Abstract
Malignant pleural mesothelioma (MPM) is an incurable surface neoplasm with peculiar pathobiology. MPM proliferates by using the tyrosine-kinase-Ras pathway. Despite representing an attractive therapeutic target, there are no standard agent(s) specifically inhibiting Ras signaling adopted in clinical settings. We posited that biologic effects of microRNA (miRNA) can disrupt this molecular network. Using patient samples, cell lines, and murine tumor xenograft models, we confirmed specific genes in the Ras pathway are targeted by an MPM-associated miRNA and then examined its therapeutic effects. We verified significant and consistent downregulation of miR-206 in MPM tissues. When miR-206 is ectopically re-expressed in MPM cells and delivered to tumor xenografts in mice, it exerted significant cell killing by suppressing multiple components of the receptor-tyrosine-kinase-Ras-cell-cycle-signaling network; some of which were prognostic when overexpressed and/or have not been druggable. Of note, we validated CDK6 as a novel target of miR-206. Overall, this miR-206-targeting mechanism manifested as induced G1/S cell cycle arrest. In addition, we identified a novel MPM therapeutic combination by adding systemic-route abemaciclib with local-route miR-206, which showed additive efficacy translating to improved survival. Our pre-clinical study suggests a potential pathophysiologic role for, and therapeutic relevance of, miR-206 in MPM.
Keywords: mesothelioma, microRNA, receptor tyrosine kinase, Ras, cell cycle
Graphical abstract

microRNAs exert profound effects when leveraged against cancer. Singh et al. demonstrate the potential utility of miR-206, which suppresses the receptor-tyrosine-kinases-KRAS-cell-cycle pathway in malignant pleural mesothelioma, an underexploited strategy. In addition, improved efficacy is possible with combinations targeting an oncogene at transcript (miR) and protein (drug) levels.
Introduction
Malignant pleural mesothelioma (MPM) is an aggressive neoplasm arising from mesothelial cells that comprise the membrane surface investment of the thoracic cavity. Contrary to historical predictions, MPM incidence continues to increase worldwide.1 MPM drug therapy has not dramatically improved upon the dismal median survival time of 12–18 months.2 Clearly, there is an ongoing and pressing need for effective treatments against this recalcitrant tumor.
Contemporary genetic profiling has largely reaffirmed known characteristics of MPM pathobiology, which entail comparatively low mutational burden with a predominance of tumor-suppressor gene losses, yet lack oncogenic driver mutations.3 Regardless of these unique features, MPM does phenotypically resemble other solid tumor types in coapting core cellular growth and survival signaling pathways mediated by the receptor tyrosine kinase (RTK) family.4,5 In most MPM cases, a multitude of RTK subfamilies are overexpressed and hyperactive, including epidermal growth factor receptor (EGFR), insulin-like growth factor receptor (IGFR), hepatocyte growth factor receptor (HGFR or c-MET), and vascular endothelial growth factor receptor (VEGFR); all of which redundantly activate Ras/mitogen-activated protein kinase (MAPK) and Ras/phosphotidylinositol-3′-kinase/Akt (PI3K/Akt) signaling.6,7 Notably, none of the components in this oncogenic pathway are mutated to confer constitutive gain-of-function in MPM.3,6 Ras proteins link growth factor signaling to the cell cycle machinery by regulating cyclin-dependent protein kinases (CDK).8 Therefore, this Ras-based signaling network represents a compelling therapeutic target in MPM.
Many of the drug strategies aimed against this pathway are classified as anti-angiogenic agents. In the first-line setting combined with chemotherapy, sorafenib (anti-VEGFR2/3, platelet-derived growth factor receptor [PDGFR], and RAF/c-KIT), imatinib (anti-BCR-ABL, c-KIT, and PDGFR), and cediranib (anti-pan-VEGFR and PDGFR) were not significantly effective against MPM to merit their clinical adoption.9 Beyond these early clinical attempts, results from randomized phase III trials have not definitively changed therapeutic paradigms in MPM. Because the Mesothelioma Avastin Cisplatin Pemetrexed study (MAPS) was not a registration trial, bevacizumab (anti-VEGFA) added to cisplatin-based chemotherapy as first-line treatment remains an unlicensed regimen, despite showing a statistically significant, improved overall survival.2 These positive findings, however, prompted the National Comprehensive Cancer Network to recommend this regimen in unresectable MPM cases.10 Therefore, drug targeting of the Ras axis in MPM is among the few clinical approaches, other than standard-of-care, to achieve a category 1 level of evidence.
Consequently, we hypothesized that a microRNA (miRNA)-based approach aimed at the Ras signaling axis in MPM could be an alternative viable method. miRNAs are short nucleotide, noncoding RNAs involved in critical biological processes. In cancers, many miRNAs are often suppressed because of their location in genomic fragile sites.11 miRNAs coherently regulate multiple gene pathways via base-pairing in the 3′ untranslated region (UTR) of mRNA transcripts and, thus, in principle, are less prone to tumor-adaptive resistance and able to exert profound intracellular, phenotypic changes upon expression or suppression in cell-specific contexts.12 These innate properties of the RNA interference machinery are, therefore, attractive as a resource for novel therapeutic agents. Currently, miRNA-based therapeutics show great potential against several tumor types;13 and for MPM, proof of concept was recently demonstrated in a phase I trial.14
Because of inherent MPM inter-tumor molecular and pathologic heterogeneity,15 multiple miRNAs may fulfill an anti-MPM role. Our interest in characterizing miR-206 stems from several lines of evidence, which have never been synthesized into a coherent therapeutic strategy until now. In our prior microarray study, miR-206 was one of the top candidates downregulated significantly in MPM tissues by 12-fold compared with normal pleura tissue.16 Our observation was further corroborated in a comprehensive analysis of The Cancer Genome Atlas (TCGA) MPM specimens, in which miR-206 was among those miRNA most differentially expressed between good and poor (lower relative expression) prognostic cases.6 In other tumor types, miR-206 has been demonstrated to directly regulate expression of mRNA transcripts, including VEGFA in lung cancer,17 KRAS in pancreas cancer,18 and CDK4 in melanoma.19
This led us to test the notion that miR-206 targets the entire RTK-Ras-MAPK-PI3K/Akt-CDK pathway in MPM, translating to potent anti-cancer effects once its expression is restored. In this study, we delineated its biological role in MPM cells. Further, in xenograft tumor models of MPM, we demonstrate the therapeutic potential of miR-206. Overall, our findings establish a preclinical rationale to develop miR-206-based therapy to treat MPM in humans.
Results
miR-206 is downregulated in mesothelioma
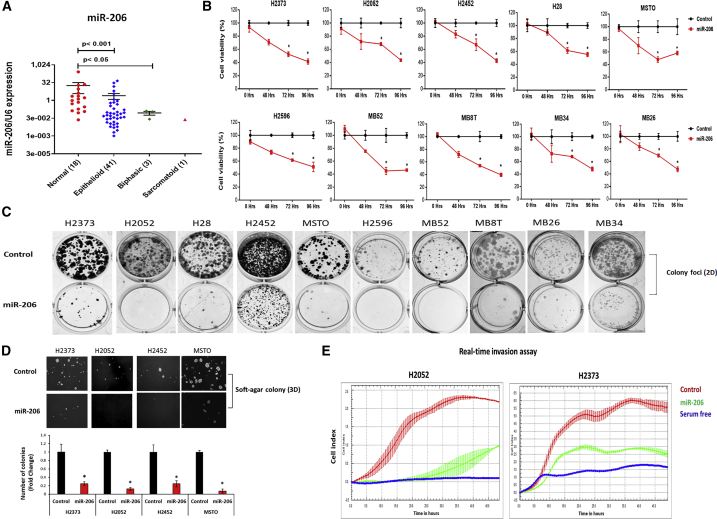
Initially, we sought to verify the status of miR-206 expression in MPM tissues because this information was still unreported from any large-sized sampling. Using quantitative real-time PCR, we confirmed the expression of miR-206 in randomly selected MPM tumors (41 epithelioid, 3 biphasic, and 1 sarcomatoid) compared with unmatched normal pleural tissues (N = 18). The mean expression level of miR-206 was significantly less, by 6.8-fold (p < 0.001), in MPM tumors versus normal pleurae (Figure 1A). This pattern of miR-206 downregulation was mirrored in a large panel of MPM cell lines (Figure S1). Overall, consistent loss of miR-206 expression in patient samples of MPM suggests that it may be clinically relevant.
