Abstract
Wheat streak mosaic virus (WSMV), a member of the genus Tritimovirus in the family Potyviridae, has been designated as a plant quarantine pathogen in South Korea. Several diagnostic methods can be applied to diagnose viral infections in plants, but polymerase chain reaction and enzyme-linked immunosorbent assay, which can identify viral species with speed and accuracy, are mainly used in Korean plant quarantine. Many variants of different viral species with highly divergent genomic sequences are constantly being discovered by high-throughput sequencing technology. This means that previously established primers may no longer be suitable for diagnostic use. In this study, we developed a reverse transcription polymerase chain reaction assay for detecting WSMV isolates/strains using all of the WSMV sequences available in NCBI GenBank. All 13 primer sets were able to produce amplicons of the expected sizes from WSMV-infected samples. To check whether nonspecific reactions occur, some closely related viruses (one tritimovirus and five potyviruses) and target imported plants (wheat, maize, oat, and proso millet) were tested. Consequently, four primer sets, which did not produce nonspecific bands, were finally selected among the 13 primer sets. Concentration-dependent amplification tests showed that the four primer sets are adequate for use in the diagnosis of WSMV in Korean plant quarantine.
Electronic supplementary material
The online version of this article (10.1007/s13337-020-00646-3) contains supplementary material, which is available to authorized users.
Keywords: WSMV, RT-PCR, Diagnosis, Quarantine
Wheat streak mosaic virus (WSMV), a member of the genus Tritimovirus in the family Potyviridae, is known to infect some monocotyledonous plants, including wheat (Triticum aestivum) [2, 13, 14]. Wheat curl mite (Aceria tosichella Keifer) is a vector involved in WSMV transmission [16]. The South Korea’s wheat cultivation area is 5224 ha, and the production is estimated to be 12,442 tons in 2020. Although the occurrence of WSMV has not yet been documented in South Korea, the infection of crops with the virus causing symptoms of chlorotic streak and mosaic leads to a significant decrease in production [2, 14]. Therefore, WSMV has been designated as one of the plant quarantine pathogens in South Korea (Animal and Plant Quarantine Agency, www.qia.go.kr). Several diagnostic methods can be applied to detect viruses [1], but polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA), which can identify viral species with speed and accuracy, are mainly used in Korean plant quarantine.
A large number of plant viral sequences have been determined over the past decade, because high-throughput sequencing (HTS) technology has become more common in research fields. It was confirmed that virus populations are more widely distributed in the natural environment than previously thought [3–5, 11, 12]. Viruses are susceptible to mutation of their nucleotide sequences during genome replication [10], and HTS technology and bioinformatic analysis have revealed the existence of a great number of genetically divergent viral isolates/strains [6–8].
Many variants of different viral species with highly divergent genomic sequences are constantly being discovered [6–8]. This means that previously established primers may no longer be suitable for diagnostic use. In this study, we developed a reverse transcription (RT)-PCR assay for the detection of WSMV isolates/strains that have been identified to date. All 175 nucleotide sequences of WSMV registered in NCBI GenBank (https://blast.ncbi.nlm.nih.gov/genbank/) were collected using CLC Main Workbench 20.0 (CLC Bio, Cambridge, MA, USA). Of the 175 sequences, 29 had complete coding sequences, and the rest of the sequences were shorter than 1800 nucleotides in length. To design primers, all of the 175 nucleotide sequences were aligned using CLC Main Workbench 6.1.1, and a graphic of the alignment showed that partial sequences, except the 29 complete genome sequences, originated from the 3′ region (corresponding to the coat protein-encoding sequence) of the genomic RNA of WSMV (data not shown). Pairwise comparison based on the alignment showed that one (WSMV strain El Batan 3; GenBank Acc. No. AF285170) out of the 29 sequences had < 80% nucleotide sequence homology to the other 28 sequences (Supplementary Fig. S1 [13]). The remaining 28 sequences had more than 88% nucleotide sequence homology to each other (Supplementary Fig. S1). A total of 18 candidate primers were designed in conserved regions across the entire WSMV genome, and 13 combinations were made using the candidate primers (Supplementary Table S1 and Supplementary Fig. S2). The expected size of fragments amplified by the 13 primer sets ranged from 300 to 800 base pairs.
