasmids. Two different variants of the integration of an MDR region were identified within the eight complete plasmid sequences. Five plasmids harbored an MDR region of 41,754 bp (p15S04714-1) to 49,700 bp (p15S0829-4) between genes encoding a hypothetical protein of the backbone (Fig. 3, p15S04714-1). The very same position of the MDR region is found in pEQ1, of equine origin, and pHCM1 (GenBank accession no. AL513383), of human origin, designated a type 2 IncHI1 plasmid based on integration sites of the MDR region inside the plasmid backbone (32). The other three plasmids contained an MDR region divided into two parts (14,708 bp and 34,143 bp) by a part of the inverted plasmid backbone of 14,228 bp (Fig. 3, p99063). This inversion was presumably mediated by two copies of IS26 flanking the backbone part (14,228 bp) and the larger, 34,143-bp part of the MDR region. This finding points to the existence of two variants of highly conserved epidemic IncHI1/ST9 plasmids differing in genetic structure, length, and the integration site of the MDR region. Besides blaCTX-M-1, each of the eight complete IncHI1/ST9 plasmids carried genes encoding resistance to tetracycline [tet(B)], trimethoprim (dfrA17), gentamicin [aac(3)-IId], sulfonamides (sul1, sul2), and streptomycin [aph(6)-Id, aph(3′')-Ib, aadA5] in the MDR region. However, they differed in the presence of the mph(A), aph-(3′)-Ia, catA1, and blaTEM-1 genes. A Mauve comparison of the MDR region variants and their rearrangements is shown in Fig. S3. The mer operon, qacEΔ1 gene, and chrA gene, encoding determinants for resistance to mercury, quaternary ammonium compounds, and chromium, respectively, were present in each complete IncHI1/ST9 plasmid, too (Fig. 2).
FIG 2.
BRIG comparison of IncHI1/ST9 plasmids including draft genome assemblies with pEQ1 as a reference. The fos operon and the blaCTX-M-1 gene are annotated in red. The origins of the samples are color-coded as follows: light blue, Czech Republic; yellow, Denmark; light green and dark green, The Netherlands (different studies); dark blue, France; purple; Germany; turquoise, Switzerland.
FIG 3.
Comparison of two variants of the MDR regions occurring in the HI1/ST9 plasmids that were studied. Diagrams of two representative plasmids are shown: p15S04714-1 (top) and p99063 (bottom). Plasmid backbone gene products are shown in blue, antimicrobial resistance gene products in red, transposases in orange, and hypothetical proteins (HP) in gray, unless they are part of the backbone (blue). An asterisk at an insertion sequence (IS) indicates <100% nucleotide identity with the reference IS.
The MDR region of the complete IncHI1/ST2 plasmid obtained using MinION (p10068) was integrated between genes encoding a hypothetical protein of the backbone, identical to the integration site in IncHI1/ST9 type 2 plasmids. Unlike IncHI1/ST9 plasmids, IncHI1/ST2 plasmids did not harbor the fos operon. Since only one complete IncHI1/ST2 plasmid was obtained, the identity of p10068 could not be properly assessed in comparison with the draft genome assemblies from other IncHI1/ST2 plasmids (n = 5) in this study. However, a BLAST Ring Image Generator (BRIG) comparison of p10068 with the draft genome assemblies of the other five IncHI1/ST2 plasmids (Fig. S4) showed low variability in the backbone and the MDR region of IncHI1/ST2 plasmids from the Czech Republic, Canada, and Switzerland, pointing toward the geographically widespread potential of the IncHI1/ST2 plasmid lineage. Plasmid p10068 harbored the blaCTX-M-1, tet(B), sul1, sul2, aac(3)-IId, aadA2, dfrA12, aph(6)-Id, and aph(3′')-Ib genes within the 40,759-bp MDR region.