Figure 1.
miR-206 expression in mesothelioma tissues and its tumor-suppressive effects in mesothelioma cells
(A) miR-206 expression in human tissues was determined by quantitative real-time PCR. This tumor cohort contained all MPM histologies (41 epithelioid, 3 biphasic, and 1 sarcomatoid). Dot plots are expressed as means ± SEM. p value in tissue samples was calculated by the Mann-Whitney test. (B) Cell viability at different time points in the MPM cell lines transfected with miR-206 compared with control mimics. Data are expressed as means ± SD of quadruplicates and are representative of three independent experiments. (C) Colony foci (2D) formation in MPM cells after control or miR-206 mimic transfection. (D) Anchorage-independent soft-agar colony (3D) formation in MPM cells on day 21 after control or miR-206 mimic transfection. (E) MPM cells transfected with miR-206 mimic compared with that of control over time showed significant decrease in cell invasion (p < 0.01). Serum-free medium was a negative control. Data are representative of three independent experiments. Bar graphs represent the means ± SEM, n = 3. ∗p < 0.05 was considered significant and was calculated by the two-tailed Student’s t test or other tests.
Re-expression of miR-206 induces tumor-suppressive effects
The biologic effects of miR-206 were characterized in MPM cells representing all three histologic subtypes. Control or miR-206 mimics were transfected into established MPM cell lines (H28, H2052, H2373, H2452, H2596, and MSTO-211H) and recently available, low-passage cell lines (MB8T, MB26, MB34, and MB52). Re-expression of miR-206 significantly inhibited the time-dependent growth (p < 0.05) of those MPM cell lines compared with control (Figure 1B) and significantly suppressed colony-forming ability (Figure 1C). Compared with control cells, miR-206 expression severely stunted anchorage-independent growth (p < 0.05) of MPM cells (Figure 1D). Furthermore, invasiveness of MPM was significantly decreased over time after miR-206 transfection into cells (Figure 1E). Of equal importance, the potential off-target effect of miR- 206 was assessed in normal mesothelial cells (recently available patient-derived cell line, NP4),20 as measured with a cell viability assay, which did not show significant change in growth compared with control cells (Figure S2). Together, these in vitro results indicate a putative tumor-suppressor role for miR-206 in MPM because its re-expression greatly diminished malignant features.
Some Ras-pathway genes targeted by miR-206 are prognostic in mesothelioma
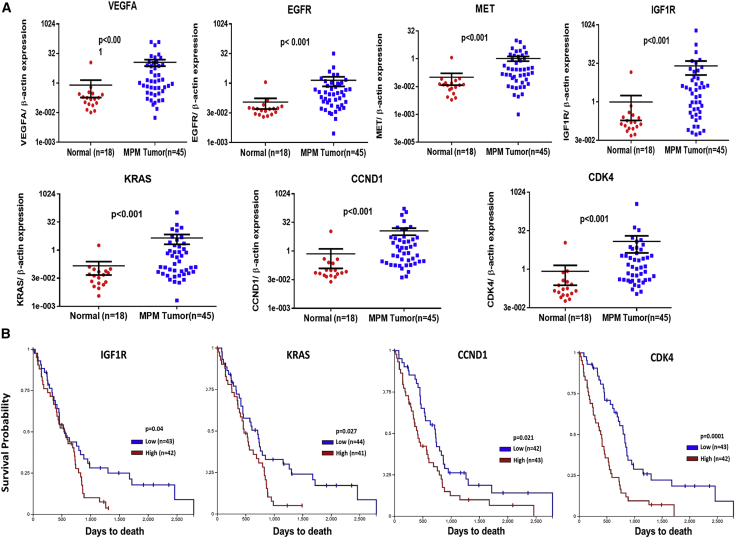
From the literature and our interrogation of miRNA databases (e.g., miRTarBase, release 7.0),21 we reviewed articles that experimentally verified the direct interaction of miR-206 in the 3′-UTR of its target transcripts to support our hypothesis that the mechanism of its anti-cancer effect in MPM is, in part, due to regulation of the RTK-Ras-MAPK-PI3K/Akt-CDK axis. From studies in human specimens, in addition to VEGFA,17 KRAS,18 and CDK4,19 miR-206 regulates c-MET in breast cancer,22 EGFR in prostate cancer,23 IGF1R24 and cyclin D2 (CCND2) in gastric cancer,25 Akt in skeletal muscle cancer,26 and cyclin D1 (CCND1) in breast cancer.27 Corresponding inversely to low miR-206 expression, we expected to observe an overabundance of these transcripts in MPM. Indeed, in randomly selected MPM specimens (N = 45), compared with normal pleurae (N = 18), mRNA levels of VEGFA, EGFR, c-MET, IGF1R, KRAS, CCND1, and CDK4 were upregulated (p < 0.05) (Figure 2A). Interestingly, from a separate cohort of MPM specimens (N = 85; Table S1) in TCGA (https://portal.gdc.cancer.gov/), relatively high expression of IGF1R, KRAS, CCND1, or CDK4 is associated with poor overall survival (p < 0.05) by Kaplan-Meier analysis (Figure 2B). Because TCGA does not include normal samples as a reference, high versus low expression of each gene in this analysis is calculated relative to its median expression among the entire MPM cohort. To support a regulatory link between miR-206 and these genes of the Ras signaling network, we reviewed their mutation and copy number status in TCGA. No significant changes were observed in the mutation and copy number of these seven genes (Figure S3), thus making plausible the loss of miR-206 expression as a contributing epigenetic mechanism. Altogether, in the context of low levels of miR-206 in MPM tissues, several predicted gene targets that contribute to Ras signaling are overexpressed, and a subset of them are associated with poor prognosis.
Figure 2.
RTKs-Ras-pathway genes regulated by miR-206 are overexpressed and prognostic in mesothelioma
(A) Quantitative real-time PCR determination of mRNA expression of VEGFA, EGFR, MET, IGF1R, KRAS, CCND1, and CDK4 in human specimens. (B) Kaplan-Meier analysis showing overall survival of patients with MPM based on dichotomized expression (relatively high or low) of IGF1R, KRAS, CCND1, and CDK4 genes. Results were derived from TCGA-Meso data. Where applicable, data are expressed as means ± SEM; p values in tissue samples were calculated by the Mann-Whitney test. p < 0.05 was considered significant.
Re-expression of miR-206 in mesothelioma cells suppresses Ras signaling
Having established a regulatory association between miR-206 and Ras-pathway genes, we performed a series of experiments to confirm a mechanistic link in MPM. Representative MPM cells were transfected with an miR-206 mimic, which induced a significant simultaneous decrease in the transcript abundance (p < 0.05) of VEGFA, EGFR, c-MET, IGF1R, KRAS, CCND1, and CDK4 genes (Figure 3A). The functional role of these miR-206-targeted genes was assessed using small interfering RNA (siRNA) knockdown, demonstrating that each one could significantly stunt the growth (p < 0.05) of MPM cells compared with that of control cells (Figure S4). Consequently, total levels of the corresponding protein to each gene noticeably decreased in response to miR-206 re-expression as demonstrated by western blot analysis (Figure 3B). From prior studies using a 3′-UTR reporter assay, it is known that miR-206 directly regulates VEGFA,17 KRAS,18 CDK4,19 c-MET,22 and CCND1.27 However, this stringent verification of the mechanism was not done for EGFR23 and IGF1R,24 despite both of these genes being recognized as miR-206 targets. So here, we queried TargetScan (release 7.2)28 and RNA22 (version 2.0)29 for predicted miR-206-binding sites in the 3′-UTR of the transcripts for EGFR and IGF1R (Table S2). Using a 3′-UTR luciferase reporter system, we confirmed both EGFR and IGF1R are specific gene targets of miR-206, with each harboring multiple 3′-UTR binding sites (Figure S5).
