Abstract
Phylogenetically, the genera Cuphophyllus, Ampulloclitocybe and Cantharocybe are treated as basal in the family Hygrophoraceae, despite weak support. However, the exact phylogenetic positions of the three genera have remained unresolved, and taxa related to these genera are poorly known. In this study, a new clitocyboid genus Spodocybe was proposed based on multigenic phylogenetic inference datasets and morphological evidence. The analyses of ITS as well as two combined datasets ITS-nrLSU-rpb2 and ITS-nrLSU-rpb1-rpb2-tef1-α-atp6 supported that (1) Spodocybe formed a well-supported monophyletic clade; and (2) sisters Spodocybe and Ampulloclitocybe, along with Cantharocybe and Cuphophyllus also formed a monophyletic lineage, as sister to the rest of the Hygrophoraceae. Meanwhile, two new species, namely S. rugosiceps and S. bispora, from southwestern China, were documented and illustrated. These results support the new proposed genus Spodocybe, and that Spodocybe, Ampulloclitocybe, Cantharocybe and Cuphophyllus should be retained in the Hygrophoraceae as a new subfamily Cuphophylloideae.
Keywords: Ampulloclitocybe , Cantharocybe , Cuphophyllus , morphological characters, phylogenetic analysis, taxonomy
Introduction
The widespread genus Clitocybe (Fr.) Staude currently encompasses large numbers of species with clitocyboid habit, sharing the features of saprophytic nutrition, funnel-shaped pileus, decurrent lamellae, a usually white, cream or pale colored spore-deposit and smooth and inamyloid spores (Singer 1986; Breitenbach and Kraenzlin 1991; Læssøe and Petersen 2019). As a consequence of the poor, broad and unrepresentative morphological characteristics, the genus appeared heterogeneous and was subsequently proven to be polyphyletic based on the phylogenetic analysis (Moncalvo et al. 2002; Harmaja 2003).
Based on phylogenetic analyses over the past 20 years, (i) many new genera within the Tricholomatoid clade were proposed to accommodate previous Clitocybe species deviating from the core Clitocybeae clade (Matheny et al. 2006), such as Cleistocybe Ammirati, A.D. Parker & Matheny (Ammirati et al. 2007), Trichocybe Vizzini (Vizzini et al. 2010), Atractosporocybe P. Alvarado, G. Moreno & Vizzini, Leucocybe Vizzini, P. Alvarado, G. Moreno & Consiglio and Rhizocybe Vizzini, G. Moreno, P. Alvarado & Consiglio (Alvarado et al. 2015); (ii) Several clitocyboid groups were reconfirmed as independent genera, for instance, Singerocybe Harmaja (Qin et al. 2014) and Infundibulicybe Harmaja (Binder et al. 2010); and (iii) some others were even transferred to the Hygrophoroid clade (Binder et al. 2010), such as Ampulloclitocybe Redhead, Lutzoni, Moncalvo & Vilgalys (Redhead et al. 2002) and Cantharocybe H.E. Bigelow & A.H. Sm. (Hosen et al. 2016). However, many clitocyboid taxa remain to be reclassified.
The molecular phylogenetic relationships among members of the Hygrophoraceae Lotsy were well studied by Lodge et al. (2014). In their work, the family was divided into subfamily Hygrophoroideae E. Larss., Lodge, Vizzini, Norvell & S.A. Redhead, Hygrocyboideae Padamsee & Lodge, Lichenomphalioideae Lücking & Redhead and Cuphophylloid grade. Meanwhile, the Cuphophylloid grade was retained in the Hygrophoraceae as the base comprising the genera Cuphophyllus (Donk) Bon, Ampulloclitocybe and Cantharocybe, despite weak phylogenetic support (Matheny et al. 2006; Binder et al. 2010; Lodge et al. 2014). Consequently, the taxonomic problem of the three genera on whether to be included or excluded in the Hygrophoraceae has remained unresolved.
Recently, some collections were shown to be closely related to Clitocybe trulliformis (Fr.) P. Karst. based on ITS-BLAST searches while at the same time they were surprisingly related to taxa of the genus Cuphophyllus based on nrLSU-BLAST searches. As far as we know, C. trulliformis and allied species were lacking taxonomic revision, especially regarding their molecular phylogenetic status. Furthermore, the phylogenetic delimitation of the Hygrophoraceae was ambiguous due to the uncertain positions of Cuphophyllus, Ampulloclitocybe and Cantharocybe. Hence, the aims of this study were (a) to propose and describe a new genus of the Hygrophoraceae for species related to C. trulliformis based on morphological and molecular analyses and (b) to reconstruct the phylogeny of the Hygrophoraceae for determining the exact phylogenetic placements of Cuphophyllus, Ampulloclitocybe and Cantharocybe with multi-gene data.
Materials and methods
Specimens
Twenty-three specimens of species similar to C. trulliformis and related species were collected from southwestern and northeastern China and western Germany, during 2007 to 2020. The fresh fruitbodies were dried using heat or silica gel. Voucher specimens were deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN-HKAS). Detail information of these specimens is given in Table 1.
Table 1.
Specimens used in phylogenetic analysis and their GenBank accession numbers. The newly generated sequences are shown in bold.