For the selection of primers suitable for use in quarantine inspections, four points should be particularly considered: (i) It should be possible to accurately detect a target virus including genetically divergent isolates/strains. (ii) There should be no nonspecific reactions from closely related viruses. (iii) There should be no nonspecific reactions in imported plants that need to be checked for viral infections. Regarding points (ii) and (iii), when a nonspecific product having a size similar to the expected size of PCR is amplified, its identity needs to be confirmed through sequencing, which takes more time. (iv) Lastly, the final selected primers should have high sensitivity to a low titer of the target virus.
To test all of the primer sets (10 pmol/each), one-step RT-PCR was carried out using positive and negative controls of WSMV (for ELISA; host plant: T. aestivum; Agdia, Elkhart, IN, USA), random N25 primer (50 pmol), and AccuPower® RT-PCR PreMix (Bioneer, Daejeon, South Korea). Random primers have been used to acquire high quality cDNA [9, 15], and it was practically effective in developing diagnostic PCR primers for various plant viruses (data not shown). Total RNA was extracted using RNeasy® Plant Mini Kit (QIAGEN, Hilden, Germany). The one-step RT-PCR began with an RT reaction at 42 °C for 1 h, followed by RT enzyme inactivation at 95 °C for 15 min. Subsequently, PCR was performed for 32 cycles using the following protocol: 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min, followed by additional extension at 72 °C for 5 min. The results of RT-PCR showed that all primer sets were able to produce amplicons with expected sizes from the positive WSMV sample (Supplementary Fig. S3).
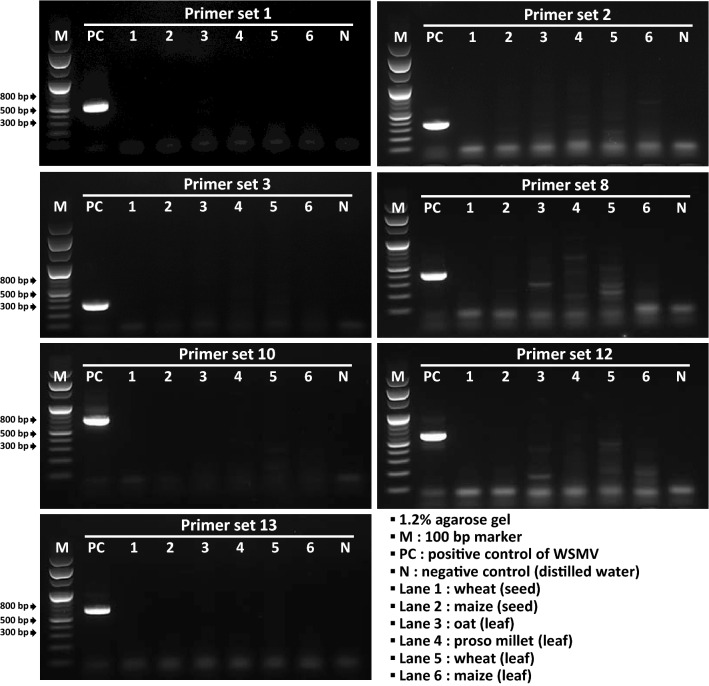
To determine whether nonspecific reactions occur in some closely related viruses belonging to the family Potyviridae, one tritimovirus (brome streak mosaic virus [host plant: Hordeum vulgare]) and five potyviruses (alstroemeria mosaic virus [host plant: Alstroemeria sp.], banana bract mosaic virus [artificial synthesis], bean common mosaic necrosis virus [host plant: Phaseolous vulgaris], chilli veinal mottle virus [ChiVMV; host plant: Nicotiana benthamiana], and tobacco etch virus [host plant: Nicotiana sp.]), which are used as positive controls for ELISA by Agdia, were tested. The five potyviruses are plant quarantine viruses along with WSMV in South Korea. In one-step RT-PCR performed on the six nontarget viruses, six primer sets (no. 4, 5, 6, 7, 9, and 11) produced nonspecific amplicons, such as having different sizes or smear bands (Fig. 1). To identify the source of nonspecific products, the amplicons were directly sequenced (Macrogen, Daejeon, South Korea). An NCBI BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that three out of four sequenced products were amplified from plants. The sequence of the other nonspecific amplicon obtained from the ChiVMV-infected sample was derived from genomic RNA of ChiVMV, and the nucleotide sequence was positioned at nucleotide 1111–1419 of the reference genome of ChiVMV (GenBank acc. no. NC_005778). The forward primer (WSMV_8F) was not specific to the ChiVMV sequence, but