Figure 3.
miR-206 re-expression suppresses RTKs-Ras signaling in mesothelioma cells
(A) Quantitative real-time PCR determined the mRNA expression of VEGFA, EGFR, MET, IGF1R, KRAS, CCND1, and CDK4 genes in MPM cells transfected with miR-206 mimic versus the control at 48 h. (B) Under similar experimental conditions, the protein levels of EGFR, MET, CCND1, CDK4, IGF1R, VEGFA, and KRAS were determined by western blot. The protein abundance of each gene (numerical value) depicted in the blot was quantified by densitometry relative to β-actin. (C) The effect of miR-206 treatment compared with that of the control was assessed on a human phospho-kinase array to evaluate its effect on kinase signaling pathways in MPM cells at 48 h. Phospho-kinase array hybridization signals are shown. Black box shows the altered phosphorylation of ERK1/2(T202/Y204), EGFR(Y1026), and Akt1/2/3(S473) with miR-206 treatment. Bar graphs represent the mean pixel density of phosphorylated proteins. (D) Western blot validation of ERK1/2(T202/Y204), EGFR(Y1026), and Akt(S473), as well as downstream Rb protein phosphorylation in MPM cells treated with miR-206 or control at 48 h. The protein abundance of each kinase (numerical value) depicted in the blot is quantified by densitometry relative to β-actin. (E) Cell cycle distribution was determined by flow cytometry in MPM cells transfected with miR-206 or the control mimic at 48 h. Bar graphs show the cell population percentage of MPM cells after miR-206 or control mimic treatment. Where applicable, data are expressed as means ± SEM. ∗∗p < 0.01; p < 0.05 was considered significant and was calculated by the two-tailed Student’s t test.
Because multiple proteins of this signaling pathway are kinases, we checked their activation status by employing a phospho-kinase array, representative of a broad collection of other signaling networks. We observed that miR-206 re-expression in MPM cells preferentially inhibited the phosphorylation of EGFR(Y1026), ERK1/2(T202/Y204), and Akt1/2/3(S473) over other kinds of kinases (Figure 3C). By western blot assay, we corroborated these results by observing decreased phosphorylation of EGFR (undoubtedly influenced by decreases in total EGFR because of miR-206-direct 3′-UTR binding), ERK, and Akt, as well as in the Rb protein further downstream of this cascade (Figure 3D). These findings indirectly suggest that the RTK-Ras-MAPK-PI3K/Akt-CDK axis is a primary target of miR-206 in MPM. Further, these transcript- and protein-level changes upon miR-206 re-expression in MPM cells led to cell cycle arrest in the G1/S-phase (Figures 3E and S6). We verified the specificity of this phenotype by noting the absence of any appreciable apoptosis in MPM cells after miR-206 re-expression (Figures S7A and S7B) and by observing expected cell senescence (Figure S7C). Collectively, our data suggest that miR-206 regulates multiple clinically important components of the Ras-pathway signaling in MPM cells, which contribute to their survival.
miR-206 suppresses mesothelioma growth in vivo
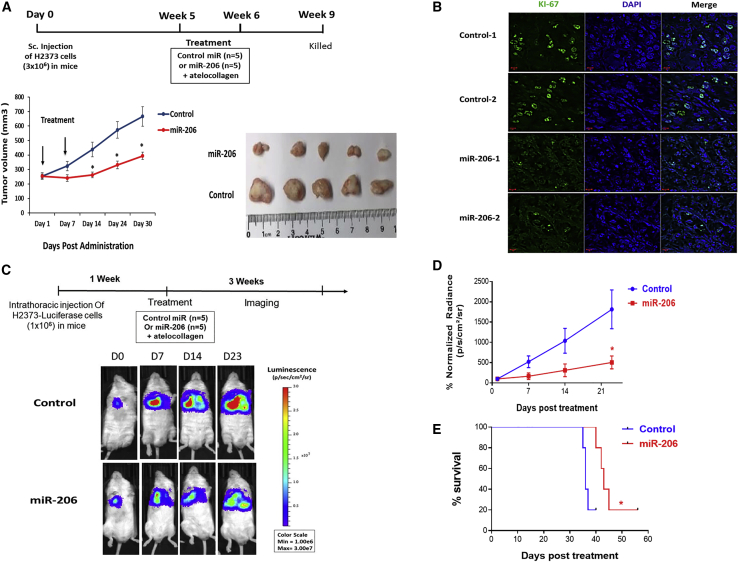
Next, we checked the anti-tumor efficacy of miR-206 when administered to MPM xenografts. For these experiments, we used H2373 cells, which represent the sarcomatoid subtype of MPM and harbor homozygous deletion of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene.30 Subcutaneous xenografts were grown in the flanks of NSG mice, according to our established protocol,31 and allowed to reach a large volume of at least 250 mm3. Control or the miR-206 mimic complexed in a atelocollagen vehicle was delivered by peritumoral (adjacent to the periphery) injection at two time points. Over a period of 4 weeks, miR-206 treatment significantly suppressed the growth (p < 0.05) of MPM tumors compared with the control treatment (Figure 4A). Immunohistofluorescence showed that miR-206 treatment significantly depressed Ki-67 levels, a marker of proliferation, consistent with its mechanism of inducing cell cycle arrest (Figure 4B). Furthermore, we confirmed successful delivery of miR-206 to the tumor by quantitating increased expression of oligonucleotide in treated xenografts compared with controls (Figure S8A) with concomitant significant decreases in the levels of miR-206-targeted genes in the Ras signaling pathway (Figures S8B and S8C).
Figure 4.
miR-206 suppresses growth of mesothelioma xenografts
(A) H2373 cells (3.0 × 106) were implanted in the flanks of NSG mice to establish subcutaneous xenografts. Once tumor volume reached an average of 250 mm3, miR-206 or control mimic complexed with atelocollagen (delivery vehicle) was injected at the peripheral edge of tumors (peritumoral injection) in mice (n = 5 in each group). A total dose of 5 nmol miR-206 was administered as treatment. Bottom panel: graph depicts the changes in tumor volume over 4 weeks with two treatment of miRNA. Image depicts the excised tumor xenografts. (B) Ki-67 levels were assessed by immunohistofluorescence in miR-206- or control-treated tumor xenografts. (C) Top panel: schematic timeline of monitoring tumor growth in mice bearing intrathoracic H2373-luciferase tumor cells followed by a single intrapleural administration of miRNA-atelocollagen complex. Bottom panel: live imaging shows tumor luminescence in mice treated with either miR-206 or control mimic at the indicated time points (n = 5 in each group). (D) Tumor growth curve of mice bearing intrathoracic H2373-Luciferase cells in miR-206 and control treatment groups based on luminescence of tumor cells. (E) Kaplan-Meier survival curve of mice treated with miR-206 or control mimic; log rank (Mantel-Cox) test was used to assess significance. Where applicable, data are presented as means ± SEM. ∗p < 0.05 was considered significant and was calculated with the two-tailed Student’s t test or other tests.
Additionally, we evaluated the efficacy of miR-206 in an orthotopic xenograft mouse model based on H2373 cells stably expressing a luciferase-reporter gene. Tumor cells were implanted in the right hemithorax of mice and allowed to mature for 7 days, according to our established protocol.31 Control or the miR-206 mimic complexed in atelocollagen vehicle was administered once intrapleurally. Over 3 weeks of monitoring luminescence (a surrogate for in vivo tumor volume) in mice, miR-206 treatment significantly inhibited tumor growth (p < 0.05) compared with control treatment (Figures 4C and 4D). Kaplan-Meier analysis showed that miR-206 treatment noticeably improved overall survival (median 43 days) compared with that of control (median 36 days) (p < 0.05) (Figure 4E). In sum, these data indicate that miR-206 re-expression suppresses the growth of tumors by functioning as a tumor suppressor in MPM.