| Species | Voucher | Locality | GenBank accession number | |||||
|---|---|---|---|---|---|---|---|---|
| ITS | nrLSU | rpb2 | rpb1 | tef1-α | atp6 | |||
| Acantholichen pannarioides | MDF352 | Costa Rica | KT429795 | KT429807 | KT429817 | |||
| Acantholichen campestris | DIC595b | Brazil | KT429798 | KT429810 | KT429818 | |||
| Acantholichen galapagoensis | MDF058 | Ecuador | KT429785 | KT429800 | KT429812 | |||
| Ampulloclitocybe clavipes | KUN-HKAS 54426 | China: Jilin | MW616462 | MW600481 | MW656471 | MW656467 | MW656461 | MW656478 |
| AFTOL-ID 542 | AY789080 | AY639881 | AY780937 | AY788848 | AY881022 | |||
| DJL06TN40 | USA | FJ596912 | KF381542 | KF407938 | ||||
| Arrhenia auriscalpium | TUB 011588 | DQ071732 | ||||||
| Arrhenia acerosa | Lueck2 | Germany | KP965766 | KP965784 | ||||
| Cantharellula umbonata | CBS 398.79 | France | MH861222 | MH872990 | ||||
| Cantharocybe gruberi | AFTOL-ID 1017 | USA | DQ200927 | DQ234540 | DQ385879 | DQ435808 | DQ059045 | |
| AH24539 | Spain | JN006422 | JN006420 | |||||
| Cantharocybe brunneovelutina | DJL-BZ-1883 | Belize | NR160458 | NG068731 | ||||
| Cantharocybe virosa | TENN63483 | India | KX452405 | JX101471 | ||||
| Iqbal-568 | Bangladesh | KX452403 | KF303143 | |||||
| Chromosera cyanophylla | AFTOL-ID 1684 | USA | DQ486688 | DQ457655 | KF381509 | |||
| Chromosera ambigua | GE18008-1 | France | MK645573 | MK645587 | MK645593 | |||
| Chromosera lilacina | GE18035 | Canada | MK645577 | MK645591 | MK645597 | |||
| Chromosera xanthochroa | GE18033 | Canada | MK645576 | MK645590 | MK645596 | |||
| Chrysomphalina chrysophylla | AFTOL-ID 1523 | USA | DQ192180 | DQ457656 | ||||
| Chrysomphalina grossula | OSC 113683 | EU644704 | EU652373 | |||||
| Clitocybe aff. costata | DJL06TN80 | USA | FJ596913 | |||||
| Clitocybe herbarum | G0171 | Hungary | MK277719 | |||||
| Clitocybe trulliformis | 14562 | Italy | JF907809 | |||||
| 4804 | Russia | MH930178 | ||||||
| Clitocybe cf. trulliformis | G0460 | Hungary | MK277728 | |||||
| Clitocybe sp. | NAMA 2015-206 | USA | MH910535 | |||||
| Clitocybe sp. | NAMA 2015-318 | USA | MH910563 | |||||
| Clitocybe sp. | Mushroom Observer 302917 | USA | MK607556 | |||||
| Cora pavonia | DIC215 | Ecuador | KF443238 | KF443261 | KF443275 | |||
| Cora aspera | DIC110 | Bolivia | KF443230 | KF443257 | KF443267 | |||
| Cora reticulifera | DIC119 | Ecuador | KF443239 | KF443262 | KF443269 | |||
| Cora squamiformis | DIC146 | Bolivia | KF443240 | KF443263 | KF443273 | |||
| Corella brasiliensis | MDF017 | Bolivia | KF443229 | KF443255 | KF443276 | |||
| Corella aff. Melvinii | MDF200 | Brazil | KJ780569 | KY861725 | ||||
| Cuphophyllus pratensis | Lueck7 | Germany | KP965771 | KP965789 | ||||
| DJL-Scot-8 | UK | KF291057 | KF291058 | |||||
| Cuphophyllus aurantius | CFMR PR-6601 | Puerto Rico | KF291099 | KF291100 | KF291102 | |||
| Cuphophyllus aff. pratensis | AFTOL-ID 1682 | USA | DQ486683 | DQ457650 | DQ435804 | |||
| Cuphophyllus sp. | KUN-HKAS 105671 | China: Tibet | MW762875 | MW763000 | MW789179 | MW789163 | ||
| Cyphellostereum galapagoense | CDS 41163 | Ecuador | NR158415 | NG068806 | ||||
| Cyphellostereum imperfectum | DIC115 | Guatemala | KF443218 | KF443243 | KF443277 | |||
| Dictyonema interruptum | Ertz 10475 | Portugal | EU825967 | KF443282 | ||||
| Dictyonema schenckianum | DIC113 | Brazil | KF443225 | KF443251 | KF443285 | |||
| Eonema pyriforme | G1063 | Poland | MK278075 | |||||
| Gliophorus psittacinus | CFMR DEN-25 | Denmark | KF291075 | KF291076 | KF291078 | |||
| Gliophorus graminicolor | TJB-10048 (CORT) | Australia | KF381520 | KF381545 | KF407936 | |||
| Gliophorus aff. laetus | CFMR PR-5408 | Puerto Rico | KF291069 | KF291070 | ||||
| Gloioxanthomyces nitidus | GDGM41710 | China: Jilin | MG712283 | MG712282 | MG711911 | |||
| Haasiella splendidissima | Herb. Roux n. 4044 | France | JN944400 | JN944401 | ||||
| Herb. Roux n. 3666 | Moldova | JN944398 | JN944399 | |||||
| Haasiella venustissima | A. Gminder 971488 | Italy | KF291092 | KF291093 | ||||
| E. C. 08191 | Italy | JN944393 | JN944394 | |||||
| Humidicutis marginata | JM96/33 | AF042580 | ||||||
| Humidicutis auratocephalus | AFTOL-ID 1727 | USA | DQ490624 | DQ457672 | DQ472720 | DQ447906 | ||
| Humidicutis dictiocephala | QCAM6000 | Ecuador | KY689661 | KY780120 | ||||
| Humidicutis sp. | CFMR BZ-3923 | Belize | KF291110 | KF291111 | ||||
| Hygroaster nodulisporus | AFTOL-ID 2020 | USA | EF561625 | |||||
| Hygroaster albellus | AFTOL-ID 1997 | Puerto Rico | KF381521 | EF551314 | ||||
| Hygrocybe conica | FO 46714 | DQ071739 | ||||||
| Hygrocybe cf. acutoconica | CFMR NC-256 | USA | KF291117 | KF291118 | KF291120 | |||
| Hygrocybe coccinea | AFTOL-ID 1715 | USA | DQ490629 | DQ457676 | DQ472723 | DQ447910 | GU187705 | |
| Hygrocybe aff. conica | AFTOL-ID 729 | AY854074 | AY684167 | AY803747 | ||||
| Hygrophorus eburneus | US97/138 | Germany | AF430279 | |||||
| GDGM70059 | USA | MT093608 | ||||||
| Hygrophorus chrysodon | KUN-HKAS 82501 | China: Tibet | MW616463 | MW600482 | MW656472 | MW656462 | MW656479 | |
| KUN-HKAS 112569 | China: Tibet | MW762876 | MW763001 | MW789180 | MW789164 | MW773440 | MW789195 | |
| Hygrophorus flavodiscus | KUN-HKAS 68013 | China: Yunnan | MW616464 | MW600483 | MW656473 | MW656468 | MW656463 | MW656480 |
| KUN-HKAS 55043 | China: Yunnan | MW616465 | MW600484 | MW656474 | MW656469 | MW656464 | MW656481 | |
| Hygrophorus gliocyclus | KUN-HKAS 79929 | China: Tibet | MW616466 | MW600485 | MW656475 | MW656465 | MW656482 | |
| Hygrophorus hypothejus | KUN-HKAS 56550 | Germany | MW616467 | MW600486 | MW656476 | MW656470 | MW656483 | |
| Hygrophorus pudorinus | AFTOL-ID 1723 | USA | DQ490631 | DQ457678 | DQ472725 | DQ447912 | GU187710 | |
| Hygrophorus sp. 1 | KUN-HKAS 112566 | China: Yunnan | MW762877 | MW763002 | MW789181 | MW789165 | MW773441 | MW789196 |