eight nucleotides at the 3′ end of the reverse primer (WSMV_11R) were identical to the ChiVMV sequence. It appears that a sufficient amount of nonspecific cDNA made by the reverse primer caused the unintended ChiVMV-derived amplicon. In this step, seven primer sets (no. 1, 2, 3, 8, 10, 12, and 13), which did not produce nonspecific bands, were selected among the 13 primer sets (Fig. 1). To check the existence of similar sequences at any other viruses including wheat-infecting viruses (such as barley yellow dwarf viruses, soil-borne wheat mosaic virus, wheat yellow mosaic virus, and wheat spindle streak mosaic virus), the BLAST searches were performed using sequences of the seven selected primer sets. As a result, similar sequences were not detected in any other viruses, except one primer (WSMV_20R). Although the primer also had sequence similarity to oat necrotic mottle virus, which belongs to the genus Tritimovirus, primer set no. 12 was included in the list for the next step due to high specificity of the other primer (WSMV_19F) only for WSMV.
Fig. 1.
Test for nonspecific reactions in six nontarget viruses. Six closely related viruses belonging to the family Potyviridae, namely, one tritimovirus (brome streak mosaic virus) and five potyviruses (alstroemeria mosaic virus, banana bract mosaic virus, bean common mosaic necrosis virus, chilli veinal mottle virus, and tobacco etch virus) were tested to check whether nonspecific reactions occur
When testing plants infected with viruses using RT-PCR, nonspecific reactions originating from the host plants may also cause confusion in making a diagnosis. To select primer sets that do not produce nonspecific products from plant samples, we tested wheat (T. aestivum), maize (Zea mays), oat (Avena sativa), and proso millet (Panicum miliaceum), which are target imported plants for WSMV infection in Korean plant quarantine. Using leaf samples of the four plants, it was determined whether a nonspecific reaction was generated; seed samples were also used for wheat and maize. Total RNA templates more than 50 ng extracted from the plant samples were used in one-step RT-PCR. Three (no. 2, 8, and 12) among the seven primer sets produced nonspecific smear bands from some plant samples; as a consequence, four primer sets (no. 1, 3, 10, and 13) were selected (Fig. 2).
Fig. 2.
Test for nonspecific reactions in four test plants. Wheat, maize, oat, and proso millet, which are target imported plants for WSMV infections in Korean plant quarantine, were tested to check whether nonspecific reactions occur
Finally, a concentration-dependent amplification test was conducted using the four primer sets. The initial amount of total RNA extracted from the positive control of WSMV was 10 ng, and a total of seven templates were prepared through 1/10 serial dilutions. In primer sets no. 1 and 10, amplified products were also observed in 0.01 ng of total RNA, and primer set no. 3 produced distinct bands up to 0.1 ng of the template (Supplementary Fig. S4). Although primer set no. 13 produced the target band only at template amounts of 10 and 1 ng (Supplementary Fig. S4), it also appeared to be sufficient for use in the diagnosis of WSMV.
