Abstract
Purpose:
Somatic variants in tumor necrosis factor receptor-associated factor 7 (TRAF7) cause meningioma, while germline variants have recently been identified in seven patients with developmental delay and cardiac, facial and digital anomalies. We aimed to define the clinical and mutational spectrum associated with TRAF7 germline variants in a large series of patients, and to determine the molecular effects of the variants through transcriptomic analysis of patient fibroblasts.
Methods:
We performed exome, targeted capture and Sanger sequencing of patients with undiagnosed developmental disorders, in multiple independent diagnostic or research centers. Phenotypic and mutational comparisons were facilitated through data exchange platforms. Whole transcriptome sequencing was performed on RNA from patient- and control-derived fibroblasts.
Results:
We identified heterozygous missense variants in TRAF7 as the cause of a developmental delay-malformation syndrome in 45 patients. Major features include a recognizable facial gestalt (characterized in particular by blepharophimosis), short neck, pectus carinatum, digital deviations and patent ductus arteriosus. Almost all variants occur in the WD40 repeats and most are recurrent. Several differentially-expressed genes were identified in patient fibroblasts.
Conclusion:
We provide the first large-scale analysis of the clinical and mutational spectrum associated with the TRAF7 developmental syndrome, and we shed light on its molecular etiology through transcriptome studies.
Keywords: TRAF7, craniofacial development, intellectual disability, blepharophimosis, patent ductus arteriosus
Introduction
The tumor necrosis factor receptor (TNF-R)-associated factor (TRAF) family contains seven members defined by shared protein domains and their involvement in mediating signal transduction from TNF-R superfamily members.1 TRAF7 contains an N-terminal RING finger domain, an adjacent TRAF-type zinc finger domain, a coiled-coil domain and seven C-terminal WD40 repeats (Figure 1). The WD40 repeats are unique to TRAF7 within the TRAF family, with all other members instead containing a C-terminal TRAF domain. In vitro studies have suggested that TRAF7 plays a role in the regulation of several transcription factors through various mechanisms. It participates in the signal transduction of cellular stress stimuli, such as TNFα stimulation, by activating pathways leading to increased transcriptional activity of AP1 and CHOP/gadd153.2–4 These effects are thought to be mediated by synergy between TRAF7 and the MAP3 kinase MEKK3, leading to the phosphorylation of JNK and p38 (regulators of AP1 and CHOP), with interaction of TRAF7 and MEKK3 occurring via the TRAF7 WD40 repeats.2,3 Depending on the context, TRAF7 can positively or negatively regulate the activity of NF-κB, through ubiquitination of pathway components p65 and NEMO.2,5,6 It also ubiquitinates p53,7 and the activity of the proto-oncogene c-Myb is negatively regulated by TRAF7 through sumoylation and consequent sequestering in the cytosol.8 In endothelial cells, TRAF7 interacts with the C-terminus of ROBO4 to suppress hyperpermeability during inflammation.9
Figure 1.
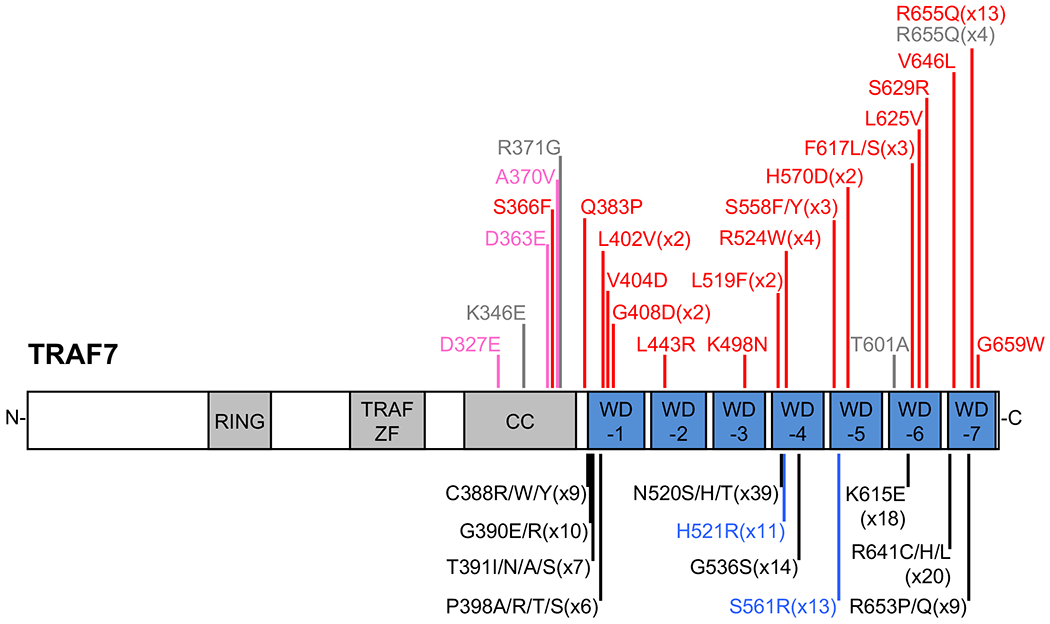
TRAF7 variants. Domain boundaries drawn approximately to scale, based on ref6. Variants causing the TRAF7 syndrome are indicated in red (reported here) or grey (previously reported18). In pink, variants of unknown significance reported here. Beneath the protein, the most recurrent somatic variants (ie, in greater than five samples) are indicated; in black, those reported in meningiomas11,12 and in blue, those in adenomatoid tumors of the genital tract16. CC; coiled-coil.
Somatic missense variants in TRAF7, concentrated within the WD40 domains and frequently recurrent, have been identified in meningiomas, mesotheliomas, intraneural perineuriomas and adenomatoid tumors of the genital tract.10–17 Heterozygous germline variants in TRAF7 have recently been reported in seven patients with a developmental disorder involving cardiac, facial, and digital anomalies and developmental delay (OMIM #618164).18 Here, we refine our understanding of the TRAF7 mutational and phenotypic spectrum through the identification of 45 previously undescribed patients, and thereby define a syndrome with a recognizable facial gestalt, specific skeletal and cardiac defects, and developmental delay/intellectual disability, which we propose to name the TRAF7 syndrome. Almost all identified variants fall in the WD40 repeats, most are recurrent and all are missense, suggestive of a gain-of-function or dominant negative mechanism, rather than haploinsufficiency. Intriguingly, somatic and germline variants do not overlap. We also present a transcriptomic analysis of fibroblasts from several patients, thereby providing insights into the pathways perturbed by TRAF7 alteration.