Additive effects by combining miR-206 with a drug inhibitor
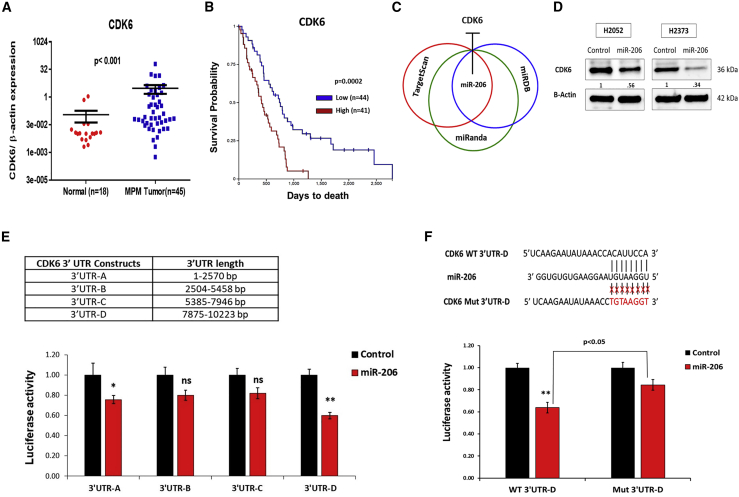
Lastly, we explored extended application of miR-206 re-expression in MPM based on the notion that simultaneous knockdown of an oncogenic transcript and inhibition of its protein activity is synergistic.32 Numerous kinds of protein-specific inhibitor drugs are available because many components of the RTK-Ras-MAPK-PI3K/Akt-CDK pathway can be oncogenic drivers. In focusing our efforts, we reasoned that drugs targeting prognostic genes in this pathway would likely be efficacious and, thus, represent top candidates for experimental verification. Of the four prognostic genes we identified in MPM (IGF1R, KRAS, CCND1, and CDK4 from Figure 2B), drugs aimed at the CCND1-CDK4 complex were the most interesting to combine with miR-206 as treatment because the CDK4/6 inhibitor abemaciclib is just entering MPM clinical trials (NCT03654833). Additional rationale to evaluate this drug included the following: (1) CDK6 mRNA is significantly overexpressed (p < 0.05) in our tissue cohort of MPM compared with normal pleurae (Figure 5A), (2) relative high expression of CDK6 in MPM tumors was associated with poor overall survival (p < 0.05) (Figure 5B), and (3) CDK6 targeting has greater potency with less side effects compared with other CDK inhibitors.35 The dual targets of abemaciclib prompted us to ascertain whether miR-206 regulates CDK6, first by querying target-prediction databases (Figure 5C). Three miRNA target-prediction databases, TargetScan,28 miRDB,33 and miRanda,34 predicted a common miR-206 binding site in the 3′-UTR of CDK6 at position 8959–8966, whereas the RNA22 database29 predicted binding sites at positions 1607–1628, 8624–8643, and 9323–9346. Using western blot analysis and a 3′-UTR luciferase reporter system coupled with site-specific mutagenesis, we verified that CDK6 is a novel target of miR-206 (Figures 5D and 5E). For target specificity, a CDK6 3′-UTR mutant reporter construct was generated in the most potent binding site (8959–8966) of miR-206 predicted by multiple databases (Figure 5F). Thus, we established that both miR-206 and abemaciclib target the CCND1-CDK4/6 cell cycle complex via different mechanisms and represent a promising therapeutic combination. As proof of concept, we tested abemaciclib to assess whether it could augment the effects of miR-206 re-expression (and vice versa) in MPM.
Figure 5.
CDK6, a novel direct target of miR-206, is prognostic in mesothelioma
(A) CDK6 mRNA expression in human specimens determined by quantitative real-time PCR. (B) Kaplan-Meier analysis shows overall survival of MPM patients based on dichotomized expression (relatively high or low) of the CDK6 gene. Data was obtained from TCGA-Meso. (C) Overlap of miR-206 binding site in the 3′-UTR of CDK6 (3′-UTR-D, see below) predicted by TargetScan,28 miRDB,33 and miRanda.34 (D) CDK6 protein levels by western blot in MPM cells transfected with miR-206 or the control mimic at 48 h. The protein abundance of CDK6 (numerical value) depicted in the blot is quantified by densitometry relative to β-actin. (E) The 3′-UTR-luciferase reporter assay confirmed the target specificity of miR-206 to the binding regions in the 3′-UTR of the CDK6 gene. Top panel: length of CDK6 3′UTR segments used for construction of four 3′-UTR-luciferase reporter vectors, 3′-UTR-A (1–2,570 bp), 3′-UTR-B (2,540–5,458 bp), 3′-UTR-C (5,385–7,946 bp), and 3′-UTR-D (7,875–10,223 bp). Bottom panel: luciferase activity was measured in H2052 cells co-transfected with wild-type CDK6 3′-UTR-luciferase reporter vectors and miR-206 or control mimic. (F) Top panel: substitution mutation at the binding site of miR-206 in CDK6 3′-UTR region (3′-UTR-D). Bottom panel: miR-206 mimic transfection in H2052 cells suppressed the luciferase activity of the wild-type construct (WT 3′-UTR-D), whereas no significant effect was observed in the luciferase activity of the mutant 3′-UTR luciferase construct (Mut 3′-UTR-D). Where applicable, data are expressed as means ± SEM. p values in tissue samples were calculated with the Mann-Whitney test. ∗p < 0.05, ∗∗p < 0.01, ns; not significant. p < 0.05 was considered significant and was calculated by the two-tailed Student’s t test or other tests.
In brief, three representative MPM cell lines were transfected with control or the miR-206 mimic and then exposed to increasing concentrations of abemaciclib over 48 h; during which, cell viability was measured. Compared with the control, those cell lines with miR-206 re-expression all exhibited lower half-maximal inhibitory concentration (IC50), demonstrating an additive MPM-cell-killing effect (Figure 6A). Next, we verified these additive effects of miRNA and drug therapy in vivo. When intrathoracic MPM luciferase-labeled xenografts had matured after 7 days, mice were randomly divided into five groups (n = 5), and each group was treated with drug vehicle, abemaciclib, control (miRNA), miR-206 (single dose), or a combination of abemaciclib (oral) and miR-206 (local intrapleural) (Figure 6B). Tumor burden in mice was monitored over 4 weeks. Notably, miR- 206 and abemaciclib mono-treatment significantly suppressed tumor growth (p < 0.05) compared with the control or the drug vehicle, whereas the combination treatment showed more-pronounced and -consistent tumor suppression at different time points (Figures 6B and 6C). Kaplan-Meier analysis revealed that the combination regimen markedly improved overall survival (median 51 days), whereas miR-206 or abemaciclib mono-therapy significantly improved overall survival with a median of 44 and 41 days, respectively, compared with the control or the vehicle (median 34 days for both) (for each comparison, p < 0.05) (Figure 6D). Our results in MPM indicate that the combination of miR-206 and abemaciclib, as one example, produced a therapeutically improved effect with overlapped targeting of the CCND1-CDK4/6 complex. Furthermore, we firmly established that miR-206 regulates the RTK-Ras-MAPK-PI3K/Akt-CDK pathway at key points (Figure 6E).