Even though IncHI1/ST9 and IncHI1/ST2 plasmids have highly conserved backbones and differ mostly within the MDR region, IncHI1/ST2 plasmids did not harbor the fos operon. A higher prevalence of IncHI1/ST2 over IncHI1/ST9 has been observed only in Swedish isolates so far (14); however, no data on the presence of the fos operon are available from that study. In summary, comparison of IncHI1/ST9 plasmids of various geographical origins showed a highly conserved structure and high nucleotide identity, highlighting the epidemic potential of IncHI1/ST9 plasmids. Similarly, plasmid lineage IncI1-Iγ, harboring Ambler class A and C genes from E. coli and Salmonella enterica of animal and human origins, has been found to be epidemic (33), and IncX3 plasmids encoding NDM-type enzymes within different enterobacterial species (34) or specific NDM-5-encoding IncX3 plasmids from E. coli strains of diverse sequence types have been obtained from five different pig farms from different geographical regions of China (35).
The fos operon is highly diffused among ST1250 strains and is carried by IncHI1/ST9 plasmids.
The collection of 79 E. coli ST1250 and ST1250-SLV/DLV isolates was screened in silico for the presence of the 8,896-bp fos operon, which is involved in the metabolism of scFOS. Fifty-eight (58/79 [73.4%]) isolates harbored the fos operon, while 1 isolate (ESC_ZA2274AA_AS) lacked the fosK gene, suggesting the presence of a nonfunctional operon. The majority (56/58 [96.6%]) of isolates harboring the fos operon also harbored an IncHI1/ST9 plasmid. A BRIG comparison including all IncHI1/ST9 draft genome assemblies from short-read sequencing data (n = 56) showed that the fos operon is highly conserved and is carried by this plasmid lineage (Fig. 2). The association of the fos operon with IncHI1/ST9 plasmids was first observed in equine isolates from the Czech Republic (6) and subsequently in The Netherlands (10) and France (16).
The broader genetic surrounding of the fos operon was examined within eight complete IncHI1/ST9 plasmids obtained using long-read sequencing. In all plasmids, the fos operon was conserved with 100% identity and surrounded by IS903B. As observed previously in pEQ1, the operon was part of a 24-kb module inserted between the repE (FIA replicon) and hns genes of the plasmid backbone (6). Only one of eight plasmids (p15S04779-4) differed within the 24-kb module; it harbored IS1203 upstream of the fos operon.
Although the association of the fos operon and the IncHI1/ST9 plasmid was evident, the fos operons of two E. coli ST1250 isolates (99783-3RV and T20) had different genetic contexts. Analysis of the genetic surroundings of the fos operon of the 99783-3RV isolate showed the presence of an IncFIB(K) replicon on the same contig, suggesting the carriage of the fos operon by plasmids other than IncHI1/ST9 plasmids. The limited information provided by the short-read sequencing did not enable us to identify the genetic context of the fos operon within the T20 strain. However, plasmid replicons IncFIB(K) and IncY were identified in T20, suggesting the possibility that the fos operon is either mediated by plasmid types other than IncHI1/ST9, as in the 99783-3RV isolate, or chromosomally encoded, as observed previously for E. coli BEN2908 (GenBank accession no. AY857617) (36).
It has been suggested that a plasmid-mediated sugar metabolic element, such as the fos operon, could play a key role in strain fitness, contributing to the successful dissemination and maintenance of the fos operon-containing MDR IncHI1/ST9 plasmids in the equine intestinal microbiota (6). We suppose that the absence of the fos operon in IncHI1/ST2 plasmids might contribute to their generally low prevalence, in contrast to IncHI1/ST9 plasmids, among equine E. coli isolates, as observed in our study, since the fos operon seems to provide isolates with a selective advantage in the presence of scFOS. The equine diet is usually rich in scFOS, which may, along with the usage of antibiotics, provide positive selective pressure for bacteria carrying this fos operon-associated plasmid lineage. However, the question of whether factors such as different amounts of scFOS in the horse feed or the geographical origin of the isolate are influencing the selection of the IncHI1/ST9 plasmid lineage needs to be clarified by further studies.