Materials and Methods
Variant identification
Genetic testing was performed according to approved ethical guidelines of the relevant institutions (Imagine Institute, GeneDx, ASST Papa Giovanni XXIII, Oslo University Hospital, Johns Hopkins University, Haukeland University Hospital, Centogene, Broad Institute, Radboud University Medical Center, UCSF Genomic Medicine Laboratory, Baylor College of Medicine, Victorian Clinical Genetics Services, Institut de Génétique Médicale (CHU Lille)) and consent was obtained from all families (including written consent to publish photographs, where applicable). Exome sequencing was performed in various independent research or diagnostic laboratories worldwide, using standard approaches (details available upon request). Targeted capture sequencing of TRAF7, in a panel of genes implicated in craniofacial malformations, was performed during clinical diagnostic screening at the Necker Hospital, using a SureSelect kit (Agilent) for capture followed by sequencing on a HiSeq machine (Illumina). Polyphen-219 was used for predicting the pathogenicity of missense variants.
Cell culture
Fibroblasts were obtained from skin biopsies of four patients (age at biopsy range: 8 to 19 years) and six controls (age at biopsy range: 17 years to adult). Corresponding informed consent and institutional ethics approval were obtained (Ethics Committee of the Universitat de Barcelona, IRB00003099), and all methods were performed in accordance with the relevant guidelines and regulations. Fibroblasts were cultured in DMEM supplemented with 10% FBS (Gibco, LifeTechnologies) and 1% penicillin-streptomycin (Gibco, LifeTechnologies) and were maintained at 37ºC and 5% CO2. When appropriate, cells were treated with 10 ng/μl recombinant human TNFα protein (R&D systems) for 6, 24 or 48 hours.
Cell viability assay
Fibroblasts were plated in 96-well plates and synchronized through serum deprivation for 24 hours. Cell viability was tested using an MTT assay, with a solution of 0.5 mg/ml Thiazolyl Blue Tetrazolium Bromide (Sigma-Aldrich) in DMEM (Gibco, LifeTechnologies). After 4 hours, formazan crystals were dissolved using DMSO (Merck Millipore) and absorbance was read at 560 nm.
Total RNA isolation and cDNA retrotranscription
RNA was extracted from fibroblasts using the High Pure RNA Isolation Kit (Roche), following the manufacturer’s instructions. Integrity and purity of the RNA was tested by agarose gel electrophoresis and 260/230 and 260/280 absorbance ratios using an ND-1000 Spectrophotometer (Nanodrop Technologies). All samples reached the quality and integrity standards for qRT-PCR. For RNA-Sequencing (RNA-Seq) analysis, the quality standards of half of the samples were tested on an Agilent Bioanalyzer 2100 (Agilent Technologies). RNA was retrotranscribed using the High-capacity cDNA Reverse Transcription kit (Applied Biosystems).
RNA-Sequencing and data analysis
RNA-Seq was performed by LEXOGEN, Inc. using the QuantSeq 3’ mRNA-Seq FWD kit for library preparation. Single-end reads were aligned to the human reference genome (GRCh37/hg19) and transcriptome using the STAR aligner. Quality metrics were obtained with tools of the RSEQC Quality control package. Differential expression analysis was performed using the R package DESeq2. The threshold to be considered as a differentially expressed gene (DEG) was set at a false discovery rate (FDR) ≤0.05 and a |log2 fold change| ≥1.
Real-time PCR
qPCR was performed using UPL probes (Roche), according to the manufacturer’s instructions. For every assay, the efficiency (E) of the reaction was calculated from a 7 point standard curve. Genomic DNA contamination was assessed and not detected in the samples. Amplification was done using the thermocycler Light Cycler 480 (Roche). Each sample was run in triplicate and the relative transcription level was quantified with the Crossing Point cycle calculation using the Light Cycler® 480 Software (release 1.5.0) (Roche). The GAPDH and PPIA genes were used as reference genes as they displayed the minimum coefficient of variation. The primer sequences and UPL probes used are available on request.
Ingenuity pathway analysis
Ingenuity Pathway Analysis (IPA, Qiagen) was performed on genes that showed an FDR ≤0.1 and |log2 fold change| ≥0.38. Separate runs were performed for each treatment condition. IPA uses the Fisher’s Exact Test to calculate statistical significance, considering associations between DEGs and annotated sets of molecules, with a p-value <0.05 (–log10 p-value >1.3) considered to be non-random.
Results
Identification of pathogenic variants in TRAF7
We performed exome sequencing, targeted capture sequencing and Sanger sequencing on individuals with undiagnosed, syndromic developmental delay/intellectual disability and dysmorphic facial features, in several independent clinical or research centers. Comparison of phenotypes and variants was facilitated by the data exchange platforms GeneMatcher20 and DECIPHER.21 We identified 45 individuals harboring missense variants in TRAF7 (Table S1). The cohort includes 36 sporadic cases in which the TRAF7 variant was de novo, one patient with low level maternal mosaicism for the TRAF7 variant, five cases with unknown inheritance and one familial case (patients 24-26) in which affected dizygotic twins inherited a TRAF7 variant from their affected mother, in whom the variant arose de novo (Figure S1). Almost all variants occurred in the WD40 repeats (Figure 1). Patients 1, 2 and 4 are sporadic cases carrying TRAF7 variants of unknown inheritance in the coiled-coil domain, and display phenotypes only partly overlapping those frequently observed in the rest of the cohort (further details below). We therefore consider these three individuals as having TRAF7 variants of unknown significance. None of the variants present in patients 1-45 have been reported in the Genome Aggregation Database (gnomAD, dataset v2.1). All affect highly conserved amino acids (based on Multiz alignments of 100 vertebrates at the UCSC Genome Browser) and all (except one coiled-coil variant) are predicted possibly or probably damaging by Polyphen-2 (Table S2). The variants in the 45 patients occur at 20 amino acid positions (Figure 1); recurrent variants occur on eight of these, with the most recurrent by far being p.(Arg655Gln) (13 index cases). The remaining recurrent variants each occur in two to four index cases, and different variants of the same residue are observed at two positions (p.(Ser558Phe) or p.(Ser558Tyr); p.(Phe617Leu) or p.(Phe617Ser)). The finding of recurrent missense variants largely restricted to the WD40 repeats suggests a disease mechanism involving specific functional changes to the mutant TRAF7 protein, rather than haploinsufficiency. In gnomAD, TRAF7 has a low probability of being loss-of-function intolerant (pLI = 0.02), further suggesting haploinsufficiency of TRAF7 does not cause severe pediatric disease.