In this study, we finally selected four primer sets (no. 1, 3, 10, and 13) for the RT-PCR assay to diagnose WSMV infection. Although the assay was developed for the plant quarantine inspection in South Korea, it would be helpful for many researchers who want to detect WSMV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Edanz (https://en-author-services.edanzgroup.com/ac) for editing the English text of a draft of this manuscript. This work was supported by a grant (Project No. B-1543086-2019-21-03) from the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not describe any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boonham N, Kreuze J, Winter S, van der Vlugt R, Bergervoet J, Tomlinson J, Mumford R. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 2014;186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Hadi BAR, Langham MAC, Osborne L, Tilmon KJ. Wheat streak mosaic virus on wheat: biology and management. J Integr Pest Manag. 2011;2(1):J1–J5. [Google Scholar]
- 3.Hodge BA, Paul PA, Stewart LR. Occurrence and high-throughput sequencing of viruses in Ohio wheat. Plant Dis. 2020;104(6):1789–1800. doi: 10.1094/PDIS-08-19-1724-RE. [DOI] [PubMed] [Google Scholar]
- 4.Lim S, Igori D, Zhao F, Yoo RH, An TJ, Lim HS, Lee SH, Moon JS. Complete genome sequence of a tentative new caulimovirus from the medicinal plant Atractylodes macrocephala. Arch Virol. 2015;160(12):3127–3131. doi: 10.1007/s00705-015-2576-y. [DOI] [PubMed] [Google Scholar]
- 5.Lim S, Kim KH, Zhao F, Yoo RH, Igori D, Lee SH, Moon JS. Complete genome sequence of a novel endornavirus isolated from hot pepper. Arch Virol. 2015;160(12):3153–3156. doi: 10.1007/s00705-015-2616-7. [DOI] [PubMed] [Google Scholar]
- 6.Lim S, Yoo RH, Igori D, Zhao F, Kim KH, Moon JS. Genome sequence of a recombinant brassica yellows virus infecting Chinese cabbage. Arch Virol. 2015;160(2):597–600. doi: 10.1007/s00705-014-2258-1. [DOI] [PubMed] [Google Scholar]
- 7.Lim S, Igori D, Zhao F, Do YS, Cho IS, Choi GS, Moon JS. Molecular detection and characterization of a divergent isolate of Plantago asiatica mosaic virus in Plantago asiatica. VirusDis. 2016;27(3):307–310. doi: 10.1007/s13337-016-0329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim S, Kwon SY, Lee JH, Cho HS, Kim HS, Park JM, Lee SH, Moon JS. Genomic detection and molecular characterization of two distinct isolates of cycas necrotic stunt virus from Paeonia suffruticosa and Daphne odora. Virus Genes. 2019;55(5):734–737. doi: 10.1007/s11262-019-01687-7. [DOI] [PubMed] [Google Scholar]
- 9.Nakaune R, Nakano M. Efficient methods for sample processing and cDNA synthesis by RT-PCR for the detection of grapevine viruses and viroids. J Virol Methods. 2006;134:244–249. doi: 10.1016/j.jviromet.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Novella IS, Presloid JB, Taylor RT. RNA replication errors and the evolution of virus pathogenicity and virulence. Curr Opin Virol. 2014;9:143–147. doi: 10.1016/j.coviro.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Roossinck MJ. Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res. 2017;239:82–86. doi: 10.1016/j.virusres.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Roossinck MJ, Martin DP, Roumagnac P. Plant virus metagenomics: advances in virus discovery. Phytopathology. 2015;105(6):716–727. doi: 10.1094/PHYTO-12-14-0356-RVW. [DOI] [PubMed] [Google Scholar]
- 13.Schubert J, Ziegler A, Rabenstein F. First detection of wheat streak mosaic virus in Germany: molecular and biological characteristics. Arch Virol. 2015;160(7):1761–1766. doi: 10.1007/s00705-015-2422-2. [DOI] [PubMed] [Google Scholar]
- 14.Singh K, Wegulo SN, Skoracka A, Kundu JK. Wheat streak mosaic virus: a century old virus with rising importance worldwide. Mol Plant Pathol. 2018;19(9):2193–2206. doi: 10.1111/mpp.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stangegaard M, Dufva IH, Dufva M. Reverse transcription using random pentadecamer primers increases yield and quality of resulting cDNA. Biotechniques. 2006;40(5):649–657. doi: 10.2144/000112153. [DOI] [PubMed] [Google Scholar]
- 16.Tatineni S, Hein GL. Genetics and mechanisms underlying transmission of Wheat streak mosaic virus by the wheat curl mite. Curr Opin Virol. 2018;33:47–54. doi: 10.1016/j.coviro.2018.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.