Abstract
It is well known that cell surface immune receptors play a critical role in regulating immune and inflammatory processes in the central nervous system (CNS). We have analyzed the function of cluster of differentiation (CD)300f immunoreceptor in a model of excitotoxic rat brain damage. First, to explore the presence of endogenous ligand(s) for this receptor we used a human CD300f‐Ig soluble protein and confocal microscopy, showing specific staining mainly in CNS white matter and on the surface of oligodendrocytes and certain astrocytes. Next, we demonstrated in a model of in vivo rat brain excitotoxic damage that the overexpression of human CD300f induced a significant reduction in the lesion volume. To validate these results, we cloned the rat ortholog of CD300f protein (rCD300f). The overexpression of rCD300f receptor had a comparable neuroprotective effect after the acute brain injury and a similar CNS staining pattern when stained with the rCD300f‐Ig soluble protein. Interestingly, when we analyzed the expression pattern of rCD300f in brain cells by quantitative polymerase chain reaction and immunohistochemistry, we detected the expression of CD300f as expected in microglial cells, but also in oligodendrocytes and neurons. These data suggest that the neuroprotective role of CD300f would be the result of a complex network of cell interactions.
Keywords: excitotoxicity, inflammation, microglia, neuroprotection, nonviral gene therapy, oligodendrocyte
INTRODUCTION
The balance of activating and inhibitory signaling produced by cell surface immune receptors contributes to determine the type and duration of inflammatory processes 18, 19, 36. In the central nervous system (CNS), immune and inflammatory reactions are involved in initiating, propagating and terminating different types of chronic and acute injuries 9, 14. It has been described that some immune receptors play a role in the maintenance of CNS homeostasis, and also participate in CNS injuries. For instance, CD200 and CD200R expressed on neurons and microglia, respectively, induce “resting” signals on microglial cells (6). Another interesting example is triggering receptor expressed on myeloid cells 2 (TREM2), which has a neuroprotective role in the CNS. The use of blocking antibodies against TREM2 in mice with experimental autoimmune encephalitis (EAE) enhances the disease, whereas the presence of myeloid cells expressing TREM2 reduces inflammation 26, 31.
CD300 immune receptors are encoded by a cluster of genes in the human chromosome 17q25.1 12, 35. The human CD300 (hCD300) family includes six molecules, expressed exclusively by cells of the myeloid lineage, with the exception of CD300a that is additionally present on different subsets of lymphoid cells (T, B and NK) (8). CD300 receptors present an extracellular V‐type immunoglobulin‐like domain with an additional pair of cystein residues, a transmembrane region and a cytoplasmic tail. Whereas CD300a (IRp60) acts exclusively as an inhibitory receptor 5, 8, 25, CD300f presents a putative functional duality. In addition to its expected inhibitory function (2), it has been recently shown to be able to deliver activating signals, although the recruitment of PI3‐Kinase and FcεRγ3, 15. Two activating receptors, CD300b and CD300e, present a transmembrane region bearing a positively charged residue that allows the recruitment of immunoreceptor tyrosine‐based activation motif (ITAM)‐bearing adaptors that contain a glutamic acid within their transmembrane sequence 1, 21. Finally, both CD300c (CMRF‐35) (16), and CD300d receptors combine a transmembrane region containing a negatively charged residue with a short cytoplasmic tail devoid of known signaling motifs. Whereas no information about the function of CD300d is available, we have shown that CD300c is a functional activating immune receptor (22). An interesting feature of this family of receptors is that CD300 members bind together extracellularly forming homo‐ and hetero‐signaling complexes (22).
At the present, few data exist regarding the roles of the human CD300 family in vivo. Very recently, it has been shown that the knockout mice for CD300f (mCD300f/CLM1) exhibits an increased neuropathology in response to the induction of EAE. Accordingly, a worsened phenotype was also noted when these authors administered a soluble mCD300f‐IgG fusion protein to wild type animals (38). Here, we aimed to characterize the presence of the unknown ligand(s) of CD300f in the CNS, the expression of CD300f in CNS cells, and the possible neuroprotective role of CD300f after an acute brain injury.
MATERIALS AND METHODS
Cloning and DNA constructs
We used a nested polymerase chain reaction (PCR) strategy to amplify rCD300f using as a template cDNA obtained by retrotranscription from total RNA from rat bone marrow or rat basophilic leukemia (RBL‐2H3) cells. PCR conditions were as follows: 94°C for 3 minutes and 30 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 2 minutes using Pfu DNA polymerase (Promega, Madison, WI, USA). Primers used in the first reaction were 5′ AAG GTG ATC CGG TGA GAG AA 3′ and 5′ CAA GGC TGA ACT CCA AGC TC 3′. For the second reaction, we used the same forward primer and the reverse primer 5′ GGT AAG GAA GCA GGC ACA GA 3′. PCR products were resolved in 1% agarose gels and visualized by ethidium bromide staining. Amplified fragments were cloned into pCDNA3.1‐TOPO vector (Invitrogen, San Diego, CA, USA) and sequenced under Big DyeTM cycling conditions on an Applied Biosystems 3730xl DNA Analyzer (Macrogen Inc., No. Macrogen Corp. Europe, Amsterdam, Netherlands). Rat CD300f extracellular domain (without signal peptide) was amplified by PCR and cloned into the BglII/SalI sites of pDisplay (Invitrogen) with primers 5′ AGA TCT TTC ACT GCT CAG GAT CCA GTC 3′ and 5′ GCC GTC GAC TCA AGG CAT GGG CCT CCT GAT 3′. Rat CD300f Sv1 extracellular domain was amplified by PCR with the pair of primers 5′GCC AAG CTT TGA TCC AGT CAC AGG TCC A 3′ and 5′GCC GGA TCC ACA TCC AGA AAC CCA TTA CC 3′ and cloned in the cassette pSecTag/mIgG2a (1) between HindIII and BamHI restriction sites. Mammalian pDisplay/hCD300f and pSecTag/hCD300f‐mIgG2a constructs were described before (2).
Real‐time PCR
RNA from cells was extracted with TRIzol reagent (Invitrogen), treated with DNAse I Amplification Grade (Invitrogen) and retrotranscribed using high‐capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Real‐time PCR was performed using TaqMan gene expression assay for rCD300f (Rn01752052_m1) and 18S amplification control for cycle normalization on an ABI‐Prism 7500 sequence detector (Applied Biosystems). Data were analyzed with 7500 SDS v1.2 Software. All quantitative PCR (Q‐PCR) reactions were set up in triplicates.