Phenotype associated with TRAF7 variants
Clinical details of all patients are provided in Table S1, and a summary of the phenotypes in the core cohort of 42 patients (i.e., excluding the three patients with coiled-coil variants of unknown significance) is provided in Table S3. Many patients presented with feeding difficulties (n=24), often requiring tube feeding in infancy. Short stature was noted in 12 cases, low weight in five and microcephaly or macrocephaly in a total of 10. All patients had some form of developmental delay; intellectual disability (n=23) and/or speech delay (n=29) occurred in all but a small minority, while motor delay occurred in the majority (n=30). Hypotonia was noted in 17 patients. Autism spectrum disorder was observed in six cases and epilepsy in seven. There was a range of nonspecific anomalies on brain MRI (most frequently, enlarged ventricles). Almost all patients presented with anomalies of the palpebral fissures; most frequently blepharophimosis (n=33), along with epicanthus (n=20), telecanthus (n=14), ptosis (n=19) and up- or downslanting palpebral fissures (n=11) (Figure 2). Hypertelorism was reported in 17 cases. Ear anomalies (n=27) most frequently consisted of low-set, posteriorly rotated and/or protruding ears. Other frequent facial features include a bulbous nasal tip (n=17), wide or flat nasal bridge (n=11), micro- or retrognathia, albeit typically mild (n=13) and a high or prominent forehead (n=11). A computational composite from multiple patient photos further highlights the facial gestalt of the syndrome (Figure S2). Other skull shape anomalies, such as trigonocephaly, dolicocephaly, plagiocephaly, brachycephaly or bitemporal narrowing, occurred in 18 cases, and craniosynostosis in three. Palatal anomalies (n=15) included submucous cleft and velopharyngeal insufficiency. Most patients presented with abnormalities of the extremities (Figure 3A). Although highly variable in nature, major anomalies of the hands were finger deviations (n=10), camptodactyly (n=10), brachydactyly (n=6) and syndactyly (n=5), and of the feet, overlapping toes (n=10), pes planus (n=10), varus or valgus abnormalities (n=10) and sandal gap (n=5). Joint limitation in the limbs, hypermobility and dislocations were occasionally present. Anomalies of the axial skeleton were frequent: short neck (n=24), pectus carinatum (n=17) and other chest shape anomalies (n=10, including barrel-shaped or narrow chest), rib anomalies (n=5), deviations of the vertebral column (n=7) and vertebral anomalies (n=14). Regarding the latter, cervical stenosis or spinal cord compression was of clinical concern in several cases. Congenital cardiac defects were also frequent: 24 patients had patent ductus arteriosus (many of which required surgical repair), nine had atrial and six had ventricular septal defects and 10 had anomalies of valves. Conductive and/or sensorineural hearing loss occurred in 21 cases. Anomalies of the eyes included refractive errors (n=10) and strabismus (n=10). Infrequent phenotypes included a range of kidney anomalies (n=10), cryptorchidism (n=7), hernias (n=11), inverted nipples (n=6) and lower limb edema (n=3). In addition to a similar facial appearance amongst many of the patients, other features contributed to an upper-body gestalt in several, i.e., short neck with sloping shoulders, pectus carinatum and relative macrocephaly (Figure 3B). Finally, amongst the few adult patients in our cohort (seven over 18 years), clinical signs of premature ageing22 were noted - two women (20 years and 44 years) had progressive hair loss, the latter also had premature atherosclerosis, and a 26 year old male had premature osteoporosis and hair loss.
Figure 2.
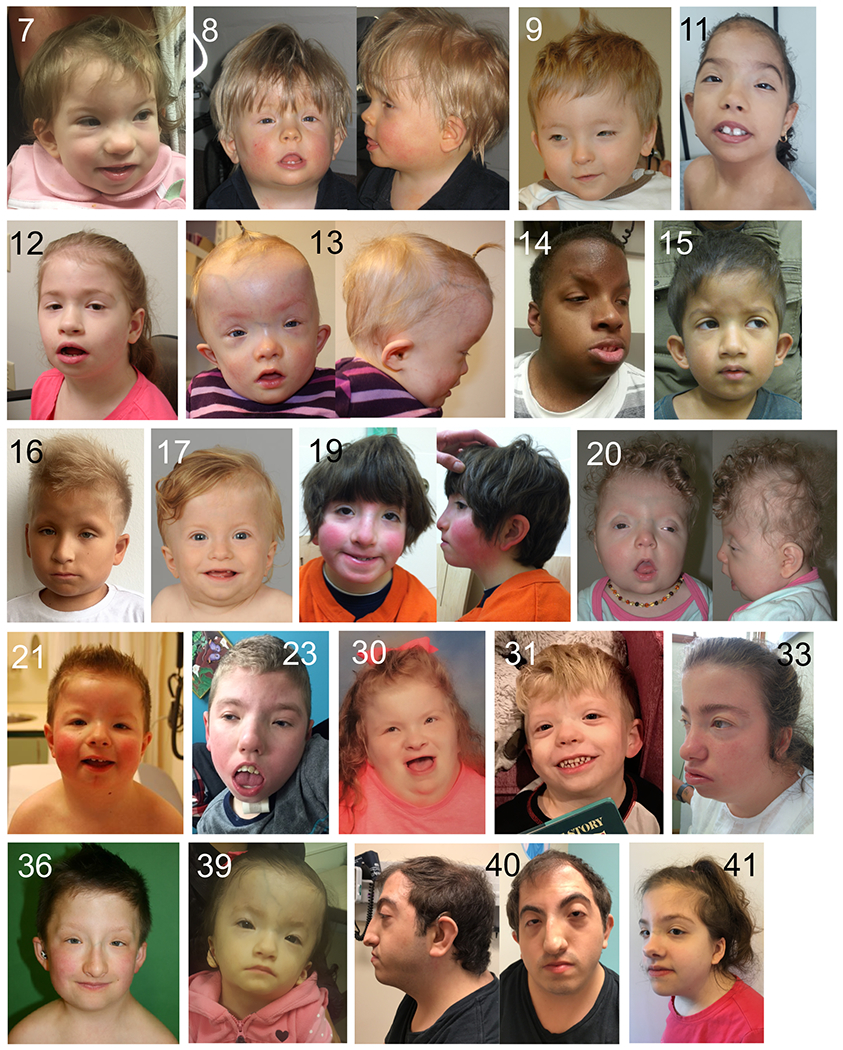
Facial features of patients with variants in TRAF7. Patient numbers are indicated at the top of each panel. See text for description of the major features.
