Plasmodium falciparum from the Greater Mekong subregion has evolved resistance to the artemisinin-based combination therapy dihydroartemisinin and the partner drug piperaquine. To monitor the potential westward spread or independent evolution of piperaquine resistance, we evaluated the in vitro susceptibility of 120 P. falciparum isolates collected at the China-Myanmar border during 2007 to 2016.
KEYWORDS: Plasmodium falciparum, drug resistance, piperaquine, pfcrt, pfmdr1, plasmepsin, gene amplification
ABSTRACT
Plasmodium falciparum from the Greater Mekong subregion has evolved resistance to the artemisinin-based combination therapy dihydroartemisinin and the partner drug piperaquine. To monitor the potential westward spread or independent evolution of piperaquine resistance, we evaluated the in vitro susceptibility of 120 P. falciparum isolates collected at the China-Myanmar border during 2007 to 2016. The parasite isolates displayed a relatively wide range of piperaquine susceptibility estimates. While 56.7% of the parasites showed bimodal drug response curves, all but five generated area-under-the-curve (AUC) estimates consistent with a susceptible phenotype. Using the piperaquine survival assay (PSA), 5.6% parasites showed reduced susceptibility. Of note, parasites from 2014 to 2016 showed the highest AUC value and the highest proportion with a bimodal curve, suggesting decreasing effectiveness in these later years. Unsupervised K-mean analysis of the combined data assigned parasites into three clusters and identified significant correlations between 50% inhibitory concentration (IC50), IC90, and AUC values. No parasites carried the E415G mutation in a putative exonuclease, new mutations in PfCRT, or amplification of the plasmepsin 2 and 3 genes, suggesting mechanisms of reduced piperaquine susceptibility that differ from those described in other countries of the region. The association of increased AUC, IC50, and IC90 values with major PfK13 mutations (F446I and G533S) suggests that piperaquine resistance may evolve in these PfK13 genetic backgrounds. In addition, the Pfmdr1 F1226Y mutation was associated with significantly higher PSA values. Further elucidation of piperaquine resistance mechanisms and continuous surveillance are warranted.
INTRODUCTION
The Greater Mekong Subregion (GMS) of Southeast Asia is one of the most threatening foci of malaria because of the emergence and spread of multidrug-resistant (MDR) Plasmodium falciparum parasites. Decades ago, parasites resistant to chloroquine (CQ) and the antifolate drug pyrimethamine arose in this location and spread to Africa, causing malaria resurgence and loss of millions of lives (1–4). Artemisinin combination therapies (ACTs) have been adopted worldwide as the frontline treatment of uncomplicated P. falciparum malaria (5) and have played an indispensable role in reducing global malaria-associated mortality and morbidity (6). However, clinical artemisinin resistance, first detected in western Cambodia (7–9), is now seen in all GMS countries due to spread and independent emergence (10–16). Since the efficacy of ACTs relies on both the fast-acting artemisinin derivatives and long-lasting partner drugs, artemisinin resistance would leave a larger parasite mass for the partner drug to clear and increase the risk of resistance development to the partner drug. This would translate into clinical failures of ACTs. Indeed, a decade or so after the deployment of ACTs, clinical resistance to artesunate-mefloquine (ATS-MFQ) and dihydroartemisinin-piperaquine (DHA-PPQ) emerged in Cambodia (17–21). More recent multisite clinical studies showed that DHA-PPQ failure rates reached high levels in several sites of the eastern GMS (22), which urged the adoption of other ACTs and consideration of triple ACTs (23). As the GMS moves toward malaria elimination, the intensity of drug selection increases, leading to rapidly adapting MDR parasites in the dwindling remnant parasite populations with different genetic backgrounds (24, 25). This situation offers a unique setting to investigate the evolution of drug resistance since important lessons could be learned for other countries moving toward elimination in the future.
Clinical artemisinin resistance in Cambodia is associated with mutations in the Kelch domain protein PfK13 (26). Molecular surveillance showed that PfK13 mutations associated with artemisinin resistance were restricted to certain areas of the GMS (16, 26, 27), but such mutations were rare in Africa (28). Within the GMS, there is also significant geographic heterogeneity in both the patterns of the PfK13 mutations and their prevalence (15, 25, 29–32), possibly reflecting regionally different drug histories and evolutionary origins of the parasites (33). Parasites in the eastern GMS harbor the predominant PfK13 C580Y mutation, whereas the most prevalent PfK13 mutation in the western GMS is F446I (26, 29, 30, 32). Consequently, PPQ resistance in Cambodia primarily arose on a PfK13 C580Y background (34–37). Using a genome-wide association (GWAS) approach, clinical PPQ resistance in western Cambodia was found to be associated with amplification of the two aspartic protease genes plasmepsin 2 and plasmepsin 3 (plasmepsin 2/3) on chromosome 14 (35, 36) and a point mutation E415G (exo-E415G) in a putative exonuclease gene (PF3D7_1362500) on chromosome 13 (35). Knockout experiments showed that disruption of plasmepsin 2/3 in the 3D7 parasite specifically sensitized the parasites to PPQ but not to other antimalarials, adding support for the roles of plasmepsin 2/3 amplification in PPQ resistance (38). However, a subsequent study showed that overexpression of these two enzymes in the 3D7 genetic background did not change the parasites’ susceptibility to PPQ, CQ, or artesunate (39), suggesting that further validation of plasmepsin 2/3 amplification in PPQ resistance is needed.
Given the potential mode of action of PPQ in blocking heme detoxification in the food vacuole (FV) (40), two FV membrane-resident transporters pfmdr1 and pfcrt have received much attention. Clinical efficacy studies revealed the reduced prevalence of multicopy pfmdr1 after switching from ATS-MFQ to DHA-PPQ (34–37), suggesting that this is due to DHA-PPQ selection of single-copy pfmdr1 or decreased use of MFQ, since pfmdr1 amplification is associated with a fitness cost (41). Interestingly, accompanying the emergence of PPQ resistance in Cambodia were several new PfCRT mutations (H97Y, F145I, M343L, and G353V) that have evolved in a Dd2 PfCRT background (74I, 75E, 76T, 220S, 271E, 326S, 356T, and 371I) (42), which were all validated by genetic studies as conferring resistance to PPQ (43). Moreover, the PfCRT F145I mutation was also associated with in vivo DHA-PPQ treatment failure and decreased ex vivo PPQ susceptibility by GWAS (44), providing further evidence that these new PfCRT mutations can serve as molecular markers for PPQ resistance.
Because the PPQ dose-response curves from the traditional in vitro or ex vivo assays to determine 50% inhibitory concentrations (IC50s) sometimes do not fit the sigmoid curve, a PPQ survival assay (PSA) was designed (42), which measures the parasite survival rates after exposure to a pharmacologically relevant dose of 200 nM PPQ for 48 h. The PSA estimate was found to be well correlated with the DHA-PPQ clinical failure phenotype. Subsequently, an in vitro study using a wide range of PPQ concentrations described a bimodal dose-response curve for some clinical PPQ-resistant parasites, showing increased parasite survival at PPQ concentrations of 100 nM to 10 μM (45). This second peak in the PPQ dose-response curve was proposed to be an alternative readout for PPQ resistance. Whereas these drug assays were developed based on clinical phenotypes observed in the GMS, their values in predicting resistance in other malaria regions remain to be evaluated.
Within the GMS, PPQ has the most prolonged use history in China, where it was first used in the early 1970 to supersede CQ as the first-line treatment for CQ-resistant P. falciparum (46). The emergence of PPQ resistance in the mid-1980s led to its diminished use in subsequent years (47–49). In a study conducted in the early 1990, the cure rates of a total dosage of 25 mg/kg PPQ dropped to ∼33% (50). DHA-PPQ was then adopted as the first-line treatment for falciparum malaria in 2005 in China. In contrast to the rapidly declining clinical efficacy of DHA-PPQ in Cambodia, recent studies along the China-Myanmar border reported that DHA-PPQ remained highly efficacious for P. falciparum malaria (51, 52). In this study, we describe the features of in vitro susceptibility to PPQ, and the associated genetic adaptions, on the China-Myanmar border. Using culture-adapted clinical parasite isolates, we compared different readouts of in vitro PPQ susceptibility assays and genotyped multiple molecular markers associated with PPQ resistance in Cambodia.
RESULTS
In vitro susceptibility of parasite isolates to PPQ.