Interestingly, the presence of the fos operon in E. coli BEN2908 was linked to an increased ability to colonize the avian intestine (17). We hypothesize that the widespread presence of the fos operon in E. coli ST1250 and ST1250-SLV/DLV isolates, as observed in our study, may compensate for the absence of known chromosomally encoded colonization factors in these strains. Further elucidation of the role of the fos operon in the colonization of the horse intestine by E. coli ST1250 would require in vivo experiments.
The genomic study of E. coli ST1250 and ST1250-SLV/DLV isolates of equine origin has shown great chromosomal diversity, which could not be linked to specific geographic regions. In comparison, the highly prevalent ESBL-encoding IncHI1/ST9 plasmids in these strains show little diversity beyond the presence of specific MDR regions and the fos operon. To summarize, E. coli ST1250 and E. coli ST1250-SLV/DLV isolates seem to be well adapted to horses as hosts, and E. coli ST1250 in particular often shows an association with IncHI1/ST9 multiresistance plasmids encoding CTX-M-type enzymes and harboring the fos operon, which may contribute to the coselection of these plasmids.
MATERIALS AND METHODS
E. coli isolates.
A total of 54 ST1250 and ST1250-SLV/DLV isolates from healthy and hospitalized horses in several countries across Europe from 2008 to 2017 were collected (Fig. S1 in the supplemental material, isolates in boldface). The isolates originated from the Czech Republic (n = 23) (5; unpublished data), The Netherlands (n = 18) (9), Germany (n = 9), Denmark (n = 3) (7), and France (n = 1) (14). Forty-two and 12 isolates were obtained by selective cultivation on MacConkey agar (MCA) with cefotaxime (1 or 2 mg/liter) and on MCA without antibiotics, respectively. The complete details regarding processing of the samples can be found in the corresponding studies mentioned above. The majority of the isolates originated from feces (42/54), various clinical materials (10/54), and environmental swabs (2/54) (Fig. S1). Susceptibility to 22 antibiotics (ampicillin [AMP], streptomycin [S], sulfonamides [S3], tetracycline [TE], trimethoprim-sulfamethoxazole [SXT], C chloramphenicol [C], cefazolin [KZ], nalidixic acid [NA], ceftazidime [CAZ], gentamicin [CN], amoxicillin-clavulanic acid [AMC], ciprofloxacin [CIP], fosfomycin [FOT], ertapenem [ETP], imipenem [IMP], meropenem [MEM], aztreonam [ATM], nitrofurantoin [F], azithromycin [AZM], colistin [TC], tigecycline [TGC], cephalothin [KF]) was tested by disk diffusion according to the CLSI (37), and the ESBL phenotype was determined by the MASTDISCS test (D68C1 AmpC & ESBL detection set; Mast Diagnostics, UK).
Short-read sequencing.
All isolates were subjected to whole-genome sequencing (WGS) with at least 50× expected coverage. Genomic DNA was extracted using the NucleoSpin Tissue kit (Macherey-Nagel, Germany) and was used for DNA library preparation with a Nextera XT Library Preparation kit. Sequencing was performed on a MiSeq system (Illumina) using 2 × 250-bp paired-end sequencing.
Publicly available data.
We searched EnteroBase (http://enterobase.warwick.ac.uk/species/index/ecoli) for sequence data for E. coli ST1250 and ST1250-SLV/DLV, and assemblies along with corresponding metadata were downloaded. Available data sets for ST1250 isolates irrespective of the plasmid content and ST1250-SLV/DLV variants harboring IncHI1 were selected. Thirteen draft genome assemblies originating in Switzerland (n = 7), Germany (n = 2), Canada (n = 1), the United States (n = 1), and unknown origin (n = 2) were retrieved from EnteroBase. Additional ST1250 and ST1250-SLV/DLV sequences from horses in The Netherlands (n = 11) and Guadeloupe (n = 1) were retrieved from BioProject (record numbers PRJEB34847 [10] and PRJNA388339 [15], respectively). In total, 25 draft genome assemblies of E. coli ST1250 and ST1250-SLV/DLV isolates were obtained from the public databases; among these, 22 originated from horses, 2 were of unknown origin, and 1 isolate was of human origin (Fig. S1).