Figure 6.
miR-206 enhances the anti-tumor efficacy of CDK4/6 inhibitor in mesothelioma
(A) miR-206 re-expression potentiates abemaciclib-induced cell killing in MPM cells. MPM cells were transfected with miR-206 or control mimic, and after 24 h, cell survival was measured over increasing concentrations of abemaciclib treatment for an additional 48 h. Data are expressed as means ± SD of quadruplicates and are representative of three independent experiments. (B) Top panel: schematic timeline of the monitoring of the tumor burden in mice bearing intrathoracic H2373-luciferase tumor cells, followed by a single intrapleural administration of miRNA-atelocollagen complex and the drug/vehicle alone or in combination with miRNA at the indicated times. When xenografts matured after 7 days, mice were randomly divided into five groups (n = 5), and each group was treated with drug vehicle, abemaciclib, control-miRNA (single dose), miR-206 (single dose), or a combination of abemaciclib (oral) and miR-206 (local intrapleural) at the indicated times. Bottom panel: live imaging shows tumor luminescence in mice treated with control-miR, miR-206, drug vehicle, abemaciclib, or a combination of abemaciclib and miR-206. (C) Tumor growth curve of mice bearing intrathoracic H2373-luciferase cells in different treatment groups based on the luminescence of tumor cells. ∗p < 0.05, ∗∗p < 0.01 versus control or drug vehicle, calculated by the two-tailed Student’s t test or Mann-Whitney test. #p < 0.05, one-way ANOVA used to determine statistical significance between the means of multiple groups. (D) Kaplan-Meier survival curve of mice in different treatment groups, log-rank (Mantel-Cox) test was used to assess significance versus control or drug vehicle. Where applicable, data are presented as means ± SEM; ∗p < 0.05 was considered significant. (E) miR-206 regulatory network in mesothelioma. Graphic summarizes the major genes directly regulated by miR-206 in MPM. Re-expression of miR-206 in MPM decreased the expression of VEGFA, EGFR, MET, IGF1R, KRAS, CCND1, CDK4, and CDK6 genes. Inhibiting the RTKs-Ras-MAPK-PI3K/Akt-CDK pathway induces cell cycle arrest.
Discussion
The precise pathogenesis of MPM remains incompletely described. Nevertheless, a crucial growth and cell survival mechanism coapted in MPM is the hyperactive signaling initiated by RTK subfamilies, many of which are overexpressed in MPM.6, 7, 8 Asbestos fibers can induce pathologic RTK signaling by autophosphorylation of EGFR (overexpressed in most MPM) as well as autophosphorylation of MAPK, inducing the subsequent pathway cascade.36 Loss of common tumor suppressors associated with MPM contribute to sustained activation of such signaling: NF237 loss influences Ras, and PTEN38 loss dysregulates PI3K/Akt. As demonstrated in breast cancer cells, hyperactive Ras signaling, not due to mutation of the gene, is mandatory for full expression of the malignant phenotype.39 A similar condition seemingly exists in MPM, and it is this state of wild-type (WT) Ras dependency that provides the rationale for pursuing pharmacologic-inhibition strategies.
Unfortunately, efficacious targeting of this Ras signaling pathway has not yet been borne out in clinical trials. Recently, the LUME-Meso study did not meet the primary progression-free endpoint with the addition of nintedanib (anti-VEGFR 1–3, PDGFR α and β, and fibroblast growth factor receptors [FGFR] 1–3, plus Src and Abl kinases).40 In an ongoing trial (NCT02863055), nintedanib is being investigated as maintenance therapy in patients with non-progressive disease after first-line chemotherapy. In addition, there are ongoing investigations of newer biosimilars, such as ramucirumab (anti-VEGFR2) in the phase II randomized, multi-center trial in a second-line setting (NCT03560973). So clearly, there remains much interest in more fully developing anti-MPM strategies based on the importance of Ras signaling.
In this study, we leveraged the gene regulatory properties of miRNA as an alternative approach. We demonstrated that miR-206 acts as a tumor suppressor against MPM by its coherent targeting of the RTK-Ras-MAPK-PI3K/Akt-CDK pathway at critical points, including KRAS directly. We confirmed in a large sample of MPM tissues the consistent loss of miR-206 expression, regardless of histologic subtype. The expression levels (inversely correlated) of multiple genes in the Ras signaling pathway that are directly regulated by miR-206 are prognostic. This group of overexpressed genes now includes CDK6, a newly verified miR-206 target. Of clinical relevance, is the prognostic expression level (higher levels correlate with worse survival) of the CCND1-CDK4/6 cell cycle complex. When miR-206 is re-expressed in MPM xenografts, it induces tumor shrinkage by arresting the cell cycle in the G1/S-phase manifested as irreversible cell senescence. In extended application, we identified a novel therapeutic MPM regimen by combining local administration of miR-206 with systemic-dosed CDK4/6 inhibition. We observed in vivo additive efficacy with this unique combination.
Our results now integrate the group of clinical studies pursuing the blockade of various RTKs at the cell surface (i.e., bevacizumab in the MAPS trial),2 with an emerging line of targeted therapy in MPM inhibiting CDK4/6. Interestingly, before any MPM-specific preclinical studies were reported, the phase II Signature program tested ribociclib (CDK4/6 inhibitor) in five patients with MPM as part of a basket trial design,41 but any results are yet to be published (NCT02187783). The first proof-of-principle study using human MPM cell lines demonstrated the anti-tumor effects of palbociclib (CDK4/6 inhibitor) alone or in sequential combination with novel PI3K inhibitors NVP-BEZ235 and NVP-BYL719.42 In addition, preliminary (abstract only) results using palbociclib alone or in combination with gemcitabine to treat subcutaneous MPM xenografts in mice was reported.43 Clearly, in MPM, the CCND1-CDK4/6 complex represents a compelling, underexploited molecular target. We have corroborated those efforts by validating that the CCND1-CDK4/6 cell cycle complex is overexpressed in MPM tissues and that this mRNA signature is prognostic of a poor outcome.
Our miRNA-based strategy of therapeutic targeting is promising for MPM. No other single agent can simultaneously inhibit multiple components of the RTK-Ras-MAPK-PI3K/Akt-CDK pathway like miR-206, which we have validated is among the most underexpressed MPM-specific miRNA. In contrast, small-molecule drugs are more limited in the total scope and extent of their mechanism(s) inhibiting this signaling pathway. It is recognized that the RTK-Ras-MAPK-PI3K/Akt-CDK axis is primed for crosstalk and is efficient in compensatory signaling when certain specific components are inhibited by traditional classes of drugs, thus explaining their lack of clinical success in mono-therapy regimens.44 miRNAs as therapeutics are attractive because they more closely mimic ideal agents targeting multiple genes and molecular networks to an extent beyond what can be covered with combinations of traditional drugs. These traits have prompted intense efforts to develop clinically practical delivery vehicles for miRNA therapies.13 Specifically, in MPM, replacement of miR-16 via systemic administration of antibody-targeted minicells14 has already progressed to phase II testing. Because of the inherent complexity and heterogeneity of MPM,15 we envision multiple miRNAs may fulfill the therapeutic role via a diverse set of delivery routes.
Using murine xenograft models, we demonstrated the efficacy of a novel combination of miR-206 delivered by local intrapleural application in the region of MPM with abemaciclib taken orally and absorbed via the circulatory system. This specific combination highlights how improved effects can be achieved by co-inhibiting oncogenes at the transcript and protein levels. This anti-cancer strategy of inhibiting dual mechanisms has been demonstrated in vitro, for example, for head and neck squamous cell carcinoma45 and glioblastoma multiforme.32 We validated this notion by showing additive, improved survival when using miRNAs and drugs in treating mice harboring intrapleural tumors. Conceivably, local delivery of therapeutic miRNAs could maximize the efficacy at the site of tumor in which the additive effect would be concentrated and improve the drug toxicity because of its lower systemic dosing requirements. Prolonged use of abemaciclib inevitably leads to drug resistance,46 so another potential benefit of combining drugs with miRNA is a practical solution of overcoming such resistance. Conversely, this novel type of therapeutic combination could extend drug sensitivity. Instead of treating only those patients with low p16 (INK4A) expression (NCT03654833), perhaps, abemaciclib could be combined with miR-206 to treat MPM, regardless of p16 status. Certainly, this is the reality borne out in breast cancer trials that demonstrated poor correlation of biomarker status and clinical benefit with CDK4/6 inhibitors.47
In summary, miR-206 is a tumor suppressor in MPM, which regulates, among multiple gene targets, the RTK-Ras-MAPK-PI3K/Akt-CDK axis. Components of the Ras pathway are associated with prognosis (higher transcript levels correlated with poorer survival) in MPM, including KRAS and the cell cycle complex. When reconstituted, miR-206 abrogated the malignant phenotype of MPM in vitro and in vivo. We identified a novel MPM therapeutic combination by adding systemic-route abemaciclib with local-route miR-206. Subsequently, dual inhibition of the CCND1-CDK4/6 cell cycle complex showed additive efficacy against MPM. Our results suggest that miRNA-based therapy should be studied and developed further in MPM, a complex surface cancer that has defied conventional treatments.