| Hygrophorus sp. 2 | KUN-HKAS 87261 | China: Jilin | MW616468 | MW600487 | MW656477 | MW656466 | MW656484 | |
| Hygrophorus sp. 3 | KUN-HKAS 112567 | China: Tibet | MW762878 | MW763003 | MW789182 | MW789166 | MW773442 | MW789197 |
| Hygrophorus sp. 4 | KUN-HKAS 112568 | China: Tibet | MW762879 | MW763004 | MW789183 | MW789167 | MW773443 | MW789198 |
| Lichenomphalia hudsoniana | GAL18249 | USA | JQ065873 | JQ065875 | ||||
| Lichenomphalia meridionalis | S-270-FB1 | Japan | LC428308 | LC428307 | ||||
| Neohygrocybe ovina | GWG H. ovina Rhosisaf (ABS) | UK | KF291233 | KF291234 | KF291236 | |||
| Neohygrocybe griseonigra | GDGM 44492 | China | MG779451 | MG786565 | ||||
| Neohygrocybe ingrata | DJL05TN62 (TENN) | USA | KF381525 | KF381558 | KF381516 | |||
| Neohygrocybe subovina | GRSM 77065 | USA | KF291140 | KF291141 | ||||
| Spodocybe bispora | KUN-HKAS 73310 | China: Yunnan | MW762880 | MW763005 | MW789184 | MW789168 | MW773444 | MW789199 |
| KUN-HKAS 73332 | China: Yunnan | MW762881 | MW763006 | MW789185 | MW789169 | MW773445 | MW789200 | |
| KUN-HKAS 112564 | China: Yunnan | MW762882 | MW763007 | MW789186 | MW789170 | MW773446 | MW789201 | |
| Spodocybe rugosiceps | KUN-HKAS 112561 | China: Yunnan | MW762883 | MW763008 | MW789187 | MW789171 | MW773447 | MW789202 |
| KUN-HKAS 81981 | China: Yunnan | MW762884 | MW763009 | MW789188 | MW789172 | MW789203 | ||
| KUN-HKAS 69830 | China: Yunnan | MW762885 | MW763010 | MW789189 | MW789173 | MW773448 | MW789204 | |
| Spodocybe rugosiceps | KUN-HKAS 71071 | China: Yunnan | MW762886 | MW763011 | MW789190 | MW789174 | MW773449 | MW789205 |
| KUN-HKAS 112562 | China: Yunnan | MW762887 | MW763012 | MW789191 | MW789175 | MW789159 | MW789206 | |
| KUN-HKAS 112563 | China: Yunnan | MW762888 | MW763013 | MW789192 | MW789176 | MW789160 | MW789207 | |
| Spodocybe sp. 1 | KUN-HKAS 112560 | China: Jilin | MW762889 | MW763014 | MW789193 | MW789177 | MW789161 | MW789208 |
| Spodocybe sp. 2 | KUN-HKAS 112565 | China: Yunnan | MW762890 | MW763015 | MW789194 | MW789178 | MW789162 | MW789209 |
| Porpolomopsis calyptriformis | CFMR ENG-3 | UK | KF291242 | KF291243 | KF291245 | |||
| Porpolomopsis aff. calyptriformis | DJL05TN80 (TENN) | USA | KF291246 | KF291247 | KF291249 | |||
| Porpolomopsis lewelliniae | TJB-10034 (CORT) | Thailand | KF291238 | KF291239 | KF291241 | |||
| Pseudoarmillariella ectypoides | AFTOL-ID 1557 | USA | DQ192175 | DQ154111 | DQ474127 | DQ516076 | GU187733 | |
| Pseudoarmillariella bacillaris | KUN-HKAS 76377 | China | KC222315 | KC222316 | ||||
| Sinohygrocybe tomentosipes | GDGM 50075 | China: Hunan | MG685873 | MG696902 | MG696906 | |||
| GDGM 43351 | China: Sichuan | MG685872 | MG696901 | MG696905 | ||||
| Amylocorticium cebennense | CFMR HHB-2808 | USA | GU187505 | GU187561 | GU187770 | GU187439 | GU187675 | |
| Aphroditeola olida | DAOM 226047 | Canada | KF381518 | KF381541 | ||||
| Macrotyphula fistulosa | IO. 14. 214 | Spain | MT232352 | KY224088 | MT242317 | MT242354 | ||
| Macrotyphula juncea | IO. 14. 177 | Sweden | MT232353 | MT232306 | MT242337 | MT242355 | ||
| Macrotyphula phacorrhiza | IO. 14. 167 | Sweden | MT232364 | MT232315 | MT242348 | MT242326 | MT242367 | |
| IO.14. 200 | France | MT232363 | MT232314 | MT242347 | MT242366 | |||
| Phyllotopsis nidulans | IO. 14. 196 | Spain | MT232308 | MT242338 | MT242319 | MT242357 | ||
| Phyllotopsis sp. | AFTOL-ID 773 | DQ404382 | AY684161 | AY786061 | DQ447933 | DQ059047 | ||
| Pleurocybella porrigens | UPS F-611822 | Sweden | MT232355 | MT232309 | MT242339 | |||
| Plicaturopsis crispa | AFTOL-ID 1924 | USA | DQ494686 | DQ470820 | GU187816 | |||
| Pterulicium echo | ZRL20151311 | LT716065 | KY418881 | KY419026 | KY418979 | KY419076 | ||
| Pterulicium gracilis | IO. 14. 142 | Sweden | MT232356 | MT232310 | MT242358 | |||
| Sarcomyxa serotina | AFTOL-ID 536 | USA | DQ494695 | AY691887 | DQ859892 | DQ447938 | GU187754 | |
| Serpulomyces borealis | CFMR L-8014 | USA | GU187512 | GU187570 | GU187782 | GU187686 | ||
| Tricholomopsis decora | AFTOL-ID 537 | DQ404384 | AY691888 | DQ408112 | DQ447943 | DQ029195 | ||
| Tricholomopsis osiliensis | ZRL20151760 | LT716068 | KY418884 | KY419029 | KY419079 | |||
| Typhula capitata | IO. 15. 122 | Spain | MT232357 | MT232312 | MT242341 | MT242321 | MT242360 | |
| Typhula incarnata | IO. 14. 92 | Sweden | MT232362 | MT232313 | MT242346 | MT242325 | ||
| Typhula micans | IO. 14. 165 | Sweden | MT232361 | KY224102 | MT242345 | MT242324 | MT242364 | |
Morphological observation
Macroscopic characters of species were described based on the raw field record data and photographs. Colors used in description referred to Kornerup and Wanscher (1978). For the microscopic structure observation, tissue sections of dried specimens were mounted in 5% KOH solution or distilled water and structures of lamellar trama, pileipellis and stipitipellis, basidia and basidiospores were observed with a light microscopy. For the description of lamellar trama structure, seven types, including regular, subregular, divergent, pachypodial, bidirectional, tri-directional and interwoven, were used following Lodge et al. (2014). Besides, Melzer’s reagent was applied to test the amyloidity of the basidiospores. In the description of basidiospores, the abbreviation [n/m/p] represent that the measurements were made on n basidiospores from m basidiomes of p collections. The range notation (a)b–c(d) stands for the dimensions of basidiospores in which b–c contains a minimum of 90% of the measured values while a and d in the brackets stand for the extreme values. In addition, a Q value show the length/width ratio of basidiospores and a Qm value for average Q ± standard deviation. All microstructures were illustrated by hand drawing.