Production of hCD300f‐IgG2a and rCD300f‐IgG2a
Chinese hamster ovary (CHO‐K1) cells were stably transfected with pSecTag/mIgG2a constructs and positive cells were selected with 250 µg/mL of Zeocin (Invivogen, San Diego, CA, USA). The chimerical protein was purified from the supernatant using a protein A‐sepharose column (GE Healthcare, Pittsburgh, PA, USA) as described before (1).
Primary cell cultures
Culture media and sera were purchased from Invitrogen and all other reagents were from Sigma–Aldrich (St. Louis, MO, USA) unless otherwise noted. Primary spinal cord and cortex mixed glial cultures and highly enriched astrocyte, oligodendrocyte or microglial cultures were prepared from 1–2‐day‐old Wistar rat pups (both sexes) according to the procedures of Saneto and De Vellis (33) with the modifications of Martínez‐Palma and collaborators (23). For oligodendrocyte differentiation, 48 h after plating, cell medium was changed for a low‐serum Dulbecco's modified Eagle's medium (DMEM) (containing 3.6 g/L N‐2‐hydroxyethylpiperazine‐N′‐2‐ethanesulfonic acid,[HEPES] 100 IU/mL penicillin and 100 mg/mL streptomycin) supplemented with conalbumin (0.1 mg/mL), putrescine (0.1 mM), insulin (5 µg/mL), sodium selenite (31 nM), progesterone (21 nM), 25 mg/mL transferrin, 30 nM tri‐iodothyronine and 2% horse serum. Differentiation was assessed by galactocerebroside‐C (Gal‐C), myelin basic protein (MBP) and glial fibrillary acidic protein (GFAP) immunoreactivity (95% purity). Only 0.5%–1% of the culture was positive for the CD11b microglial marker. Purified cortical and hippocampal neuron cultures were prepared from E18 Wistar rat embryos according to Peluffo and collaborators (30).
Histological, immunocytochemical and immunohistochemical procedures
Three days after the lesion was performed, rats were anesthetized and perfused intracardially with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). Brains were post‐fixed (2 h), cryoprotected in 30% sucrose, and frozen with dry CO2. Parallel cryostat coronal sections (32 µm) of the entire brain were used. Sections and cultured cells were processed for astroglial labeling by using rat anti‐GFAP (1:1800; Dakopatts‐Z0334; Dakopatts, Glostrup, Denmark), for microglial labeling by using mouse anti‐rat CD11b (1:50, MCA275G, AbD Serotec, Raleigh, NC, USA) and goat anti‐Iba1 (1:200, ab5076, abcam, Cambridge, MA, USA) , for oligodendroglial labeling by using anti‐2′,3′‐cyclic nucleotide 3′‐phosphohydrolase (CNPase) (1:50, HPA023266, Sigma‐Aldrich), anti‐GalC (1:150, G9152, Sigma‐Aldrich), mouse anti‐MBP (1:300, Oncogene, Cambridge, MA, USA) and mouse anti‐adenomatous polyposis coli (APC) (1:200, OP80 CC‐1, EMD4‐Bioscience‐Calbiochem, Gibbstown, NJ, USA), or for neuronal labeling by using mouse anti‐β‐3‐tubulin (1:3000, Promega‐G7121). For detection of CD300f expression, we used the following primary antibodies: rat anti‐CD300f (1:50, 132701, BioLegend, San Diego, CA, USA), mouse anti‐mouse myeloid‐associated immunoglobulin‐like receptor five (MAIRV)/CMRF‐35‐like molecule 1 (CLM1) (1:20, MAB27881 R&D Systems, Minneapolis, MN, USA). As a control negative staining for CD300f transfectants, the rat anti‐mCD300b/CLM7 antibody (MAB2580, R&D Systems) was used. Alexa‐labeled biotin or appropriate secondary antibodies (Invitrogen) were used to detect specific staining of primary antibodies and fusion proteins. Controls were made to rule out nonspecific staining by incubation without primary antibody, and in the case of double immunofluorescence with two mouse primary antibodies, the second primary antibody was omitted. Microglia/macrophages and blood vessels were also demonstrated using biotinylated Lycopersicon esculentum (tomato) lectin (6 µg/mL; L9389; Sigma‐Aldrich).
Animals, in vivo excitotoxic lesions and transgene overexpression
All experimental work was approved by the UDELAR university ethical commission and conducted according to directives of the Federation of Laboratory Animal Science Associations (FELASA). Adult (8–10 weeks) heterozygote yellow fluorescent protein H (YFP‐H) mice [B6.Cg‐Tg (Thy1‐YFPH)2Jrs/Jackson Laboratories, Bar Harbor, ME, USA]s (13) of both sexes were used for co‐localization studies. Adult male Wistar rats (280–300 g) were submitted to excitotoxic lesions by injection of 120 nmol N‐methyl‐D‐aspartate (NMDA, Sigma‐Aldrich) in 1 µL saline solution (0.9% NaCl) into the striatum (coordinates A: +0.5; L: −3.5; V: −4.0) using a stereotaxic frame under isoflurane (Abbott, Abbott Park, IL, USA) anesthesia and a nanoinjector (0.2 µL/minute; Quintessential Stereotaxic Nanoinjector, Stoelting CO. Wood Dale, IL, USA). Four hours later, 5 µL of either vehicle (NaCl 0.9%) or NLSCt vector accommodating an expression plasmid (0.8 µg/µL NLSCt + 0.024 µg/µL plasmid) were injected at 0.5 µL/minute (2.5 µL at the same coordinates as the lesion and 2.5 µL 2 mm above the lesion point) as previously described (28). The needle was left in place for an additional 10 minutes to allow diffusion into the brain parenchyma. The expression plasmids used were pEGFP‐C1 (Clontech, Mountain View, CA, USA), the pDisplay‐hCD300f or the pDisplay‐rCD300f. All transgenes were under the cytomegalovirus (CMV) promoter regulation. In vivo studies were designed to inject six animals and two different treatments in each surgical procedure. Treatments were randomly distributed (17 animals NLSCt/GFP, 17 animals NLSCt/hCD300f and 12 animals NLSCt/rCD300f). Parallel coronal cryostat sections (32 µm thick) of the entire brain, separated by 352 µm (every 12th section) were stained for Nissl and used for the quantification of the lesioned area and the total area of the hemisphere. Quantification of the lesioned Nissl pale area was performed using the Image J 44 software (National Institutes of Health, USA, http://rsbweb.nih.gov/ij/index.html) with parallel microscope observation (using 100× and 200× magnification) by two independent researchers blinded for the treatment administration as described previously (29). The lesion area and the total lesioned hemisphere area were used to calculate the lesion volume and the total hemisphere volume, and data were expressed as “% of lesioned hemisphere” to correct for the possible edema effect.