Long-read sequencing.
PacBio sequencing was used to obtain complete high-quality reference chromosomal and plasmid sequences. Subsequently, sequences of randomly selected IncHI1 plasmids were obtained using MinION sequencing. For PacBio sequencing, genomic DNA was extracted using a NucleoSpin Microbial DNA kit (Macherey-Nagel, Germany). Libraries were prepared according to the microbial multiplexing protocol. Shearing was performed using g-tubes (Covaris, USA), and no size selection was done during library preparation. Sequencing was performed with the Sequel I platform (Pacific Biosciences, Menlo Park, CA, USA). For Oxford Nanopore Technologies (ONT) sequencing, the DNA was extracted using a Gentra Puregene Yeast/Bact. kit and a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Libraries were prepared using a 1D ligation barcoding kit (catalog no. SQK-LSK109 and EXP-NBD104; ONT, Oxford, UK) and were sequenced in R9.4.1 and Flongle flow cells with a MinION Mk1B (ONT).
Sequence data analysis.
Raw data from short-read sequencing were adaptor and quality (Q ≤ 20) trimmed using Trimmomatic (38), v 0.36. De novo assembly was performed using SPAdes (39), v3.13.1, with the “careful” option. For PacBio data, HGAP4 (40) in SMRT Link, v.6, was used to perform the assemblies with a minimum seed coverage of 30. Long reads from MinION were demultiplexed and converted from fast5 to fastq format using Guppy, v3.2.1 (ONT). The adaptors and low-quality ends (Q ≤ 8) were trimmed using Porechop, v0.2.2 (https://github.com/rrwick/Porechop) and BBDuk (https://sourceforge.net/projects/bbmap/), respectively. De novo hybrid assembly using short reads from Illumina and long reads from MinION was performed with Unicycler (41), v0.4.7. The complete chromosomal and plasmid sequences were annotated using RASTtk (42) and were manually curated according to GenBank in Geneious, v9.0.5.
Genomic characterization.
The draft genome assemblies from WGS and EnteroBase were analyzed for the presence of antibiotic resistance genes (ARGs), virulence genes, plasmid replicons, and serotypes. For this purpose, ABRicate (https://github.com/tseemann/abricate) was used with thresholds set to 90% identity and 95% query coverage against the databases ResFinder (43), VFDB (44), PlasmidFinder (45), and EcOH (46), respectively. Plasmid multilocus sequence typing (pMLST) was performed using pMLST by the Center for Genomic Epidemiology (45). MLST profiles were verified by mlst (https://github.com/tseemann/mlst), and the phylogroups were assessed using ClermonTyping (47).
Phylogenetic analysis.
The phylogenetic relatedness of isolates was evaluated. The open reading frames were predicted from draft genome assemblies using Prokka (48), and GFF3 files were used for core genome alignment in Roary (49) with default settings (the percentage of isolates a gene must be in to be considered a core gene is 99%). The alignment was then used in RAxML (50) for calculating the phylogenetic tree under the GTR+GAMMA model supported by 1,000 bootstraps. The tree was visualized in iTOL, v4 (51), and GrapeTree (52). The nucleotide similarity between the isolates was determined by assessing the number of single nucleotide polymorphisms using snp-dists (https://github.com/tseemann/snp-dists) with the core genome alignment as an input.
Comparative genomics.
Comparison of four high-quality annotated complete chromosomal E. coli ST1250 sequences obtained from PacBio sequencing with E. coli K-12 as a reference (GenBank accession no. U00096) was performed using progressiveMauve (53). E. coli K-12 was selected especially because of the lack of virulence genes and the high level of characterization its genetic content. Therefore, the comparison allowed us to see if there are virulence factors or any other genes that could offer an advantage for E. coli ST1250, since E. coli K-12 is a nonpathogenic strain. The complete annotated plasmid sequences were compared with respect to their plasmid replicon and plasmid ST using BLAST Ring Image Generator (54). The structure of the fos operon (nucleotides 61904 to 70800 of pEQ1 [GenBank accession no. KF362121]) was examined and compared to the sequence within pEQ1.