Materials and methods
Reagents
The CDK4/6 inhibitor abemaciclib mesylate (LY2835219) was purchased from Selleckchem.
Tissues and cell culture
Specimen collection followed our institutional review board (IRB)-approved protocols. De-identified surgical specimens were stored at −80°C. We selected 45 MPM tumors of all three histologies and 18 non-patient-matched, non-malignant pleurae based on the amounts of usable tissue available (Table S1). National Cancer Institute (NCI)-derived MPM cell lines (H28, H226, H2052, H2373, H2596, H2452, H2691, and MSTO-211H) were obtained from ATCC. Low-passage MPM cell lines, nos. 19 (MB19), 24 (MB24), 26 (MB26), 34 (MB34), 52 (MB52), and 8T (MB8T), were obtained from MesobanK, UK, an international bioresource.48 These cell lines represent all three histologies of MPM (Table S3). All MPM cell lines were cultured as monolayers at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) plus 1% penicillin/streptomycin solution. The normal mesothelial cell line, LP-9, was purchased from Coriell Cell Repository and cultured according to the manufacturer or a recently available pleural mesothelial cell line, NP4,20 was used accordingly. All media, media supplements, and FBS were purchased from Thermo Fisher Scientific.
Cell transfection
The miR-206 mimic and the mirVana miRNA mimic negative-control no. 1 were transfected into cell lines at a final concentration of 20 nM using Lipofectamine RNAiMAX, according to manufacturer’s (Thermo Fisher Scientific) instructions. For transient gene silencing, chemically modified siRNAs (SMARTpool: ON-TARGETplus siRNA) and scrambled negative controls (Dharmacon) were transfected at a final concentration of 50 nM into the indicated MPM cell lines using Lipofectamine RNAiMAX.
Quantitative real-time PCR
Total RNA was extracted from tissues and cells using TRIzol. RNA was quantified using a NanoDrop 2000 spectrophotometer. For miRNA analysis, reverse transcription was performed with TaqMan advanced miRNA cDNA synthesis kit. For mRNA analysis, total RNA was reverse transcribed with the high-capacity cDNA reverse transcription kit. Expression of mRNA or miRNA was determined by TaqMan analysis on a QuantStudio 6 Flex PCR system.49 The expression of gene and miRNAs was normalized to β-actin and U6 (RNU6B) small-nucleolar RNA, respectively. Quantitative real-time PCR primers used for the analysis of genes and miRNA expression were available from Applied Biosystems (Table S4). All quantitative real-time PCR reactions were performed independently in triplicate. All reagents and equipment are from Thermo Fisher Scientific.
Cell viability
Consistently, 3.0 × 103 MPM cells/well were seeded into 96-well plates. Under several experimental conditions, cell viability was measured using the CellTiter-Glo Assay (Promega). Studies were performed in three independent cell preparations.
Clonogenicity
Colony-foci formation (two dimensional [2D]) was assessed in vitro. After miRNA transfection (16 h), MPM cells were trypsinized, and 1.0 to 1.5 × 103 cells/well were seeded in 12-well plates. Media was replenished every third day. After 10–14 days, foci were stained with crystal violet (0.5%) and imaged using the Bio-Rad gel imager.
Anchorage-independent growth
Anchorage-independent growth (three dimensional [3D]) was assessed in soft agarose. Six-well plates were pre- coated with 0.5% (w/v) agarose in appropriate cell media and allowed to set. Approximately 5.0 × 103 MPM cells/well were resuspended in a soft agarose medium consisting of 0.4% (w/v) agarose (Sigma-Aldrich), 10% FBS, and RPMI 1640 media before being seeded on a 0.5% (w/v) agarose base. After 3–4 weeks of culture, colonies were imaged under a microscope after being stained with crystal violet solution (0.05%).
Invasion assay
According to manufacturer instructions, experiments were performed on the real-time cell analyzer dual-purpose system, an electronically integrated 16-well Boyden chamber apparatus with a membrane pore size of 8 μm (ACEA Biosciences). Briefly, MPM cells were serum starved overnight after miRNA transfection. Then, 5.0 × 104 cells/well were seeded into the upper chamber of wells coated with Matrigel, whereas the lower chambers contained RPMI growth medium supplemented with 10% FBS.50 Impedance change generated from migrating MPM cells was recorded in real time and expressed as the cell index (CI). All cellular-invasion experiments (three independent runs) were run for 48 h, and CI was monitored every 10 min.
Western blot analysis
MPM cells were lysed in radio-immunoprecipitation buffer, and their protein concentration was measured using the bicinchoninic acid protein assay kit (Thermo Fisher Scientific). This material was fractionated on 4%–15% polyacrylamide gels and transferred onto nitrocellulose (Bio-Rad). Primary monoclonal antibodies used were the following: β-actin (no. 8457), KRAS (no. 8955), CDK4 (no. 12790), CCND1 (no. 2978), MET (no. 8198), EGFR (no. 4267), phospho-Akt (Ser473) (no. 4060), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (no. 9101), phospho-EGFR (Y1086) (no. 2220), phospho-Rb (Ser807/811) (no. 8516), cleaved caspase-3 (no. 9661), and PARP (no. 9542), all at 1:1,000 dilution (Cell Signaling Technology); in addition, VEGFA (ab46154), CDK6 (ab124821), and IGF1R (ab182408), all at 1:1,000 dilution (Abcam), were used. Secondary antibodies anti-rabbit and anti- mouse, conjugated with horseradish peroxidase, were used for the detection of protein signal (Abcam). All protein experiments were performed in triplicate. Blots were quantified with ImageJ software.
Phosphorylation status
A proteome profiler human phospho-kinase array kit (R&D Systems, no. ARY003B) was used to detect the phosphorylation of 43 kinases in MPM cells treated with miR-206 or with the control mimic. In brief, we used 2.0 × 106 MPM cells per 10-cm cell culture dish, and 24 h later, the cells were transfected with miR-206 or the control mimic. After 48 h, cells were washed with PBS, and the lysate was prepared. For each array panel, 300 μg of protein lysates were incubated, and array membranes were developed according to manufacturer’s protocols. Protein levels that changed with miR-206 treatment were validated by western blot analysis.
Cell cycle analysis
Flow cytometric analysis of DNA content in fixed MPM cells was performed with the FxCycle PI/RNase staining solution according to manufacture instructions (Thermo Fisher Scientific). Into each well of a 6-well plate, 2.0 × 105 MPM cells were seeded and transfected 24 h later with miR-206 or the control mimic. Data were acquired and analyzed on a BD FACSCalibur platform according to manufacturer’s recommendations (BD Biosciences). Assays were performed in triplicate.
Apoptosis analysis
MPM cells, at a density of 2.0 × 105 cells/well, were transfected with control or the miR-206 mimic (20 nM) in six-well plates for 72 h. Adherent and floating cells were collected. Apoptotic or dead cells were detected by fluorescence-activated cell sorting (FACS) analysis using the Annexin V-FITC apoptosis detection kit (Abcam) according to manufacturer’s protocols.
Cell senescence
In brief, 1.0 × 105 MPM cells/well were transfected with control or miR-206 mimic (20 nM) in 12-well plates. After 72 h, senescent cells were detected with the senescence β-galactosidase staining kit (Cell Signaling Technology) according to manufacturer’s protocols.