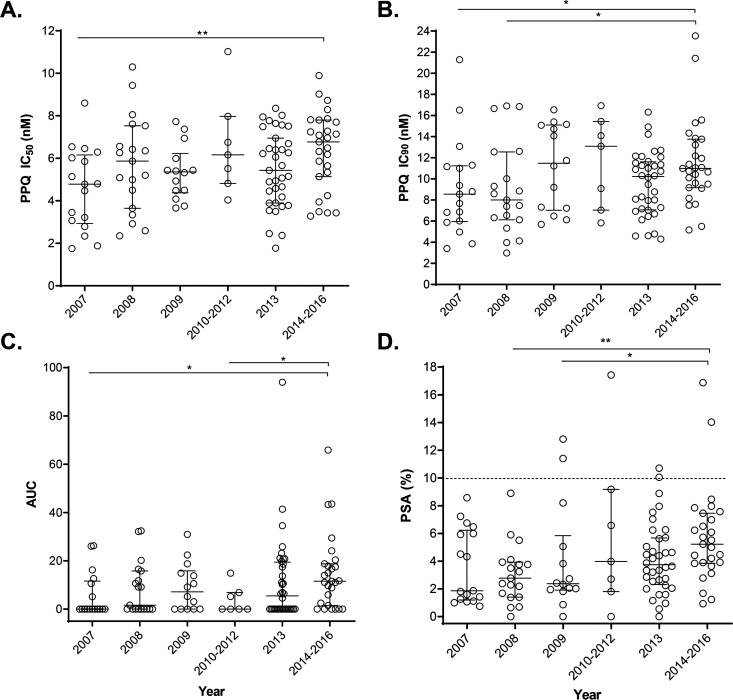
We tested in vitro susceptibilities of 120 archived P. falciparum isolates to PPQ using recently developed in vitro assays. These parasites are monoclonal, culture-adapted clinical isolates collected from the China-Myanmar border area during 2007 to 2016, which represent parasite populations after the adoption of DHA-PPQ as the frontline treatment for uncomplicated P. falciparum malaria in 2005. We first performed a modified 72-h standard drug assay using 24 dilutions of PPQ with the highest concentration being 100 μM to ensure complete parasite killing. Consistent with an earlier study (45), the drug response curves in 56.7% (68/120) of the parasite isolates exhibited a bimodal pattern with a second peak at PPQ concentrations of 100 nM to 100 μM (Fig. 1). We used the whole range of the drug concentrations to estimate the IC50 and IC90 values after removing the outliers following the strategy adopted in a previous ex vivo study (53). Overall, we found that the parasites were susceptible to PPQ with a median IC50 value of 5.6 nM (interquartile range [IQR], 4.1 to 7.1 nM) and a median IC90 value of 10.1 nM (IQR, 7.1 to 12.6 nM) (Fig. 2A and B). Despite this, the most and the least sensitive parasites had >6-fold differences in IC50 (1.8 to 11.0 nM) and ∼8-fold differences in IC90 (3.0 to 23.5 nM), indicating the presence of parasites with reduced susceptibility to the drug. Over the 9 years, annual median PPQ IC50 varied between 4.8 and 6.8 nM (Table 1), but a significant difference was only identified between the 2014-2016 and 2007 parasites (Fig. 2A). Similarly, the annual median IC90 value fluctuated between 8.0 and 13.1 nM (Table 1). The IC90 in parasites from the most recent years (2014 to 2016) was significantly higher than the IC90s in the earliest samples from years 2007 and 2008, suggesting declining PPQ susceptibility over the years (Fig. 2B). With regard to the fluctuation in the annual IC50 and IC90 data, the Mann-Kendall trend test did not detect a clear trend of increase in IC50 or IC90 (P > 0.05).
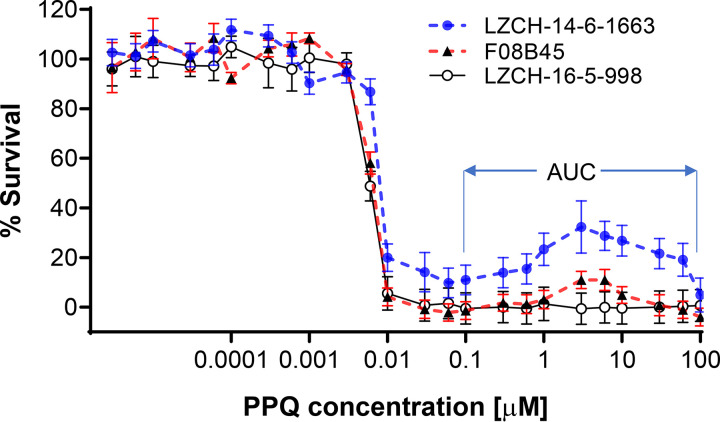
FIG 1.
Piperaquine drug response curves of representative parasite isolates from the China-Myanmar border using the modified SYBR green I assay with 24 drug concentrations. One parasite isolate (LZCH-16-5-998) showed a typical sigmoid curve, while the other two isolates (LZCH-14-6-1663 and F08B45) showed a second peak in PPQ concentrations of 1 to 10 μM. The concentration range used to calculate the AUC is indicated.
FIG 2.
Dot plots of in vitro susceptibilities to piperaquine of longitudinally collected parasites from the China-Myanmar border area during 2007 to 2016. (A) IC50 values. (B) IC90 values. (C) AUC values. (D) PSA values. The dotted line represents the 10% survival rate cutoff that distinguishes piperaquine-resistant (≥10%) from piperaquine-sensitive (<10%) parasites in PSA. For samples from each year, the medians and interquartile ranges are shown. Asterisks indicate significant differences in drug sensitivity between 2 years. * and **, P < 0.05 and < 0.01, respectively (Mann-Whitney U test).
TABLE 1.
In vitro susceptibilities of Plasmodium falciparum strains from the China-Myanmar border area to piperaquinea
| Period (yr) | No. | Median (IQR) |
No. (%) |
Median AUC (IQR)d | |||
|---|---|---|---|---|---|---|---|
| IC50 (nM) | IC90 (nM) | PSA (%) | PSA ≥ 10%b | Bimodalc | |||
| 2007 | 17 | 4.8 (2.9–6.2) | 8.6 (6.0–11.2) | 1.9 (1.2–6.2) | 0 | 6 (35.3) | 0.0 (0–11.6) |
| 2008 | 19 | 5.9 (3.7–7.5) | 8.0 (6.1–12.6) | 2.8 (1.4–3.9) | 0 | 10 (52.6) | 1.7 (0–15.8) |
| 2009 | 14 | 5.4 (4.4–6.2) | 11.5 (7.0–15.1) | 2.4 (1.9–5.8) | 2 (14.3) | 9 (64.3) | 7.2 (0–15.9) |
| 2010–2012 | 7 | 6.2 (4.8–8.0) | 13.1 (7.0–15.5) | 4.0 (1.8–9.2) | 1 (14.3) | 3 (42.9) | 0.0 (0–6.9) |
| 2013 | 36 | 5.4 (3.9–7.0) | 10.2 (7.1–11.6) | 3.7 (2.3–5.7) | 2 (5.6) | 19 (52.8) | 5.5 (0–19.5) |
| 2014–2016 | 27 | 6.8 (5.1–7.8) | 11.0 (9.2–13.8) | 5.2 (3.8–7.5) | 2 (7.4) | 21 (77.8) | 11.2 (1.5–18.8) |
| Totale | 120 | 5.6 (4.1–7.1)* | 10.1 (7.1–12.6)** | 3.8 (2.0–6.1)† | 7 (5.8) | 68 (56.7) | 5.6 (0–16.5)‡ |
AUC, area under the curve; IQR, interquartile range; PSA, piperaquine survival assay score.
Number and percentage of parasites with a PSA of ≥10%.
Number and percentage of parasites with bimodal PPQ response curves (67.4%).
AUC for the drug response curve at PPQ concentrations of 102 to 105 nM.
A Kruskal-Wallis test was used to compare results between years (*, P = 0.0557; **, P = 0.1103; †, P = 0.0450; ‡, P = 0.1246). A Mann-Kendall trend test was used to evaluate the presence of annual trends for IC50, IC90, PSA, and AUC values (all of which were nonsignificant).
