Abstract
ARIEL2 (NCT01891344) is a single-arm, open-label phase 2 study of the PARP inhibitor (PARPi) rucaparib in relapsed high-grade ovarian carcinoma. In this post hoc exploratory biomarker analysis of pre- and post-platinum ARIEL2 samples, RAD51C and RAD51D mutations and high-level BRCA1 promoter methylation predict response to rucaparib, similar to BRCA1/BRCA2 mutations. BRCA1 methylation loss may be a major cross-resistance mechanism to platinum and PARPi. Genomic scars associated with homologous recombination deficiency are irreversible, persisting even as platinum resistance develops, and therefore are predictive of rucaparib response only in platinum-sensitive disease. The RAS, AKT, and cell cycle pathways may be additional modulators of PARPi sensitivity.
Subject terms: Ovarian cancer, Tumour biomarkers
The identification of biomarkers of response to PARP inhibitors can enable selection of appropriate ovarian cancer patients for treatment. In this study, the authors report clinical results and exploratory biomarker analyses from the ARIEL2 phase 2 clinical trial on the safety and efficacy of the PARP inhibitor rucaparib in patients with recurrent ovarian cancers
Introduction
Rucaparib is an inhibitor of poly(ADP-ribose) polymerase (PARP) 1, PARP2, and PARP3, DNA damage repair enzymes in the base excision repair pathway1. Homologous recombination deficiency (HRD) sensitizes neoplasms to rucaparib and other DNA-damaging agents (e.g., platinum-based chemotherapy)2,3, and platinum sensitivity in high-grade ovarian carcinoma (HGOC) is a strong clinical predictor of benefit from PARP inhibitors (PARPi)4–7.
Genetic, epigenetic, and genomic biomarkers can suggest the presence of HRD and help identify patients most likely to respond to PARPi8–10. Germline and somatic BRCA1 or BRCA2 (BRCA) mutations are well-defined biomarkers for PARPi response for a number of cancer types, including breast, ovarian, pancreatic, and prostate11–16. Alterations in other homologous recombination repair (HRR) pathway genes, including PALB2, RAD51C, and RAD51D, have been associated with improved responses to rucaparib and other PARPi17–19. However, the extent to which many HRR genes contribute to PARPi sensitivity remains unclear20. BRCA1 and RAD51C promoter methylation results in transcriptional silencing21 and commonly leads to HRD in HGOC22. In preclinical studies, BRCA1 or RAD51C methylation led to increased sensitivity to PARPi and platinum23–25, but establishing a consistent association of methylation with clinical responses has been elusive26,27.
Given the variety of mechanisms that can result in HRD, methods for identifying HRD cancers independent of the mechanisms involved are desirable. HRD cancers exhibit high genomic instability, characterized by deletions of large genomic segments (genome-wide loss of heterozygosity [LOH]), among other genomic aberrations28,29. Next-generation sequencing (NGS) can identify multiple patterns of genomic change, including copy number variations, single-nucleotide variations, insertions/deletions, rearrangements characteristic of HRD29,30, and others31–34. HGOCs with a BRCA mutation or BRCA1/RAD51C methylation have high levels of genomic LOH25. Patients with BRCA wild-type (BRCAwt) HGOC with high LOH (≥16%) show greater clinical benefit from rucaparib than those with low LOH7. High LOH is also correlated with improved response to platinum and other PARPi6,35. However, genomic scars accumulate in HRD cancer cells over time and do not disappear when HRR is restored or other PARPi resistance mechanisms develop, making them imperfect biomarkers of PARPi sensitivity36. Therefore, understanding the mechanisms leading to HRD and how they might change during prior therapies is important to predict outcomes with PARPi.
ARIEL2 is an international, open-label, 2-part, phase 2 study assessing the safety and efficacy of rucaparib as active treatment in patients with relapsed HGOC. Part 1 enrolled patients with platinum-sensitive disease; clinical results and molecular data from this portion were published17,25,37,38. Part 2 enrolled patients who had received three to four prior chemotherapies, including patients with platinum-sensitive or platinum-resistant/refractory disease. Here we present clinical results from Part 2 and post hoc exploratory biomarker analyses using the rich dataset of archival tissue samples and screening biopsies required from patients in both Parts 1 and 2.
Results
Part 2 efficacy and safety
Between October 2013 and August 2016, 491 patients were enrolled and received rucaparib in ARIEL2 (Part 1, n = 204; Part 2, n = 287; Supplementary Fig. S1). Baseline demographics and disease characteristics are presented in Table 1 and Supplementary Table S1. The protocol-prespecified primary endpoints classified patients’ HGOC into one of three HRD groups (BRCA-mutant [BRCAmut], BRCAwt/LOH-high, and BRCAwt/LOH-low) using mutations and genome-wide LOH estimates provided by targeted panel NGS of neoplastic tissue (Online Methods)24. Among heavily pretreated patients in Part 2, confirmed objective response rates (ORR), the study’s primary endpoint, were 31.0% (95% confidence interval [CI], 21.3–42.0), 6.8% (95% CI, 2.3–15.3), and 5.6% (95% CI, 2.1–11.8), respectively, in patients with BRCAmut, BRCAwt/LOH-high, and BRCAwt/LOH-low HGOC (Supplementary Table S2), with durable responses seen across HRD subgroups (Supplementary Fig. S2a). Data from secondary efficacy endpoints, including ORR by Response Evaluation Criteria In Solid Tumors version 1.1 (RECIST)/Gynecological Cancer InterGroup (GCIG) cancer antigen 125 (CA-125) criteria, progression-free survival (PFS), and overall survival are presented in Supplementary Table S2 and Supplementary Fig. S2b, c.
Table 1.
Select baseline demographics and disease characteristics.
| ARIEL2 Part 1 (n = 204) | ARIEL2 Part 2 (n = 287) | |
|---|---|---|
| Age, median (range), years | 64.5 (31.0–86.0) | 63.0 (35.0–91.0) |
| ECOG PS, n (%) | ||
| 0 | 134 (65.7) | 134 (46.7) |
| 1 | 70 (34.3) | 151 (52.6) |
| ≥2 | 0 | 2 (0.7) |
| Cancer type, n (%) | ||
| Epithelial ovarian cancer | 164 (80.4) | 234 (81.5) |
| Primary peritoneal cancer | 24 (11.8) | 28 (9.8) |
| Fallopian tube cancer | 16 (7.8) | 25 (8.7) |
| Histology, n (%)a | ||
| Serous | 197 (96.6) | 269 (93.7) |
| Endometrioid | 4 (2.0) | 12 (4.2) |
| Mixed | 3 (1.5) | 6 (2.1) |
| No. of prior chemotherapy regimens, median (range) | 1 (1–6) | 3 (2–5) |
| No. of prior chemotherapy regimens, n (%) | ||
| 1 | 119 (58.3) | 0 |
| 2 | 52 (25.5) | 2 (0.7)b |
| 3 | 24 (11.8) | 186 (64.8) |
| 4 | 5 (2.5) | 97 (33.8) |
| ≥5 | 4 (2.0) | 2 (0.7) |
| No. of platinum-based regimen, median (range) | 1 (1–5) | 3 (1–4) |
| Platinum status, n (%) | ||
| Sensitive | 202 (99.0) | 81 (28.2) |
| Resistant | 2 (1.0) | 158 (55.1) |
| Refractory | 0 | 48 (16.7) |
| No. of non-platinum regimens following last platinum regimen, n (%) | ||
| 0 | 204 (100.0) | 81 (28.2) |
| 1 | 0 | 150 (52.3) |
| 2 | 0 | 52 (18.1) |
| 3 | 0 | 4 (1.4) |
ECOG Eastern Cooperative Oncology Group, PS performance status.
aHistology based on information provided at enrollment. Pathology re-review triggered by molecular findings reclassified 11 cases as non-high-grade serous or Grade 2/3 endometrioid histology (see Supplementary Table S12).
bReceipt of <3 prior chemotherapy regimens was considered a protocol deviation.
The toxicity profile of rucaparib in the heavily pretreated patients in Part 2 (Supplementary Tables S3–S5) was consistent with that previously reported in patients with HGOC7,37,39.
Molecular subgroup profiling across Parts 1 and 2
Taking advantage of the large sample size with varying degree of platinum sensitivity and number of prior treatments, we analyzed the association between molecular characteristics and clinical outcomes across both parts of ARIEL2 to identify molecular mechanisms resulting in rucaparib sensitivity and resistance.
Molecular analyses were used to characterize HGOC based on mutation, methylation, and molecular subgroup status. In these post hoc analyses, molecular subgroups were determined using a 16% LOH cutoff for LOH-high in the BRCAwt subgroup; this refined cutoff was validated within ARIEL2 Part 1 (ref. 40) and was prospectively tested in the maintenance setting in ARIEL3 (NCT01968213)7. To define the BRCAmut subgroup comprehensively, we utilized both available local testing data and an updated BRCA missense mutation classifier to reclassify the HGOC from 14 patients as BRCAmut, beyond those initially identified by targeted NGS (Supplementary Fig. S3, Online Methods). In total, 138 HGOC were classified as BRCAmut, 156 were BRCAwt/LOH-high, 168 were BRCAwt/LOH-low, and 29 were BRCAwt/LOH-unclassified.