Cell transefection
In the presence of 20 µg of pDisplay/rCD300f linearized construct, 20 × 106 RBL‐2H3 cells were electroporated at 250 V, 960 µF and 100 Ω in a Gene Pulser Electroporator (Bio‐Rad, Hercules, CA, USA). Transfectant was selected and maintained in culture with G418 (BioWhittaker, Walkersville, MO, USA). Positive cells were further selected by immunostaining with the appropriate antibodies and sorting with magnetic Dynabeads® M‐450 coated with Sheep anti‐Mouse IgG (Invitrogen).
Flow cytometry
Cell surface expression of the desired molecules was tested by indirect immunofluorescence following standard techniques. Analysis was performed using a FACScalibur instrument and CellQuest software (BD Biosciences, Franklin Lakes, NJ, USA).
Data processing and statistical analysis
All results are expressed as mean ± standard error of the mean (SEM). Student t‐test was used to determine significant differences (P < 0.05) between two experimental groups after determination of the normal distribution of the ssamples and variance analysis. One‐way analysis of variance (ANOVA) followed by Tukey's multiple comparison test was used to determine significant differences (P < 0.05) for experimental settings with more than two experimental groups.
RESULTS
Expression of hCD300f ligand(s) in vitro and in vivo
Because of the elevated homology among the extracellular domains of human, mouse and rat CD300f, we tested if hCD300f would be able to recognize the ligand for mouse and rat CD300f. We performed immunocytochemical studies using a soluble fusion protein containing the extracellular domain of hCD300f fused to the Fc region of the IgG2a mouse heavy chain. We analyzed both, CNS primary glial cultures and rat and mouse brains. In culture, we found that oligodendrocytes and a subset of GFAP positive astrocytes showed in their surface a positive punctate pattern for hCD300f‐IgG2a fusion protein (Figure 1A–D). In cell cultures, two populations of astrocytes can be observed by morphological criteria, fibrous star‐shaped cells and protoplasmic polygonal cells (10). Only the subset of fibrous astrocytes were positive for the fusion protein staining (Figure 1D). This staining was not observed in control conditions consisting in incubation with an identical concentration of mouse IgG2a (not shown). Regarding oligodendrocytes, both small undifferentiated cells (GalC+/MBP−, Figure 1A) and large highly ramified differentiated cells (GalC+/MBP+, Figure 1B,C) were positive for this staining. Interestingly, polygonal GFAP positive cells were always negative for the staining (Figure 1D), whereas two populations of fibrous GFAP‐positive cells were observed, some of which were positive for the ligand(s), and some of which were negative. Microglial cells were negative for the staining with hCD300f‐IgG2a (Figure 1E).
Figure 1.

Localization of the putative ligand(s) of hCD300f in vitro and in vivo. Primary cultures of the different glial central nervous system (CNS) cell types (A–E) or sections from mouse and rat brain (mouse brain, F–I) were used for co‐localization studies of hCD300f‐IgG2a (10 µg/mL) fusion protein and different cell‐specific markers. Small undifferentiated oligodendrocytes with few ramifications (A, GalC) and large highly ramified differentiated oligodendrocytes (B, GalC; C, myelin basic protein [MBP]) showed specific punctate staining with the fusion protein. Glial fibrillary acidic protein‐(GFAP) positive polygonal protoplasmic astrocytes did not show specific staining with the fusion proteins (D, arrow), being only positive the ramified fibrous GFAP‐stained cells. Microglial cells were negative for the staining with the fusion protein (E). A similar punctuate staining pattern was observed by confocal microscopy in the white matter of the brain stained with the fusion protein, which did not co‐localized with myelin markers (stained with CNPase, F, corpus callosum) but co‐localized with the oligodendrocyte soma and proximal projections (stained with APC, arrows in G–I). Scale bars: F–I 10 µm.
In vivo, rat and mouse brains also showed a punctate pattern of staining when incubated with hCD300f‐IgG2a, which was mainly present in the brain white matter (1, 2). Co‐localization studies by confocal microscopy showed that the punctate staining did not co‐localized with myelin markers such as CNPase (Figure 1F) or MBP (not shown). Contrary, the signal co‐localized with the oligodendrocyte marker APC (Figure 1G–I), which stains exclusively the cell soma and proximal projections but not the myelin sheet.
Figure 2.
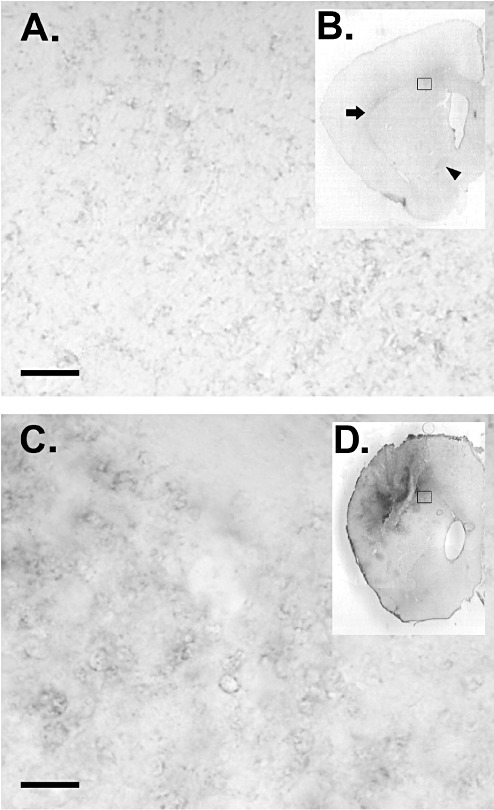
Increased staining for the ligand(s) of CD300f in lesioned rat brain. Normal brain (A,B) showed a punctate staining with hCD300f‐IgG2a fusion protein mainly in white matter areas like corpus callosum, external capsule (B: arrow) or the anterior commisure (B: arrowhead). Brains subjected to intra‐striatal injection of N‐methyl‐D‐aspartate (NMDA) showed an increased hCD300f‐IgG2a staining in the lesion core (C,D) in comparison with the noninjured brain (A,B). Representative higher magnification images from the sections in B and D (open squares) are shown in A and C. No staining was observed when sections were incubated with control mouse IgG2a. Scale bar: 10 µm.