DNA extraction, PCR and sequencing
Total genomic DNA was extracted using the Ezup Column Fungi Genomic DNA Purificaton Kit (Sangon Biotech, Shanghai, China) according to the manual. For the PCR amplification, (1) Primers ITS5 and ITS4 (White et al. 1990) were used for the internal transcribed spacer (ITS); (2) LROR and LR5 (Vilgalys and Hester 1990) for the nuclear ribosomal large subunit (nrLSU); (3) EF1-983F and EF1-1953R (Matheny et al. 2007), designed primers SPO-TEF1-F (5’-ATTGCYGGYGGTACYGGTGA-3’) and SPO-TEF1-R (5’-TCVAGDGATTTACCTGTHCGRC-3’) or another pair of designed primers HYG-TEF1-F (5’-CTTGCCTTYACTCTYGGYGTCC-3’) and HYG-TEF1-R (5’-GCGAACTTGCASGCAATGTG-3’) for the translation elongation factor 1-α (tef1-α); (4) RPB1-Af and RPB1-Cr (Matheny et al. 2002) or designed primers SPO-RPB1-F (5’-ACGAGGTTGYGTGGTGAAAT-3’) and SPO-RPB1-R (5’-GGAGGNGGDACHGGCATNA-3’) for the DNA-directed RNA polymerase II second largest subunit 1 (rpb1); (5) RPB2-6F and RPB2-7.1R (Matheny 2005) for the DNA-directed RNA polymerase II second largest subunit 2 (rpb2); and (6) ATP6-3 and ATP-6 (Kretzer and Bruns 1999) for ATP synthase subunit 6 (atp6).
The PCR mixtures contained 1× PCR buffer, 1.5mM MgCl2, 0.2mM dNTPs, each primer at 0.4 μM, 1.25U of Taq polymerase (Sangon Biotech, Shanghai, China), and 1 μL of DNA template in a total volume of 25 μL. Reactions were performed with the following program: initial denaturation at 94 °C for 5 min, 35 cycles at 94 °C for 30 s, 50 °C (atp6), 52 °C (nrLSU, tef1-α, rpb1 and rpb2) or 54 °C (ITS) for 30 s, and 72 °C for 30 s (ITS and atp6), 50 s (nrLSU and rpb2) or 75 s (tef1-α and rpb1), and for terminal elongation, the reaction batches were incubated at 72 °C for 10 min. All PCR products were detected by 2% agarose gel electrophoresis and then sent to the Kunming branch of Tsingke Biological Technology Co., Ltd. (Beijing, China) for sequencing.
Phylogenetic tree construction
Sequences used for phylogenetic analysis (presented in Table 1) were aligned by using MAFFT v7.471 (Katoh and Standley 2016) and then manually adjusted by using BIOEDIT v7.2.5 (Hall 1999). The intron regions of tef1-α, rpb2 and rpb1 were excluded except the conserved rpb1-intron2. Three datasets of ITS-nrLSU-rpb2, ITS-nrLSU-rpb1-rpb2-tef1-α-atp6 and ITS (Suppl. materials 1, 2 and 3) were used to construct phylogenetic trees. The two multi-gene matrixes were generated by SEQUENCEMATRIX 1.7.8 (Vaidya et al. 2011). GTR + I + G was inferred as the best-fit model for the three matrixes selected according to the AIC in MRMODELTEST v2.4 (Nylander 2004). Maximum likelihood (ML) trees with 1000 bootstrap replicates and Bayesian inferences were generated with RAXML v8.0.20 (Stamatakis 2006) and MRBAYES v3.2.7 (Ronquist and Huelsenbeck 2003), respectively.
Results
Molecular phylogenetic analysis
As shown in Table 1, a total of 393 sequences (109 ITS, 110 nrLSU, 40 tef1-α, 38 rpb1, 74 rpb2 and 22 atp6) from 118 samples were used in the phylogenetic analyses, 131 (23 ITS, 23 nrLSU, 20 tef1-α, 20 rpb1, 23 rpb2 and 22 atp6) of which were newly generated in the present study.
The combined dataset ITS-nrLSU-rpb2 comprised 221 sequences from 88 samples with a total of 3135 positions. In the three-gene tree (Fig. 1), 11 specimens from four novel Spodocybe species collected in this study, C. cf. trulliformis and C. herbarum formed a strongly supported monophyletic clade (BP = 100%, PP = 1.0), as sister to Ampulloclitocybe (BP = 63%, PP = 0.98). The phylogenetic analysis showed that the new proposed genus Spodocybe should be placed within the Hygrophoraceae, although intergeneric branched orders among Spodocybe, Ampulloclitocybe, Cantharocybe and Cuphophyllus were unstable with low support values.
Figure 1.
ML analysis of Hygrophoraceae combined ITS, nrLSU and rpb2 sequence data, with Macrotyphula juncea, Macrotyphula phacorrhiza and Phyllotopsis sp. as outgroups. Bootstrap values (BP) ≥ 50% from ML analysis and Bayesian posterior probabilities (PP) ≥ 0.90 are shown at nodes. The newly generated sequences are shown in bold.
In order to accurately determine the position of Spodocybe in the family Hygrophoraceae and better clarify the phylogenetic relationships of Spodocybe, Ampulloclitocybe, Cantharocybe and Cuphophyllus, a further six-gene matrix ITS-nrLSU-rpb1-rpb2-tef1-α-atp6 composed of 179 sequences from 54 samples with 5405 positions was used to rebuild the Hygrophoraceae tree. As revealed by the six-gene phylogenetic analysis (Fig. 2), the branch support level of the six-gene tree was obviously improved, compared with that of the previous three-gene tree. The monophyly of Spodocybe clade was strongly supported (BP = 100%, PP = 1.00), including Spodocybe rugosiceps (BP = 100%, PP = 1.00), S. bispora (BP = 100%, PP = 1.00) and two unnamed Spodocybe species. Spodocybe and Ampulloclitocybe were sister clades (BP = 78%, PP = 0.99), then further clustered with Cantharocybe (BP = 59%, PP = 0.97) and finally together with Cuphophyllus formed an independent lineage (BP = 85%, PP = 1.00). Meanwhile, this lineage (Cuphophylloideae) comprising the four genera was well-supported (BP = 83%, PP = 1.00) as sister to the rest of the Hygrophoraceae.
Figure 2.