Efficacy based on molecular subgroup and platinum status
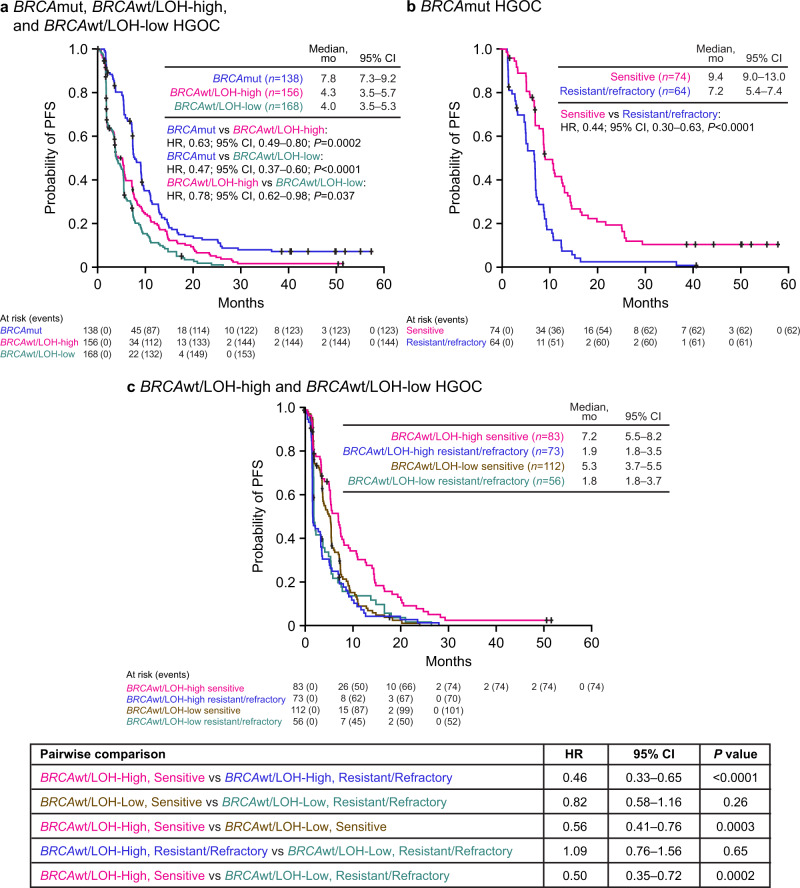
PFS and ORR differed significantly among molecular subgroups. Patients with BRCAmut HGOC had superior outcomes, with a median PFS of 7.8 months and ORR of 45.7% (95% CI, 37.2–54.3). In contrast, among patients with BRCAwt/LOH-high and BRCAwt/LOH-low HGOC, median PFS was 4.3 months and 4.0 months and ORR was 16.7% (95% CI, 11.2–23.5) and 7.7% (95% CI, 4.2–12.9), respectively (Figs. 1a and 2 and Supplementary Table S6).
Fig. 1. PFS by molecular subgroup and platinum-sensitivity status.
a PFS in patients with BRCAmut (blue), BRCAwt/LOH-high (magenta), and BRCAwt/LOH-low (teal) HGOC. b PFS in patients with BRCAmut HGOC that are platinum resistant/refractory (blue) or platinum sensitive (magenta). c PFS in patients with BRCAwt/LOH-high HGOC that are platinum resistant/refractory (blue) or platinum sensitive (magenta) and PFS in patients with BRCAwt/LOH-low HGOC that are platinum resistant/refractory (teal) or platinum sensitive (brown). P values were computed using a Cox proportional hazard model. The interaction between molecular subgroup and platinum status was also tested in the Cox proportional hazard model and found to be significant (P < 0.05). BRCA BRCA1 or BRCA2, CI confidence interval, HGOC high-grade ovarian carcinoma, HR hazard ratio, LOH loss of heterozygosity, mut mutated, PFS progression-free survival, wt wild type.
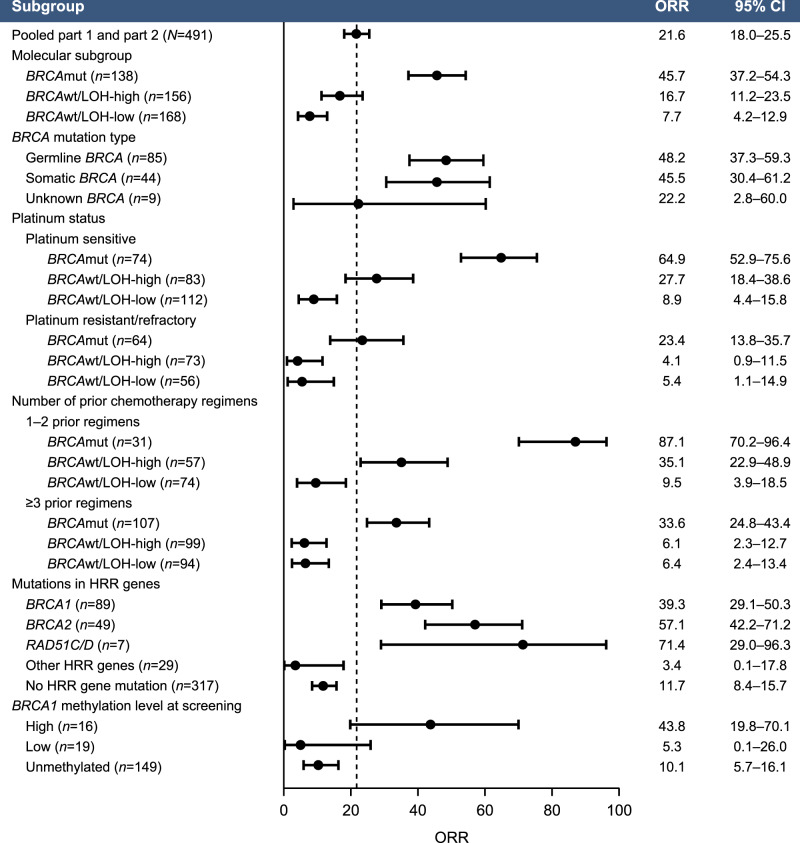
Fig. 2. ORR based on post hoc molecular subgroups and baseline clinical characteristics.
Data are plotted as ORR (dots) with the corresponding two-sided 95% CI (error bars) based on the Clopper–Pearson method. BRCA BRCA1 or BRCA2, CI confidence interval, HRR homologous recombination repair, LOH loss of heterozygosity, mut mutated, ORR objective response rate, wt wild type.
We hypothesized that cross-resistance mechanisms that emerge during prior lines of platinum treatment affect rucaparib sensitivity. Among patients with BRCAmut HGOC, those who were sensitive to the most recent line of platinum therapy (platinum-free interval [PFI] >6 months) had a median PFS of 9.4 months and ORR of 64.9% (95% CI, 52.9–75.6), whereas those with platinum-resistant (PFI <6 months) or platinum-refractory (progression on prior platinum) disease performed significantly worse, with a median PFS of 7.2 months (hazard ratio [HR], 0.44; 95% CI, 0.30–0.63; P < 0.0001) and ORR of 23.4% (95% CI, 13.8–35.7; P < 0.0001, Fisher’s exact test) (Figs. 1b and 2 and Supplementary Table S6). Among patients with BRCAwt HGOC, the LOH-high, platinum-sensitive subgroup performed better (median PFS, 7.2 months; ORR, 27.7% [95% CI, 18.4–38.6]) than the LOH-high, platinum-resistant/refractory subgroup (median PFS, 1.9 months [HR, 0.46; 95% CI, 0.33–0.65]; ORR, 4.1% [95% CI, 0.9–11.5]) and the LOH-low subgroups regardless of platinum status (Fig. 1c). Genomic scarring is, therefore, a good predictor of rucaparib sensitivity before the emergence of cross-resistance, while LOH-low HGOC lacks PARPi-sensitizing HRD mechanisms, and therefore shows little rucaparib efficacy even with limited prior platinum exposure. Indeed, rucaparib efficacy in BRCAmut and BRCAwt/LOH-high but not BRCAwt/LOH-low HGOC was better for patients with platinum-sensitive disease who had received one or two prior therapies (Part 1) than those who had three or four prior chemotherapy regimens with predominantly platinum-resistant/refractory disease (Part 2) (Fig. 2 and Supplementary Table S6). These observations suggest that platinum sensitivity and fewer lines of prior platinum treatments are both linked to better outcomes on rucaparib in HRD-associated HGOC; this finding is further supported by multivariate analyses that identified platinum status and number of prior chemotherapy regimens as significant predictors for ORR (Supplementary Tables S7 and S8). Other patient demographics and baseline disease characteristics (age, body mass index, race, Eastern Cooperative Oncology Group performance status, type of ovarian cancer) also tested in multivariate analysis were not identified as significant predictors for response.
HRD-associated genetic events leading to rucaparib sensitivity
In view of the association between BRCA mutation status and PARPi response7,37, we examined the relationship between rucaparib response and HRR gene mutation status. Deleterious germline or somatic BRCA mutations were detected in HGOC from 28% of patients (138/491; n = 89 BRCA1, n = 49 BRCA2). Consistent with prior analyses of rucaparib treatment in patients with BRCAmut HGOC, similar ORRs were observed between patients with HGOC associated with germline or somatic BRCA mutations (48.2% [95% CI, 37.3–59.3] and 45.5% [30.4–61.2]; Fig. 2). All somatic BRCA mutations were present at similar variant allele fractions to concurrent TP53 mutations, consistent with a driver mutation, and no subclonal somatic BRCA mutations were identified. BRCA reversion mutations restoring the open reading frame were enriched in patients with platinum-resistant/refractory disease and were associated with poor responses to rucaparib38. None of the 10 patients who had a reversion mutation before initiating rucaparib achieved a confirmed response (6 progressive disease [PD]; 4 stable disease [SD]). Reversion mutations accounted for 6/16 (37.5%) of BRCAmut HGOC whose best response on rucaparib was PD.