Considering the presence of CD300f ligand(s) in rat and mouse brains, we wondered whether the staining observed was increased after an acute lesion. We performed a well‐established model of excitotoxic injury by intra‐striatal injection of NMDA in rats (11). Interestingly, 3 days after the lesion, the staining with hCD300f‐IgG2a appeared more intense in the lesioned area than in the rest of the brain (Figure 2). This result suggested that CD300f would be involved in CNS inflammatory processes.
Overexpression of hCD300f reduce the lesion volume after an acute brain injury
In order to approach the in vivo function of hCD300f, we induced an acute excitotoxic injury in rats by injecting NMDA in the striatum and cortex. Four hours later, we introduced at the same location a modular recombinant gene therapy vector bound to plasmids encoding for hCD300f or EGFP. The chosen vector, termed NLSCt, was previously described to induce the transient expression of transgenes under similar experimental conditions 28, 29. The NMDA injection induced a well‐delimitated lesion both in the striatum and the surrounding dorsal cortex. Interestingly, the delayed overexpression of hCD300f caused a significant reduction (≅40%) in the lesion volume when compared 3 days post‐intervention to the enhanced green fluorescent protein (EGFP) transgene (Figure 3).
Figure 3.

Overexpression of hCD300f is neuroprotective after and acute brain injury. Striatal injection of N‐methyl‐D‐aspartate (NMDA) induced a well‐delimitated lesion in the striatum and the cortex when observed by Nissl staining 3 days post‐intervention (A,B). At this time point, the measurement of the % of lesioned hemisphere (C, black lines in A,B) showed that the overexpression of hCD300f using the NLSCt vector induced a significant neuroprotection (P < 0.05) when compared with the overexpression of a control transgene (EGFP).
Cloning of rat CD300f
In order to further analyze the role of the endogenous CD300f in the rat brain, we cloned the rat ortholog of this molecule. We used the hCD300f sequence to blast the Ensembl rat database (http://www.ensembl.org) and we identified a predicted cDNA homologous to the human sequence. We designed primers and cloned a cDNA using RBL‐2H3 cells RNA as template in a two‐round nested PCR strategy. The nucleotide sequence obtained (GeneBank accession GU057984) was 994 bp in length and contained an open reading frame of 618 nucleotides. The cDNA encoded for a polypeptide of 301 amino acids with a predicted molecular weight of 33.5 kDa (Figure 4A). As expected, the cloned molecule presented a high homology with hCD300f (64% similarity) and conserved the most important residues within the extracellular domain as well as in the cytoplasmic tail (Figure 4B). The conservation of the five tyrosine‐based motifs within the intracellular region strongly suggested that both molecules would share the same signaling mechanisms and consequently a similar function. Indeed, using a three‐hybrid assay in yeast, we found out that rCD300f was able to recruit SH2 domain‐containing protein phosphatase 1 (SHP1), Grb2 and the p85 subunit of PI3‐kinase similarly as described for hCD300f (data not shown) 2, 3.
Figure 4.

Cloning of Rattus Norvegicus CD300f. A. Nucleotide and predicted amino acid sequences of rCD300f (GU057984). The predicted amino acid sequence is shown below the nucleotide sequence. The putative signal peptide is double underlined, the Ig‐like domain is in bold type, the transmembrane domain is underlined (dotted line), and the consensus immunoreceptor tyrosine‐based inhibitory motif (ITIM)‐like sequences are underlined (single line). The N‐glycosylation site is boxed, and cysteine residues involved in the Ig‐like fold are circled. B. Protein sequence alignment of human and rat CD300f. Identical amino acids are shown on black background and similar residues are on gray background.
Additionally, to the full length rCD300f receptor, we have cloned two alternatively spliced forms of rCD300f. The rCD300f Sv1 cDNA was amplified from RNA obtained from rat bone marrow cells, while rCD300f Sv2 was cloned from RNA from the mast cell line RBL‐2H3. The sequence for rCD300Sv1 was almost identical to rCD300f, except for the lack of 20 amino acids in the stem region, located between the Ig extracellular and the transmembrane domains. The rCD300Sv2 cDNA encoded for a putative soluble protein as a result of an exon skipping process in the productive mRNA (not shown).
Expression of rCD300f in CNS cells
Recently, CLM1 was reported to be absent from mouse spinal cord (38). Here, we analyzed the expression of the rat ortholog of this molecule in rat CNS cultures. Highly purified cultures of microglia, astrocytes, oligodendrocytes and neurons were subjected to Q‐PCR. The transcript for rCD300f was detected in microglia, immature oligondendroglia, differentiated oligondendroglia (not shown), and unexpectedly, at a low level in neurons, but not in astrocytes (Figure 5A). Interestingly, the mRNA for rCD300f increased in microglia after a stimulus mimicking an inflammatory process (IL1β + lipopolysaccharide [LPS]). The expression of rCD300f was also confirmed by immunocytochemistry using an anti‐CLM1 antibody. First, we analyzed whether the anti‐mouse CD300f antibody cross‐reacted with the rat molecule. In a flow cytometry assay, only the R&D Systems anti‐CLM1 antibody recognized the endogenous rCD300f expressed on the surface of RBL‐2H3 cells (Figure 5B). Accordingly, in rCD300f‐transfected RBL‐2H3 cells the anti‐CLM1 staining was more intense compared with the parental cell line (Figure 5B). Moreover, cells in both mixed and highly enriched primary cultures of microglial cells (Figure 5C,D), undifferentiated and differentiated oligodendrocytes (Figure 5E,F), and cortical (not shown) or hippocampal neurons (Figure 5G) were immunoreactive to this antibody, whereas astrocytes were not (Figure 5C, arrows). In vivo, we failed to detect positive cells in CNS or spleen cryosections using two different commercial monoclonal antibodies.
Figure 5.