ML analysis of Hygrophoraceae combined ITS, nrLSU, rpb1, rpb2, tef1-α and atp6 sequence data, with representatives of Amylocorticiaceae, Pterulaceae and the Hygrophoroid clade (Aphroditeola, Macrotyphula, Phyllotopsis, Pleurocybella, Sarcomyxa, Tricholomopsis and Typhula) as outgroups. Bootstrap values (BP) ≥ 50% from ML analysis and Bayesian posterior probabilities (PP) ≥ 0.90 are shown at nodes. Branches with BP ≥ 75% and PP ≥ 0.95 are bolded. The newly generated sequences are shown in bold. Lamellar trama type B for bidirectional, D for divergent, I for interwoven, P for pachypodial, R for regular, S for subregular, T for tri-directional. Lamellar trama types of specimens collected in this study were identified by ourselves and others referred to Lodge et al. (2014) and Hosen et al. (2016).
In addition, an ITS dataset (23 sequences, 1053 positions) was applied to phylogenetic analysis for displaying the relationships among Spodocybe species from this study and species of Clitocybe treated from GenBank. In the ITS tree (Fig. 3), Spodocybe species formed a highly supported monophyletic clade with C. trulliformis and related species (BP = 100%, PP = 1.00), which was also a sister clade to Ampulloclitocybe with strong support (BP = 91%, PP = 0.99).
Figure 3.
Phylogram showing the phylogenetic relationships among Spodocybe species and species of Clitocybe treated from Genbank based on ITS sequence data, with representatives of Ampulloclitocybe, Cuphophyllus and Hygrophorus as outgroups (rooted with Hygrophorus eburneus). Bootstrap values (BP) ≥ 50% from ML analysis and Bayesian posterior probabilities (PP) ≥ 0.90 are shown at nodes. The newly generated sequences are shown in bold. EA, NA and SE refer to East Asia, North America and South Europe, respectively.
Taxonomy
Cuphophylloideae
Z. M. He & Zhu L. Yang subf. nov.
C96C3465-EDF8-5E8F-B6E1-C1FDD9144C40
839377
Diagnosis.
Characterized generally by clitocyboid basidiomes, convex to funnel-shaped pileus, decurrent lamellae, absence of veils, inamyloid basidiospores and presence of clamps.
Etymology.
From the type genus Cuphophyllus.
Type genus.
Cuphophyllus (Donk) Bon.
Description.
Basidiomes small, medium-sized to large, mostly clitocyboid, rarely omphalinoid or mycenoid; veils absent. Pileus convex, applanate to funnel-shaped; surface usually dry, smooth, lubricous or rarely viscid. Lamellae decurrent to deeply decurrent. Basidiospores ellipsoid, oblong or subglobose, thin-walled and inamyloid. Pileipellis usually a cutis, sometimes ixocutis or trichoderm. Lamellar trama regular, subregular, interwoven or bidirectional. Clamp connections present.
Habitat, ecology and distribution.
Usually gregarious or caespitose on ground, rarely on wood; widespread in temperate and tropical regions.
The genera Ampulloclitocybe, Cantharocybe, Cuphophyllus and Spodocybe are included in the subfamily Cuphophylloideae, which is in correspondence with Cuphophylloid grade of Lodge et al. (2014) plus Spodocybe.
Spodocybe
Z. M. He & Zhu L. Yang gen. nov.
F15F630C-60D4-54EE-9717-AA0BF80B6081
839050
Diagnosis.
Differs from Ampulloclitocybe by its small basidiomes and subregular lamellar trama rather than medium-sized basidiomes and bidirectional lamellar trama. Differs from Cuphophyllus in the ratio of basidia to basidiospore length less than 5, and lamellar trama subregular rather than interwoven. Differs from Cantharocybe in its absence of cheilo- and caulocystidia, having small basidiomes rather than large ones and having subregular lamellar trama rather than regular one.
Etymology.
Spodo- refers to grey; -cybe refers to head; that is a Clitocybe-like genus with gray pileus.
Type species.
Spodocybe rugosiceps Z. M. He & Zhu L. Yang.
Description.
Basidiomes small, clitocyboid. Pileus convex, applanate to infundibuliform; surface dry, greyish (2B1), grey-brown (5C4) to dark grey-brown (5E4); center depressed with age. Lamellae decurrent to deeply decurrent, white (1A1) to cream (1A2), thin, moderately crowded, sometimes furcate and interveined. Stipe central, subcylindrical, concolorous with pileus. Basidiospores ellipsoid, oblong to cylindrical, colourless, hyaline, smooth, thin-walled, inamyloid; ratio of basidia to basidiospore length less than 5. Pileipellis and stipitipellis a cutis. Lamellar trama subregular. Clamp connections abundant, present in all parts of basidiome.
Habitat, ecology and distribution.
Saprophytic, usually gregarious or caespitose on the ground of coniferous or coniferous and broad-leaved mixed forest; distributed in the temperate and subtropical zones from June to November.
Spodocybe rugosiceps
Z. M. He & Zhu L. Yang sp. nov.
CCB3B84F-91A5-5DAD-B5A5-E6CC5355CF68
839052
Figure 4.
Basidiomes of described Spodocybe species. A, BSpodocybe rugosiceps (KUN-HKAS 112563, KUN-HKAS 112562, respectively) C, DSpodocybe bispora (KUN-HKAS 73332, KUN-HKAS 112562, respectively). Scale bars: 1 cm.
Figure 5.
Microscopic features of Spodocybe rugosiceps (KUN-HKAS 112563, holotype) a basidiospores b basidia c pileipellis. Scale bars: 10 μm.
Diagnosis.
Differs from S. bispora in having a rugose pileus, smaller basidiospores and 4-spored rather than 2-spored basidia. Differs from C. trulliformis in having smaller basidiospores and a rugose rather than felty-squamulose pileus.
Etymology.
rugosiceps refers to the rugose pileus.
Type.
China. Yunnan Province: Kunming City, near Yeya Lake, at 25.136658°N, 102.873027°E, alt. 2000 m, 11 Aug 2020, Z. M. He 72 (KUN-HKAS 112563, holotype).
Description.
Basidiomes small, clitocyboid. Pileus 0.5–2 cm in diam, at first nearly applanate, then concave; surface dry and rugose, gray-brown (5E2-4) to gray-black (4F2-4) in the center and gray-brown (5C2-4) or gray (5B1-2) towards margin; center often slightly umbonate; margin straight and undulating; context thin and white (1A1) to cream (1A2). Lamellae deeply decurrent, white (1A1) to cream (1A2), thin (up to 2 mm high), crowded, sometimes forked and intervenose. Stipe 2.5–6 × 0.2–0.4 cm, central, narrowly cylindrical to subcylindrical, sometimes flexuous, hollow; surface dry and nearly smooth, concolorous with pileus; context white (1A1).