Data availability.
The draft genome assemblies, the complete plasmid and chromosomal sequences, and the raw data from short-read sequencing have been deposited in GenBank and SRA, respectively, under BioProject number PRJNA638007. The complete nucleotide sequences of the plasmids have been deposited in GenBank under accession numbers MT586601 to MT586609.
Supplementary Material
ACKNOWLEDGMENTS
We thank Katerina Rehakova, Iva Kutilova and Martina Masarikova for their excellent work in the laboratory. We thank Alois Cizek, the staff from the Equine Clinic of the University of Veterinary and Pharmaceutical Sciences Brno, and the terrain veterinarians for collecting and providing the samples or bacterial isolates. We are also grateful to Stefan Börjesson for his cooperation.
The study was funded mainly by the Czech Science Foundation (award 18-23532S to M.D.). P.S. received funds from the Internal Grant Agency of the University of Veterinary and Pharmaceutical Sciences Brno (award 204/2020/FVHE). J.H., I.B., and PacBio sequencing were funded by the Charles University Research Fund PROGRES (project Q39), by National Sustainability Program I (NPU I) grant LO1503, and by project CZ.02.1.01/0.0/0.0/16_019/0000787, “Fighting Infectious Diseases,” provided by the Ministry of Education, Youth, and Sports of the Czech Republic. Isolates from the Czech Republic were collected and partially characterized within the project funded by the Internal Grant Agency of the University of Veterinary and Pharmaceutical Sciences Brno (grant 116/2015/FVL). MinION sequencing was funded by Horizon 2020 (award 773830), OH-EJP-H2020-JRP-AMR-2-ARDIG, and internal funding from Utrecht University.
We have no potential conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. 2012. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 18:646–655. 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 2.El Salabi A, Walsh TR, Chouchani C. 2013. Extended spectrum β-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. Crit Rev Microbiol 39:113–122. 10.3109/1040841X.2012.691870. [DOI] [PubMed] [Google Scholar]
- 3.McNulty C, Lecky DM, Xu-McCrae L, Nakiboneka-Ssenabulya D, Chung KT, Nichols T, Thomas HL, Thomas M, Alvarez-Buylla A, Turner K, Shabir S, Manzoor S, Smith S, Crocker L, Hawkey PM. 2018. CTX-M ESBL-producing Enterobacteriaceae: estimated prevalence in adults in England in 2014. J Antimicrob Chemother 73:1368–1388. 10.1093/jac/dky007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. 2019. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control 8:39. 10.1186/s13756-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolejska M, Duskova E, Rybarikova J, Janoszowska D, Roubalova E, Dibdakova K, Maceckova G, Kohoutova L, Literak I, Smola J, Cizek A. 2011. Plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding centre. J Antimicrob Chemother 66:757–764. 10.1093/jac/dkq500. [DOI] [PubMed] [Google Scholar]
- 6.Dolejska M, Villa L, Minoia M, Guardabassi L, Carattoli A. 2014. Complete sequences of IncHI1 plasmids carrying blaCTX-M-1 and qnrS1 in equine Escherichia coli provide new insights into plasmid evolution. J Antimicrob Chemother 69:2388–2393. 10.1093/jac/dku172. [DOI] [PubMed] [Google Scholar]
- 7.Damborg P, Marskar P, Baptiste KE, Guardabassi L. 2012. Faecal shedding of CTX-M-producing Escherichia coli in horses receiving broad-spectrum antimicrobial prophylaxis after hospital admission. Vet Microbiol 154:298–304. 10.1016/j.vetmic.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Jakobsen L, Bortolaia V, Bielak E, Moodley A, Olsen SS, Hansen DS, Frimodt-Møller N, Guardabassi L, Hasman H. 2015. Limited similarity between plasmids encoding CTX-M-1 β-lactamase in Escherichia coli from humans, pigs, cattle, organic poultry layers and horses in Denmark. J Glob Antimicrob Resist 3:132–136. 10.1016/j.jgar.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Apostolakos I, Franz E, van Hoek AHAM, Florijn A, Veenman C, Sloet-van Oldruitenborgh-Oosterbaan MM, Dierikx C, van Duijkeren E. 2017. Occurrence and molecular characteristics of ESBL/AmpC-producing Escherichia coli in faecal samples from horses in an equine clinic. J Antimicrob Chemother 72:1915–1921. 10.1093/jac/dkx072. [DOI] [PubMed] [Google Scholar]