We examined whether alterations in 28 HRR genes beyond BRCA (Supplementary Table S9) correlated with rucaparib sensitivity. Patients with BRCAwt HGOC associated with any deleterious HRR gene mutation (36/491; 7.3%) had a median PFS of 5.7 months and ORR of 16.7% (95% CI, 6.4–32.8), which was not different from that of patients with BRCAwt HGOC not associated with an HRR gene mutation (median PFS, 3.7 months [HR, 0.83; 95% CI, 0.58–1.20; P = 0.32]; ORR, 11.7% [95% CI, 8.4–15.7]; Supplementary Fig. S4a, Supplementary Table S10 and Fig. 2).
Given strong evidence linking mutations in PALB2, RAD51C, and RAD51D to PARPi sensitivity17,37,41, we examined separately how HGOC harboring mutations in any of these three genes responded. Seven patients had HGOC with a deleterious RAD51C/D mutation (n = 4 RAD51C, n = 3 RAD51D), and none had a PALB2 mutation. The response rate among the seven patients with RAD51C/D-mutated HGOC was high (5/7; 71.4%; 95% CI, 29.0–96.3; Fig. 2). With the exception of one responder with an NBN mutation (Supplementary Table S10), all responders with HGOC harboring a non-BRCA HRR gene mutations had a RAD51C or a RAD51D alteration; results from the multivariate analysis also identified RAD51C/D mutation as a significant predictor of ORR (Supplementary Tables S7 and S8). Median PFS among patients with RAD51C/D-mutated HGOC was similar to that of patients with BRCAmut HGOC (11.0 and 7.8 months, respectively; HR, 1.52; 95% CI, 0.67–3.44; P = 0.32; Supplementary Fig. S4b). Although 6/7 RAD51C/D-mutated HGOC were platinum sensitive, one patient had platinum-resistant disease and achieved a partial response to rucaparib, with a PFS of 13.0 months. Additionally, 4/7 patients with RAD51C/D-mutated HGOC had received three or more lines of prior chemotherapy regimens, suggesting that as long as cross-resistance is not present, rucaparib can be highly effective in HGOC with RAD51C/D mutations, even in late lines of treatment.
HRD-associated epigenetic events leading to rucaparib sensitivity
We assessed the presence of both BRCA1 and RAD51C promoter methylation by methylation-specific polymerase chain reaction (MSP)25 in available biopsies obtained from archival HGOC tissues (n = 321), and at screening prior to initiating rucaparib (n = 230). Consistent with published estimates (11–15% for BRCA1 and 1–3% for RAD51C)22,42, ARIEL2 methylation frequencies were 16.8% for BRCA1 and 1.6% for RAD51C in archival tissues, and 13.5% for BRCA1 and 2.6% for RAD51C at screening. None of the BRCAmut HGOC included in the MSP analysis (n = 79) exhibited BRCA1 promoter methylation, suggesting that BRCA mutations and BRCA1 promoter methylation are mutually exclusive HRD mechanisms.
Interestingly, we saw no difference in PFS based on either archival or screening methylation status in patients with BRCAwt HGOC, suggesting that the mere presence of methylation at the two promoters is not a biomarker for rucaparib outcomes (Supplementary Fig. S5).
MSP analysis detects the presence of methylation but does not estimate what fraction of the neoplastic cells is methylated or if all alleles are silenced25. To determine the fraction of methylated BRCA1 and RAD51C copies, we employed methylation-sensitive digital droplet polymerase chain reaction (MS-ddPCR). Initial results from Part 1 platinum-sensitive samples analyzed for quantitative BRCA1 methylation have been published25. Here, we report MS-ddPCR analysis of all ARIEL2 patients with BRCA1 or RAD51C promoter methylation detectable by MSP in at least one time point or for whom MSP data were inconclusive. Of the 99 samples submitted (56 archival; 43 screening), 82 were identified as methylated or unmethylated by both methods; four samples that were unmethylated by MSP showed very low levels of methylation by MS-ddPCR (all <2%), suggesting that their methylation levels were below the level of detection of MSP. Methylation in two samples was detected by MSP but was not confirmed by MS-ddPCR. The remaining samples analyzed by MS-ddPCR did not have MSP data available. Only two RAD51C methylated cases, as determined by MSP, were available for MS-ddPCR analysis. Quantitative analysis confirmed low (<70%) RAD51C methylation in the screening and archival samples for both; however, due to this small sample size, RAD51C methylated cases were excluded from quantitative methylation analysis.
Using a cutoff of 70% to define high methylation25, we observed lower BRCA1 RNA expression among samples with high BRCA1 methylation compared with unmethylated samples (P < 0.0001, Wilcoxon rank-sum test) (Supplementary Fig. S6a). However, samples with low methylation had intermediate BRCA1 expression similar to unmethylated cases (P = 0.079, Wilcoxon rank-sum test). This analysis is consistent with BRCA1 promoter methylation repressing gene expression, with high and low methylation resulting in different levels of BRCA1 suppression and, therefore, differential impact on HRR activity.
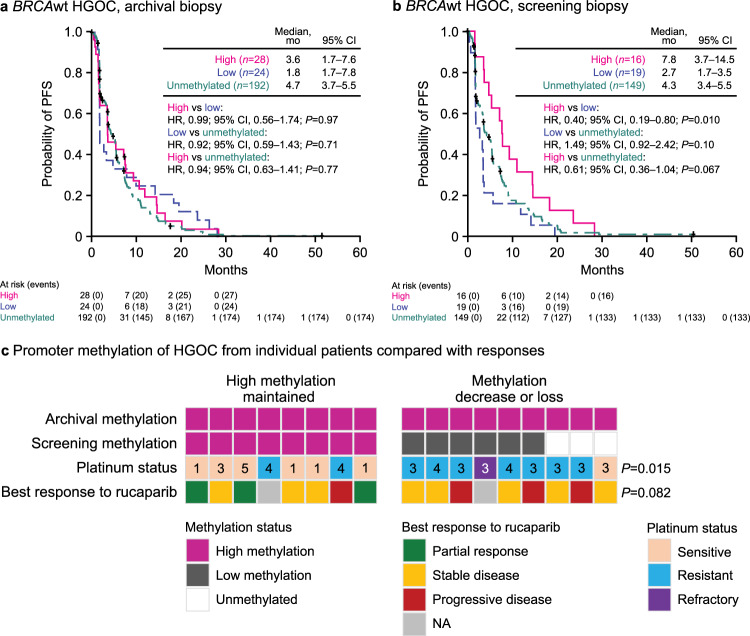
Although archival BRCA1 methylation levels were not associated with differential PFS with rucaparib treatment (Fig. 3a), PFS was better in patients with BRCAwt HGOC containing high BRCA1 methylation immediately prior to initiating rucaparib than those with unmethylated or low levels of methylation (Fig. 3b). Median PFS in patients with high methylation HGOC immediately prior to initiating rucaparib was similar to that of patients with BRCAmut HGOC without reversion mutations (7.8 vs 9.0 months; HR, 0.84; 95% CI, 0.50–1.42; P = 0.52), whereas patients with HGOC with low methylation had a similar median PFS as patients with HGOC with BRCA reversion mutations (2.7 vs 1.8 months; HR, 0.73; 95% CI, 0.33–1.62; P = 0.44) (Supplementary Fig. S6b). The ORR was 43.8% (95% CI, 19.8–70.1) in patients with high methylation HGOC, 5.3% (95% CI, 0.1–26.0) in patients with low methylation HGOC, and 10.1% (95% CI, 5.7–16.1) in patients with unmethylated HGOC (Fig. 2). This difference in response rates indicates that decreased BRCA1 methylation may be an acquired resistance mechanism leading to lower rucaparib efficacy, similar to BRCA reversion mutations. Consistent with these findings, methylation status was identified as a significant predictor of ORR in addition to BRCA and RAD51C/D mutation, in a multivariate analysis (Supplemental Tables S7 and S8).
Fig. 3. High BRCA1 methylation levels at screening are associated with better outcomes.
a Kaplan–Meier plot showing PFS in ARIEL2 patients with BRCAwt HGOC having high (magenta), low (blue), or no methylation (unmethylated, teal) in archival biopsy. b Kaplan–Meier plot showing PFS in ARIEL2 patients with BRCAwt HGOC having high (magenta), low (blue), or no methylation (unmethylated, green) in screening biopsy. P values in panels a and b are based on Cox proportional hazard model. c Methylation status at screening as compared to the archival sample, platinum status, and best response to rucaparib of 17 HGOC with high BRCA1 methylation levels in the archival sample and an available matched screening biopsy; numbers indicate number prior lines of chemotherapy treatment; P values are based on a two-sided Fisher’s exact test testing the proportion of patients with platinum status of sensitive and best response of partial response between HGOC that maintained methylation vs decrease or loss of methylation. BRCA BRCA1 or BRCA2, CI confidence interval, HGOC high-grade ovarian carcinoma, HR hazard ratio, NA not applicable, PFS progression-free survival, wt wild type.