Figure 3.
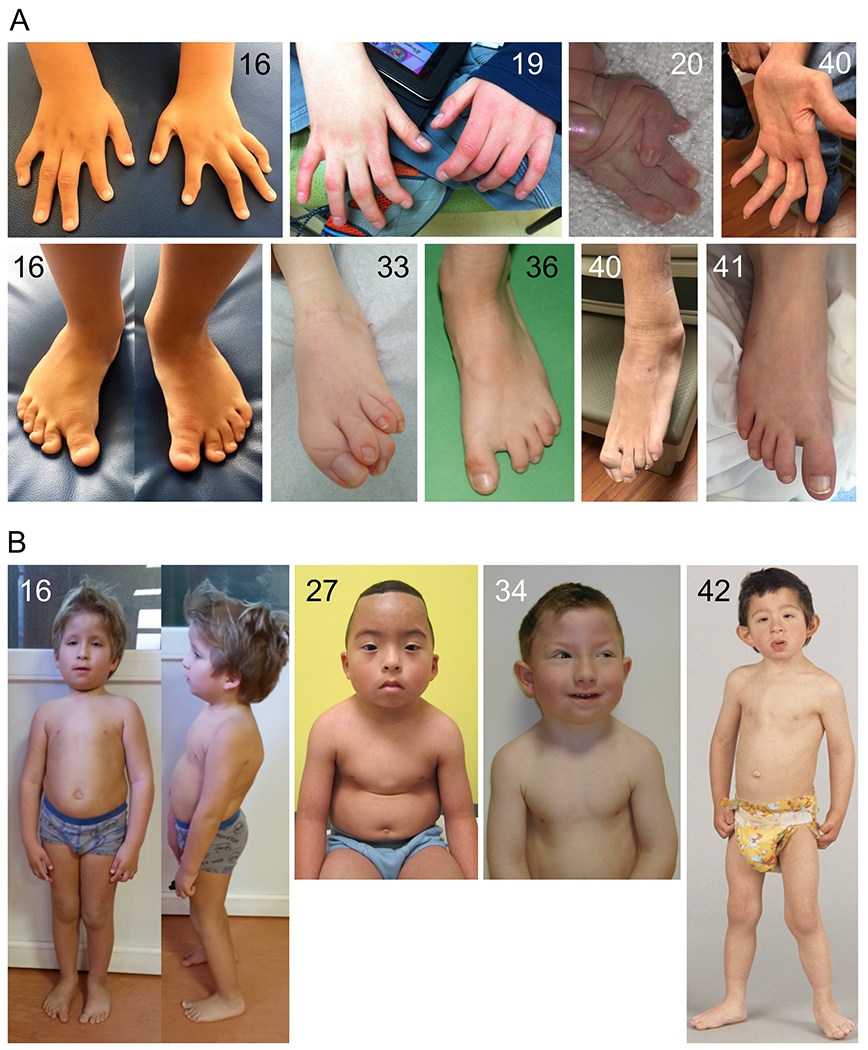
Anomalies of the extremities (A) and upper body appearance (B) of patients with variants in TRAF7. Patient numbers are indicated at the top of each panel. See text for description of the major features.
Amongst the differential diagnoses in our cohort, an Ohdo-related syndrome (OMIM 249620) was suspected in several patients, with KAT6B sequencing performed for five individuals prior to exome, highlighting that the TRAF7 syndrome overlaps with the group of blepharophimosis-mental retardation syndromes23. Also, Noonan/RAS-MAPK pathway gene panels were tested in eight patients, suggesting similarities to rasopathies. Although almost all TRAF7 variants were identified through trio exome sequencing, inclusion of TRAF7 on an NGS panel of genes mutated in neurocristopathy and craniofacial malformation syndromes led to the identification of two further individuals through diagnostic screening (patients 5 and 29). In patients 38 and 42, a strong clinical suspicion of TRAF7 syndrome, based on comparison with our cohort at the time, led to identification of a TRAF7 variant through Sanger sequencing or reanalysis of a previously unresolved singleton exome, respectively, highlighting the recognizability of this syndrome.
As noted above, three individuals (1, 2 and 4) harbored variants of unknown inheritance in the coiled-coil domain of TRAF7 (pink variants in Figure 1). These patients presented with neurodevelopmental defects, with seizures in two. Their facial features were not reminiscent of those of patients with variants in the WD40 repeats. Interestingly, patient 3, the only case with a de novo coiled-coil variant, had typical TRAF7 syndrome facial features (photographs were reviewed but permission to publish was denied). Of note, patient 4 had an endometrioid adenocarcinoma, diagnosed at 36 years.
Syndromic TRAF7 missense variants affect gene expression
As a first approach to begin to explore the effects of syndromic TRAF7 missense variants, the expression of 17 candidate target genes was assessed by qRT-PCR using skin fibroblast RNA from three patients bearing different variants in the WD40 repeats [p.(Leu402Val), p.(Leu519Phe), p.(Arg655Gln); patient numbers 6, 13, 33, respectively] versus fibroblast RNA from six control individuals. Of these, nine genes (EEF1A2, FLNB, IGFBP4, IGFBP7, LASS2, MMP2, NFKBIA, NOTCH3, SQSTM1) were selected because their expression appeared to be altered in a previous global transcriptome analysis performed in TRAF7-silenced cells,6 and because of their known involvement in human developmental disorders or putative role in developmental events relevant to the TRAF7 syndrome. The expression of JUN (encoding a subunit of the AP1 transcription factor) was tested because although AP1 activity can be stimulated by TRAF7, the effect of TRAF7 on transcription of AP1 components is unknown. TRAF7 itself was tested for auto-regulation. The rest are known NF-κB target genes (CFLAR (cFLIP), CCL2 (MCP1), VEGFA, MYC, BCL2, PTGS2 (COX2)).24 The above genes were also tested after TNFα stimulation.
We found NOTCH3 (ENSG00000074181) expression decreased by half in untreated patient cells (Figure 4), matching the direction of differential expression reported in TRAF7-silenced cells. In contrast, two other genes displayed an expression profile in patients opposite to that observed in TRAF7-silenced cells: FLNB (ENSG00000136068) expression levels in patients were over 2-fold higher than in controls, whereas IGFBP7 (ENSG00000163453) RNA was 4-fold lower in patients. We also found differences in BCL2 (ENSG00000171791), with lower expression levels in untreated patient cells than in controls, and PTGS2 (ENSG00000073756), whose expression was reduced to one third in untreated patient cells. Regarding TRAF7 itself, none of the three heterozygous missense variants tested altered its expression levels. In the same way, JUN, EEF1A2, IGFBP4, NFKBIA, LASS2, MMP2, SQSTM1, CFLAR, CCL2, VEGFA and MYC did not show any variation in their expression levels between patients and controls (data not shown).