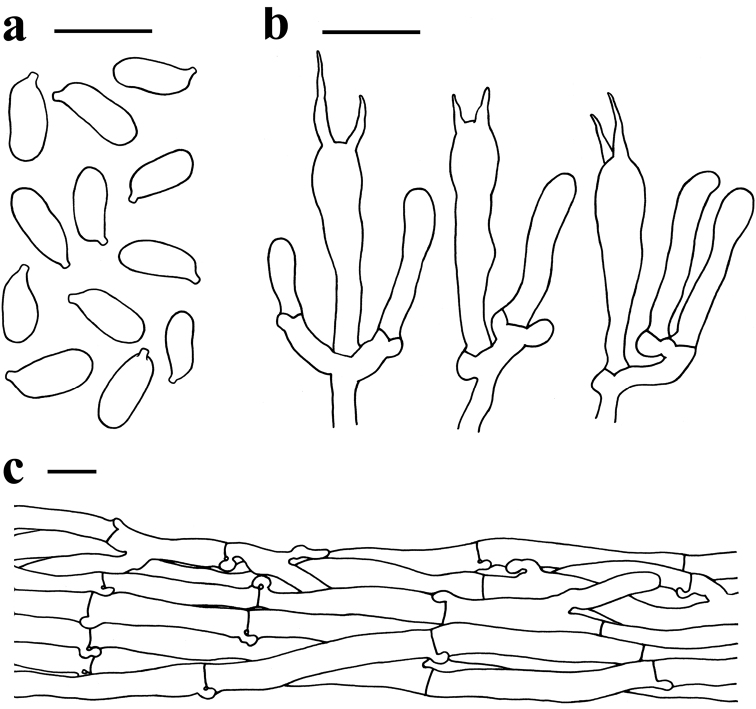
Basidiospores [60/3/3] 5–6 (6.5) × (2.5)3–3.5(4) μm, Q = (1.38)1.55–1.95(2), Qm = 1.73 ± 0.14, elongate, colorless, hyaline, smooth, thin-walled, inamyloid. Basidia 20–24 × 5–6 μm, clavate, 4-spored, colorless, hyaline, thin-walled; sterigmata up to 4 μm long; ratio of basidia to basidiospore length values about 3–5. Cystidia absent. Lamellar trama subregular; hyphae colorless, hyaline, cylindrical, thin-walled, 3–10 µm wide. Pileipellis a cutis, but in places upright or trichodermial in appearance, made up with thin-walled cylindrical hyphae 3–9 µm wide. Stipitipellis a cutis, composed of thin-walled cylindrical hyphae 3–10 μm wide. Clamp connections present in all parts of basidiome.
Habitat, ecology and distribution.
Gregarious or caespitose, growing saprotrophically in forest litter, often under conifers, on the ground, known from subtropical zone of Yunnan, China; from July to October.
Additional specimens examined.
China. Yunnan Province: Dali Bai Autonomous Prefecture, Yunlong Country, Tianchi National Nature Reserve, at 25.850365°N, 99.274236°E, alt. 2509 m, 28 Sep 2019, X. H. Wang 7471 (KUN-HKAS 112561); Kunming City, Fangwang Tree Farm, at 25.063737°N, 102.870690°E, alt. 2262 m, 22 Sep 2011, Z. L. Yang 5586 (KUN-HKAS 71071); Kunming City, Kunming Institute of Botany, at 25.147081°N, 102.748855°E, alt. 1990 m, 24 Aug 2020, Z. L. Yang 6391 (KUN-HKAS 112562); Kunming City, Qiongzhu Temple, at 25.071304°N, 102.630934°E, alt. 1900 m, 28 Jul 2013, T. Guo 779 (KUN-HKAS 81981); Yulong Country, Lashi Village, at 26.883902°N, 100.234594°E, alt. 2655 m, 31 Jul 2011, L. P. Tang 1369 (KUN-HKAS 69830).
Spodocybe bispora
Z. M. He & Zhu L. Yang sp. nov.
EB4F3677-029E-51F4-A106-7B5D99A2D117
839054
Figure 6.
Microscopic features of Spodocybe bispora (KUN-HKAS 73310, holotype) a basidiospores b basidia c pileipellis. Scale bars: 10 μm.
Diagnosis.
Differs from S. rugosiceps in having a nearly smooth pileus, larger basidiospores and 2-spored rather than 4-spored basidia. Differs from C. trulliformis in having a nearly smooth rather than felty-squamulose pileus.
Etymology.
Bispora refers to 2-spored.
Type.
China. Yunnan Province: Baoshan City, Longyang District, Shuizhai Village, at 25.273967°N, 99.306216°E, alt. 2400 m, 12 Aug 2011, J. Qin 324 (KUN-HKAS 73310, holotype).
Description.
Basidiomes small, clitocyboid. Pileus 1.5–3 cm in diam, plano-convex to funnel-shaped; surface dry and nearly smooth, greyish-brown (4B2-3) to grey-brown (4E3-5); center depressed, usually with a low umbo, somewhat darker; margin generally straight and undulating, incurved when old; context thin and white (1A1). Lamellae deeply decurrent, white (1A1) to cream (1A2), thin, 1–2 mm high, relatively crowded, sometimes forked and intervenose. Stipe 1–3 × 0.2–0.4 cm, central, subcylindrical, hollow; surface dry and nearly smooth, concolorous with pileus; context white (1A1).
Basidiospores [60/3/3] (7)7.5–10.5(11.5) × 3–4 μm, Q = (2.05)2.11–3(3.33), Qm = 2.56 ± 0.3, cylindrical, colorless, hyaline, smooth, thin-walled, inamyloid. Basidia 20–30 × 4–5.5 μm, clavate, 2-spored, colorless, hyaline, thin-walled; sterigmata up to 10 μm long; ratio of basidia to basidiospore length less than 5 (about 2–4). Cystidia absent. Lamellar trama subregular, colorless, hyaline, made up of thin-walled cylindrical hyphae with 3–10 µm wide. Pileipellis a cutis, composed of thin-walled cylindrical hyphae 3–11 µm wide. Stipitipellis a cutis, composed of thin-walled cylindrical hyphae 3–10 μm wide. Clamp connections in all parts of basidiomes.
Habitat, ecology and distribution.
Saprophytic, usually gregarious on the ground of coniferous or coniferous and broad-leaved mixed forest, known from Yunnan, China; July to September.
Additional specimens examined.
China. Yunnan Province: Kunming City, Qipan Mountain, at 26.060020°N, 102.576823°E, alt. 1900 m, 25 Jul 2020, Z. M. He 35 (KUN-HKAS 112564); Nujiang City, Lanping Country, No. 311 Provincial Highway, at 26.636613°N, 99.557809°E, alt. 2660 m, 14 Aug 2011, J. Qin 346 (KUN-HKAS 73332).