To examine whether a decrease in methylation correlates with resistance to prior therapies and poor response to rucaparib, we focused on patients with BRCAwt HGOC whose archival samples showed high methylation and who had a matched screening biopsy prior to rucaparib treatment (n = 17) (Fig. 3c). HGOCs that maintained high methylation across archival and screening biopsies were enriched for platinum-sensitive disease (6/8), while those with a decrease or loss in methylation were predominantly platinum resistant/refractory (8/9) (P = 0.015, Fisher’s exact test). Loss/decrease of methylation was also associated with higher number (≥3) of prior lines of chemotherapy treatment (P = 0.029, Fisher’s exact test; Fig. 3c).
The ORR among the eight patients with HGOC that maintained high methylation was 38%, whereas no responses were observed among the nine with HGOC with methylation decrease or loss (Fig. 3c), indicating that demethylation may be associated with reduced rucaparib efficacy (P = 0.082, Fisher’s exact test). Although based on a small number of cases, this analysis suggests that methylation plasticity is an important factor in developing cross-resistance to both platinum agents and PARPi via de-repression of BRCA1 expression and reactivation of HRR. BRCA1 methylation, both high and low, is highly enriched in BRCAwt/LOH-high HGOC compared with other molecular subgroups (Supplementary Table S11; P < 0.0001, Chi-square test), confirming an association between methylation and accumulation of genome scars43. In ARIEL2, 14% of BRCAwt/LOH-high HGOC (13/94) had high BRCA1 methylation at screening, and 18% (17/94) had low BRCA1 methylation. Outcomes in the two groups were vastly different with ORRs of 46.2% (95% CI, 19.2–74.9) and 5.9% (95% CI, 0.1–28.7; P = 0.025, Fisher’s exact test) in patients with BRCAwt/LOH-high HGOC with high vs low screening methylation, respectively, and median PFS of 9.3 vs 3.3 months (HR, 0.44; 95% CI, 0.21–0.92; P = 0.015). Thus, acquired resistance by demethylation before initiating rucaparib may account for a substantial fraction of PARPi resistance among BRCAwt/LOH-high HGOC, a population expected to be PARPi responsive.
Non-HRR gene alterations may modulate response to rucaparib
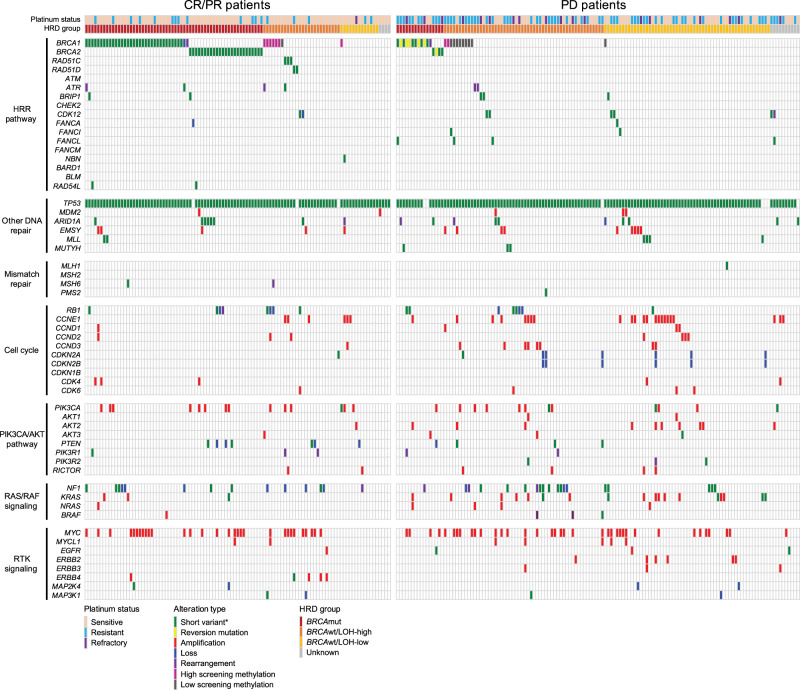
Apart from the HRR pathway, alterations in other pathways may also affect rucaparib response. Mutation and copy number data for 315 genes based on targeted carcinoma NGS were available for 484 of 491 patients. The genetic and epigenetic alterations of HGOC from patients who achieved a confirmed complete or partial response (n = 103) and those whose best response was PD (n = 136) are shown in Fig. 4; patients who achieved a best response of SD are summarized in Supplementary Fig. S7. Genes and pathways commonly altered in ovarian cancer were included. TP53 was the most frequently mutated gene (92.8%), consistent with published data22,44. We also observed frequent alterations in genes involved in non-HRR DNA repair, cell cycle regulation, PI3K/AKT signaling, RAS pathway, and receptor tyrosine kinase signaling.
Fig. 4. Genetic and epigenetic alteration landscape of HGOC with confirmed best response of CR/PR (left) or PD (right).
Methylation levels shown are at screening. *Short variant include nonsense, missense, frameshift, and splice site alterations. All reported alterations are deleterious or likely deleterious. BRCA BRCA1 or BRCA2, CR complete response, HGOC high-grade ovarian carcinoma, HRR homologous recombination repair, LOH loss of heterozygosity, mut mutated, PD progressive disease, PR partial response, RTK receptor tyrosine kinase, wt wild type.
Although events in most non-HRR genes are individually rare and did not show significantly different alteration rates between responders and nonresponders, a few interesting observations can be noted. CCNE1 amplification was mutually exclusive with both BRCA mutations, as previously reported22,45, and BRCA1 methylation. KRAS and NRAS amplifications were enriched among BRCAwt HGOC with PD (14/120, 11.7%); in contrast, no amplification was identified in BRCAwt responders (0/43, P = 0.023, Fisher’s exact test), suggesting that dysregulated RAS signaling may lead to poorer outcomes in BRCAwt HGOC.
Several patients (n = 14) had a KRAS, NRAS, or BRAF activating mutation in the absence of a TP53 mutation, a genetic profile associated with low-grade serous carcinoma, mucinous carcinoma, and mesonephric-like adenocarcinoma46–48. A blinded pathology re-review confirmed the presence of non-high-grade serous or non-grade 2/3 endometrioid histologies in 11 of the 14 cases, while all cancers with KRAS, NRAS, or BRAF mutations and a TP53 alteration (n = 7) were confirmed to be high-grade serous (Supplementary Table S12 and Supplementary Fig. S8). Eleven of 12 cancers with KRAS, NRAS, or BRAF activating mutations in TP53 wild-type background that could be classified into molecular subgroups were LOH-low, consistent with the observation that low grade and mesonephric-like histologies are not driven by HRD49. Although none of the 14 patients achieved a clinical response to rucaparib, indicating that PARPi are not likely to be active in these rare histologies, some experienced PFS longer than 200 days, an observation that may be indicative of slower overall tumor growth47,50.
Amplification and overexpression in CCNE1/cell cycle and AKT pathway genes have been previously associated with platinum resistance42,51. Among platinum-resistant/refractory patients in ARIEL2, patients with BRCAwt HGOC and alterations (predominantly via copy number change) either in the AKT1/2/3 genes or cell cycle pathway at screening had poorer PFS outcomes compared with other patients with BRCAwt HGOC (AKT: HR, 0.45; 95% CI, 0.24–0.85; P = 0.013; cell cycle: HR, 0.66; 95% CI, 0.44–1.00; P = 0.050; Supplementary Fig. S9), consistent with the hypothesis that certain platinum resistance mechanisms may modulate responses to other DNA-damaging agents, including PARPi.
Discussion
ARIEL2 assessed the safety and efficacy of rucaparib in an unselected HGOC population. Platinum sensitivity was a strong clinical predictor of rucaparib response in this PARPi-naïve population, especially in the BRCAmut and BRCAwt/LOH-high molecular subgroups. As HRD is a common mechanism leading to both platinum and PARPi sensitivity1, this finding suggests that cross-resistance mechanisms arising during platinum therapies also impact responses to PARPi. Here, we describe HRD mechanisms leading to both platinum and rucaparib sensitivity (BRCA mutation, RAD51C/D alterations, and high BRCA1 promoter methylation) and summarize two important cross-resistance mechanisms: BRCA reversion mutations, which we have previously shown38, and loss of BRCA1 methylation described here for the first time using archival and screening clinical specimens. Other cross-resistance mechanisms to PARPi have been previously proposed, including BRCA1 alternative splicing52, 53BP1 loss53,54, increased expression of BRCA hypomorphs55–57, and ABCB1 gene fusions42,58. These mechanisms may also play a role in ARIEL2 but were not assessed here due to insufficient patient samples.
Alterations in RAD51C and RAD51D correlated with meaningful clinical activity of rucaparib similar to that of BRCAmut HGOC. Therefore, we propose utilizing panels incorporating RAD51C/D when considering targeted therapies. Importantly, we previously showed that reversion mutations also occur in RAD51C/D as a resistance mechanism17, supporting their essential role in generating synthetic lethality with PARPi.