Figure 4.
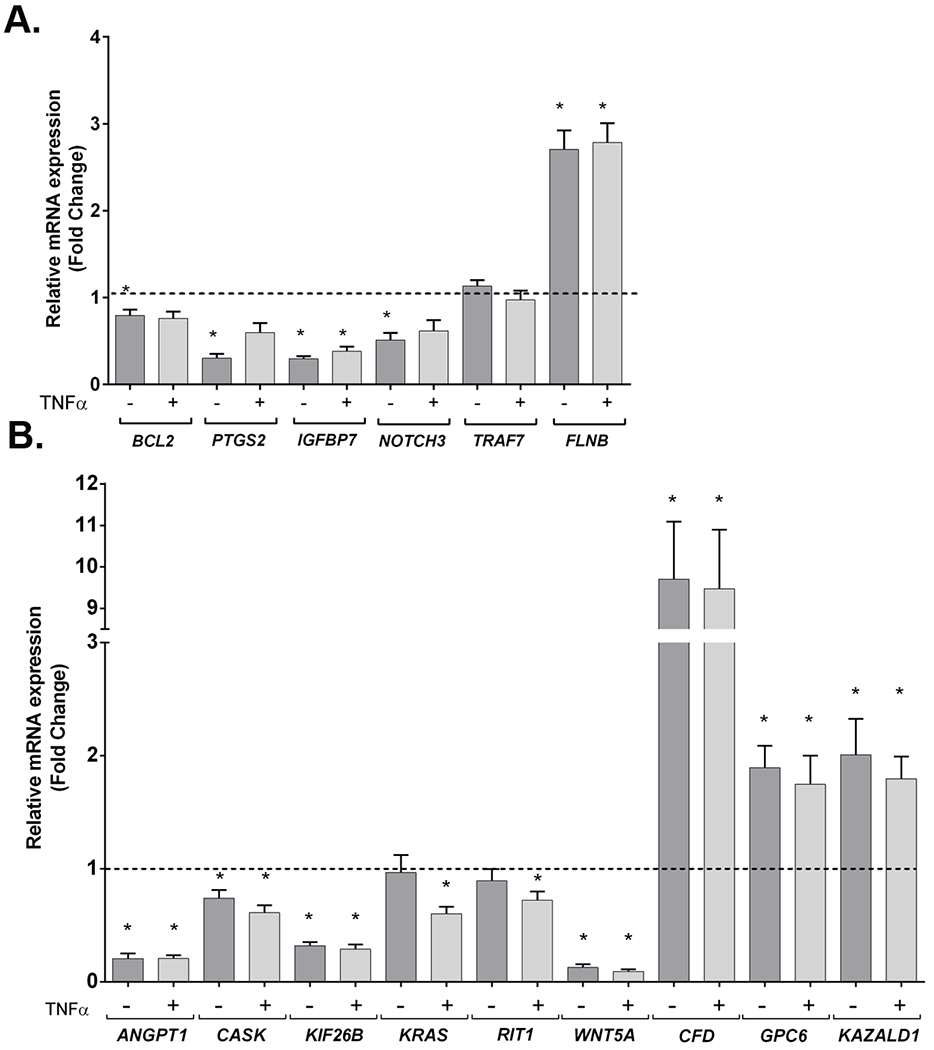
qRT-PCR quantification of mRNA levels in TRAF7 syndrome patient fibroblasts. (A) qRT-PCR of BCL2, PTGS2, IGFBP7, NOTCH3, TRAF7 and FLNB in fibroblasts from three patients, with or without TNFα treatment. Relative mRNA level was normalized to the mean of six controls in each treatment condition (dotted line). GAPDH was used as a reference gene (the use of PPIA gave similar results). Data shown represents the mean ± SEM of the three patients, in six independent experiments. Asterisks indicate significant differences (p<0.05) between patients and controls in the corresponding condition (treated or untreated). (B) qRT-PCR validation of DEGs identified by RNA-Seq: ANGPT1, CASK, KIF26B, KRAS, RIT1, WNT5A, CFD, GPC6 and KAZALD1. Relative mRNA level was normalized to the mean of six controls in each treatment condition (dotted line). GAPDH was used as a reference gene (the use of PPIA gave similar results). Data shown represents the mean ± SEM of the four patients, in two independent experiments. Asterisks indicate significant differences (p<0.05) between patients and controls in the corresponding condition (treated or untreated).
To further characterize the effects that syndromic TRAF7 missense variants could have on gene expression levels in a more unbiased fashion, we performed an mRNA whole transcriptomic analysis of skin fibroblasts. We used samples from the three patients and the six controls, with and without TNFα treatment. We considered DEGs as presenting an adjusted p-value <0.05 and |log2 fold change| in expression ≥1 (Table S4). We identified 76 DEGs in basal conditions (51.31% up-regulated and 48.68% down-regulated) and 90 DEGs after TNFα treatment (40% up-regulated and 60% down-regulated) (Figure 5A–5B). A substantial overlap in several DEGs was detected between the untreated and treated conditions, as 16 DEGs were up-regulated and 19 down-regulated independently of treatment (Figure 5C). In addition, 55 DEGs were identified only in the treatment condition, suggesting that TRAF7 syndromic variants may cause an alteration of the signaling pathway that is activated as a response to TNFα ligand.
Figure 5.