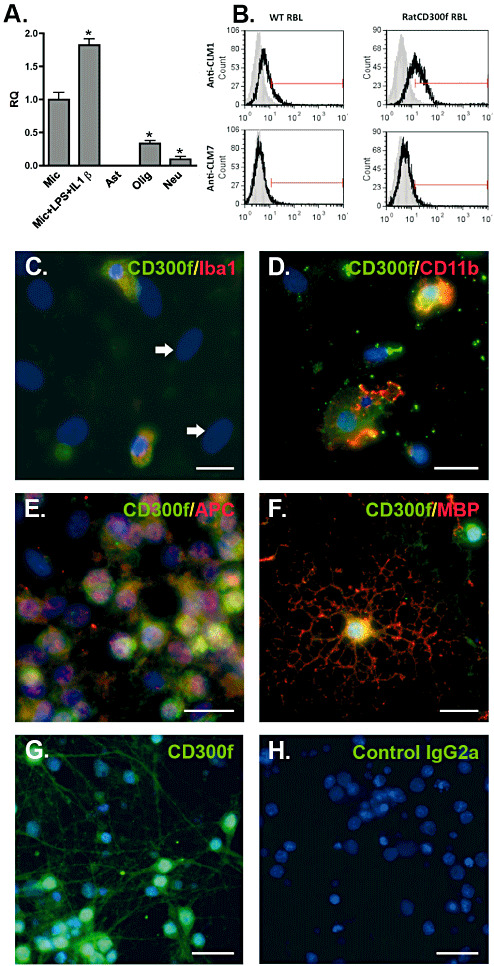
Expression of rCD300f in CNS primary cultures. Real–time polymerase chain reaction (PCR) (A) fluorescence‐activated cell sorting (FACS) (B) and immunocytochemistry (C–H) of different central nervous system (CNS) primary cultures were performed to analyze the expression of rCD300f. Microglial cells (Mic), undifferentiated oligodendrocytes (Olig) and hippocampal neurons (Neu) expressed mRNA for rCD300f while astrocytes (Ast) did not. Treatment of microglia with lipopolysaccharide (LPS) (1 µg/mL) plus IL1β (10 ng/mL) induced an increase in the mRNA for rCD300f (A, P < 0.05). Its expression was also detected in parental and rCD300f transfected RBL‐2H3 cells by FACS staining, whereas staining with an anti‐CLM7 antibody was negative. Isotypic antibody (grey histograms), anti‐CLM1/CLM7 (white histograms) (B). Microglia (stained with C: Iba1 or D: CD11b), differentiated oligodendrocytes (stained with E: GalC or F: myelin basic protein [MBP]) and hippocampal neurons (G) were positive by immunocytochemistry. Astrocytes were negative (big ovoid nuclei, arrows in B). No staining was observed in the absence of primary antibody (H, control IgG2a). Scale bars: 20 µm.
Expression of rCD300f ligand(s) in vitro and in vivo
By generating a rCD300f‐IgG2a fusion protein we investigated the in vitro and in vivo presence of the putative ligand(s) for rCD300f in the CNS. A very similar punctate staining pattern was overall observed between the rCD300f‐IgG2a and the hCD300f‐IgG2a both in vitro and in rat and mouse brains (Figure 6A,B). In culture, besides oligodendrocytes and fibrous GFAP‐positive astrocytes, hippocampal (not shown) and cortical neurons were also positive for the staining with rCD300f‐IgG2a, although the staining was fainter and mainly observed in the cell soma (Figure 6H). In vivo, the staining with rCD300f‐IgG2a was also mainly localized in white matter tracts, and co‐localized with the oligodendrocyte marker APC (Figure 6C–E), but it did not co‐localize with GFAP (not shown), tomato lectin‐positive microglia or blood vessels (Figure 6G). Finally, to evaluate the putative in vivo neuronal expression of rCD300f and hCD300f ligand(s), brain and spinal cord sections from Thy1‐YFP‐H mice were used (13). These mice constitute an excellent model to co‐localize molecules within neuronal cells, as they express constitutively YFP in a small proportion of neurons, allowing an unambiguously visualization of cell somas and their projections. Surprisingly, no co‐localization was observed between neuronal soma, dendrites or axons and the fusion protein rCD300f‐IgG2a in the brain (Figure 6I–K) or in the spinal cord (Figure 7).
Figure 6.

Localization of the putative ligand(s) of rCD300f in vitro and in vivo. Sections from mouse and rat brain were incubated with the fusion proteins rCD300f‐IgG2a for the localization of the putative ligand(s) of CD300f, and confocal co‐localization studies were performed with cell‐specific markers (C–H) or with the Thy1‐YFP‐H mice (I–K). Staining with the fusion proteins was observed mainly in the brain white matter as in the external capsule (A, external capsule (EC); Cx = neocortex; St = striatum; V = ventricle) or the fiber tracts of the striatum (B, St.). The punctuate staining pattern co‐localized with the oligodendrocyte soma and proximal projections (stained with APC, arrows in C–E, corpus callosum) but not with myelin itself (stained with CNPase, F, striatum). No colocalization could be observed with microglia or blood vessels (stained with TL, G). Cortical neuron cultures were positive for rCD300f‐IgG2a staining (stained with β3‐tubulin, H). For extensive in vivo neuronal labeling, Thy1‐YFP mice were used, and no co‐localization was detected between YFP neuronal soma, dendrites, spines or axons and the fusion proteins (I: striatum; J: neocortex, red staining is nonspecific capillaries staining; K: corpus callosum). Scale bars: A,B: 50 µm, C–K: 10 µm.
Figure 7.
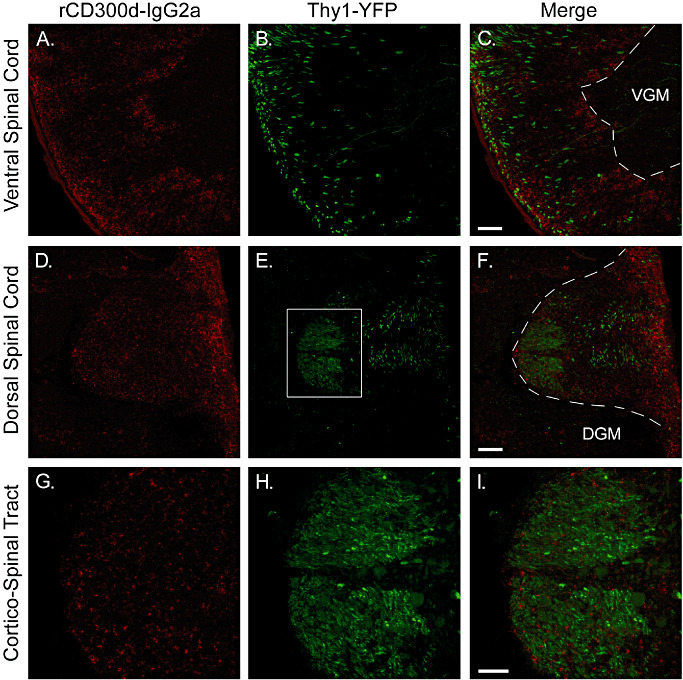
Localization of the putative ligand(s) of rCD300f in spinal cord in vivo. Spinal cord sections from Thy1‐YFP mice were stained with rCD300f‐IgG2a fusion protein, and confocal microscopy was used to visualize specific staining. Corticospinal tract axons can be visualized in green. The red staining for rCD300f‐IgG2a showed a punctate pattern similar to that observed in the brain, and was also mainly present in the white matter tracts of the spinal cord. G–I are higher magnification images from the dorsal corticospinal tract (square area shown in E). No co‐localization was observed between green and red staining. VGM = ventral grey matter; DGM = dorsal grey matter.