- 10.Hordijk J, Farmakioti E, Smit LAM, Duim B, Graveland H, Theelen MJP, Wagenaar JA. 2020. Fecal carriage of extended-spectrum-β-lactamase/AmpC-producing Escherichia coli in horses. Appl Environ Microbiol 86:e02590-19. 10.1128/aem.02590-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isgren CM, Edwards T, Pinchbeck GL, Winward E, Adams ER, Norton P, Timofte D, Maddox TW, Clegg PD, Williams NJ. 2019. Emergence of carriage of CTX-M-15 in faecal Escherichia coli in horses at an equine hospital in the UK; increasing prevalence over a decade (2008–2017). BMC Vet Res 15:268. 10.1186/s12917-019-2011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother 69:1224–1230. 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 13.Kaspar U, von Lützau K, Schlattmann A, Rösler U, Köck R, Becker K. 2019. Zoonotic multidrug-resistant microorganisms among non-hospitalized horses from Germany. One Health 7:100091. 10.1016/j.onehlt.2019.100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupo A, Haenni M, Saras E, Gradin J, Madec JY, Börjesson S. 2018. Is blaCTX-M-1 riding the same plasmid among horses in Sweden and France? Microb Drug Resist 24:1580–1586. 10.1089/mdr.2017.0412. [DOI] [PubMed] [Google Scholar]
- 15.Sadikalay S, Reynaud Y, Guyomard-Rabenirina S, Falord M, Ducat C, Fabre L, Le Hello S, Talarmin A, Ferdinand S. 2018. High genetic diversity of extended-spectrum β-lactamases producing Escherichia coli in feces of horses. Vet Microbiol 219:117–122. 10.1016/j.vetmic.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 16.de Lagarde M, Larrieu C, Praud K, Lallier N, Trotereau A, Sallé G, Fairbrother JM, Schouler C, Doublet B. 2020. Spread of multidrug resistance IncHI1 plasmids carrying ESBL gene blaCTX-M-1 and metabolism operon of prebiotic oligosaccharides in commensal Escherichia coli from healthy horses, France. Int J Antimicrob Agents 55:105936. 10.1016/j.ijantimicag.2020.105936. [DOI] [PubMed] [Google Scholar]
- 17.Schouler C, Taki A, Chouikha I, Moulin-Schouleur M, Gilot P. 2009. A genomic island of an extraintestinal pathogenic Escherichia coli strain enables the metabolism of fructooligosaccharides, which improves intestinal colonization. J Bacteriol 191:388–393. 10.1128/JB.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Respondek F, Myers K, Smith TL, Wagner A, Geor RJ. 2011. Dietary supplementation with short-chain fructo-oligosaccharides improves insulin sensitivity in obese horses. J Anim Sci 89:77–83. 10.2527/jas.2010-3108. [DOI] [PubMed] [Google Scholar]
- 19.Sonnenborn U, Schulze J. 2009. The non-pathogenic Escherichia coli strain Nissle 1917 — features of a versatile probiotic. Microb Ecol Health Dis 21:122–158. 10.3109/08910600903444267. [DOI] [Google Scholar]
- 20.Qi B, Han M. 2018. Microbial siderophore enterobactin promotes mitochondrial iron uptake and development of the host via interaction with ATP synthase. Cell 175:571–582.e11. 10.1016/j.cell.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 21.Girón JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol 44:361–379. 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 22.Qin X, Hu F, Wu S, Ye X, Zhu D, Zhang Y, Wang M. 2013. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS One 8:e61169. 10.1371/journal.pone.0061169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talmi-Frank D, Altboum Z, Solomonov I, Udi Y, Jaitin DA, Klepfish M, David E, Zhuravlev A, Keren-Shaul H, Winter DR, Gat-Viks I, Mandelboim M, Ziv T, Amit I, Sagi I. 2016. Extracellular matrix proteolysis by MT1-MMP contributes to influenza-related tissue damage and mortality. Cell Host Microbe 20:458–470. 10.1016/j.chom.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Francetic O, Pugsley AP. 1996. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol 178:3544–3549. 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francetic O, Belin D, Badaut C, Pugsley AP. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J 19:6697–6703. 10.1093/emboj/19.24.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bébien M, Kirsch J, Méjean V, Verméglio A. 2002. Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology (Reading) 148:3865–3872. 10.1099/00221287-148-12-3865. [DOI] [PubMed] [Google Scholar]