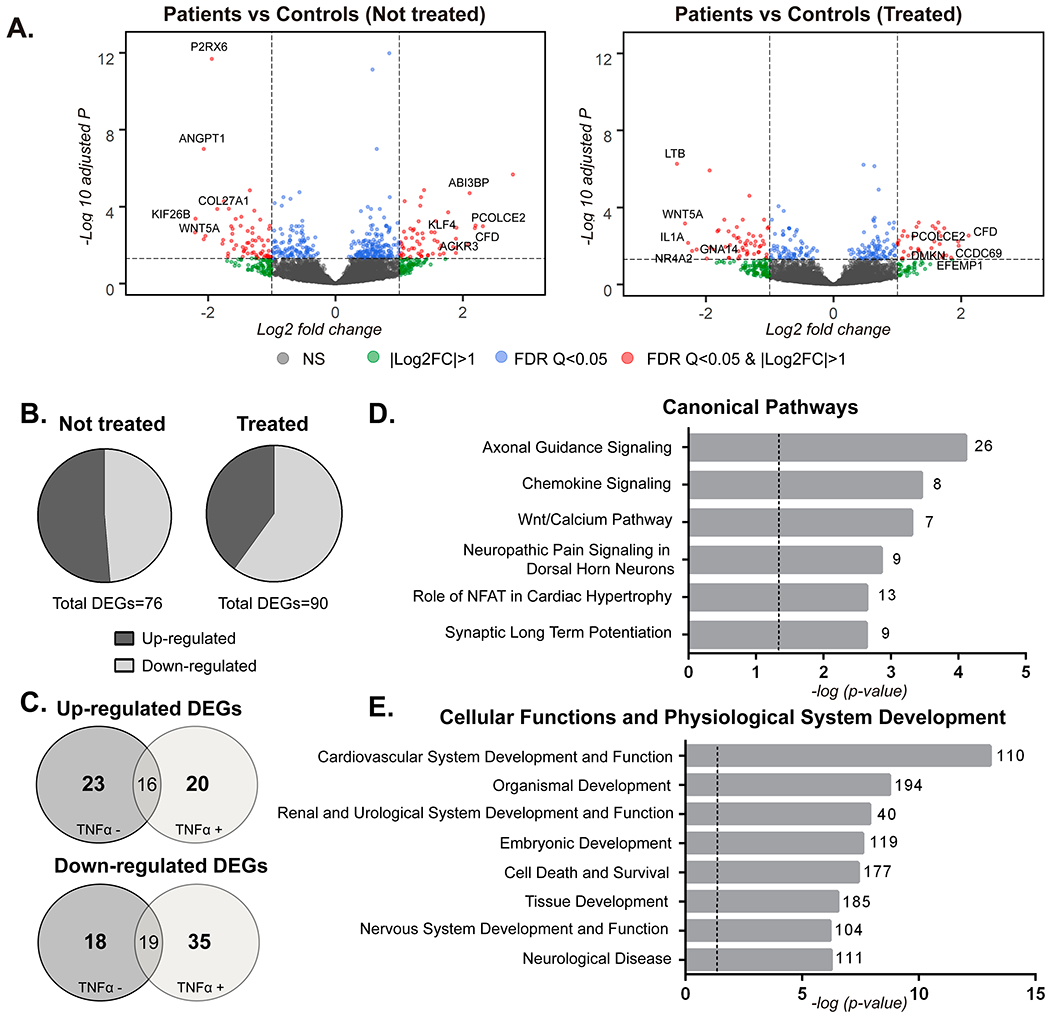
Transcriptome analysis of fibroblasts bearing missense TRAF7 variants. (A) Volcano plots representing differentially expressed genes (DEGs) in each condition (untreated or treated with TNFα (10 ng/ml) for 6 hours), measuring changes in expression (log2 fold change) and their significance (FDR; -log10 adjusted p-value). (B) Proportions of up- and down-regulated transcripts in untreated or treated patient fibroblasts. (C) Venn diagrams representing overlapping up- and down-regulated DEGs. (D and E) Ingenuity Pathway Analysis of DEGs in TRAF7 syndrome patient fibroblasts. (D) Selected over-represented Canonical Pathways. The number of DEGs included in each pathway is specified and a −log10 p-value >1.3 was considered significant. (E) Selected over-represented categories of “Cellular Functions and Physiological System Development”. The number of DEGs associated with each category is specified and a −log10 p-value >1.3 was considered significant.
Twelve DEGs from the RNA-Seq data were selected for qRT-PCR validation based on their high log2 fold change and/or functional criteria (i.e., for most, involvement in a human developmental disorder or relevant phenotype in an animal model – see Supplementary Discussion for details) (Table S5). For this step, an additional patient [number 36, bearing the variant p.(Arg655Gln)] was included. Results for seven out of the 12 were confirmatory (ANGPT1, CASK, KIF26B, WNT5A, CFD, GPC6, KAZALD1) (Figure 4B). KRAS, which was slightly upregulated in the RNA-Seq analysis, appeared downregulated in treated patient cells by qRT-PCR, while RIT1 was slightly downregulated only in untreated cells by RNA-Seq and only in treated cells by qRT-PCR. Finally, three genes (FOXP1, SPTAN1, MAPK11) whose log2 fold changes and significance values were below the general threshold to be considered as DEGs, but which had been selected based on functional criteria, were not validated.
To explore the different pathways and biological functions that might be affected, we performed IPA on the 726 DEGs identified under basal conditions using less stringent criteria than the preceding analyses (Table S6). The “Axonal guidance signaling” canonical pathway was significantly enriched, as 26 genes within this pathway showed alterations in transcript levels between patients and controls. Other significantly enriched canonical pathways included “Wnt/Ca2+ pathway” and “Role of NFAT in Cardiac Hypertrophy” (Figure 5D). IPA results also showed significant enrichment amongst the DEGs for genes involved in the development and function of the cardiovascular and nervous systems (Figure 5E). Similar results were obtained under TNFα-treated conditions (data not shown).
Syndromic TRAF7 missense variants have mild effects on cell viability
Putative differences in cell viability between patient and control fibroblasts were assessed, both in untreated and TNFα-treated conditions, during 24 or 48 hours. Although a slight tendency for increased cell viability was observed in patient fibroblasts under all conditions, significant differences were only observed at 24 hours of treatment with TNFα (Figure S3).
Discussion
In the present study, we have characterized the clinical features associated with germline variants in TRAF7, through analysis of 45 patients. In a cohort of seven patients, Tokita et al reported speech and motor delay, a range of dysmorphic facial features (including epicanthal folds, ptosis and dysmorphic ears), variable cardiac defects and anomalies of the extremities (digit deviations and variant creases) as principal phenotypes associated with TRAF7 variants.18 The large size of the cohort described here allowed us to further refine the phenotypic spectrum and to highlight several major phenotypes which were not emphasized previously: blepharophimosis, short neck, pectus carinatum and other thoracic defects, vertebral anomalies, patent ductus arteriosus and hearing loss. All the TRAF7 variants in our cohort are missense and several are recurrent, with one major mutational hotspot; p.(Arg655Gln). Previously, Tokita et al reported TRAF7 variants at only four positions (also missense); two in the coiled-coil domain and two in the WD40 repeats (including p.(Arg655Gln)).18 Our results reveal a highly skewed variant distribution along the protein, that was not evident in the prior study (Figure 1), and strongly implicate alteration of the WD40 repeats as the central disease mechanism.