Overexpression of rCD300f reduce the lesion volume after an acute brain injury
A comparative study was performed to assess the effect of the rat ortholog of CD300f after an excitotoxic injury. Under the same experimental paradigm described previously, we observed that the delayed overexpression of rCD300f after an excitotoxic injury induced a significant reduction (≅35%) in the lesion volume when compared 3 days post‐intervention to the GFP transgene (Figure 8).
Figure 8.
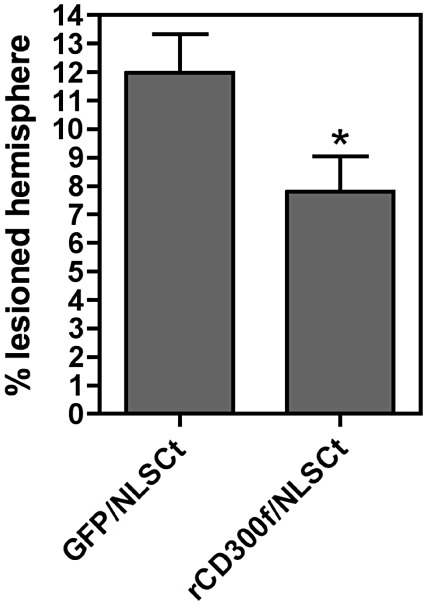
Overexpression of rCD300f is neuroprotective after and acute brain injury. Striatal injection of N‐methyl‐D‐aspartate (NMDA) induced a well‐delimitated lesion when observed by Nissl staining 3 days post‐intervention. At this time point, the measurement of the % of lesioned hemisphere showed that the overexpression of rCD300f using the NLSCt vector induced a significant neuroprotection (P < 0.05) when compared with the overexpression of a control transgene (EGFP).
DISCUSSION
In the present work, we have demonstrated that the overexpression of CD300f using the modular recombinant NLSCt vector has a neuroprotective role in a model of acute rat brain damage produced by injection of the glutamic acid analog NMDA. The excitotoxic process is a well‐known mechanism underlying several acute injuries such as stroke or trauma with important toxicity toward neurons and oligodendrocytes, and thus has been widely used as an acute model for CNS damage (32). Two differentiated spatio‐temporal lesioned areas can be established upon focal traumatisms or ischemic episodes caused by hypoxia: the core and the penumbra zone. The core degenerates rapidly and consequently is very difficult to rescue, but the secondary adjacent area engages a delayed proinflammatory program leading to cell death that can be modulated. Accordingly, most of the therapies are directed to overcome the neuropathological events in the penumbra zone.
Our initial hypothesis was that CD300f, an inhibitory immune receptor found on myeloid cells, could be expressed by microglial cells, and thus, could participate in the regulation of their activity. To test this hypothesis, we decided to overexpress the protein using a nonviral gene therapy vector. The NLSCt vector described by Aris and collaborators (4) has proven earlier to be a safe gene therapy vector for in vivo use as it does not induce acute inflammation or immunological response in the normal brain (28). The NLSCt modular vector was very efficient in conducting widespread expression after an excitotoxic damage, transfecting both neurons and reactive astroglial and microglial cells in the lesion zone (28). Our data showing a neuroprotective role for both human and rat CD300f in the NMDA excitotoxic model seemed to confirm our hypothesis. We assumed initially that the detected neuroprotective effect was due to the overexpression of CD300f in microglial cells, and that the overexpression of CD300f in other cell types would have no relevance in the reduction in the lesion volume. In that scenario, CD300f would inhibit microglia activation, decreasing the overall inflammatory process and thus reducing the lesioned area. But our data demonstrated that in addition to microglia, oligodendrocytes and neurons could, at least in vitro, express this molecule. Considering this circumstances, the overexpression of CD300f in these cell types could also be relevant for the detected neuroprotection. The presence of immune receptors in neurons is not unusual. For instance, neurons in the adult brain express some receptors from the adaptive immune system like CD200 (37) or TLR3 (7). It has also been described that neurons are able to transiently express high levels of toll‐like receptor 8 (TLR8) during the development but they decline in the adult brain (20). The primary neurons used in our studies have their origin in 18‐day‐old rat embryos. Consequently, the CD300f expression detected could be the endogenous for the developing brain or alternatively, the result of upregulation because of tissue damage or stress. However, we cannot discard that CD300f could be expressed constitutively by neurons. New antibodies that detect CD300f in vivo will be necessary to answer these questions.
Here, we detected the expression of rCD300f in microglial cells in vitro, which is coincident with the expression of CLM1/mCD300f in microglia/macrophages in EAE lesions (38). Interestingly, the LPS + IL1β inflammatory stimulus induced an increase in the mRNA for rCD300f in microglial cultures, contrary to what has been reported for other microglial immune receptors such as TREM2 or CD200R, which are downregulated after the in vitro treatment with LPS + IFNγ(34) or intraperitoneal LPS injection (24), respectively. We also show that besides microglial cells and neurons, oligodendrocytes in culture express CD300f. In accordance, oligodendrocytes have been shown to express both TREM2 and DNAX‐activating protein (DAP)12 (17), two important immune regulatory molecules.
We also report here the presence of the putative ligand(s) of CD300f in cultured CNS cells and in the brain and spinal cord. The staining was also present in peripheral nerves with an equivalent pattern to the one described for the brain and spinal cord (not shown). The in vitro and in vivo punctuate pattern observed with both human and rat CD300f‐IgG2a fusion proteins is intriguing. In primary cultures, oligodendrocytes, neurons and some astrocytes were positive for CD300f‐IgG2a staining, whereas no signal was detected in microglia. The in vitro hippocampal and cortical neuron expression of the ligand(s) for hCD300f/rCD300f contrasted with the absence of its expression in vivo. The Thy‐1‐YFP‐H mice display YFP‐positive neurons, but no co‐localization was detected by confocal microscopy when assessing hCD300f and rCD300f ligand(s) distribution. Precise co‐localization studies after a lesion need to be performed to investigate in detail the scenario in which neurons express CD300f ligand, both in physiological and pathological conditions.