We then measured the AUC at PPQ concentrations 0.1 to 100 μM in the drug response curves. In different years, 35.3 to 77.8% of parasites displayed bimodal drug response curves (Table 1), and parasites collected in 2014 to 2016 had the highest proportion (77.8%). Similar to the IC50 and IC90 values, parasites from 2014 to 2016 had significantly higher AUC values than parasites from 2007 and from 2010 to 2012 (Fig. 2C). However, compared to the AUC range defined earlier (45), only five parasite isolates had the intermediate AUC values (35 to 100), whereas no parasites had an AUC of >100, a cutoff value found to correspond to PSA value of >10% (Fig. 2C).
We used PSA to assess the parasite’s resistance to PPQ, which measures the parasite survival rate after exposure to 200 nM PPQ for 48 h (42). The overall parasite survival rate was low (3.8%), whereas parasites from 2014 to 2016 had significantly higher PSA values than parasites from 2008 and 2009 (Fig. 2D). There seemed to be a trend of gradual increase over the years (from 1.9% in 2007 to 5.2% in 2014 to 2016), albeit the trend was not significant (Table 1, P > 0.05, Mann-Kendall trend test). In total, there were only 7 (5.6%) parasite isolates displaying PSA values of >10% (Table 1 and Fig. 2D), the proposed cutoff value for PPQ resistance (42), and the highest PSA value was 17.4%. For the two earlier years, 2007 and 2008, all 36 parasite isolates tested had PSA values lower than 10%.
Comparison among different in vitro PPQ sensitivity assays.
The PSA was developed to better differentiate recrudescent from nonrecrudescent P. falciparum cases after treatment with DHA-PPQ in Cambodia (42). Using the cutoff value of 10%, we compared the IC50, IC90, and AUC values between the 113 parasite isolates with PSA values <10% and the seven isolates with PSA values of ≥10%. The results showed no significant differences between the two groups in IC50, IC90, or AUC values (see Fig. S1 in the supplemental material), indicating that PSA and other parameters measured different aspects of PPQ susceptibilities.
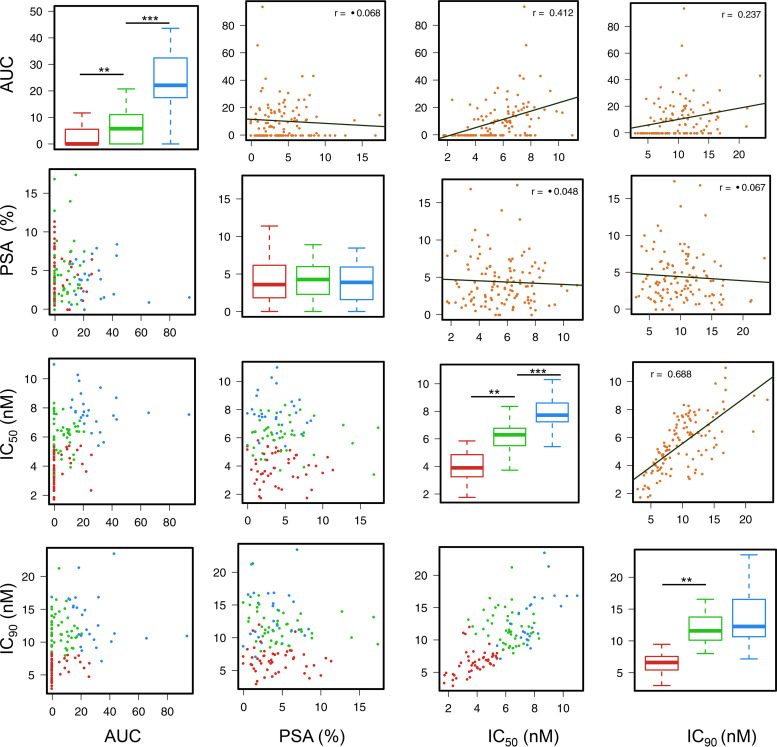
To discover the underlying patterns of the in vitro assay data for PPQ and properly group the parasites based on their IC50, IC90, AUC, and PSA values, we subjected the in vitro assay results to analysis using the unsupervised K-means clustering algorithm. This clustering analysis assigned the parasites in three groups of 44, 46, and 26 parasites, respectively (Fig. 3). While this grouping identified significantly different clusters in AUC, IC50, and IC90 values, PSA results remained indistinguishable among the three clusters (Table 2 and Fig. 3). Group 1 (44 parasites) contained mostly parasites having typical sigmoid in vitro drug response curves (median AUC = 0), with low IC50 (median, 3.9 nM) and IC90 (median, 6.6 nM) values. Group 2 (46 parasites) had intermediate AUC value (median, 5.8), but relatively high IC50 (median, 6.3 nM) and IC90 (median, 11.6 nM) values. Group 3 (26 parasites) had the highest AUC value (median, 22.1) and highest IC50 (median, 7.7 nM) and IC90 (median, 12.3 nM) values (Fig. 3). The results showed that IC50 and IC90 values were highly correlated (r = 0.688, P < 0.05, Pearson correlation), while they also exhibited significant correlations with the AUC results (Fig. 3, upper diagonal). However, results from the 72-h assays showed no correlation with the PSA results.
FIG 3.
Clustering of parasite populations into three groups based on unsupervised K-means clustering analysis of piperaquine drug assay data. The diagonal panels show comparisons in AUC, PSA, IC50, and IC90 values among the three groups. Groups 1, 2, and 3 are indicated in red, green, and blue, respectively. Panels below the diagonal are scatterplots showing the distribution of values of samples, which are color-coded based on the three K-mean groups. Panels above the diagonal are scatterplots showing the correlation between each pair of experimental data. Pearson correlation coefficient values are shown.
TABLE 2.
PPQ susceptibilities categorized based on IC50, IC90, and the presence of bimodal curves and AUCa
| Parasite group or P value | Median (IQR) |
PSA ≥ 10%, no. (%) | Median PSA (IQR) | ||
|---|---|---|---|---|---|
| AUC | IC50 (nM) | IC90 (nM) | |||
| Groups | |||||
| 1 (red) | 0.0 (0.0–5.7) | 3.9 (3.2–4.9) | 6.6 (5.4–7.6) | 3 (6.3) | 3.6 (1.8–6.2) |
| 2 (green) | 5.8 (0.0–11.1) | 6.3 (5.5–6.8) | 11.6 (10.1–13.8) | 4 (8.7) | 4.3 (2.2–6.1) |
| 3 (blue) | 22.1 (17.2–33.0) | 7.7 (7.2–8.6) | 12.3 (10.6–16.6) | 0 (0.0) | 3.9 (1.6–6.0) |
| P valuesb | |||||
| * | P < 0.0001 | P < 0.0001 | P < 0.0001 | ND | P = 0.6450 |
| ** |
P = 0.0034 (1 vs 3) P < 0.0001 (2 vs 3) P = 0.4108 (1 versus 2) |
P < 0.0001 (1 vs 2, 1 vs 3, 2 vs 3) |
P < 0.0001 (1 vs 2, 1 vs 3) P = 0.1230 (2 versus 3) |
ND | ND |
AUC, area under curve; IQR, interquartile range; PSA, piperaquine survival assay score; ND, not done.
*, Comparison among three groups (Kruskal-Wallis test); **, comparison between two groups (Mann-Whitney U test).
Association with known resistance markers.
PPQ resistance has been associated with the E415G mutation in a putative exo-nuclease (35), and new mutations (H97Y, F145I, M343L, and G353V) in PfCRT (43, 44), and plasmepsin 2/3 amplification (35, 36). Genotyping the 120 isolates from the China-Myanmar border by PCR and sequencing found no E415G mutation (data not shown). Also, real-time PCR analysis did not identify any parasites carrying more than one copy of plasmepsin 2/3 (see Fig. S2). In order to determine mutations in the pfcrt gene, we used RT-PCR to circumvent problems associated with multiple AT-rich introns. Sequencing of the full-length pfcrt genes showed that the overwhelming majority (115/120) of the parasites carried the Dd2 haplotype (M74I, N75E, K76T, A220S, Q271E, N326S, I356T, and R371I). Compared with the Dd2 haplotype, one isolate carried an M74T mutation, one was wild-type (WT) at N326, one carried two additional mutations at L243S and K284E, and one had an additional mutation at F78S (Table 3). For the four new mutations described in Cambodia, only one parasite carried an H97L mutation in the Dd2 haplotype background.
TABLE 3.