The effect of mutations in other HRR genes on rucaparib sensitivity remains unclear given the low relative frequency of each gene in the ARIEL2 cohort. Although it has been hypothesized that ATM mutations are correlated with sensitivity to PARP inhibition, in a recent phase 3 trial, there was no survival benefit seen for patients with prostate cancer associated with ATM mutations who were treated with olaparib vs androgen-receptor targeted therapy (HR, 0.93; 95% CI, 0.53–1.75)59. In ARIEL2, no responses were observed among five patients with HGOC harboring an ATM mutation. However, three of these patients had platinum-resistant/refractory disease, and only one of the remaining two was evaluable for response. Therefore, with the limited data, the impact of ATM on rucaparib sensitivity in HGOC remains inconclusive.
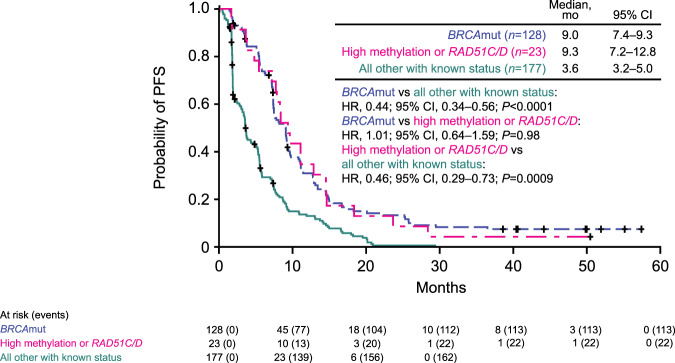
Through quantitative MS-ddPCR assessment of BRCA1 methylation in archival and screening biopsies, we show that high-level methylation of the BRCA1 promoter is a strong biomarker of rucaparib sensitivity. Patients with carcinomas harboring this modification or a RAD51C/D mutation had PFS similar to that of patients with BRCAmut HGOC treated with rucaparib, even after cases with BRCA reversion mutations are removed from comparison (Fig. 5). Further supporting this finding, each of these characteristics (methylation status, BRCA mutation, or RAD51C/D mutation) were identified as a significant predictor for ORR in multivariate analysis. Given strong preclinical data linking BRCA1 promoter methylation to HRD, and platinum and PARPi sensitivity23,24, the previous lack of definitive clinical evidence associating BRCA1 methylation and therapy response has been perplexing60,61. The TNT trial (NCT00532727) found no association between BRCA1 methylation and response to carboplatin vs docetaxel in patients with triple-negative breast cancer26. Discrepancies between these findings and our own may have both biological and technical explanations. First, most studies analyze methylation in samples obtained at diagnosis, thus missing methylation loss that may have occurred since sample collection. Second, previous studies only report the binary presence or absence of BRCA1 promoter methylation without considering the levels of methylation observed. All BRCA1 alleles likely need to be methylated to establish complete gene silencing and subsequent HRD26. Importantly, we show that methylation should be quantitatively assessed immediately prior to treatment to ensure high enough levels to result in sufficient promoter silencing, especially in later-line settings.
Fig. 5. High BRCA1 methylation at pretreatment or the presence of a RAD51C/D mutation result in similar PFS as the presence of BRCA mutations.
PFS of patients with BRCAmut HGOCs, excluding cases with reversion mutations (blue), patients with HGOCs harboring a RAD51C/D mutation or high methylation at pretreatment (magenta), and all other patients with known mutation and methylation status, including cases with reversion mutations (teal). P values were computed using a Cox proportional hazard model. BRCA BRCA1 or BRCA2, CI confidence interval, HGOC high-grade ovarian carcinoma, HR hazard ratio, mut mutated, PFS progression-free survival.
Relying on genomic scarring as evidence of HRD in later lines of treatment poses significant disadvantages. Once established, genomic scars persist and do not provide a real-time predictor of sensitivity after multiple treatment lines. In the platinum-sensitive ARIEL2 Part 1 cases, high LOH was associated with a higher ORR among BRCAwt HGOC. But among the more heavily pretreated Part 2 patients, high LOH did not predict rucaparib sensitivity. Notably, all carcinomas with acquired platinum resistance (i.e., BRCA reversion mutation and BRCA1 promoter demethylation) remained LOH-high. Thus, accumulation of genomic scarring is an irreversible process, persisting even as cancers re-acquire functional HRR. In the QUADRA trial of the PARPi niraparib after three lines of therapy, which used the myChoice HRD test to determine HRD status, BRCAmut HGOC were included in the HRD group, driving much of the 15% ORR62. When only considering the BRCAwt cases in QUADRA, HRD was associated with a higher ORR only for platinum-sensitive disease (20% vs 4% for non-HRD) and was not predictive for platinum-resistant cases (2.4% vs 3% non-HRD), similar to our findings. Therefore, although stratifying BRCAwt HGOC for PARPi treatment based on evidence of accumulated genomic scarring is useful early in the disease course, these tests lose utility in platinum-resistant/refractory cases.
We also examined alterations in non-HRR pathways that may indirectly modulate response to rucaparib. Cell cycle gene alterations, including amplifications of cyclin E, cyclin D, and CDK4/6 genes and deletions in the cell cycle inhibitors CDKN2A/B and CDKN1B, were common in BRCAwt HGOC in ARIEL2 and have been associated with platinum resistance22,63. AKT signaling has also been linked to platinum resistance, and AKT inhibition appears to reverse resistance in certain models64. In ARIEL2, platinum-resistant/refractory BRCAwt HGOC with alterations in the cell cycle pathway and the AKT1/2/3 genes showed poorer outcomes on rucaparib, implying that these pathways may be relevant to responses to multiple DNA-damaging agents. Further in vitro and in vivo studies would be essential to confirm these clinical observations, describe the mechanistic connections between these pathways and PARPi, and determine if combination therapies with PARPi and cell cycle or AKT inhibitors may be beneficial for certain late-stage HGOC patients. Most of the cancers with KRAS, NRAS, or BRAF activating mutations in the absence of a TP53 mutation were subsequently found to be non-HGOC, and the lack of response to rucaparib in these cancers indicates the importance of confirming histological subtype when these mutations are detected.
Overall, our analysis highlights significant overlap between molecular mechanisms resulting in platinum and PARPi sensitivity and the extent of cross-resistance that exists between these two drug classes. In addition to BRCA mutations, RAD51C/D mutations and high-level BRCA1 promoter methylation are strong predictors of sensitivity to a PARPi. Given its general tolerability and efficacy, especially in patients with BRCAmut and BRCAwt/LOH-high HGOC, and data supporting the use of PARPi as maintenance following primary therapy8–10,65, we propose that administration of rucaparib, as active treatment, should be considered in earlier lines of therapy, before the emergence of platinum resistance. Such an approach would increase the likelihood of patients experiencing significant clinical benefit while maintaining the improved quality of life associated with targeted vs systemic therapy.
Methods
Study design and participants
ARIEL2 (CO-338-017; NCT01891344) was an international, multicenter, two-part, phase 2 open-label study conducted across 64 sites in Australia, Canada, France, Spain, the United Kingdom, and the United States. The study design and clinical results from Part 1 were published previously37. In ARIEL2, eligible patients were aged 18 years or older with histologically confirmed, relapsed, high-grade serous or Grade 2 or Grade 3 endometroid epithelial ovarian, fallopian tube, or primary peritoneal cancer. Patients were required to have measurable disease according to RECIST, an Eastern Cooperative Oncology Group Performance Status of 0 or 1, and adequate organ function. Part 1 enrolled patients with relapsed HGOC who had received at least one prior platinum-based regimen and had platinum-sensitive disease (disease progression ≥6 months after last platinum). Part 2 enrolled patients with relapsed HGOC who had received three to four prior chemotherapies and had a treatment-free interval of more than 6 months following first-line chemotherapy. Patients in Part 2 could be platinum sensitive, platinum resistant (disease progression <6 months after last platinum, with best response other than PD), or platinum refractory (best response of PD on last platinum with progression-free interval <2 months). Both Part 1 and Part 2 enrolled patients regardless of their HRD status, with the exception that Part 1 had a cap on the number of patients with a known germline BRCA mutation (n = 15) allowed, in order to enrich for the BRCAwt population.
Patients were ineligible if they previously received a PARPi in either the treatment setting or as a maintenance therapy, had an active second malignancy, had central nervous system metastases, or received anticancer therapy 14 days or fewer before receiving their first dose of rucaparib.
The study was approved by national or local institutional review boards, as appropriate at each site (see Supplementary Information for full list), and was carried out in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines of the International Conference on Harmonization. Patients provided written informed consent before participation.
Procedures
Patients were treated with oral rucaparib at 600 mg twice daily until disease progression, unacceptable toxicity, or death. Supportive care (e.g., antiemetics or analgesics for pain control) was permitted at the investigator’s discretion. Treatment interruptions and dose reductions (in decrements of 120 mg formulation in Part 1 and 100 mg in Part 2) were permitted if a patient had a grade 3 or greater adverse event. Treatment was discontinued if a dose interruption occurred for more than 14 consecutive days (longer dose interruptions were permitted with sponsor approval).