One important question is through which mechanisms CD300f induce a neuroprotective effect in our model of excitotoxic damage. CD300f was initially identified as an inhibitory receptor able to recruit the tyrosine phosphatase SHP1 and efficiently block the signaling of activating receptors (2). However, CD300f presents a putative functional duality, as it has been recently shown to deliver activating signals 3, 15. Although our initial hypothesis was that the inhibitory capability of CD300f was responsible for the detected effect, the idea of an activating receptor having a neuroprotective effect should also be considered. For instance, the association of CD300f with the regulatory subunit of PI3‐kinase could provide additional survival signals that protect against cell death. TREM2, an immune‐activating receptor expressed by microglia, recruits the ITAM‐bearing adaptor DAP12 and has a protective role in mouse EAE (31). Whatever the signaling outcome may be, the neuroprotective effect achieved by the overexpression of CD300f was most probably mediated by its activation by the endogenous ligand(s), which we have shown is present in the brain. Nevertheless, it is also possible that the unknown CD300f ligand(s) could neuroprotect itself by signaling after the engagement with CD300f. This back‐to‐back signaling has been described for other ligand‐receptors systems (27).
The overall coincident pattern of staining observed both in rat and mice with hCD300f‐IgG2a and rCD300f‐IgG2a suggests that the ligand or ligands for hCD300f and rCD300f might be highly conserved, and most probably be the same molecule(s). Further studies identifying the ligand(s) of these molecules will be of interest, raising the tempting possibility of activating endogenous CD300f for neuroprotection against acute CNS injuries or neurodegenerative disorders.
In this work, we have injected the NLSCt vector carrying the transgene for CD300f 4 h after the lesion. Although the extent of the therapeutic window has to be investigated, the success of this delayed neuroprotective treatment suggests that the CD300f molecule in combination with the NLSCt gene therapy vector constitutes a robust candidate for neuroprotective drug formulations. This may be especially true for acute localized traumatic or ischemic injures in humans, which may require decompressing‐scull interventions, as a direct injection of the NLSCt vector containing the hCD300f transgene might be possible.
Taken together all the data, we have demonstrated that the overexpression of CD300f in a model of acute brain injury could have therapeutic applications, diminishing the secondary extension of the lesion. The fact that both the endogenous CD300f receptor and its physiological ligand(s) are found in diverse types of cells in brain suggests that the neuroprotective role of CD300f is the result of a complex network of cell‐to‐cell interactions.
ACKNOWLEDGMENTS
This work was supported by grants from Fondo Clemente Estable (FCE_197) ANII, Ministerio de Educación y Cultura, Uruguay; Comisión Sectorial de Investigación Científica (CSIC) UDELAR, Uruguay; Fondo de Investigaciones Sanitarias (PI080366), Ministerio de Ciencia e Innovación (ACI2009‐0919), and Agencia de Gestio d'Ajuts Universitaris i de Recerca de Catalunya (AGAUR) (2009 SGR 493 and 2009 SGR 108). A.E‐O. is supported by a contract Juan de la Cierva from the Ministerio de Ciencia e Innovación, E.C‐C. by a fellowship from AGAUR and J.S. by a contract Miguel Servet from Fondo de Investigaciones Sanitarias from Instituto de Salud Carlos III (CP06/00058). Thanks to Agustín Correa (UPR), Gonzalo Obal (UBP) and Soledad Astrada from the Institut Pasteur de Montevideo for their excellent technical help.
REFERENCES
- 1. Aguilar H, Alvarez‐Errico D, Garcia‐Montero AC, Orfao A, Sayos J, Lopez‐Botet M (2004) Molecular characterization of a novel immune receptor restricted to the monocytic lineage. J Immunol 173:6703–6711. [DOI] [PubMed] [Google Scholar]
- 2. Alvarez‐Errico D, Aguilar H, Kitzig F, Brckalo T, Sayos J, Lopez‐Botet M (2004) IREM‐1 is a novel inhibitory receptor expressed by myeloid cells. Eur J Immunol 34:3690–3701. [DOI] [PubMed] [Google Scholar]
- 3. Alvarez‐Errico D, Sayos J, Lopez‐Botet M (2007) The IREM‐1 (CD300f) inhibitory receptor associates with the p85alpha subunit of phosphoinositide 3‐kinase. J Immunol 178:808–816. [DOI] [PubMed] [Google Scholar]
- 4. Aris A, Villaverde A (2003) Engineering nuclear localization signals in modular protein vehicles for gene therapy. Biochem Biophys Res Commun 304:625–631. [DOI] [PubMed] [Google Scholar]
- 5. Bachelet I, Munitz A, Moretta A, Moretta L, Levi‐Schaffer F (2005) The inhibitory receptor IRp60 (CD300a) is expressed and functional on human mast cells. J Immunol 175:7989–7995. [DOI] [PubMed] [Google Scholar]
- 6. Barclay AN, Wright GJ, Brooke G, Brown MH (2002) CD200 and membrane protein interactions in the control of myeloid cells. Trends Immunol 23:285–290. [DOI] [PubMed] [Google Scholar]
- 7. Cameron JS, Alexopoulou L, Sloane JA, DiBernardo AB, Ma Y, Kosaras B et al (2007) Toll‐like receptor 3 is a potent negative regulator of axonal growth in mammals. J Neurosci 27:13033–13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cantoni C, Bottino C, Augugliaro R, Morelli L, Marcenaro E, Castriconi R et al (1999) Molecular and functional characterization of IRp60, a member of the immunoglobulin superfamily that functions as an inhibitory receptor in human NK cells. Eur J Immunol 29:3148–3159. [DOI] [PubMed] [Google Scholar]
- 9. Carson MJ, Thrash JC, Walter B (2006) The cellular response in neuroinflammation: the role of leukocytes, microglia and astrocytes in neuronal death and survival. Clin Neurosci Res 6:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cassina P, Peluffo H, Pehar M, Martinez‐Palma L, Ressia A, Beckman JS et al (2002) Peroxynitrite triggers a phenotypic transformation in spinal cord astrocytes that induces motor neuron apoptosis. J Neurosci Res 67:21–29. [DOI] [PubMed] [Google Scholar]