Haplotypes of the pfcrt gene in the studied parasite population and association with altered piperaquine susceptibility
| Isolate |
pfcrt positiona |
Frequency, no. (%) | AUC | Pb | PSAc (%) | P | IC50 (nM) | P | IC90 (nM) | P | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 74 | 75 | 76 | 78 | 97 | 220 | 243 | 271 | 284 | 326 | 356 | 371 | ||||||||||
| 3D7 | M | N | K | F | H | A | L | Q | K | N | I | R | 0 | 0 | <0.0001 | 1.19 | <0.0001 | 5.47 | <0.0001 | 7.05 | <0.0001 |
| Dd2 type | I | E | T | F | H | S | L | E | K | S | T | I | 115 (95.8) | 6.14 | 3.84 | 5.66 | 9.55 | ||||
| F09N29 | I | E | T | F | H | S | L | E | K | N | T | I | 1 (0.97) | 0 | <0.0001 | 0.87 | <0.0001 | 5.32 | 0.0985 | 15.08 | <0.0001 |
| F07-29 | I | E | T | F | H | S | S | E | E | S | T | I | 1 (0.97) | 0 | <0.0001 | 1.86 | <0.0001 | 3.45 | <0.0001 | 6.87 | <0.0001 |
| NB-14-6-3 | I | E | T | F | L | S | L | E | K | S | I | I | 1 (0.97) | 15.02 | <0.0001 | 6.52 | <0.0001 | 7.81 | <0.0001 | 15.58 | <0.0001 |
| F09N6 | I | E | T | S | H | S | L | E | K | S | T | I | 1 (0.97) | 5.22 | <0.0001 | 1.82 | <0.0001 | 4.5 | <0.0001 | 14.72 | <0.0001 |
| NB13-1827 | T | E | T | F | H | S | L | E | K | S | T | I | 1 (0.97) | 4.59 | <0.0001 | 4.47 | 0.1795 | 5.07 | 0.0037 | 12.62 | <0.0001 |
aMutations in the Dd2 parasite are highlighted in boldface; new mutations are underlined.
bAll P values were determined using a Wilcoxon matched-pairs signed-rank test, compared to the Dd2 type.
cPSA, piperaquine survival assay score.
PPQ-resistant parasites have evolved mainly in the background of PfK13 mutations and loss of amplification of the pfmdr1 gene (34, 35, 42, 54). For PfK13, the predominant mutation in the China-Myanmar border area is the F446I, but in 2014 to 2016, the new mutation G533S appeared and reached 44% prevalence (55). Parasites with the NN insertion at amino acids 136 to 137 of PfK13 continuously increased in frequency and reached 100% in 2014 to 2016. For pfmdr1, gene amplification was rare in the parasite population from the China-Myanmar border (56). Only Y184F was consistently present throughout most of the years and slightly increased in the 2013-2016 samples. Whereas N1042D was only found in the 2009 samples, the N86Y, D90H, and E130K mutations appeared in samples collected in later years (2013 to 2016) at very low frequencies. It is noteworthy that the F1226Y mutation appeared from 2013 onward, but its frequency reached 44.4% in the 2014-2016 samples.
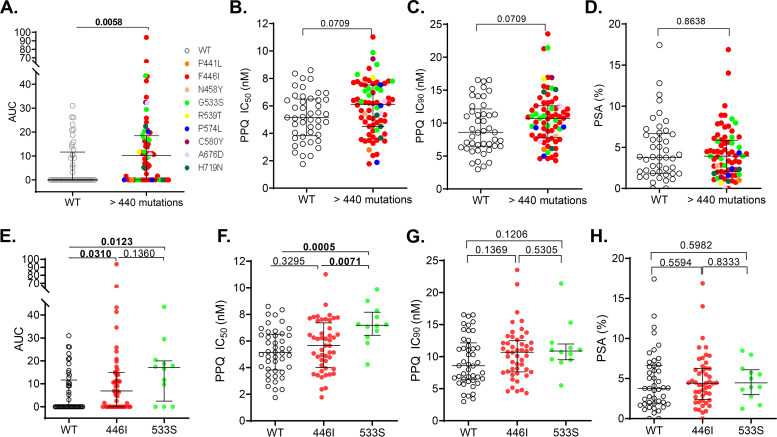
To determine whether any of these mutations were associated with altered PPQ susceptibility, we performed Lasso regression analysis, assessing all mutations regardless of their prevalence. The PfK13 G533S and two rare mutations PfK13 C580Y and Pfmdr1 D90H were significantly associated with PPQ IC50 values (see Fig. S3A). For the mutations associated with increased PSA values, only the Pfmdr1 Y184F and PfCRT I356T mutations were relatively prevalent. In contrast, the PfK13 mutations and Pfmdr1 N86Y and E130K were very rare in the study parasite population (see Fig. S3B). We then used the Mann-Whitney U test to determine whether the relatively abundant (≥5%) mutations or haplotypes were associated with altered susceptibilities to PPQ. When parasites with PfK13 mutations in the propeller domain (>440 amino acids) were compared to the PfK13 WT parasites, we found that those with PfK13 mutations had significantly elevated AUC values (Fig. 4A, P < 0.01). Although these parasites with the PfK13 mutations also had higher IC50 and IC90 values than the WT parasites, the differences were only marginally significant (Fig. 4B and C, P = 0.0709). When comparing the two most predominant mutations F446I and G533S, we found that parasites carrying either of these two mutations had significantly higher AUC values than the WT parasites (Fig. 4E). In addition, parasites with the G533S mutation also had significantly higher PPQ IC50 values than the WT parasites (P < 0.0001) and parasites with the F446I mutation (Fig. 4F, P < 0.01). Compared to the WT parasites, the parasites with PfK13 mutations in the propeller domain (amino acids >440) or the predominant mutations showed no significant differences in their PSA values (Fig. 4D and H).
FIG 4.
Comparisons of in vitro piperaquine susceptibility (AUC [A and E], IC50 [B and F], IC90 [C and G], and PSA [D and H]) between parasites with PfK13 mutations (colored dots) and wild-type parasites (WT, open circles). PfK13 mutations were indicated by different colors. AUC, area under curve. Horizontal lines indicate medians ± IQRs. P values are indicated, and significance is highlighted in boldface (Mann-Whitney U test).
For the Pfmdr1 Y184F and F1226Y mutations, 1226Y was associated with significantly increased PPQ IC50 values, while 184F was associated with significantly higher PSA values (see Fig. S4). For the haplotypes, Y184/1226Y had significantly higher IC50 than the WT (Y184/F1226) and the 184F/F1226 haplotype (P < 0.05, Mann-Whitney U test). The 184F/F1226 haplotype had a significantly higher PSA value than the WT parasites (see Fig. S4, P < 0.05).
DISCUSSION
Surveillance for antimalarial drug resistance and elucidation of resistance mechanisms are essential for guiding effective treatment of malaria cases. This is especially true for eliminating malaria in the GMS, where parasites have developed resistance to all commonly used antimalarial drugs, especially the frontline ACT drugs ATS-MFQ and DHA-PPQ. Since the deployment of DHA-PPQ in Cambodia in 2008, failure rates to this ACT rapidly increased, which is associated with the increased prevalence of parasites with PfK13 C580Y mutation and plasmepsin 2/3 gene amplification (22). Population genomics studies identified the expansion of an MDR colineage KEL1/PLA1 carrying these mutations in eastern GMS, which diversified into multiple subgroups with the emergence of novel pfcrt mutations (57). To evaluate the evolution of drug resistance in the pre-elimination setting of the GMS, we assessed the in vitro PPQ susceptibility of parasites collected from the China-Myanmar border. Despite the historical detection of clinical PPQ resistance in southern China when PPQ was used as monotherapy after the mid-1980s (47–50), our study suggests the lack of PPQ resistance in this region or that PPQ resistance in this parasite population may involve additional mechanisms. While the lack of clinical outcome data prevents us from connecting the in vitro assay data with the in vivo efficacy result, this assumption is nonetheless consistent with the excellent clinical efficacy of DHA-PPQ from recent studies in this region (51, 52).