Tumor response was assessed by the investigators using RECIST, with computed tomography scans at screening and every 8 weeks (±4 days) during treatment and after treatment for patients who discontinued for any reason other than disease progression. Patients who had been on study at least 18 months could have the frequency of disease assessment decreased to every 16 weeks (±2 days). Assessment continued until confirmed disease progression, death, start of subsequent treatment, or loss to follow-up. Serum CA-125 measures were taken at screening, day 1 of each cycle, and the end of treatment, or when clinically indicated. Hematology, serum chemistry, and safety assessments were done at screening, day 1 and day 15 of cycle 1, and day 1 of any subsequent cycles. Adverse events were classified in accordance with the Medical Dictionary for Drug Regulatory Activities classification system version 19.1 and graded for severity in accordance with the National Cancer Institute’s Common Terminology Criteria for Adverse Event version 4.03 (https://nciterms.nci.nih.gov/ncitbrowser/start.jsf). Part 1 patients were treated and followed up until disease progression, death, or discontinuation of treatment due to other reasons. Part 2 patients were followed for survival, subsequent therapy, and secondary malignancy every 12 weeks until death, loss to follow-up, withdrawal of consent from study, or study closure, whichever happened first.
Molecular subgroup classification
Biomarker analysis was performed following the REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) guidelines66. A known HRR gene mutation was not required for enrollment in ARIEL2. Patients were required to have adequate formalin-fixed paraffin-embedded (FFPE) archival tumor tissue available and to have undergone a pretreatment (screening) biopsy (optional in patients with HGOC having a known deleterious germline BRCA mutation in Part 2). For the ARIEL2 Part 1 and Part 2 prespecified analyses, tumor BRCA status was determined centrally using the FoundationOneTM NGS assay (Supplementary Fig. S3)37,67.
Local BRCA test results were collected when available (Supplementary Fig. S3). For our post hoc analysis of the pooled ARIEL2 patient population, we classified a patient as BRCAmut if a deleterious BRCA alteration was detected by either central tumor testing or local testing (Supplementary Fig. S3). The majority of BRCA alterations that were detected by local testing but not by tissue testing were large rearrangements, known to be challenging to detect by NGS.
FoundationOneTM was also used to calculate the percentage of genomic LOH in archival and screening biopsies and to identify alterations in genes other than BRCA37,67.
To classify BRCAwt patients into LOH-high and LOH-low subgroups, we used a prespecified cutoff of 14% for patients in Part 1 and a prespecified cutoff of 18% for patients in Part 2 (Supplementary Fig. S3). For the post hoc, exploratory molecular subgroup analyses, both the local and central test information for BRCA was utilized. For BRCAwt, we utilized an optimized cutoff of 16%, which was shown to be the optimal cutoff in ARIEL2 Part 1 (ref. 40) and was prospectively evaluated in the phase 3 ARIEL3 study (Supplementary Fig. S3)7.
BRCA1 and BRCA2 germline/somatic status was determined by a combination of methods. Blood samples from all ARIEL2 Part 1 BRCAmut samples and a subset of ARIEL2 Part 2 BRCAmut cases (n = 74) were analyzed using the BROCA homologous recombination sequencing assay (University of Washington, Seattle WA, USA). Any alteration detected by FoundationOne™ NGS but not BROCA was considered somatic. The remainder of the samples were assigned germline/somatic status based on local testing data provided by the study sites (n = 29) or computational inference method using the targeted NGS data (n = 26)68. Computational inference was not attempted for samples with tumor purity >80%; those samples were listed as Unknown. Nine BRCAmut cases remained with unknown germline/somatic status. Agreement between germline/somatic status determined through germline sequencing (BROCA and local testing) and the computational inference method was very high for samples that had both data available (n = 67, 95.5% agreement, kappa = 0.88, P < 0.0001, Cohen’s Kappa statistics).
HRR gene mutation subgroup was based on alterations in the genes listed in Supplementary Table S6. Germline/somatic status for ARIEL2 Part 1 non-BRCA HRR genes was determined by the BROCA homologous recombination sequencing assay (University of Washington, Seattle, WA, USA). Any alteration detected by FoundationOne™ NGS but not BROCA was considered somatic. BRCAwt ARIEL2 Part 2 samples were not analyzed by central germline testing and germline/somatic status for these cases was determined by computational inference as described above68. The same computational approach was used to determine zygosity for all non-BRCA HRR gene alterations68.
The detection of BRCA reversion mutations has been described in detail previously38. In short, for reversion mutations present in cell-free DNA (cfDNA), plasma samples collected from ARIEL2 patients were analyzed using the Guardant360 (ref. 69) or FoundationACT70 cfDNA assays. Central tumor tissue NGS data were also re-analyzed to determine if reversion mutations were present. BRCA reversion mutations were defined as: (i) a base substitution that changed a nonsense mutation to a missense mutation, (ii) an insertion/deletion that restored the ORF, or (iii) a larger intragenic deletion that deleted the primary deleterious mutation. Reversion mutations were detected in 10/112 (8.9%) BRCAmut HGOC pretreatment samples: nine by Guardant360 plasma analysis (eight previously described38 and one identified in post-publication analysis) and one by central tumor tissue NGS analysis.
Patients were classified as having a cell cycle pathway alteration if an alteration in any of the following genes was detected in their screening cancer sample: CCNE1, CCND1, CCND2, CCND3, CDKN2A, CDKN2B, CDKN1B, CDK4, or CDK6. Patients were classified as having an AKT alteration if they had an alteration in AKT1, AKT2, or AKT3 in their screening cancer sample. Amplifications reported were high-copy gains (CN > 5).
Methylation analysis
The presence of methylation at the BRCA1 and RAD51C promoters was detected using MSP, as previously described37. In summary five 10 μm sections of FFPE tissue were deparaffinized, rehydrated, and digested with Proteinase K (Zymo Research, Irvine, CA, USA) overnight and bisulfite conversion of 10 μL of supernatant was performed in duplicates using the EZ DNA Methylation-Direct kit (Zymo Research). Following bisulfite conversion, the samples underwent desulfonation and cleanup, and 2 μL of bisulfite-converted DNA was evaluated with MSP for BRCA1 and RAD51C using primers listed in Supplementary Table S13.
Quantification of BRCA1 and RAD51C methylation levels in HGOC from ARIEL2 Part 1 and Part 2 patients was performed by quantitative MS-ddPCR methodology as previously described25: DNA extracted from FFPE-preserved tissue sections was bisulfite converted using the EZ DNA Methylation-Lightning kit (Zymo Research). BRCA1 primers were designed for a 72 bp amplicon in the untranslated region (UTR) (Supplementary Table S13). RAD51C methylation, primers were designed for a 142 bp amplicon in the RAD51C UTR. Minor groove binder probes hybridizing to the fully unmethylated (2′-chloro-7′phenyl-1,4-dichloro-6-carboxyfluorescein [VIC] labeled) and the fully methylated sequences (6-carboxyfluorescein [FAM] labeled) were used. The ddPCR was performed on the Bio-Rad QX-200 system. Methylation frequencies determined by MS-ddPCR were normalized to neoplastic purity and BRCA1/RAD51C copy number estimates using the following formula: RM × (CN × TP + (100 − TP) × 2)/(TP × CN)/100, where RM is the raw fraction methylated copies detected, TP is the tumor purity, and CN is the BRCA1 copy number25. The tumor purity and copy number were estimated based on the targeted NGS data68. Patient samples were classified as having high or low methylation levels based on a predefined cutoff of 70%25. In the absence of contradicting MS-ddPCR data, biopsies with 0% BRCA1 promoter methylation by MS-ddPCR, or ones determined to be unmethylated by MSP, were labeled as unmethylated.
BRCA1 expression analysis
For a subset of the samples analyzed for BRCA1 promoter methylation status, we assessed the BRCA1 RNA expression levels by NanoString71.
Pathology analysis and histology reclassification
For all patients, hematoxylin and eosin (H&E) staining was performed following standard procedures on archival and/or screening biopsy tissue corresponding to the tissue that was submitted for central NGS testing. The H&E slides for the patients harboring activating mutations in the KRAS/NRAS/BRAF genes were re-reviewed by gynecologic pathologists (J.A.E. and D.I.L.), and a consensus diagnosis was rendered. At time of re-review, the pathologists were blinded to the genomic findings for these patients. Both archival and screening tissue H&E slides were available for at least two patients each for mesonephric-like carcinomas, low-grade serous carcinomas, and endometrioid adenocarcinomas, and the reclassification was confirmed in both samples. Representative fields were selected to describe features concidered for reclassification.
Statistical analysis
All analyses were performed using the safety population, which included all patients who received at least one dose of rucaparib. The primary endpoint in Part 1 was PFS by predefined HRD subgroups. PFS was defined as the number of days from the first dose of study drug to disease progression by RECIST, as determined by the investigator, or death due to any cause, whichever occurred first. Patients without a documented event of progression were censored on the date of their last adequate cancer assessment (i.e., radiologic assessment) or the date of the first dose of the study drug if no cancer assessments were performed. In Part 2, the primary endpoint was ORR, defined as the proportion of patients achieving a best response of complete or partial response according to RECIST as assessed by the investigator by predefined HRD subgroups. Secondary endpoints included the proportion of patients achieving an objective response (according to RECIST and GCIG CA-125 criteria), duration of response (according to RECIST), and overall survival. Response endpoints were summarized with frequencies and percentage using Clopper–Pearson methodology to calculate 95% CIs. Rates of response were compared with pair-wise comparison using Fisher’s exact test. The response (partial or complete response) by RECIST needed to be confirmed by a second assessment after at least 4 weeks. Duration of confirmed response (complete or partial response) was calculated from the initial date a response was detected to the first date of PD. Patients without a documented event of progression were censored on the date of their last adequate cancer assessment (i.e., radiologic assessment) or date of response if no cancer assessments were performed. Overall survival was defined as the number of days from the date of first dose of study drug to the date of death (due to any cause). Patients without a known date of death were censored on the date the patient was last known to be alive. Duration of response, PFS, and overall survival were summarized with Kaplan–Meier methodology, including median estimates and 95% CIs using log–log distribution. In addition, a Cox proportional hazard model was used to summarize these endpoints and make comparisons between molecular subgroups. Here, we also present post hoc exploratory molecular biomarker analyses of PFS and ORR outcomes in order to further explore clinical benefit. Most of the post hoc analyses are based on univariate comparisons. In addition, a stepwise multivariate logistics regression model was used to identify predictors of confirmed response (PR or CR) using baseline and molecular characteristic variables as predictors in the model (Supplementary Tables S7 and S8). For all time-to-event analyses, the proportional hazards assumption was assessed visually with log–log plots (Supplementary Fig. S10); as the assumption of the model was valid in each instance, we provide HR and 95% CI for these endpoints. All P values for the post hoc exploratory analyses of molecular subgroups are presented for descriptive purposes only.
