We have read with interest the article by Crinier et al. describing a comprehensive single-cell RNA sequencing dataset of human bone marrow natural killer (NK) cells from eight healthy donors.1 Their unbiased analysis resulted in the identification of three NK cell populations in bone marrow: hNK_Bm1, hNK_Bm2 and hNK_Bm3. These populations correspond to the NK cell populations that the authors previously described in human spleen: hNK_Spl1, hNK_Sp2 and hNK_Sp3.2 The authors compared the gene signatures of the three bone marrow populations to the conventional CD56bright and CD56dim NK cells in the blood and concluded that hNK_Bm1 cells resemble CD56dim NK cells, hNK_Bm2 cells correspond to CD56bright NK cells, and hNK_Bm3 represent a CD56bright tissue-resident NK cell population that is not present in the blood.1
Previously, we described a tissue-resident CD56bright CD69+CXCR6+ NK cell population in human lymphoid tissues and reported bulk RNA sequencing data on this lymphoid tissue NK (ltNK) population in bone marrow together with conventional CD56dim and CD56bright NK cells from bone marrow and blood.3,4 ltNK cells typically express CD69 and CXCR6, two hallmarks of tissue residency.4–7 These markers are also reported among the top ten membrane markers enriched in hNK_Bm2 cells.1 This prompted us to investigate the possibility that hNK_Bm2 cells, rather than hNK_Bm3 cells, are the tissue-resident NK cell population.
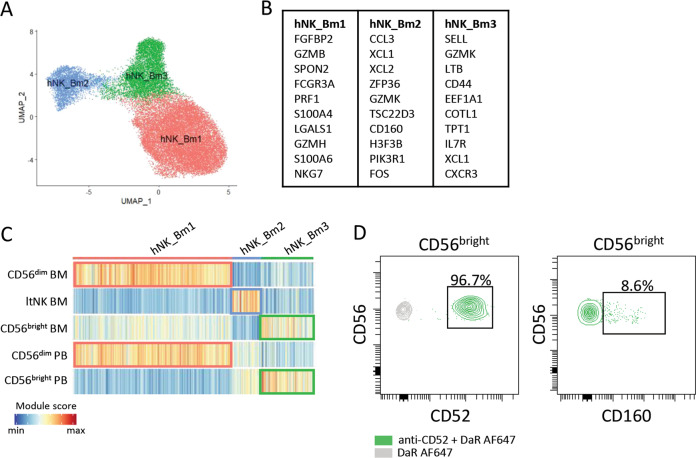
To study this hypothesis, we reanalyzed the raw data uploaded to the GEO repository (GSE159624) and performed the same data preprocessing and quality control steps as reported by Crinier et al.1 Data integration was performed by applying MNN correction to eliminate donor-to-donor variation.8 Louvain clustering with a low resolution parameter revealed three clusters (Fig. 1A). Based on the differentially expressed genes, these clusters were comparable to those described by Crinier et al. (Fig. 1B).1 We then compared the transcriptomes of hNK_Bm1-3 cells with the bulk gene signature of sorted blood- and bone marrow-derived CD56dim and CD56bright NK cells and bone marrow ltNK cells3 by applying the AddModuleScore function of Seurat, as used by Crinier et al.1 This demonstrated a strong correlation between hNK_Bm1 and CD56dim cells in blood and bone marrow, between hNK_Bm2 and ltNK cells in bone marrow and between hNK_Bm3 cells and CD56bright NK cells in blood and bone marrow (Fig. 1C).1,3 To further validate this finding, we performed flow cytometry on blood NK cells, which demonstrated that nearly all circulating CD56bright NK cells have the CD52+CD160- hNK_Bm3 phenotype (Fig. 1D).
Fig. 1.
Reanalysis of single-cell RNA sequence data of human bone marrow NK cells. A Louvain clustering and UMAP were performed on the integrated dataset of eight healthy donors (data: GEO repository GSE15962, Crinier et al.1). B The top ten most differentially expressed genes per cluster are shown. Ribosomal protein coding genes were excluded. C Module score analysis was performed to compare the gene signatures of the clusters (Crinier et al.1) to the gene signatures determined by bulk RNA sequencing of CD56bright, CD56dim and ltNK cells (Melsen et al.3). D The CD52 and CD160 expression of CD56bright NK cells of one representative blood donor is shown. As a control for the two-step staining protocol of CD52, staining with the secondary antibody (DaR) only is shown in gray. BM = bone marrow, PB = peripheral blood, ltNK = lymphoid tissue NK, DaR = donkey anti-rat
We agree with the conclusion of Crinier et al. that NK cells in human bone marrow can be categorized into three subpopulations and that the large hNK_Bm1 population corresponds to CD56dim NK cells. However, our re-evaluation supports the hypothesis that hNK_Bm2 cells represent a tissue-resident NK cell population: the transcriptome of hNK_Bm2 cells is most similar to the transcriptome of the tissue-resident ltNK cell population from human bone marrow that we previously described.3,4 Moreover, the CD69+CD52- phenotype of hNK_Bm2 cells (Fig. 3D & 3F in Crinier et al.) is suggestive of tissue residency.4,6,7,9 The fact that hNK_Bm2 cells bear a strong resemblance to hNK_Sp2 cells in the human spleen (Fig. 3C in Crinier et al.)1,2 suggests that the hNK_Sp2 population might also represent a tissue-resident NK cell population.
On the other hand, hNK_Bm3 cells show strong similarities with the CD56bright NK cells in the blood. This is in line with Fig. 3B and Supplementary Fig. 2B in Crinier et al., in which the authors compared the transcriptomes of hNK_Bm2 and hNK_Bm3 populations with those of circulating CD56bright NK cells as published by Hanna et al.10 Taken together, these findings support the hypothesis that hNK_Bm3 cells represent conventional CD56bright NK cells, whereas hNK_Bm2 cells represent the tissue-resident NK cell population in human bone marrow.
Competing interests
The authors declare no competing interests.
References
- 1.Crinier, A. et al. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell. Mol. Immunol.18, 1290–1304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crinier, A. et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity49, 971–986.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melsen, J. E. et al. Human Bone Marrow-Resident Natural Killer Cells Have a Unique Transcriptional Profile and Resemble Resident Memory CD8+ T Cells. Front. Immunol.9, 1829 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugthart, G. et al. Human Lymphoid Tissues Harbor a Distinct CD69 + CXCR6 + NK Cell Population. J. Immunol.197, 78–84 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Wein, A. N. et al. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med. 216, 2748–2762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiow, L. R. et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature440, 540–544 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Dogra, P. et al. Tissue Determinants of Human NK Cell Development, Function, and Residence. Cell180, 749–763.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol.36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hotta, R. et al. CD52-Negative NK Cells Are Abundant in the Liver and Less Susceptible to Alemtuzumab Treatment. PLoS ONE11, e0161618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna, J. et al. Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling. J. Immunol.173, 6547–6563 (2004). [DOI] [PubMed] [Google Scholar]