The three assays, measuring the IC50, IC90, AUC, and PSA as the parasite’s response to PPQ, present a complicated picture for identifying in vitro surrogates of clinical PPQ resistance. PSA measures the survivorship of parasites after exposure to 200 nM PPQ for 48 h (42). Using clinical DHA-PPQ efficacy data, a PSA value of ≥10% was found to be a relevant value to define PPQ resistance, and the PSA results were used to identify plasmepsin 2/3 gene amplification (36). Using the PSA, we determined that 5.8% of the 120 parasites collected from the China-Myanmar border had PSA values of higher than 10%, conforming to the definition of PPQ resistance. However, the lack of parasites with plasmepsin 2/3 amplification and new pfcrt mutations suggests that they may not represent the clinical PPQ resistance previously described. Thus, without clinical and molecular data as backups, the usefulness of PSA outside eastern GMS remains to be further evaluated. In evaluating the bimodal curve-based assay, Bopp et al. found that the PSA survival rates (0 to 30%) in a small set of nine representative isolates were significantly correlated with the AUC values, and an AUC of >100, corresponding to a PSA value of >10%, was considered to be PPQ resistant (45). In this study, comparing the results from these different assays did not identify any correlation between PSA and IC50, IC90, or AUC values. The relatively low IC50 and IC90 values and the finding that only five parasites had AUC values in the intermediate range of 35 to 100 are both consistent with the excellent DHA-PPQ efficacy of the China-Myanmar border parasite population (51, 52). This study identified correlations among the IC50, IC90, and AUC estimates, suggesting that the use of a wide range of PPQ concentrations with the high concentrations able to kill all parasites would allow a more realistic determination of these in vitro assay values (53). Furthermore, given that the use of the ex vivo IC50 or IC90 estimates also allowed the identification of the plasmepsin 2/3 amplification as associated with clinical PPQ resistance by GWAS (35, 44), we consider that the IC50 or IC90 values are useful parameters for predicting PPQ resistance.
Plasmepsin 2/3 amplification has previously been associated with clinical PPQ resistance and was a useful marker for predicting DHA-PPQ clinical failures in Cambodia and eastern GMS. However, validation of these two genes in the 3D7 background using genetic inactivation and overexpression produced conflicting results, suggesting that the effect of plasmepsin 2/3 amplification on PPQ resistance may vary depending on the parasites’ genetic backgrounds. This also undermines the use of plasmepsin 2/3 amplification for monitoring PPQ resistance outside the eastern GMS. Novel pfcrt mutations were found to be associated with PPQ resistance, and there is genetic evidence showing that these novel mutations mediate PPQ resistance in the genetic background without plasmepsin 2/3 amplification (43). Although we did not observe any of the new pfcrt mutations (H97Y, F145I, M343L, and G353V) described in the DHA-PPQ-resistant populations in Cambodia (42–44), one parasite collected in 2014 had an H97L mutation. This parasite had a significantly elevated CQ IC50 value (826.9 nM) compared to parasites carrying the Dd2 haplotype pfcrt (259.2 nM). It also had higher PPQ IC50, IC90, PSA, and AUC estimates than parasites carrying the Dd2 haplotype pfcrt (Table 3), but the limited data from a single parasite isolate carrying the H97L mutation warrant future studies. Of note, this mutation was identified in a single parasite from a set of 183 samples collected in ACT efficacy studies in Cambodia, but it was not associated with PPQ IC90 (44).
The PPQ resistance occurring in western Cambodia probably reflects a two-step selection process. The spread of artemisinin resistance has led to the limited genetic diversity of the parasite populations with most parasites carrying the PfK13 mutations (e.g., C580Y), and on top of these genetic backgrounds, PPQ resistance has emerged (34, 42). Within a short time period, parasite lineages harboring the PfK13 C580Y mutation and plasmepsin 2/3 amplification rapidly spread in eastern GMS in the fashion of a hard selective sweep (37, 57). At the China-Myanmar border, P. falciparum had PfK13 alleles similar to those of parasites from northern Myanmar, with a predominance of the F446I mutation (29, 30). This mutation, when introduced into the 3D7 background, did not confer artemisinin resistance as measured by the in vitro ring-stage survival assay, but the parasites had a prolonged ring stage and showed no fitness cost (58). When we assessed the connection between the PfK13 mutations in the parasites studied here with PPQ in vitro assay results, we found that the total K13 mutations after the position 440 or the two predominant mutations F446I and G553S were all associated with increased IC50, IC90, and AUC values compared to the WT parasites. The F446I and G553S parasites developed significantly higher AUC values than WT, and the G553S parasites showed significantly higher IC50 values than WT and F446I parasites. Of note, both of these predominant PfK13 mutations were associated with significantly higher ring survival rates than the WT parasites (55). We speculate that they may not be causal for the elevated PPQ AUC and IC50 values, but, like the C580Y mutation in Cambodia, may represent the PfK13 background mutations on which PPQ resistance emerges. GWAS may reveal the genetic changes underlying the altered susceptibility to PPQ.
The ATS-MFQ combination has previously selected for increased copy number of pfmdr1 in the eastern GMS, whereas DHA-PPQ used to replace ATS-MFQ appears to have selected for the opposite. Consistent with the extensive deployment of DHA-PPQ at the China-Myanmar border, parasites from this region mostly had a single copy pfmdr1. In these parasite isolates, we found that the Y184F mutation had an overall frequency of 36%, while the F1226Y mutations appeared after 2013 and reached a high frequency of 44% in the 2014-2016 samples. Whereas the Y184F mutation was not significantly associated with changes in PSA in the Cambodian parasites, we found that it was associated with an increased PSA value in the current work. Also, we found that the newly acquired F1226Y mutation in later years was associated with an increased PPQ IC50 value. It would be interesting to determine whether the F1226Y mutation was selected by the extensive use of DHA-PPQ and whether it indeed mediates PPQ resistance.
We did not find clear evidence of PPQ resistance in the parasites from the China-Myanmar border area, nor did we observe temporally declining PPQ susceptibility. Alternatively, the fluctuating PPQ susceptibility over the years suggests that PPQ resistance emerged one or more times, but did not spread significantly. Despite this, we found that parasites collected in 2014 to 2016 had significantly higher IC50, IC90, AUC, and PSA values than those collected earlier (Fig. 2). These samples also appeared to be genetically distinct from those collected in earlier years for the pfk13 gene. The F446I mutation showed a gradual increase in the frequency between 2007 and 2013, whereas the G533S mutation only emerged in the 2014-2016 parasite population and reached 44% (55). It would be interesting to test whether this parasite population was the result of intensive selection by DHA-PPQ, the most popular ACT deployed in this area, or the result of the spread of parasites from elsewhere in the GMS. Regardless of the origins, the drug resistance situation requires continued surveillance efforts using a combination of complementary tools to monitor clinical efficacy, in vitro drug susceptibility, and molecular epidemiology of resistance-associated markers. We did not identify clear evidence of PPQ resistance from in vitro assays or association with known molecular markers, consistent with the known sustained clinical efficacy of DHA-PPQ in the region.
MATERIALS AND METHODS
Parasite collection and culture adaption.
Uncomplicated P. falciparum malaria was diagnosed by microscopy in patients attending local clinics located along the China-Myanmar border during 2007 to 2016. P. falciparum-infected blood samples were collected and cryopreserved in liquid nitrogen for laboratory culture adaption. Informed consent was obtained from all patients, and the study was approved by the institutional review boards of Kachin Health Bureau, Kunming Medical University, and Pennsylvania State University. A total of 120 parasite isolates (17 in 2007, 19 in 2008, 14 in 2009, 7 in 2010 to 2012, 36 in 2013, and 27 in 2014 to 2016), determined to be monoclonal infections by genotyping three polymorphic antigen markers—msp1, msp2, and glutamate-rich protein—were culture adapted and used for in vitro drug assay and genotyping (56). Parasites were cultured in type O+ human red blood cells (RBCs) in a complete medium at 37°C in an incubator under 5% O2, 5% CO2, and 90% N2.
Drug assays.
PPQ was obtained from Chongqing Kangle Pharmaceutical Co., Ltd. (Chongqing, China). The stock solution of PPQ (10 mM) was prepared in a mixture of 90% methanol and 10% HCl. In vitro susceptibilities of the parasite isolates to PPQ were determined using the standard SYBR green I assay (66). Twenty-four dilutions of the drug were added to 96-well microplates, which cover both the high-range (100, 60, 30, 10, 6, 3, 1, 0.6, and 0.3 μM) and low-range concentrations (100, 60, 30, 10, 6, 3, 1, 0.6, 0.3, 0.1, 0.06, 0.03, 0.01, 0.006, and 0.003 nM). Parasite cultures were synchronized with 5% d-sorbitol treatment (59), and the drug assay was performed using 0.5% synchronized parasites at the ring stage and 2% hematocrit (60). A drug assay was carried out with three technical replicates and two biological replicates. For consistency and comparison, 3D7 was included throughout the study as an internal control.