Discussion
The new genus Spodocybe
In our current study, the new clitocyboid species were clustered into a monophyletic lineage (BP = 100%, PP = 1.00) in the Hygrophoraceae according to the multi-gene phylogenetic analysis (Figs 1, 2). As a result, the new generic name Spodocybe is proposed here to accommodate the new lineage, which is irrelevant to Clitocybeae of the Tricholomatoid clade (Matheny et al. 2006; Alvarado et al. 2015). The three-gene tree of the Hygrophoraceae (Fig. 1) in this study presented basically consistent topological structure with Lodge et al. (2014), and showed that Spodocybe was a sister to Ampulloclitocybe located within the family Hygrophoraceae and further confirmed by a six-gene tree (Fig. 2).
Besides the molecular analyses, morphological data also support its separation from the relative genera. Spodocybe shares clitocyboid basidiomes, decurrent lamellae, inamyloid basidiospores and the presence of clamps with the other genera Ampulloclitocybe, Cuphophyllus and Cantharocybe. However, the genus Ampulloclitocybe, typified by A. clavipes, differs from Spodocybe in having medium-sized basidiomes and bidirectional lamellar trama (Harmaja 2002; Lodge et al. 2014). Afterwards, Cuphophyllus differs from Spodocybe in having long basidia, typically 7−8 (rarely 5−6) times the length of the basidiospores, highly interwoven lamellar trama, rarely subregular (Voitk et al. 2020). Finally, Cantharocybe differs from Spodocybe in having large basidiomes, broad lamellae, cheilo- and caulocystidia, clamps but not on all hyphal septa or at the base of every basidium and more regular lamellar trama (Ovrebo 2011; Hosen et al. 2016). In view of the four genera above with different structures in lamellar trama (Fig. 2), the type of lamellar trama can become a good distinguishing microscopic character for them.
For a long time, C. trulliformis has been placed in the genus Clitocybe based on the clitocyboid feature and habit since 1879 (Karsten 1879). However, C. trulliformis shares many morphological characteristics with Spodocybe, such as the small basidioma with applanate to infundibuliform pileus, grey-brown pileus and stipe, decurrent and whitish lamellae, and smooth and inamyloid basidiospores (Bas et al. 1995). Besides, the ITS phylogenetic analysis in our study (Fig. 3) showed that C. trulliformis and related Clitocybe species were involved in the Spodocybe clade as well, indicating that C. trulliformis and related species should be placed with Spodocybe. In consequence, it is foreseeable that C. trulliformis and other related clitocyboid species will eventually be moved to Spodocybe. Accordingly, more taxonomic work is needed in future.
The placements of Spodocybe, Cuphophyllus, Ampulloclitocybe and Cantharocybe
In previous studies, Cuphophyllus, Ampulloclitocybe and Cantharocybe were treated as basal in Hygrophoraceae (Lodge et al. 2014), but their phylogenetic placements were not resolved. In a six-gene phylogenetic analysis by Binder et al. (2010) and a three-gene analysis by Wang et al. (2018), Ampulloclitocybe and Cantharocybe were located between Cuphophyllus and the rest of the Hygrophoraceae, but without support. While two four-gene analyses by Lodge et al. (2014) showed that Ampulloclitocybe and Cantharocybe were sister clades as basal to Cuphophyllus along with the rest of the Hygrophoraceae with weak support. However, in our six-gene analysis (Fig. 2), the new proposed genus Spodocybe and Ampulloclitocybe were sisters (BP = 78%, PP = 0.99) and they clustered with Cantharocybe followed by Cuphophyllus, forming a supported monophyletic sister clade to the rest of the Hygrophoraceae (BP = 83%, PP = 1.00). Hence, Spodocybe, Ampulloclitocybe, Cantharocybe and Cuphophyllus should be retained in Hygrophoraceae, and a new subfamily, Cuphophylloideae, is proposed to accommodate the lineage.
Supplementary Material
Acknowledgements
The authors are very grateful to their colleagues at Kunming Institute of Botany, Chinese Academy of Sciences, including Drs. Xiang-Hua Wang, Jiao Qin, Bang Feng, Qi Zhao and Master students Hua Qu and Si-Peng Jian for collecting and providing specimens; and Drs. Gang Wu, Yang-Yang Cui, Qing Cai for providing help on morphological observation and phylogenetic analysis. This study was financed by Yunnan Ten-Thousand-Talents Plan – Yunling Scholar Project and Postdoctoral Directional Training Foundation of Yunnan Province.
Citation
He Z-M, Yang ZL (2021) A new clitocyboid genus Spodocybe and a new subfamily Cuphophylloideae in the family Hygrophoraceae (Agaricales). MycoKeys 79: 129–148. https://doi.org/10.3897/mycokeys.79.66302
Funding Statement
Yunnan Ten-Thousand-Talents Plan - Yunling Scholar Project and Postdoctoral Directional Training Foundation of Yunnan Province
Supplementary materials
Alignment of ITS-LSU-RPB2 dataset used in the three-gene phylogenetic analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zheng-Mi He, Zhu L. Yang
Data type
fasta file
Explanation note
ITS: 1-1380, LSU: 1381–2356, RPB2: 2357–3135.