- 27.Minazzato G, Gasparrini M, Amici A, Cianci M, Mazzola F, Orsomando G, Sorci L, Raffaelli N. 2020. Functional characterization of COG1713 (YqeK) as a novel diadenosine tetraphosphate hydrolase family. J Bacteriol 202:e00053-20. 10.1128/jb.00053-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederix M, Hütter K, Leu J, Batth TS, Turner WJ, Rüegg TL, Blanch HW, Simmons BA, Adams PD, Keasling JD, Thelen MP, Dunlop MJ, Petzold CJ, Mukhopadhyay A. 2014. Development of a native Escherichia coli induction system for ionic liquid tolerance. PLoS One 9:e101115. 10.1371/journal.pone.0101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins LS, Nunn WD. 1987. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol 169:2096–2102. 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeger C, Poulsen C, Dandanell G. 1995. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol 177:5506–5516. 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koga M, Otsuka Y, Lemire S, Yonesaki T. 2011. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187:123–130. 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cain AK, Hall RM. 2013. Evolution of IncHI1 plasmids: two distinct lineages. Plasmid 70:201–208. 10.1016/j.plasmid.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, Guerra B, Rodríguez I, van Essen-Zandbergen A, Brouwer M, Mevius D. 2015. Characterization of epidemic IncI1-Iγ plasmids harboring Ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother 59:5357–5365. 10.1128/AAC.05006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Tong MK, Chow KH, Cheng VCC, Tse CWS, Wu AKL, Lai RWM, Luk WK, Tsang DNC, Ho PL. 2018. Occurrence of highly conjugative IncX3 epidemic plasmid carrying blaNDM in Enterobacteriaceae isolates in geographically widespread areas. Front Microbiol 9:2272. 10.3389/fmicb.2018.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho PL, Wang Y, Liu MCJ, Lai ELY, Law PYT, Cao H, Chow KH. 2018. IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob Agents Chemother 62:e00295-17. 10.1128/AAC.02295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chouikha I, Germon P, Brée A, Gilot P, Moulin-Schouleur M, Schouler C. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J Bacteriol 188:977–987. 10.1128/JB.188.3.977-987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.CLSI. 2018. Performance standards for antimicrobial susceptibility testing; twenty-eighth informational supplement. CLSI document M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Bolger AM, Marc L, Bjoern U. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 41.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingle DJ, Valcanis M, Kuzevski A, Tauschek M, Inouye M, Stinear T, Levine MM, Robins-Browne RM, Holt KE. 2015. EcOH: in silico serotyping of E. coli from short read data. BioRxiv:e032151. 10.1101/032151. [DOI] [PMC free article] [PubMed]
- 47.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. 2018. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genomics 4:7. 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carriço JA, Achtman M. 2018. Grapetree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The draft genome assemblies, the complete plasmid and chromosomal sequences, and the raw data from short-read sequencing have been deposited in GenBank and SRA, respectively, under BioProject number PRJNA638007. The complete nucleotide sequences of the plasmids have been deposited in GenBank under accession numbers MT586601 to MT586609.