PSA was performed as described earlier (42). Briefly, tightly synchronized 0- to 3-h ring-stage parasites in 2 ml of culture media, diluted to 2% hematocrit and 0.5% parasitemia, were exposed to 200 nM PPQ or distilled water as the control in 24-well plates for 48 h. The drug was then washed off with RPMI 1640, and parasites were cultured for an additional 24 h. Thin smears were stained with Giemsa, and the number of surviving parasites in 20,000 RBCs were independently enumerated by two investigators. A third independent assessment was conducted if the difference between the first two counts was >20%. Survival rates were calculated by determining the proportion of viable parasites (second-generation rings or trophozoites with normal morphology) at the 72-h time point in PPQ-treated versus control cultures. A survival rate of 10% was used as the cutoff value for PPQ resistance (42).
Single nucleotide polymorphisms and copy number variations.
The E415G mutation in PF3D7_1362500 was determined by PCR and sequencing. For this, parasite genomic DNA was extracted from freshly cultured parasites using a genomic DNA extraction kit according to the manufacturer’s instructions (Roche). The exo-E415G fragment was amplified in 25 μl of reaction mixture, which contained 12.5 μl of Premix Taq (TaKaRa Taq version 2.0), 1 μl of primers (0.1 μM) (5′-TTTCCTTCTGACCCCTTT-3′ and 5′-TCCCATTCGATATCTATACCTAT-3′), and 50 ng of genomic DNA. PCR was performed under the following conditions: 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 53°C for 30 s, and 68°C for 1 min. For the year 2013 samples, the full-length pfk13 gene and the two pfmdr1 fragments covering codons 86, 184, 1042, and 1246 were amplified and sequenced as described previously (29, 61). For other samples, the pfk13 and pfmdr1 sequences were obtained from earlier studies (61, 62).
CNVs of the plasmepsin 2/3 genes were evaluated by real-time PCR following an established procedure (35). Briefly, each 20-μl PCR contained 10 μl of Bestar SybrGreen qPCR master mix, 0.25 μM concentrations of each forward and reverse primer for the plasmepsin 2/3 gene or the β-tubulin gene as an internal control, 0.4 μl of 5× ROX reference dye, and 50 ng of the parasite DNA. Real-time PCR was performed with an initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 10 s, 56°C for 30 s, and 72°C for 30 s and then a final extension for 5 min at 72°C. A copy number ratio of ≥1.5 was considered an increased copy number.
The full-length coding sequence of the pfcrt gene consisting of 424 codons was obtained by RT-PCR. Total RNA was isolated from 1 ml of cultured parasites at 1% parasitemia using the TRIzol reagent (Life Technologies). cDNA was synthesized using a RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, 1 μg of total RNA was mixed with 1 μl of Oligo(dT)18 primer, 1 μl of RiboLock RNase Inhibitor, 2 μl of 10 mM dNTP mix, and 1 μl of RevertAid M-MuLV reverse transcriptase in a total volume of 20 μl, and the reaction mixture was incubated at 42°C for 60 min. The pfcrt gene was amplified using primers located in the 5′ untranslated region (5′ UTR; CCGTTAATAATAAATACACGCAG) and the 3′ UTR (ATTCCTTATAAAGTGTAATGCGA) of the pfcrt gene (63). PCR was performed in 25 μl, including 12.5 μl of PrimeSTAR Max Premix (2×) (TaKaRa PrimeSTAR Max DNA polymerase), 1 μl of each primer (0.1 μM), and 2 μl of cDNA. PCR conditions were initial denaturing at 95°C for 3 min, followed by 30 cycles of 95°C for 10 s, 55°C for 15 s, and 72°C for 1 min. PCR products were sequenced in both directions using the dideoxy method, and sequences were aligned to the reference gene in 3D7 using ClustalW to identify the single nucleotide polymorphisms.
Statistical analysis.
To calculate IC50 and IC90 values, drug response data in the PPQ concentration range of 0.03 to 100 nM were fit to a sigmoid curve after removing the outliers (64). Outliers were determined using Grubb’s test as implemented in GraphPad Prism 6.0. If a parasite isolate had 3 or more of the 10 data values in the 0.1 to 100 μM PPQ range exceeding 3 × standard deviations of the baseline value (based on the value at 100 μM PPQ, approximating 100% growth inhibition), it was considered to have a bimodal PPQ response curve. The area under the curve (AUC) for the drug response curves at 100 nM to 100 μM was calculated using GraphPad Prism 6.0. Statistical analyses were performed using GraphPad Prism 6.0. Medians and IQRs were calculated, given that the data were not normally distributed. IC50s and PSA survival rates between the groups were compared by the Kruskal-Wallis test. Correlations were determined using the Pearson’s test. For the grouping of samples, we used an unsupervised machine-learning algorithm K-means to cluster the AUC, PSA, IC50, and IC90 values of all the samples after the data were normalized by their z-scores. All possible pairs of AUC, PSA, IC50, and IC90 values were plotted in scatterplots, and the regression lines were built using a least-squares approach. To identify the mutation markers associated with PPQ susceptibility data, the Lasso regression, a widely used method with proven better performance than linear regression and stepwise regression (65), was used first to analyze the 30 mutation markers in 120 samples. Each model was trained by 10-fold cross-validation, and the final parameters are listed in Table S1 in the supplemental material. The shrunk coefficient returned from Lasso regression usually indicates the association between a mutation marker and PPQ susceptibility. For further validation, a Wilcoxon test was performed. In addition, the predominant mutations in PfK13 or all propeller domain mutations combined, as well as the major haplotypes of pfmdr1, were analyzed by the Mann-Whitney U test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shiling Xu and Jinting Geng for their assistance with the drug assays. L.C. received funding (U19 AI089672) from the National Institute for Allergy and Infectious Diseases, The National Institutes of Health, USA. Z.Y. was supported by the National Science Foundation of China (31860604 and U1802286), International Science and Technology Cooperation Yunnan (202003AE140004), and the Major Science and Technology Project of Yunnan Province (2018ZF0081).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320–323. 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 2.Wellems TE, Plowe CV. 2001. Chloroquine-resistant malaria. J Infect Dis 184:770–776. 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 3.Trape JF, Pison G, Preziosi MP, Enel C, Desgrees Du Lou A, Delaunay V, Samb B, Lagarde E, Molez JF, Simondon F. 1998. Impact of chloroquine resistance on malaria mortality. C R Hebd Seances Acad Sci III 321:689–697. 10.1016/s0764-4469(98)80009-7. [DOI] [PubMed] [Google Scholar]
- 4.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 5.Ashley EA, White NJ. 2005. Artemisinin-based combinations. Curr Opin Infect Dis 18:531–536. 10.1097/01.qco.0000186848.46417.6c. [DOI] [PubMed] [Google Scholar]
- 6.Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, Battle KE, Moyes CL, Henry A, Eckhoff PA, Wenger EA, Briet O, Penny MA, Smith TA, Bennett A, Yukich J, Eisele TP, Griffin JT, Fergus CA, Lynch M, Lindgren F, Cohen JM, Murray CL, Smith DL, Hay SI, Cibulskis RE, Gething PW. 2015. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526:207–211. 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaratunga C, Sreng S, Suon S, Phelps ES, Stepniewska K, Lim P, Zhou C, Mao S, Anderson JM, Lindegardh N, Jiang H, Song J, Su XZ, White NJ, Dondorp AM, Anderson TJ, Fay MP, Mu J, Duong S, Fairhurst RM. 2012. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect Dis 12:851–858. 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium . 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359:2619–2620. 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 10.Hien TT, Thuy-Nhien NT, Phu NH, Boni MF, Thanh NV, Nha-Ca NT, Thai Le H, Thai CQ, Toi PV, Thuan PD, Long Le T, Dong Le T, Merson L, Dolecek C, Stepniewska K, Ringwald P, White NJ, Farrar J, Wolbers M. 2012. In vivo susceptibility of Plasmodium falciparum to artesunate in Binh Phuoc Province, Vietnam. Malar J 11:355. 10.1186/1475-2875-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustos MD, Wongsrichanalai C, Delacollette C, Burkholder B. 2013. Monitoring antimalarial drug efficacy in the Greater Mekong Subregion: an overview of in vivo results from 2008 to 2010. Southeast Asian J Trop Med Public Health 44(Suppl 1):201–230. [PubMed] [Google Scholar]