Comparisons of proportions between methylation subgroups were summarized using Fisher’s exact test or chi-square tests; for the analysis of number prior lines of chemotherapy treatment, patients were grouped in 1–2 and ≥3 prior lines subgroups. The distribution of BRCA1 gene expression levels was analyzed using a nonparametric Wilcoxon rank-sum test to compare molecular subgroups. Agreement analyses were performed using non-weighted Cohen’s Kappa statistics.
Data analysis was performed using SAS v9.4 (SAS Institute, Cary, NC, USA) and Excel 2016 (Microsoft Corporation, Redmond, WA, USA).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Funding from the National Breast Cancer Foundation of Australia (to A.D.) and the Department of Defense Ovarian Cancer Research Program (OC12056, to E.M.S. and S.H.K.) and a Stand Up To Cancer-Ovarian Cancer Research Fund Alliance-National Ovarian Cancer Coalition Dream Team Translational Cancer Research Grant (SU2C-AACR-DT16-15, to E.M.S. and S.H.K.). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. We thank Elaina Mann, Jennifer Borrow, Jen Lenore, Jeff Isaacson, and Lindsey Rolfe for clinical development, statistical guidance, and operational support of the ARIEL2 study. In addition, we thank Vivian Chen and Mary Joyce for assistance in manuscript preparation. ARIEL2 was designed by the funder and a subgroup of investigators. Data presented and herein were collected by the funder; the funder and all authors interpreted and analyzed the data. This article was written by the authors, with medical writing and copy editing support paid for by the funder and provided by Nathan Yardley and Frederique H. Evans of Ashfield MedComms, an Ashfield Health company. All authors had full access to all trial data and had final responsibility for the decision to submit for publication.
Author contributions
J.A.E. and D.I.L. conducted pathologist re-review of histology type of tumors with KRAS/NRAS/BRAF mutations with no TP53 mutations. E.M.S., C.L.S., K.K.L., and I.A.M. were involved in the study conception. E.M.S., S.H.K., C.L.S., K.K.L., and I.A.M. were involved in the study design. E.M.S. and S.H.K. acquired funding. E.M.S., K.K.L., and I.A.M. were involved in the protocol development. E.M.S., T.T.K., K.K.L., and I.A.M. co-wrote the first draft of the manuscript. E.M.S., A.M.O, A.V.T., I.R.-C., A.O., R.L.C., C.A., G.E.K., D.M.O., A.L., D.P., S.W., L.-m.C., A.E.W.H., L. Ma, P.G., R.S.K., O.D., A.M., S.H.K., S.K.C., A.D., C.L.S., and I.A.M. treated patients, acquired data, and obtained samples. E.M.S., T.T.K., I.R.C., J.A.E., D.I.L., C.L.S., K.K.L., and I.A.M. interpreted the data. E.M.S., T.T.K., J.A.E., D.I.L., and K.K.L. analyzed data. E.M.S., T.T.K., A.M.O., A.V.T., I.R.C., A.O., R.L.C., C.A., G.E.K., D.M.O., A.L., D.P., S.W., L.-m.C., A.E.W.H., L.Ma, P.G., R.S.K., O.D., A.M., S.H.K., J.A.E., D.I.L, S.K.C., E.D., L.-T.V., S.G., L. Maloney, H.G., T.H., A.D., C.L.S., K.K.L., and I.A.M. contributed to manuscript revisions and approved the final draft for submission.
Data availability
Consent was not obtained from patients to allow posting of the data to public repositories. Requests for de-identified datasets for the results reported in this publication will be made available to qualified researchers following submission of a methodologically sound proposal to medinfo@clovisoncology.com. Data will be made available for such requests following online publication of this article and for 1 year thereafter in compliance with applicable privacy laws, data protection, and requirements for consent and anonymization. Data will be provided by Clovis Oncology. The redacted protocol for the ARIEL2 clinical study is available on thelancet.com: https://ars.els-cdn.com/content/image/1-s2.0-S1470204516305599-mmc1.pdf. Clovis Oncology does not share identified participant data or a data dictionary.
Competing interests
E.M.S. has received funding for clinical trials from Clovis Oncology and TESARO (paid to her institution). T.T.K., E.D., L.-T.V., S.G., L. Maloney, H.G., T.H., and K.K.L. are employees of Clovis Oncology and may own stock or have stock options in that company. A.M.O. has served on advisory boards for Clovis Oncology, Amgen, Immunovaccine, and Verastem; received support for travel or accommodation from AstraZeneca; and received honoraria from WebRx. A.V.T. has served on an advisory board for and received grants from AstraZeneca. I.R.-C. has served on advisory boards for Clovis Oncology, AstraZeneca, Genmab/Seattle Genetics, ImmunoGen, Merck Sharpe Dohme, PharmaMar, Roche, and Tesaro/GlaxoSmithKline and received support for travel or accommodation from AstraZeneca, GlaxoSmithKline, and Roche. A.O. has served on advisory boards for Clovis Oncology, AstraZeneca, ImmunoGen, Genmab/Seattle Genetics, PharmaMar, Roche, and Tesaro and received support for travel or accommodation from AstraZeneca, PharmaMar, Roche, and Tesaro. R.L.C. reports grants from Clovis Oncology, AbbVie, AstraZeneca, Esperance, Janssen, Merck, Millennium, OncoMed, and Roche/Genentech and has served as an advisor to Clovis Oncology, AbbVie, AstraZeneca, Bayer, Esperance, GamaMabs, Genmab, Gradalis, Janssen, Millennium, Merck, OncoMed, Pfizer, Roche/Genentech, and Tesaro. C.A. has served on steering committees for Clovis Oncology and Mateon Therapeutics; served on advisory boards for Clovis Oncology, Bayer, Cerulean Pharma, Tesaro, and VentiRx, and received honoraria from Clovis Oncology, Bayer, Cerulean Pharma, Mateon Therapeutics, Tesaro, and VentiRx. G.E.K. has received research funding (to the University of California outside the scope of this work) from Lilly, Merck, and Pfizer, and received honorarium from Clovis Oncology, AstraZeneca, and Tesaro. D.M.O. has served on advisory boards for Clovis Oncology, AstraZeneca, Gynecologic Oncology Group, Janssen, Myriad, and Tesaro; has served on steering committees for Clovis Oncology, Amgen, and ImmunoGen; has served as a consultant to AbbVie, Ambry, AstraZeneca, Health Analytics, and Tesaro, and his institution has received research support from Clovis Oncology, Agenus, Ajinomoto, Array BioPharma, AstraZeneca, Bristol-Myers Squibb, ERGOMED Clinical Research, Exelixis, Genentech, GlaxoSmithKline, Gynecologic Oncology Group, ImmunoGen, INC Research, inVentiv Health Clinical, Janssen Research and Development, Ludwig Institute for Cancer Research, Novartis Pharmaceuticals, PRA International, Regeneron Pharmaceuticals, Serono, Stemcentrx, Tesaro, and TRACON Pharmaceuticals. A.L. has served on advisory boards for Clovis Oncology, Ability, AstraZeneca, Biocad, Merck Serono, MSD, Pfizer, PharmaMar, Seattle Genetics, and Tesaro PharmaMar; reports institutional research grant support from GamaMabs, Inivata, Merus, and Sanofi, and reports boarding and travel expenses for congress activities from Clovis Oncology, AstraZeneca, Roche, and Tesaro. D.P. has served on advisory boards for AstraZeneca and GlaxoSmithKline, and received support for travel from AstraZeneca. S.W. has served on advisory boards for AstraZeneca and Roche; reports institutional research support from Clovis Oncology, AstraZeneca, Merck, Regeneron, and Tesaro, and has received honoraria from AstraZeneca. P.G. has served on an advisory board for AstraZeneca, GSK, and Merck, and received support for travel expenses for congress activities from AstraZeneca. R.S.K. has served on advisory boards for Clovis Oncology, Roche, and Tesaro. O.D. has served on advisory boards for Clovis Oncology, IMV, Tesaro, and Merck, and has served on the speaker bureau for AstraZeneca and Tesaro. J.A.E. and D.I.L. are employees of Foundation Medicine, Inc., which is a wholly-owned subsidiary of Roche, and may own stock or have stock options in Roche. A.D. has served on advisory boards for AstraZeneca Australia and has received travel support from Bio-Rad. C.L.S. has served on advisory boards for Clovis Oncology, AstraZeneca, Takeda, Roche Australia, MSD, Eisai Co., has received travel support from AstraZeneca, has received in kind research support from Clovis Oncology, Roche, Beigene and Funded research support from Sierra oncology and Eisai Co. I.A.M. has served on advisory boards for Clovis Oncology, AstraZeneca, Roche, Takeda, and Tesaro and receives institutional funding from AstraZeneca. The remaining authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Elizabeth M. Swisher, Tanya T. Kwan.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-22582-6.