Alignment of ITS-LSU-RPB1-RPB2-TEF1-ATP6 dataset used in the six-gene phylogenetic analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zheng-Mi He, Zhu L. Yang
Data type
fasta file
Explanation note
ITS: 1–1217, LSU: 1218–2158, RPB1: 2159–3358, RPB2: 3359–4089, TEF1: 4090–4967, ATP6: 4968–5405
Alignment of ITS dataset used in the single-gene phylogenetic analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zheng-Mi He, Zhu L. Yang
Data type
fasta file
References
- Alvarado P, Moreno G, Vizzini A, Consiglio G, Manjón JL, Setti L. (2015) Atractosporocybe, Leucocybe and Rhizocybe: three new clitocyboid genera in the Tricholomatoid clade (Agaricales) with notes on Clitocybe and Lepista. Mycologia 107(1): 123–136. 10.3852/13-369 [DOI] [PubMed] [Google Scholar]
- Ammirati JF, Parker AD, Matheny PB. (2007) Cleistocybe, a new genus of Agaricales. Mycoscience 48(5): 282–289. 10.1007/S10267-007-0365-5 [DOI] [Google Scholar]
- Bas C, Kuyper TW, Noordeloos ME, Vellinga EC. (1995) Flora agaricina neerlandica 3: critical monographs on families of agarics and boleti occurring in the Netherlands. A. A. Balkema, Rotterdam, 183 pp. [Google Scholar]
- Binder M, Larsson KH, Matheny PB, Hibbett DS. (2010) Amylocorticiales ord. nov. and Jaapiales ord. nov.: early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia 102(4): 865–880. 10.3852/09-288 [DOI] [PubMed] [Google Scholar]
- Breitenbach J, Kraenzlin F. (1991) Fungi of Switzerland 3: Boletales and Agaricales. Mykologia, Luzern, 361 pp. [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Harmaja H. (2002) Amylolepiota, Clavicybe and Cystodermella, new genera of the Agaricales. Karstenia 42(2): 39–48. 10.29203/ka.2002.386 [DOI] [Google Scholar]
- Harmaja H. (2003) Notes on Clitocybe s. lato (Agaricales). Annales Botanici Fennici 40(3): 213–218. [Google Scholar]
- Hosen MI, Li TH, Lodge DJ, Rockefeller A. (2016) The first ITS phylogeny of the genus Cantharocybe (Agaricales, Hygrophoraceae) with a new record of C. virosa from Bangladesh. Mycokeys 14: 37–50. 10.3897/mycokeys.14.9859 [DOI] [Google Scholar]
- Karsten PA. (1879) Rysslands, Finlands och den Skandinaviska halföns Hattsvampar. Förra Delen: Skifsvampar. Bidrag till Kännedom av Finlands Natur och Folk 32: 1–571. [Google Scholar]
- Katoh K, Standley DM. (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32(13): 1933–1942. 10.1093/bioinformatics/btw108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978) Methuen handbook of colour (3rd edn). Eyre Methuen, London, 252 pp. [Google Scholar]
- Kretzer AM, Bruns TD. (1999) Use of atp6 in fungal phylogenetics: an example from the Boletales. Molecular Phylogenetics and Evolution 13(3): 483–492. 10.1006/mpev.1999.0680 [DOI] [PubMed] [Google Scholar]
- Læssøe T, Petersen JH. (2019) Fungi of temperate Europe. Princeton University Press, Princeton, 1708 pp. [Google Scholar]
- Lodge DJ, Padamsee M, Matheny PB, Aime MC, Cantrell SA, Boertmann D, Kovalenko A, Vizzini A, Dentinger BTM, Kirk PM, Ainsworth AM, Moncalvo JM, Vilgalys R, Larsson E, Lücking R, Griffith GW, Smith ME, Norvell LL, Desjardin DE, Redhead SA, Ovrebo CL, Lickey EB, Ercole E, Hughes KW, Courtecuisse R, Young A, Binder M, Minnis AM, Lindner DL, Ortiz-Santana B, Haight J, Læssøe T, Baroni TJ, Geml J, Hattori T. (2014) Molecular phylogeny, morphology, pigment chemistry and ecology in Hygrophoraceae (Agaricales). Fungal Diversity 64: 1–99. 10.1007/s13225-013-0259-0 [DOI] [Google Scholar]
- Matheny PB. (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Molecular Phylogenetics and Evolution 35(1): 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Liu YJ, Ammirati JF, Hall BD. (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany 89(4): 688–698. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Hofstetter V, Aime MC, Moncalvo JM, Ge ZM, Yang ZL, Slot JC, Ammirati JF, Baroni TJ, Bougher NL, Hughes NW, Lodge DJ, Kerrigan R, Seidl MT, Aanen DK, DeNitis M, Daniele GM, Desjardin DE, Kropp BR, Norvell LL, Parker A, Vellinga EC, Vilgalys R, Hibbett DS. (2006) Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia 98(6): 982–995. 10.1080/15572536.2006.11832627 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson H, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, Langer E, Langer G, McLaughlin DJ, Wilson AW, Frøslev T, Ge ZW, Kerrigan RW, Slot JC, Yang ZL, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vauras J, Hibbett DS. (2007) Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 43(2): 430–451. 10.1016/j.ympev.2006.08.024 [DOI] [PubMed] [Google Scholar]
- Moncalvo JM, Vilgalys R, Redhead SA, Johnson JE, James TY, Aime MC, Hofstetter V, Verduin SJW, Larsson E, Baroni TJ, Thorn RG, Jacobsson S, Clémençon H, Miller Jr OK. (2002) One hundred and seventeen clades of euagarics. Molecular Phylogenetics and Evolution 23(3): 357–400. 10.1016/S1055-7903(02)00027-1 [DOI] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Ovrebo CL. (2011) A new Cantharocybe from Belize with notes on the type of Cantharocybe gruberi. Mycologia 103(5): 1102–1109. 10.3852/10-360 [DOI] [PubMed] [Google Scholar]
- Qin J, Feng B, Yang ZL, Li YC, Ratkowsky D, Gates G, Takahashi H, Rexer KH, Kost GW, Karunarathna SC. (2014) The taxonomic foundation, species circumscription and continental endemisms of Singerocybe: evidence from morphological and molecular data. Mycologia 106(5): 1015–1026. 10.3852/13-338 [DOI] [PubMed] [Google Scholar]
- Redhead SA, Lutzoni F, Moncalvo JM, Vilgalys R. (2002) Phylogeny of agarics: partial systematics solutions for core omphalinoid genera in the Agaricales (euagarics). Mycotaxon 83: 19–57. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Singer R. (1986) The Agaricales in Modern Taxonomy (4th edn). Koeltz Scientific Books, Koenigstein, 981 pp. [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21): 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) Sequencematrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzini A, Musumeci E, Murat C. (2010) Trichocybe, a new genus for Clitocybe puberula (Agaricomycetes, Agaricales). Fungal Diversity 42: 97–105. 10.1007/s13225-010-0030-8 [DOI] [Google Scholar]
- Voitk A, Saar I, Lodge DJ, Boertmann D, Berch SM, Larsson E. (2020) New species and reports of Cuphophyllus from northern North America compared with related Eurasian species. Mycologia 112(2): 438–452. 10.1080/00275514.2019.1703476 [DOI] [PubMed] [Google Scholar]
- Wang CQ, Zhang M, Li TH, Liang XS, Shen YH. (2018) Additions to tribe Chromosereae (Basidiomycota, Hygrophoraceae) from China, including Sinohygrocybe gen. nov. and a first report of Gloioxanthomyces nitidus. Mycokeys 38: 59–76. 10.3897/mycokeys.38.25427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of ITS-LSU-RPB2 dataset used in the three-gene phylogenetic analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zheng-Mi He, Zhu L. Yang
Data type
fasta file
Explanation note
ITS: 1-1380, LSU: 1381–2356, RPB2: 2357–3135.
Alignment of ITS-LSU-RPB1-RPB2-TEF1-ATP6 dataset used in the six-gene phylogenetic analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zheng-Mi He, Zhu L. Yang
Data type
fasta file
Explanation note
ITS: 1–1217, LSU: 1218–2158, RPB1: 2159–3358, RPB2: 3359–4089, TEF1: 4090–4967, ATP6: 4968–5405
Alignment of ITS dataset used in the single-gene phylogenetic analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zheng-Mi He, Zhu L. Yang
Data type
fasta file