- 12.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M. et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, Ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NP, White NJ, Anderson TJ, Nosten F. 2012. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379:1960–1966. 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyaw MP, Nyunt MH, Chit K, Aye MM, Aye KH, Aye MM, Lindegardh N, Tarning J, Imwong M, Jacob CG, Rasmussen C, Perin J, Ringwald P, Nyunt MM. 2013. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One 8:e57689. 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, Nyunt MM, Adams M, Zhou S, Xia Z, Ringwald P, Bustos MD, Tang L, Plowe CV. 2015. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis 212:1629–1635. 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Nhien NT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercereau-Puijalon O, Menard D, Newton PN, Khanthavong M, Hongvanthong B, Starzengruber P, Fuehrer HP, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MH, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. 2015. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 211:670–679. 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wongsrichanalai C, Meshnick SR. 2008. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis 14:716–719. 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. 2016. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 16:357–365. 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders DL, Vanachayangkul P, Lon C, Royal Cambodian Armed Forces . 2014. Dihydroartemisinin-piperaquine failure in Cambodia. N Engl J Med 371:484–485. 10.1056/NEJMc1403007. [DOI] [PubMed] [Google Scholar]
- 20.Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, Kim S, Witkowski B, Duru V, Domergue A, Khim N, Ringwald P, Menard D. 2015. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in Western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother 59:4719–4726. 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-Ngam P, Kuntawunginn W, Arsanok M, Buathong N, Chaorattanakawee S, Gosi P, Ta-Aksorn W, Chanarat N, Sundrakes S, Kong N, Heng TK, Nou S, Teja-Isavadharm P, Pichyangkul S, Phann ST, Balasubramanian S, Juliano JJ, Meshnick SR, Chour CM, Prom S, Lanteri CA, Lon C, Saunders DL. 2015. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 15:683–691. 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 22.van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, Jittamala P, Hanboonkunupakarn B, Chutasmit K, Saelow C, Runjarern R, Kaewmok W, Tripura R, Peto TJ, Yok S, Suon S, Sreng S, Mao S, Oun S, Yen S, Amaratunga C, Lek D, Huy R, Dhorda M, Chotivanich K, Ashley EA, Mukaka M, Waithira N, Cheah PY, Maude RJ, Amato R, Pearson RD, Goncalves S, Jacob CG, Hamilton WL, Fairhurst RM, Tarning J, Winterberg M, Kwiatkowski DP, Pukrittayakamee S, Hien TT, Day NP, Miotto O, White NJ, Dondorp AM. 2019. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis 19:952–961. 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Pluijm RW, Tripura R, Hoglund RM, Pyae Phyo A, Lek D, Ul Islam A, Anvikar AR, Satpathi P, Satpathi S, Behera PK, Tripura A, Baidya S, Onyamboko M, Chau NH, Sovann Y, Suon S, Sreng S, Mao S, Oun S, Yen S, Amaratunga C, Chutasmit K, Saelow C, Runcharern R, Kaewmok W, Hoa NT, Thanh NV. et al. 2020. Triple artemisinin-based combination therapies versus artemisinin-based combination therapies for uncomplicated Plasmodium falciparum malaria: a multicentre, open-label, randomised clinical trial. Lancet 395:1345–1360. 10.1016/S0140-6736(20)30552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Duong S, Nguon C, Chuor CM, Saunders D, Se Y, Lon C, Fukuda MM, Amenga-Etego L, Hodgson AV, Asoala V, Imwong M, Takala-Harrison S, Nosten F. et al. 2013. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45:648–655. 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng W, Bai Y, Wang M, Wang Z, Deng S, Ruan Y, Feng S, Yang Z, Cui L. 2017. Significant divergence in sensitivity to antimalarial drugs between neighboring Plasmodium falciparum populations along the eastern border of Myanmar. Antimicrob Agents Chemother 61:e01689-16. 10.1128/AAC.01689-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyunt MH, Hlaing T, Oo HW, Tin-Oo LL, Phway HP, Wang B, Zaw NN, Han SS, Tun T, San KK, Kyaw MP, Han ET. 2015. Molecular assessment of artemisinin resistance markers, polymorphisms in the k13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis 60:1208–1215. 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- 28.Menard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen JH, Collet L, Cui L, Thakur GD, Dieye A, Djalle D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino FE, Fandeur T, Ferreira-da-Cruz MF. et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Shrestha S, Li X, Miao J, Yuan L, Cabrera M, Grube C, Yang Z, Cui L. 2015. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007–2012. Malar J 14:168. 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, Dhorda M, Cheah PY, Pukrittayakamee S, Ashley EA, Anderson TJ, Nair S, McDew-White M, Flegg JA, Grist EP, Guerin P, Maude RJ, Smithuis F, Dondorp AM, Day NP, Nosten F, White NJ, Woodrow CJ. 2015. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis 15:415–421. 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putaporntip C, Kuamsab N, Kosuwin R, Tantiwattanasub W, Vejakama P, Sueblinvong T, Seethamchai S, Jongwutiwes S, Hughes AL. 2016. Natural selection of K13 mutants of Plasmodium falciparum in response to artemisinin combination therapies in Thailand. Clin Microbiol Infect 22:285 e281–285 e288. [DOI] [PubMed] [Google Scholar]
- 32.Ye R, Hu D, Zhang Y, Huang Y, Sun X, Wang J, Chen X, Zhou H, Zhang D, Mungthin M, Pan W. 2016. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci Rep 6:20100. 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, Kachur PS, Wongsrichanalai C, Satimai W, Barnwell JW, Udhayakumar V. 2015. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog 11:e1004789. 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parobek CM, Parr JB, Brazeau NF, Lon C, Chaorattanakawee S, Gosi P, Barnett EJ, Norris LD, Meshnick SR, Spring MD, Lanteri CA, Bailey JA, Saunders DL, Lin JT, Juliano JJ. 2017. Partner-drug resistance and population substructuring of artemisinin-resistant Plasmodium falciparum in Cambodia. Genome Biol Evol 9:1673–1686. 10.1093/gbe/evx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17:164–173. 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, Eam R, Khean C, Ken M, Loch K, Bouillon A, Domergue A, Ma L, Bouchier C, Leang R, Huy R, Nuel G, Barale JC, Legrand E, Ringwald P, Fidock DA, Mercereau-Puijalon O, Ariey F, Menard D. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 17:174–183. 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, Smithuis FM, Hlaing TM, Tun KM, van der Pluijm RW, Tripura R, Miotto O, Menard D, Dhorda M, Day NPJ, White NJ, Dondorp AM. 2017. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis 17:491–497. 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee A, Gagnon D, Wirth DF, Richard D. 2018. Inactivation of plasmepsins 2 and 3 sensitizes Plasmodium falciparum to the antimalarial drug piperaquine. Antimicrob Agents Chemother 62:e02309-17. 10.1128/AAC.02309-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loesbanluechai D, Kotanan N, de Cozar C, Kochakarn T, Ansbro MR, Chotivanich K, White NJ, Wilairat P, Lee MCS, Gamo FJ, Sanz LM, Chookajorn T, Kumpornsin K. 2019. Overexpression of plasmepsin II and plasmepsin III does not directly cause reduction in Plasmodium falciparum sensitivity to artesunate, chloroquine and piperaquine. Int J Parasitol Drugs Drug Resist 9:16–22. 10.1016/j.ijpddr.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warhurst DC, Craig JC, Adagu IS, Guy RK, Madrid PB, Fivelman QL. 2007. Activity of piperaquine and other 4-aminoquinoline antiplasmodial drugs against chloroquine-sensitive and resistant blood-stages of Plasmodium falciparum: role of beta-haematin inhibition and drug concentration in vacuolar water- and lipid-phases. Biochem Pharmacol 73:1910–1926. 