References
- 1.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dal Molin GZ, Omatsu K, Sood AK, Coleman RL. Rucaparib in ovarian cancer: an update on safety, efficacy and place in therapy. Ther. Adv. Med. Oncol. 2018;10:1758835918778483. doi: 10.1177/1758835918778483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennington KP, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rafii S, et al. Baseline clinical predictors of antitumor response to the PARP inhibitor olaparib in germline BRCA1/2 mutated patients with advanced ovarian cancer. Oncotarget. 2017;8:47154–47160. doi: 10.18632/oncotarget.17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledermann JA, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 2016;17:1579–1589. doi: 10.1016/S1470-2045(16)30376-X. [DOI] [PubMed] [Google Scholar]
- 6.del Campo JM, et al. Niraparib maintenance therapy in patients with recurrent ovarian cancer after a partial response to the last platinum-based chemotherapy in the ENGOT-OV16/NOVA trial. J. Clin. Oncol. 2019;37:2968–2973. doi: 10.1200/JCO.18.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman RL, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman RL, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N. Engl. J. Med. 2019;381:2403–2415. doi: 10.1056/NEJMoa1909707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Martin A, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 10.Ray-Coquard I, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N. Engl. J. Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 11.Robson M, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 12.Litton JK, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golan T, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateo J, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bono J, et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 16.Abida W, et al. Non-BRCA DNA damage repair gene alterations and response to the PARP Inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the phase 2 TRITON2 study. Clin. Cancer Res. 2020;28:2487–2496. doi: 10.1158/1078-0432.CCR-20-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondrashova O, et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017;7:984–998. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikkila J, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat. Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotsopoulos J, et al. Frequency of germline PALB2 mutations among women with epithelial ovarian cancer. Fam. Cancer. 2017;16:29–34. doi: 10.1007/s10689-016-9919-z. [DOI] [PubMed] [Google Scholar]
- 20.Castaneda A, et al. BRIP1 mutation does not confer sensitivity to PARP inhibition. Gynecol. Oncol. 2019;154:87. doi: 10.1016/j.ygyno.2019.04.205. [DOI] [Google Scholar]
- 21.Kawazu M, et al. Integrative analysis of genomic alterations in triple-negative breast cancer in association with homologous recombination deficiency. PLoS Genet. 2017;13:e1006853. doi: 10.1371/journal.pgen.1006853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ter Brugge, P. et al. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J. Natl. Cancer Inst. 108, 10.1093/jnci/djw148 (2016). [DOI] [PubMed]
- 24.AlHilli MM, et al. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol. Oncol. 2016;143:379–388. doi: 10.1016/j.ygyno.2016.08.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondrashova O, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018;9:3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tutt A, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat. Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lheureux S, et al. Long-term responders on olaparib maintenance in high-grade serous ovarian cancer: clinical and molecular characterization. Clin. Cancer Res. 2017;23:4086–4094. doi: 10.1158/1078-0432.CCR-16-2615. [DOI] [PubMed] [Google Scholar]
- 28.Abkevich V, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies H, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polak P, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menghi F, et al. The tandem duplicator phenotype is a prevalent genome-wide cancer configuration driven by distinct gene mutations. Cancer Cell. 2018;34:197–210.e195. doi: 10.1016/j.ccell.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macintyre G, et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018;50:1262–1270. doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telli ML, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swisher EM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 38.Lin KK, et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9:210–219. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 39.Oza AM, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: integrated analysis of data from Study 10 and ARIEL2. Gynecol. Oncol. 2017;147:267–275. doi: 10.1016/j.ygyno.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 40.Coleman RL, et al. Refinement of prespecified cutoff for genomic loss of heterozygosity (LOH) in ARIEL2 part 1: a phase II study of rucaparib in patients (pts) with high grade ovarian carcinoma (HGOC) J. Clin. Oncol. 2016;34:5540. doi: 10.1200/JCO.2016.34.15_suppl.5540. [DOI] [Google Scholar]
- 41.Goodall J, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7:1006–1017. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patch AM, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 43.Patel JN, et al. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br. J. Cancer. 2018;119:1060–1066. doi: 10.1038/s41416-018-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed AA, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Etemadmoghadam D, et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc. Natl Acad. Sci. USA. 2013;110:19489–19494. doi: 10.1073/pnas.1314302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emmanuel C, et al. Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver. Clin. Cancer Res. 2014;20:6618–6630. doi: 10.1158/1078-0432.CCR-14-1292. [DOI] [PubMed] [Google Scholar]
- 47.Kaldawy A, et al. Low-grade serous ovarian cancer: a review. Gynecol. Oncol. 2016;143:433–438. doi: 10.1016/j.ygyno.2016.08.320. [DOI] [PubMed] [Google Scholar]
- 48.Mirkovic J, et al. Targeted genomic profiling reveals recurrent KRAS mutations in mesonephric-like adenocarcinomas of the female genital tract. Am. J. Surg. Pathol. 2018;42:227–233. doi: 10.1097/PAS.0000000000000958. [DOI] [PubMed] [Google Scholar]
- 49.Gershenson DM, et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecol. Oncol. 2009;114:48–52. doi: 10.1016/j.ygyno.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Monk BJ, et al. MILO/ENGOT-ov11: binimetinib versus physician’s choice chemotherapy in recurrent or persistent low-grade serous carcinomas of the ovary, fallopian tube, or primary peritoneum. J. Clin. Oncol. 2020;38:3753–3762. doi: 10.1200/JCO.20.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahne JC, et al. Downregulation of AKT reverses platinum resistance of human ovarian cancers in vitro. Oncol. Rep. 2012;28:2023–2028. doi: 10.3892/or.2012.2041. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, et al. The BRCA1-delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76:2778–2790. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hurley RM, et al. 53BP1 as a potential predictor of response in PARP inhibitor-treated homologous recombination-deficient ovarian cancer. Gynecol. Oncol. 2019;153:127–134. doi: 10.1016/j.ygyno.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drost R, et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J. Clin. Invest. 2016;126:2903–2918. doi: 10.1172/JCI70196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park PH, et al. Amplification of the mutation-carrying BRCA2 allele promotes RAD51 loading and PARP inhibitor resistance in the absence of reversion mutations. Mol. Cancer Ther. 2020;19:602–613. doi: 10.1158/1535-7163.MCT-17-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson N, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl Acad. Sci. USA. 2013;110:17041–17046. doi: 10.1073/pnas.1305170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christie EL, et al. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat. Commun. 2019;10:1295. doi: 10.1038/s41467-019-09312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hussain M, et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020;383:2345–2357. doi: 10.1056/NEJMoa2022485. [DOI] [PubMed] [Google Scholar]
- 60.Ignatov T, et al. BRCA1 promoter methylation is a marker of better response to anthracycline-based therapy in sporadic TNBC. Breast Cancer Res. Treat. 2013;141:205–212. doi: 10.1007/s10549-013-2693-9. [DOI] [PubMed] [Google Scholar]
- 61.Bernards SS, et al. Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated ovarian carcinoma. Gynecol. Oncol. 2018;148:281–285. doi: 10.1016/j.ygyno.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 62.Moore KN, et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019;20:636–648. doi: 10.1016/S1470-2045(19)30029-4. [DOI] [PubMed] [Google Scholar]
- 63.Noel EE, et al. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am. J. Pathol. 2010;176:2607–2615. doi: 10.2353/ajpath.2010.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whicker ME, Lin ZP, Hanna R, Sartorelli AC, Ratner ES. MK-2206 sensitizes BRCA-deficient epithelial ovarian adenocarcinoma to cisplatin and olaparib. BMC Cancer. 2016;16:550. doi: 10.1186/s12885-016-2598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore K, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 66.McShane LM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br. J. Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun JX, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol. 2018;14:e1005965. doi: 10.1371/journal.pcbi.1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lanman RB, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clark TA, et al. Analytical validation of a hybrid capture–based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J. Mol. Diagn. 2018;20:686–702. doi: 10.1016/j.jmoldx.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Talhouk A, et al. Single-patient molecular testing with NanoString nCounter data using a reference-based strategy for batch effect correction. PLoS ONE. 2016;11:e0153844. doi: 10.1371/journal.pone.0153844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Consent was not obtained from patients to allow posting of the data to public repositories. Requests for de-identified datasets for the results reported in this publication will be made available to qualified researchers following submission of a methodologically sound proposal to medinfo@clovisoncology.com. Data will be made available for such requests following online publication of this article and for 1 year thereafter in compliance with applicable privacy laws, data protection, and requirements for consent and anonymization. Data will be provided by Clovis Oncology. The redacted protocol for the ARIEL2 clinical study is available on thelancet.com: https://ars.els-cdn.com/content/image/1-s2.0-S1470204516305599-mmc1.pdf. Clovis Oncology does not share identified participant data or a data dictionary.