The germline TRAF7 variants reported here are strikingly mutually exclusive to the somatic variants previously identified in tumors (lower part of Figure 1). This is underscored by the presence of recurrent variants restricted to each disease; p.(Arg655Gln) identified in 13 index cases here but never previously reported in a tumor, and variants at Asn520 reported 39 times in meningioma11,12 but not in syndromic patients. This suggests differences in activity of the mutant protein in each disease (for example, disruption of different protein-protein interactions or differences in degree of the same activity). It is possible that when present in the germline, the somatic variants may have a more severe phenotypic effect, perhaps not compatible with life. Interestingly, even amongst somatic variants, there may also be a trend for certain variants to be more frequent in particular tumor types; p.(His521Arg) and p.(Ser561Arg) (in blue in Figure 1) are highly recurrent in adenomatoid tumors of the genital tract,16 but have not been reported in meningiomas.10–13 In a few cases, the same amino acid is mutated to a different residue in each disease; for example, the syndromic variant p.(Leu519Phe) and the meningioma variants p.(Leu519Pro) and p.(Leu519Arg) (the latter two are not included in Figure 1). To the best of our knowledge, there is only one syndromic variant, p.(Arg524Trp) (de novo or maternal mosaic in four unrelated cases here) that has also been reported in a tumor sample.11 However, this meningioma also harbored a SMO variant, p.(Ala459Val), known to cause increased activity of SMO;25 the oncogenicity of the TRAF7 variant in this case is therefore unclear. One patient in our series with a variant of unknown significance in the coiled-coil domain had an endometrioid adenocarcinoma, while no tumors were reported in the patients with variants in the WD40 domain (although only a few are adults). One patient (43 years of age) in the cohort of Tokita et al18 had a meningioma. Meningiomas harboring a TRAF7 variant typically also contain a variant in KLF4, AKT1 or PIK3CA,10–13 suggesting a second hit may be required for development of TRAF7-associated tumors. Careful monitoring of aging syndromic patients will be required in order to determine whether they have a greater risk of developing tumors.
WD40 repeats are typically protein- or nucleic acid-interaction surfaces,26 and the WD40 domain of TRAF7 is known to interact with MEKK32,3 and with the DNA-binding domain of c-Myb.8 Whether alteration of these or other interactions underlies TRAF7 syndrome is unknown. Interestingly, Mekk3 is required for early cardiovascular development in mice.27 Also, phosphorylation of ERK1/2, which are MAP kinases downstream of MEKK3, is reduced in cells overexpressing TRAF7 syndromic variants,18 and loss of Erk2 in mice causes craniofacial and cardiac malformations and neurogenesis defects.28,29 In vitro overexpression of TRAF7 harboring WD40 domain variants identified in adenomatoid tumors of the genital tract leads to activation of the NF-κB pathway,16 and although dysregulation of this pathway has not typically been associated with congenital malformations in humans,30 knock-out of Ikka (a central component of the NF-κB pathway) in mice results in craniofacial and skeletal defects.31 The TRAF7 mutation distribution identified here, with clustering of recurrent missense variants in the WD40 repeats, is more consistent with a gain-of-function or dominant negative effect rather than haploinsufficiency. Structural studies suggest TRAF proteins can form trimers via the coiled-coil domain,32 and co-immunoprecipitation experiments have shown that TRAF7 can interact with itself2 and with TRAF6.33 An interesting possibility is that TRAF7 harboring syndromic variants could dominantly interfere not only with wildtype TRAF7 molecules but also with other TRAF proteins during development. TRAF7 loss of function animal models have not been reported. Amongst TRAF family members, Traf4 knock-out mice have congenital malformation phenotypes, including tracheal ring disruption, spina bifida and axial skeletal defects;34,35 the latter is of particular interest given the high frequency of costo-sternal and vertebral anomalies in TRAF7 syndrome patients.
Tokita et al reported two TRAF7 syndrome patients with variants in the coiled-coil domain (p.(Lys346Glu) and p.(Arg371Gly)), although these had no or less negative effect on ERK1/2 phosphorylation compared to the two WD40 repeat variants they identified (p.(Thr601Ala) and p.(Arg655Gln)). We report four individuals with coiled-coil domain variants, of which only one was confirmed to have a facial gestalt similar to patients with WD40 repeat variants. This suggests that some coiled-coil domain variants can have a similar molecular effect as those that perturb the WD40 domain. On the other hand, it has been reported that the coiled-coil domain of TRAF7 is required for interaction with NEMO,6 and in combination with the zinc finger domain for homodimerization and subcellular localization,2 raising the possibility that the molecular consequences of some coiled-coil domain variants may be different from those in the WD40 repeats. Note that none of the fibroblast samples used here for transcriptomic studies were from patients with coiled-coil variants, but comparison of transcriptomes from patients with WD40 versus coiled-coil domain variants may be informative. Confirmation of the causality of all TRAF7 coiled-coil domain variants, and the associated phenotypic spectrum, will require further functional studies and analysis of a larger number of individuals in this mutational subset.
To gain a better understanding of the pathogenic role of syndromic TRAF7 variants, patient fibroblasts were compared with controls at a transcriptomic level. This initially involved directly testing the expression of a small number of candidate genes previously found to be altered in TRAF7-silenced cells, and subsequently a global analysis of the gene expression landscape by RNA-Seq. Several of the identified DEGs are especially interesting due to the effects that their alteration are known to produce in human disease or animal models, and their dysregulation therefore plausibly contributes to the pathogenesis of the TRAF7 syndrome. In this context, the following DEGs are discussed further in the Supplementary Discussion: FLNB, IGFBP7, NOTCH3, BCL2, PTGS2, ANGPT1, WNT5A, KIF26B, CASK, GPC6, KAZALD1 and CFD.
In conclusion, through analysis of a large series of patients, we have defined the phenotypic spectrum associated with germline TRAF7 variants. The major features in our series are intellectual disability, motor delay, a recognizable facial gestalt including blepharophimosis, short neck, pectus carinatum, digital deviations, hearing loss and patent ductus arteriosus. TRAF7 syndrome patients typically require assisted learning and may be at risk of cervical stenosis. Older patients may benefit from monitoring for development of premature ageing phenotypes and tumors, although at this stage we cannot conclude whether there is an increased risk of the latter. We have shown there is a strong bias for TRAF7 syndrome-associated variants to occur in the WD40 repeats, and a major avenue for future investigation will involve determining whether there are different consequences on direction or strength of downstream signaling between these germline variants versus those previously reported in various cancers. Our transcriptomic studies of TRAF7 syndrome patient fibroblasts revealed a large number of DEGs. The fact that the expression of some genes (but not all) is only affected after TNFα treatment indicates that TRAF7 function in this pathway is disturbed, but that this is not the only pathway affected by the syndromic variants. Several identified DEGs are involved in cardiovascular, skeletal or nervous system development or function, and are therefore relevant to the phenotypes observed in the patients. Further exploration of the link between TRAF7 and these putative transcriptional targets is warranted in an animal model of the TRAF7 syndrome.