10.1016/j.bcp.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Preechapornkul P, Imwong M, Chotivanich K, Pongtavornpinyo W, Dondorp AM, Day NP, White NJ, Pukrittayakamee S. 2009. Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. AAC 53:1509–1515. 10.1128/AAC.00241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, Ke S, Chy S, Eam R, Khean C, Loch K, Ken M, Lek D, Beghain J, Ariey F, Guerin PJ, Huy R, Mercereau-Puijalon O, Witkowski B, Menard D. 2015. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med 13:305. 10.1186/s12916-015-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kumpornsin K, Takala-Harrison S, Witkowski B, Fairhurst RM, Ariey F, Menard D, Fidock DA. 2018. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 9:3314. 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal S, Moser KA, Morton L, Cummings MP, Parihar A, Dwivedi A, Shetty AC, Drabek EF, Jacob CG, Henrich PP, Parobek CM, Jongsakul K, Huy R, Spring MD, Lanteri CA, Chaorattanakawee S, Lon C, Fukuda MM, Saunders DL, Fidock DA, Lin JT, Juliano JJ, Plowe CV, Silva JC, Takala-Harrison S. 2017. Association of a novel mutation in the Plasmodium falciparum chloroquine resistance transporter with decreased piperaquine sensitivity. J Infect Dis 216:468–476. 10.1093/infdis/jix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, Dhorda M, Amaratunga C, Woodrow CJ, Ashley EA, White NJ, Dondorp AM, Fairhurst RM, Ariey F, Menard D, Wirth DF, Volkman SK. 2018. Plasmepsin II-III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun 9:1769. 10.1038/s41467-018-04104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis TM, Hung TY, Sim IK, Karunajeewa HA, Ilett KF. 2005. Piperaquine: a resurgent antimalarial drug. Drugs 65:75–87. 10.2165/00003495-200565010-00004. [DOI] [PubMed] [Google Scholar]
- 47.Wu Z. 1985. [Resistance to piperaquine phosphate in Hainan islanders with pernicious malaria]. Zhonghua Yi Xue Za Zhi 65:483–484. [PubMed] [Google Scholar]
- 48.Huang JZ, Lan XH, Xu WZ. 1985. Sensitivity of Plasmodium falciparum to piperaquine in Baoting County, Hainan Island. Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 3:276–277. (In Chinese.) [PubMed] [Google Scholar]
- 49.Lan CX, Lin X, Huang ZS, Chen YS, Guo RN. 1989. In vivo sensitivity of Plasmodium falciparum to piperaquine phosphate assayed in Linshui and Baisha counties, Hainan Province. (In Chinese.) Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 7:163–165. [PubMed] [Google Scholar]
- 50.Guo XB. 1993. Randomized comparison on the treatment of falciparum malaria with dihydroartemisinin and piperaquine. Zhonghua Yi Xue Za Zhi 73:602–604, 638. []. [PubMed] [Google Scholar]
- 51.Wang Y, Yang Z, Yuan L, Zhou G, Parker D, Lee MC, Yan G, Fan Q, Xiao Y, Cao Y, Cui L. 2015. Clinical efficacy of dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria at the China-Myanmar border. Am J Trop Med Hyg 93:577–583. 10.4269/ajtmh.15-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H, Yang HL, Tang LH, Li XL, Huang F, Wang JZ, Li CF, Wang HY, Nie RH, Guo XR, Lin YX, Li M, Wang J, Xu JW. 2015. In vivo monitoring of dihydroartemisinin-piperaquine sensitivity in Plasmodium falciparum along the China-Myanmar border of Yunnan Province, China, from 2007 to 2013. Malar J 14:47. 10.1186/s12936-015-0584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaorattanakawee S, Lon C, Jongsakul K, Gawee J, Sok S, Sundrakes S, Kong N, Thamnurak C, Chann S, Chattrakarn S, Praditpol C, Buathong N, Uthaimongkol N, Smith P, Sirisopana N, Huy R, Prom S, Fukuda MM, Bethell D, Walsh DS, Lanteri C, Saunders D. 2016. Ex vivo piperaquine resistance developed rapidly in Plasmodium falciparum isolates in northern Cambodia compared to Thailand. Malar J 15:519. 10.1186/s12936-016-1569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in vitro and ex vivo drug-response studies. Lancet Infect Dis 13:1043–1049. 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Li N, Siddiqui FA, Xu S, Geng J, Zhang J, He X, Zhao L, Pi L, Zhang Y, Li C, Chen X, Wu Y, Miao J, Cao Y, Cui L, Yang Z. 2019. In vitro susceptibility of Plasmodium falciparum isolates from the China-Myanmar border area to artemisinins and correlation with K13 mutations. Int J Parasitol Drugs Drug Resist 10:20–27. 10.1016/j.ijpddr.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng H, Zhang R, Yang H, Fan Q, Su X, Miao J, Cui L, Yang Z. 2010. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob Agents Chemother 54:4306–4313. 10.1128/AAC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamilton WL, Amato R, van der Pluijm RW, Jacob CG, Quang HH, Thuy-Nhien NT, Hien TT, Hongvanthong B, Chindavongsa K, Mayxay M, Huy R, Leang R, Huch C, Dysoley L, Amaratunga C, Suon S, Fairhurst RM, Tripura R, Peto TJ, Sovann Y, Jittamala P, Hanboonkunupakarn B, Pukrittayakamee S, Chau NH, Imwong M, Dhorda M, Vongpromek R, Chan XHS, Maude RJ, Pearson RD, Nguyen T, Rockett K, Drury E, Goncalves S, White NJ, Day NP, Kwiatkowski DP, Dondorp AM, Miotto O. 2019. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis 19:943–951. 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siddiqui FA, Boonhok R, Cabrera M, Mbenda HGN, Wang M, Min H, Liang X, Qin J, Zhu X, Miao J, Cao Y, Cui L. 2020. Role of Plasmodium falciparum Kelch 13 protein mutations in Plasmodium falciparum populations from northeastern Myanmar in mediating artemisinin resistance. mBio 11:e01134-19. 10.1128/mBio.01134-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65:418–420. 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- 60.Hao M, Jia D, Li Q, He Y, Yuan L, Xu S, Chen K, Wu J, Shen L, Sun L, Zhao H, Yang Z, Cui L. 2013. In vitro sensitivities of Plasmodium falciparum isolates from the China-Myanmar border to piperaquine and association with polymorphisms in candidate genes. Antimicrob Agents Chemother 57:1723–1729. 10.1128/AAC.02306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai Y, Zhang J, Geng J, Xu S, Deng S, Zeng W, Wang Z, Ngassa Mbenda HG, Zhang J, Li N, Wu Y, Li C, Liu H, Ruan Y, Cao Y, Yang Z, Cui L. 2018. Longitudinal surveillance of drug resistance in Plasmodium falciparum isolates from the China-Myanmar border reveals persistent circulation of multidrug resistant parasites. Int J Parasitol Drugs Drug Resist 8:320–328. 10.1016/j.ijpddr.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S, Xu S, Geng J, Si Y, Zhao H, Li X, Yang Q, Zeng W, Xiang Z, Chen X, Zhang Y, Li C, Kyaw MP, Cui L, Yang Z. 2020. Molecular surveillance and in vitro drug sensitivity study of Plasmodium falciparum isolates from the China-Myanmar border. Am J Trop Med Hyg 103:1100–1106. 10.4269/ajtmh.20-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gadalla NB, Malmberg M, Adam I, Oguike MC, Beshir K, Elzaki SE, Mukhtar I, Gadalla AA, Warhurst DC, Ngasala B, Martensson A, El-Sayed BB, Gil JP, Sutherland CJ. 2015. Alternatively spliced transcripts and novel pseudogenes of the Plasmodium falciparum resistance-associated locus pfcrt detected in East African malaria patients. J Antimicrob Chemother 70:116–123. 10.1093/jac/dku358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaorattanakawee S, Saunders DL, Sea D, Chanarat N, Yingyuen K, Sundrakes S, Saingam P, Buathong N, Sriwichai S, Chann S, Se Y, Yom Y, Heng TK, Kong N, Kuntawunginn W, Tangthongchaiwiriya K, Jacob C, Takala-Harrison S, Plowe C, Lin JT, Chuor CM, Prom S, Tyner SD, Gosi P, Teja-Isavadharm P, Lon C, Lanteri CA. 2015. Ex vivo drug susceptibility testing and molecular profiling of clinical Plasmodium falciparum isolates from Cambodia from 2008 to 2013 suggest emerging piperaquine resistance. Antimicrob Agents Chemother 59:4631–4643. 10.1128/AAC.00366-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayers KL, Cordell HJ. 2010. SNP selection in genome-wide and candidate gene studies via penalized logistic regression. Genet Epidemiol 34:879–891. 10.1002/gepi.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004. Simple and inexpensive fluorescence-based technique for high-through put antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806. 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.