Immunohistofluorescence
Tissues sections (5 μm) were deparaffinized and serially rehydrated in ethanol (100%, 95%, 70%, and 50%, each for 5 min). Antigen retrieval was performed with heated citrate buffer (Vector Laboratories, H3300) according to manufacturer’s instructions. Protein signal of Ki-67 was monitored using a standard immunolabeling protocol. The following was used: primary antibody against Ki-67 (no. 9027, Cell Signaling Technology) and secondary antibody Alexafluor488-conjugated, goat anti-rabbit (111-545-144, Jackson ImmunoResearch). Tissue sections were mounted with a medium containing DAPI (Vector Laboratories) and imaged using a Zeiss LSM 710 NLO confocal microscope.
Luciferase reporter assay for miRNA target validation
To construct luciferase reporter vectors, the 3′-UTR segments of EGFR, IGF1R, and CDK6 were cloned downstream to the firefly luciferase open reading frame (ORF) into the pEZX-MT06 vector (GeneCopoeia). Because of the large size (>6.01 kb) of the EGFR 3′-UTR, two 3′-UTR (UTR A, and B) luciferase reporter constructs were generated that harbored binding sites for miR-206. Similarly, three 3′-UTR luciferase reporter vectors were constructed (UTR A, B, and C) for long IGF1R 3′-UTR (>7.08 kb) from three segments of IGF1R 3′-UTR. The CDK6 3′-UTR luciferase reporter vectors were constructed from four segments of the CDK6 3′-UTR (UTR A, B, C, and D) inserted into the pEZX-MT06 vector. Use of four 3′-UTR segments was required because of the long length (>10.2 kb) of the CDK6 3′-UTR, which contains the binding region for miR-206. To verify the target specificity of miR-206, the mutant 3′-UTR luciferase reporter construct of CDK6, with substitution mutations within the target sequence of miR-206, was generated by site-directed mutagenesis (GeneCopoeia). In brief, MPM cells were seeded into six-well plates at a density of 2.0 × 105 cells/well. After 24 h, these cells were co-transfected with each gene-3′-UTR-luciferase reporter construct and control or miR-206 mimic using Lipofectamine 2000 with Opti-MEM (Thermo Fisher Scientific). After 48 h of transfection, cells were harvested, and the relative luciferase activity (firefly/Renilla) was measured by Duo-Luciferase assay kit 2.0 (GeneCopoeia).
Mice xenografts
All animal experiments were approved by our Animal Care and Use Committee in accordance with NIH Guidelines. H2373 cells (3.0 × 106) were injected subcutaneously into the right flank of NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (6–8 weeks old).51 Xenografts reached an average of 250 mm3 before experimentation. To study the anti-tumor efficacy of miR-206 treatment in vivo, control or the miR-206 mimic (10 μM) was complexed with atelocollagen (AteloGene local use, Koken Co.) according to our established protocol.31 In the intrapleural xenograft models, 1.0 to 2.0 × 106 H2373 cells stably expressing the luciferase reporter vector pGL4.51[luc2/CMV/Neo] (Promega) were implanted into the pleural space of NSG mice (6–8 weeks old). Xenografts matured over 7 days before experimentation. Next, control or the miR-206 mimic (10 μM) was complexed with atelocollagen before intrathoracic delivery, and in vivo studies were conducted as previously described.31 In addition, separate cohorts of mice were treated with miR-206 (intrathoracic delivery) or abemaciclib (oral gavage 50 mg/kg/day over 14 days) compared with mice treated with the combination of drug and miRNA in the context of appropriate treatment controls (drug vehicle or miRNA mimic). Abemaciclib was formulated in 1% hydroxyethyl cellulose and 20 mM phosphate buffer (pH 2.0).
Statistics
Means and standard error of the mean (SEM) or standard deviation (SD) were calculated from numerical data. Fold or percentage changes indicate the difference between experimental and control samples. As indicated, values in graphs are presented as the means ± SEM or ± SD. Two-tailed, unpaired Student’s t test assessed significance between the two conditions. Nonparametric Mann-Whitney test compared the differences in mRNA or miRNA expression between normal pleura and MPM tumor specimens. One-way analysis of variance (ANOVA) was used to determine statistical significance between the means of multiple groups. For survival analysis, Kaplan-Meier and the log-rank test were applied. p <0.05 was considered statistically significant. GraphPad Prism version 7.05 software was used for statistical calculations.
Acknowledgments
We would like to thank Shivani Dixit for assistance with critical proofreading of the manuscript. This work was supported by the NIH Intramural Research Program with funding (ZIA BC 011657) provided to C.D.H.
Author contributions
Data curation, analysis, and writing of the manuscript, A.S.; data curation and interpretation, N.P., R.P., and A.M.; collection of clinical specimens and editing of the manuscript, D.S.; conceptualization, study design and direction, data interpretation, writing and editing of the manuscript, and project administration, C.H.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.04.001.
Supplemental information
References
- 1.Beckett P., Edwards J., Fennell D., Hubbard R., Woolhouse I., Peake M.D. Demographics, management and survival of patients with malignant pleural mesothelioma in the National Lung Cancer Audit in England and Wales. Lung Cancer. 2015;88:344–348. doi: 10.1016/j.lungcan.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Zalcman G., Mazieres J., Margery J., Greillier L., Audigier-Valette C., Moro-Sibilot D., Molinier O., Corre R., Monnet I., Gounant V., French Cooperative Thoracic Intergroup (IFCT) Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet. 2016;387:1405–1414. doi: 10.1016/S0140-6736(15)01238-6. [DOI] [PubMed] [Google Scholar]
- 3.Bueno R., Stawiski E.W., Goldstein L.D., Durinck S., De Rienzo A., Modrusan Z., Gnad F., Nguyen T.T., Jaiswal B.S., Chirieac L.R. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 4.Menges C.W., Chen Y., Mossman B.T., Chernoff J., Yeung A.T., Testa J.R. A phosphotyrosine proteomic screen identifies multiple tyrosine kinase signaling pathways aberrantly activated in malignant mesothelioma. Genes Cancer. 2010;1:493–505. doi: 10.1177/1947601910375273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S., Liu L., Li H., Eilers G., Kuang Y., Shi S., Yan Z., Li X., Corson J.M., Meng F. Multipoint targeting of the PI3K/mTOR pathway in mesothelioma. Br. J. Cancer. 2014;110:2479–2488. doi: 10.1038/bjc.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hmeljak J., Sanchez-Vega F., Hoadley K.A., Shih J., Stewart C., Heiman D., Tarpey P., Danilova L., Drill E., Gibb E.A., TCGA Research Network Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–1565. doi: 10.1158/2159-8290.CD-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel M.R., Jacobson B.A., De A., Frizelle S.P., Janne P., Thumma S.C., Whitson B.A., Farassati F., Kratzke R.A. Ras pathway activation in malignant mesothelioma. J. Thorac. Oncol. 2007;2:789–795. doi: 10.1097/JTO.0b013e31811f3aab. [DOI] [PubMed] [Google Scholar]
- 8.Ingham M., Schwartz G.K. Cell-cycle therapeutics come of age. J. Clin. Oncol. 2017;35:2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantini L., Hassan R., Sterman D.H., Aerts J.G.J.V. Emerging treatments for malignant pleural mesothelioma: where are we heading? Front. Oncol. 2020;10:343. doi: 10.3389/fonc.2020.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Comprehensive Cancer Network . 2020 edition. NCCN; 2020. Malignant Pleural Mesothelioma Guidelines. [Google Scholar]