Supplementary Material
Acknowledgements
We thank the families for their participation. This work was supported by the Agence Nationale de la Recherche (CranioRespiro and ANR-10-IAHU-01), MSD Avenir (Devo-Decode), the Morton S. and Henrietta K. Sellner Professorship in Human Genetics (J.W.I.), JPB Foundation and the Simons Foundation SFARI program (W.K.C.), German Research Foundation (DFG; LE 4223/1; to D.L.), BC Children’s Hospital Foundation and Genome BC (CAUSES Study), PG23 / FROM 2017 Call for Independent Research as part of the Rapid Analysis for Rapid carE project (M.I., A.C.), the Victorian Government’s Operational Infrastructure Support Program, the NHGRI (UM1 HG008900, U01HG009599, UM1 HG006542), an NIH Common Fund grant (U01HG00769) and the Health Innovation Challenge Fund (DDD study; grant number HICF-1009-003). See Table S7 for supplementary acknowledgements.
Footnotes
Disclosure
M.T.C. and S.Y. are employees of GeneDx, Inc.
References
- 1.Zotti T, Scudiero I, Vito P, Stilo R. The Emerging Role of TRAF7 in Tumor Development. J Cell Physiol. 2017;232(6):1233–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. [DOI] [PubMed] [Google Scholar]
- 3.Xu L-G, Li L-Y, Shu H-B. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J Biol Chem. 2004;279(17):17278–17282. [DOI] [PubMed] [Google Scholar]
- 4.Scudiero I, Zotti T, Ferravante A, et al. Tumor necrosis factor (TNF) receptor-associated factor 7 is required for TNFα-induced Jun NH2-terminal kinase activation and promotes cell death by regulating polyubiquitination and lysosomal degradation of c-FLIP protein. J Biol Chem. 2012;287(8):6053–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsikitis M, Acosta-Alvear D, Blais A, et al. Traf7, a MyoD1 transcriptional target, regulates nuclear factor-κB activity during myogenesis. EMBO Rep. 2010;11(12):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zotti T, Uva A, Ferravante A, et al. TRAF7 protein promotes Lys-29-linked polyubiquitination of IkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J Biol Chem. 2011;286(26):22924–22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Wang L, Zhang S, et al. Downregulation of ubiquitin E3 ligase TNF receptor-associated factor 7 leads to stabilization of p53 in breast cancer. Oncol Rep. 2013;29(1):283–287. [DOI] [PubMed] [Google Scholar]
- 8.Morita Y, Kanei-Ishii C, Nomura T, Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol Biol Cell. 2005;16(11):5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirakura K, Ishiba R, Kashio T, et al. The Robo4-TRAF7 complex suppresses endothelial hyperpermeability in inflammation. J Cell Sci. 2019;132(1). [DOI] [PubMed] [Google Scholar]
- 10.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark VE, Harmancı AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuss DE, Piro RM, Jones DTW, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125(3):351–358. [DOI] [PubMed] [Google Scholar]
- 13.Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro-oncology. 2016;18(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. [DOI] [PubMed] [Google Scholar]
- 15.Klein CJ, Wu Y, Jentoft ME, et al. Genomic analysis reveals frequent TRAF7 mutations in intraneural perineuriomas. Ann Neurol. 2017;81(2):316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goode B, Joseph NM, Stevers M, et al. Adenomatoid tumors of the male and female genital tract are defined by TRAF7 mutations that drive aberrant NF-kB pathway activation. Mod Pathol. 2018;31(4):660–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevers M, Rabban JT, Garg K, et al. Well-differentiated papillary mesothelioma of the peritoneum is genetically defined by mutually exclusive mutations in TRAF7 and CDC42. Mod Pathol. 2019;32(1):88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokita MJ, Chen C-A, Chitayat D, et al. De Novo Missense Variants in TRAF7 Cause Developmental Delay, Congenital Anomalies, and Dysmorphic Features. Am J Hum Genet. 2018;103(1):154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lessel D, Kubisch C. Hereditary Syndromes with Signs of Premature Aging. Dtsch Arztebl Int. 2019;116(29-30):489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verloes A, Bremond-Gignac D, Isidor B, et al. Blepharophimosis-mental retardation (BMR) syndromes: A proposed clinical classification of the so-called Ohdo syndrome, and delineation of two new BMR syndromes, one X-linked and one autosomal recessive. Am J Med Genet A. 2006;140(12):1285–1296. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Ma J, Wang KS, et al. Baicalein inhibits TNF-α-induced NF-κB activation and expression of NF-κB-regulated target gene products. Oncol Rep. 2016;36(5):2771–2776. [DOI] [PubMed] [Google Scholar]
- 25.Sharpe HJ, Pau G, Dijkgraaf GJ, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schapira M, Tyers M, Torrent M, Arrowsmith CH. WD40 repeat domain proteins: a novel target class? Nat Rev Drug Discov. 2017;16(11):773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Boerm M, McCarty M, et al. Mekk3 is essential for early embryonic cardiovascular development. Nat Genet. 2000;24(3):309–313. [DOI] [PubMed] [Google Scholar]
- 28.Newbern J, Zhong J, Wickramasinghe RS, et al. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc Natl Acad Sci USA. 2008;105(44):17115–17120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuels IS, Karlo JC, Faruzzi AN, et al. Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J Neurosci. 2008;28(27):6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-κB: A Blossoming of Relevance to Human Pathobiology. Cell. 2017;168(1-2):37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sil AK, Maeda S, Sano Y, Roop DR, Karin M. IkappaB kinase-alpha acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature. 2004;428(6983):660–664. [DOI] [PubMed] [Google Scholar]
- 32.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398(6727):533–538. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida H, Jono H, Kai H, Li J-D. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem. 2005;280(49):41111–41121. [DOI] [PubMed] [Google Scholar]
- 34.Régnier CH, Masson R, Kedinger V, et al. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc Natl Acad Sci USA. 2002;99(8):5585–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiels H, Li X, Schumacker PT, et al. TRAF4 deficiency leads to tracheal malformation with resulting alterations in air flow to the lungs. Am J Pathol. 2000;157(2):679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