- 11. Castillo‐Ruiz MM, Campuzano O, Acarin L, Castellano B, Gonzalez B (2007) Delayed neurodegeneration and early astrogliosis after excitotoxicity to the aged brain. Exp Gerontol 42:343–354. [DOI] [PubMed] [Google Scholar]
- 12. Clark GJ, Ju X, Azlan M, Tate C, Ding Y, Hart DN (2009) The CD300 molecules regulate monocyte and dendritic cell functions. Immunobiology 214:730–736. [DOI] [PubMed] [Google Scholar]
- 13. Feng G, Mellor RH, Bernstein M, Keller‐Peck C, Nguyen QT, Wallace M et al (2000) Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28:41–51. [DOI] [PubMed] [Google Scholar]
- 14. Griffiths MR, Gasque P, Neal JW (2010) The regulation of the CNS innate immune response is vital for the restoration of tissue homeostasis (repair) after acute brain injury: a brief review. Int J Inflam 2010:151097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Izawa K, Kitaura J, Yamanishi Y, Matsuoka T, Kaitani A, Sugiuchi M et al (2009) An activating and inhibitory signal from an inhibitory receptor LMIR3/CLM‐1: LMIR3 augments lipopolysaccharide response through association with FcRgamma in mast cells. J Immunol 183:925–936. [DOI] [PubMed] [Google Scholar]
- 16. Jackson DG, Hart DN, Starling G, Bell JI (1992) Molecular cloning of a novel member of the immunoglobulin gene superfamily homologous to the polymeric immunoglobulin receptor. Eur J Immunol 22:1157–1163. [DOI] [PubMed] [Google Scholar]
- 17. Kiialainen A, Hovanes K, Paloneva J, Kopra O, Peltonen L (2005) Dap12 and Trem2, molecules involved in innate immunity and neurodegeneration, are co‐expressed in the CNS. Neurobiol Dis 18:314–322. [DOI] [PubMed] [Google Scholar]
- 18. Lanier LL (2009) DAP10‐ and DAP12‐associated receptors in innate immunity. Immunol Rev 227:150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Long EO (2008) Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev 224:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma Y, Li J, Chiu I, Wang Y, Sloane JA, Lu J et al (2006) Toll‐like receptor 8 functions as a negative regulator of neurite outgrowth and inducer of neuronal apoptosis. J Cell Biol 175:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez‐Barriocanal A, Sayos J (2006) Molecular and functional characterization of CD300b, a new activating immunoglobulin receptor able to transduce signals through two different pathways. J Immunol 177:2819–2830. [DOI] [PubMed] [Google Scholar]
- 22. Martinez‐Barriocanal A, Comas‐Casellas E, Schwarzt SJ, Martín M, Sayós J (2010) CD300 heterocomplexes, a new and family‐restricted mechanism for myeloid cell signaling regulation. J Biol Chem 285:41781–41794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martinez‐Palma L, Pehar M, Cassina P, Peluffo H, Castellanos R, Anesetti G et al (2003) Involvement of nitric oxide on kainate‐induced toxicity in oligodendrocyte precursors. Neurotox Res 5:399–406. [DOI] [PubMed] [Google Scholar]
- 24. Masocha W (2009) Systemic lipopolysaccharide (LPS)‐induced microglial activation results in different temporal reduction of CD200 and CD200 receptor gene expression in the brain. J Neuroimmunol 214:78–82. [DOI] [PubMed] [Google Scholar]
- 25. Munitz A, Bachelet I, Eliashar R, Moretta A, Moretta L, Levi‐Schaffer F (2006) The inhibitory receptor IRp60 (CD300a) suppresses the effects of IL‐5, GM‐CSF, and eotaxin on human peripheral blood eosinophils. Blood 107:1996–2003. [DOI] [PubMed] [Google Scholar]
- 26. Neumann H, Takahashi K (2007) Essential role of the microglial triggering receptor expressed on myeloid cells‐2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol 184:92–99. [DOI] [PubMed] [Google Scholar]
- 27. Nolte MA, van Olffen RW, van Gisbergen KP, van Lier RA (2009) Timing and tuning of CD27‐CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev 229:216–231. [DOI] [PubMed] [Google Scholar]
- 28. Peluffo H, Aris A, Acarin L, Gonzalez B, Villaverde A, Castellano B (2003) Nonviral gene delivery to the central nervous system based on a novel integrin‐targeting multifunctional protein. Hum Gene Ther 14:1215–1223. [DOI] [PubMed] [Google Scholar]
- 29. Peluffo H, Acarin L, Aris A, Gonzalez P, Villaverde A, Castellano B, Gonzalez B (2006) Neuroprotection from NMDA excitotoxic lesion by Cu/Zn superoxide dismutase gene delivery to the postnatal rat brain by a modular protein vector. BMC Neurosci 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peluffo H, González P, Arís A, Acarin L, Villaverde A, Castellano B, González B (2007) RGD domains neuroprotect the immature brain by a glial dependent mechanism. Ann Neurol 62:251–256. [DOI] [PubMed] [Google Scholar]
- 31. Piccio L, Buonsanti C, Mariani M, Cella M, Gilfillan S, Cross AH et al (2007) Blockade of TREM‐2 exacerbates experimental autoimmune encephalomyelitis. Eur J Immunol 37:1290–1301. [DOI] [PubMed] [Google Scholar]
- 32. Rothman SM, Olney JW (1995) Excitotoxicity and the NMDA receptor–still lethal after eight years. Trends Neurosci 18:57–58. [DOI] [PubMed] [Google Scholar]
- 33. Saneto RP, De Vellis J (1987) Neuronal and glial cells: cell culture of the central nervous system. In: Neurochemistry: A Practical Approach. Turner AJ, Brachelard HS (eds), pp. 27–63. IRL Press: Washington, DC. [Google Scholar]
- 34. Schmid CD, Sautkulis LN, Danielson PE, Cooper J, Hasel KW, Hilbush BS et al (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells‐2 on adult murine microglia. J Neurochem 83:1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Speckman RA, Wright Daw JA, Helms C, Duan S, Cao L, Taillon‐Miller P et al (2003) Novel immunoglobulin superfamily gene cluster, mapping to a region of human chromosome 17q25, linked to psoriasis susceptibility. Hum Genet 112:34–41. [DOI] [PubMed] [Google Scholar]
- 36. Veillette A, Latour S, Davidson D (2002) Negative regulation of immunoreceptor signaling. Annu Rev Immunol 20:669–707. [DOI] [PubMed] [Google Scholar]
- 37. Webb M, Barclay AN (1984) Localisation of the MRC OX‐2 glycoprotein on the surfaces of neurones. J Neurochem 43:1061–1067. [DOI] [PubMed] [Google Scholar]
- 38. Xi H, Katschke KJ Jr, Helmy KY, Wark PA, Kljavin N, Clark H et al (2010) Negative regulation of autoimmune demyelination by the inhibitory receptor CLM‐1. J Exp Med 207:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