- 11.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah M.Y., Ferrajoli A., Sood A.K., Lopez-Berestein G., Calin G.A. microRNA therapeutics in cancer—an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 14.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 15.Oehl K., Vrugt B., Opitz I., Meerang M. Heterogeneity in malignant pleural mesothelioma. Int. J. Mol. Sci. 2018;19:1603. doi: 10.3390/ijms19061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y., Zheng M., Merritt R.E., Shrager J.B., Wakelee H.A., Kratzke R.A., Hoang C.D. miR-1 induces growth arrest and apoptosis in malignant mesothelioma. Chest. 2013;144:1632–1643. doi: 10.1378/chest.12-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H., Yu X., Yang F., Zhang Z., Shen J., Sun J., Choksi S., Jitkaew S., Shu Y. Reprogramming of normal fibroblasts into cancer-associated fibroblasts by miRNAs-mediated CCL2/VEGFA signaling. PLoS Genet. 2016;12:e1006244. doi: 10.1371/journal.pgen.1006244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keklikoglou I., Hosaka K., Bender C., Bott A., Koerner C., Mitra D., Will R., Woerner A., Muenstermann E., Wilhelm H. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene. 2015;34:4867–4878. doi: 10.1038/onc.2014.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgantas R.W., 3rd, Streicher K., Luo X., Greenlees L., Zhu W., Liu Z., Brohawn P., Morehouse C., Higgs B.W., Richman L. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res. 2014;27:275–286. doi: 10.1111/pcmr.12200. [DOI] [PubMed] [Google Scholar]
- 20.Pruett N., Singh A., Shankar A., Schrump D.S., Hoang C.D. Normal mesothelial cell lines newly derived from human pleural biopsy explants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020;319:L652–L660. doi: 10.1152/ajplung.00141.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Leva G., Gasparini P., Piovan C., Ngankeu A., Garofalo M., Taccioli C., Iorio M.V., Li M., Volinia S., Alder H. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson R.S., Yi M., Esposito D., Watkins S.K., Hurwitz A.A., Yfantis H.G., Lee D.H., Borin J.F., Naslund M.J., Alexander R.B. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren J., Huang H.J., Gong Y., Yue S., Tang L.M., Cheng S.Y. MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci. 2014;4:26. doi: 10.1186/2045-3701-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Liu X., Jin H., Guo X., Xia L., Chen Z., Bai M., Liu J., Shang X., Wu K. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 2013;332:94–101. doi: 10.1016/j.canlet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 26.Margolis L.M., McClung H.L., Murphy N.E., Carrigan C.T., Pasiakos S.M. Skeletal muscle myomiR are differentially expressed by endurance exercise mode and combined essential amino acid and carbohydrate supplementation. Front. Physiol. 2017;8:182. doi: 10.3389/fphys.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliman S.J., Howley B.V., Mehta D.S., Fearnhead H.O., Kemp D.M., Barkley L.R. Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis. 2014;3:e113. doi: 10.1038/oncsis.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M., Lim B., Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Klorin G., Rozenblum E., Glebov O., Walker R.L., Park Y., Meltzer P.S., Kirsch I.R., Kaye F.J., Roschke A.V. Integrated high-resolution array CGH and SKY analysis of homozygous deletions and other genomic alterations present in malignant mesothelioma cell lines. Cancer Genet. 2013;206:191–205. doi: 10.1016/j.cancergen.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A., Bhattacharyya N., Srivastava A., Pruett N., Ripley R.T., Schrump D.S., Hoang C.D. MicroRNA-215-5p treatment suppresses mesothelioma progression via the MDM2-p53-signaling axis. Mol. Ther. 2019;27:1665–1680. doi: 10.1016/j.ymthe.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plaisier C.L., O’Brien S., Bernard B., Reynolds S., Simon Z., Toledo C.M., Ding Y., Reiss D.J., Paddison P.J., Baliga N.S. Causal mechanistic regulatory network for glioblastoma deciphered using systems genetics network analysis. Cell Syst. 2016;3:172–186. doi: 10.1016/j.cels.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K. Differences of cyclin-dependent kinase 4/6 inhibitor, palbociclib and abemaciclib, in breast cancer. Jpn. J. Clin. Oncol. 2019;49:993–998. doi: 10.1093/jjco/hyz151. [DOI] [PubMed] [Google Scholar]
- 36.Zanella C.L., Posada J., Tritton T.R., Mossman B.T. Asbestos causes stimulation of the extracellular signal-regulated kinase 1 mitogen-activated protein kinase cascade after phosphorylation of the epidermal growth factor receptor. Cancer Res. 1996;56:5334–5338. [PubMed] [Google Scholar]
- 37.Cui Y., Groth S., Troutman S., Carlstedt A., Sperka T., Riecken L.B., Kissil J.L., Jin H., Morrison H. The NF2 tumor suppressor merlin interacts with Ras and RasGAP, which may modulate Ras signaling. Oncogene. 2019;38:6370–6381. doi: 10.1038/s41388-019-0883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cedrés S., Montero M.A., Martinez P., Martinez A., Rodríguez-Freixinós V., Torrejon D., Gabaldon A., Salcedo M., Ramon Y Cajal S., Felip E. Exploratory analysis of activation of PTEN-PI3K pathway and downstream proteins in malignant pleural mesothelioma (MPM) Lung Cancer. 2012;77:192–198. doi: 10.1016/j.lungcan.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 39.Galiè M. RAS as supporting actor in breast cancer. Front. Oncol. 2019;9:1199. doi: 10.3389/fonc.2019.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scagliotti G.V., Gaafar R., Nowak A.K., Nakano T., van Meerbeeck J., Popat S., Vogelzang N.J., Grosso F., Aboelhassan R., Jakopovic M. Nintedanib in combination with pemetrexed and cisplatin for chemotherapy-naive patients with advanced malignant pleural mesothelioma (LUME-Meso): a double-blind, randomised, placebo-controlled phase 3 trial. Lancet Respir. Med. 2019;7:569–580. doi: 10.1016/S2213-2600(19)30139-0. [DOI] [PubMed] [Google Scholar]
- 41.Kang B.P., Slosberg E., Snodgrass S., Lebedinsky C., Berry D.A., Corless C.L., Stein S., Salvado A. The signature program: bringing the protocol to the patient. Clin. Pharmacol. Ther. 2015;98:124–126. doi: 10.1002/cpt.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonelli M.A., Digiacomo G., Fumarola C., Alfieri R., Quaini F., Falco A., Madeddu D., La Monica S., Cretella D., Ravelli A. Combined inhibition of CDK4/6 and PI3K/AKT/mTOR pathways induces a synergistic anti-tumor effect in malignant pleural mesothelioma cells. Neoplasia. 2017;19:637–648. doi: 10.1016/j.neo.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aliagas E., Martinez-Iniesta M., Hernandez M., Alay A., Cordero D., Sole X., Rivas F., Urena A., Vilarino N., Munoz-Pinedo C. MA23.02 CDK4/6 inhibitors show antitumor effects in preclinical models of malignant pleural mesothelioma. J. Thorac. Oncol. 2019;14:S343. [Google Scholar]
- 44.Beckwith H., Yee D. Minireview: were the IGF signaling inhibitors all bad? Mol. Endocrinol. 2015;29:1549–1557. doi: 10.1210/me.2015-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y.L., Yuksel Durmaz Y., Nör J.E., ElSayed M.E. Synergistic combination of small molecule inhibitor and RNA interference against antiapoptotic Bcl-2 protein in head and neck cancer cells. Mol. Pharm. 2013;10:2730–2738. doi: 10.1021/mp4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Q., Guo X., Wang M., Li Y., Sun X., Li Q. The application and prospect of CDK4/6 inhibitors in malignant solid tumors. J. Hematol. Oncol. 2020;13:41. doi: 10.1186/s13045-020-00880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sobhani N., D’Angelo A., Pittacolo M., Roviello G., Miccoli A., Corona S.P., Bernocchi O., Generali D., Otto T. Updates on the CDK4/6 inhibitory strategy and combinations in breast cancer. Cells. 2019;8:321. doi: 10.3390/cells8040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rintoul R.C., Rassl D.M., Gittins J., Marciniak S.J., MesobanK collaborators MesobanK UK: an international mesothelioma bioresource. Thorax. 2016;71:380–382. doi: 10.1136/thoraxjnl-2015-207496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Tong W.H., Sourbier C., Kovtunovych G., Jeong S.Y., Vira M., Ghosh M., Romero V.V., Sougrat R., Vaulont S., Viollet B. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell. 2011;20:315–327. doi: 10.1016/j.ccr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.