Abstract
The present work considers how connectome-wide differences in brain organization might distinguish good and poor readers. The connectome comprises a ‘rich-club’ organization in which a small number of hub regions play a focal role in assisting global communication across the whole brain. Prior work indicates that this rich-club structure is associated with typical and impaired cognitive function although no work so far has examined how this relates to skilled reading or its disorders. Here we investigated the rich-club structure of brain’s white matter connectome and its relationship to reading subskills in 64 children with and without reading disabilities. Among three types of white matter connections, the strength of feeder connections that connect hub and non-hub nodes was significantly correlated with word reading efficiency and phonemic decoding. Phonemic decoding was also positively correlated with connectivity between connectome-wide hubs and nodes within the left-hemisphere reading network, as well as the local efficiency of the reading network. Exploratory analyses also identified sex differences indicating these effects were stronger in girls. This work highlights the independent roles of connectome-wide structure and the more narrowly-defined reading network in understanding the neural bases of skilled and impaired reading in children.
Keywords: Reading disabilities, Rich-club, Reading network, Connectome, Diffusion tensor imaging
1. Introduction
Skilled readers are able to recognize and map phonological, orthographic and semantic components of language quickly and accurately. However, around 10 % of children have reading disabilities (RD), which entails dysfluent and inaccurate reading performance (Lyon et al., 2003). One predominant theory proposes phonological deficits are the main cause of RD (Boets et al., 2013, 2011; Ramus, 2003; Snowling, 2000). Neuroimaging studies have demonstrated links between poor reading or phonology and structural anomalies in RD. However, a clear picture describing neural and cognitive substrates of RD still requires further exploration as these studies reported inconsistent results across measures of white matter tracts and structural morphology (Ramus et al., 2018).
Magnetic resonance imaging (MRI) studies have identified a specialized leftward lateralized reading network in the brain supporting reading functions (Price and Devlin, 2011; Pugh et al., 2000). Several white matter pathways within the reading network are proposed to support specific reading functions, with dorsal pathways supporting the orthographic-phonology mapping and ventral pathways supporting orthographic-semantic mapping (Jobard et al., 2003; Schlaggar and Mccandliss, 2007). The connectivity of white matter tracts is usually evaluated via diffusion tensor imaging (DTI), which quantifies the orientation and integrity of white matter fibers based on the direction and degree of diffusion of water molecules within voxels (Basser, 1995; Basser et al., 1994). Studies using DTI have reported altered connectivity in reading-related white matter pathways, and their associations with reading performance in RD (Deutsch et al., 2005; Klingberg et al., 2000; Lou et al., 2019; Niogi and McCandliss, 2006; Richards et al., 2008; Rimrodt et al., 2010; Steinbrink et al., 2008; Su et al., 2018; Vandermosten et al., 2012a; Zhao et al., 2016). Although diverse major white matter pathways, such as the arcuate fasciculus and superior longitudinal fasciculus, have been generally found to differ in poor readers, results have been somewhat inconsistent with respect to the involvement of other white matter pathways and widespread brain regions linked with those tracts. This suggests that reading-related white matter pathways may contribute to the individual variance of reading abilities via interactions among each other instead of each supporting particular cognitive function independently. However, the manner in which the alterations of these connections constrain reading performance as a global system has not been well explored. Analytic strategies that consider connectome-wide white matter structure represent a potentially useful model to investigate anatomical structure at large-scale network level.
The term ‘connectome’ describes the human brain as a network with nodes representing cortical regions, and edges representing white matter tracts that connect them into a matrix (Sporns et al., 2005). The connectome has been proposed as the elementary building block of cognitive architecture (Petersen and Sporns, 2015), and it has been investigated in a range of cognitive functions (for review see Medaglia et al., 2015). Hypothesised connectivity alterations in neurological disorders are reflected by both convergence and divergence in the types of connectome disruptions across various clinical populations (van den Heuvel and Sporns, 2019).
The human connectome follows a hierarchical connection principle that generally divides regions into a small number of hubs and a larger number of peripheral regions. Hubs are more centrally embedded in the connectome, such that they have a higher than usual level of connectivity with other regions (Gong et al., 2008; Hagmann et al., 2008; Sporns et al., 2007; van den Heuvel and Sporns, 2011). Hubs are also more likely to connect to each other, thereby forming “rich-club” connections in which hub regions represent the primary route to interregional signal transmission, representing a central backbone for global brain communication (van den Heuvel et al., 2012; van den Heuvel and Sporns, 2011). Crossley et al. (2014), using massive open source DTI data with 26 brain disorders, demonstrated that hubs in the white matter connectome were more likely to present anatomical anomalies than peripheral regions in various neurological disorders. Altered rich-club-wise connectivity has also been implicated in various disorders such as autism spectrum disorder (Ray et al., 2014; Hong et al., 2019), ADHD (Ray et al., 2014), schizophrenia (Griffa et al., 2015; van den Heuvel et al., 2013), and bipolar disorder (Roberts et al., 2018).
As a developmental disorder with neurological origins, RD has also been associated with alterations at the connectome level in different modalities (Bailey et al., 2018; Finn et al., 2014; Liu et al., 2015; Lou et al., 2019; Qi et al., 2016). Specifically, Lou et al. (2019) recently examined the structure of the white matter connectome in children with developmental dyslexia, reporting a subnetwork consisting of brain regions which overlapped with the reading network and connections among them. Results showed fewer white matter streamlines within the subnetwork in the left hemisphere of children with RD (Lou et al., 2019). Moreover, significant correlations between global topological properties, which quantify the structure of the white matter connectome, and reading performance have been illustrated in children with RD (Lou et al., 2019) and children with poor reading performance in school (Bathelt et al., 2018). Those studies support the view that the connectome is related to reading and RD at both a global and local level. Reading requires not only processing various language components within individual brain regions, but also mapping the components via long-distance connections among those regions. Therefore, complex cognitive tasks like reading might be expected to take advantage of this rich-club structure. However, only one study to date has examined the relationship between topological properties of hub regions and reading performance in the white matter connectome (Bathelt et al., 2018). This study found critical associations between topological properties of brain network hubs and reading and math achievement in school-age children. However, the concept of rich-club provides a more intuitive view of connections extending from hub regions, raising the question of whether rich-club-wise correlates of specific reading mechanisms are important to reading disabilities. Similarly, the association between the connectomic hubs and the better-established reading network has not been well explored yet.
Associations between white matter connectivity and either word or pseudoword reading have been reported in many prior studies of earlier reading disability (Deutsch et al., 2005; Klingberg et al., 2000; Lou et al., 2019; Niogi and McCandliss, 2006; Rimrodt et al., 2010; Odegard et al., 2009; Steinbrink et al., 2008; Zhao et al., 2016). White matter connectivity was also related to rapid automatized naming and comprehension skills (Deutsch et al., 2005; Carter et al., 2009). Here, we hypothesized that associations between white matter and reading subskills could also be reflected at the larger-scale white matter network level.
The present study aims to investigate the relationship between rich-club structure and these four important subskills of reading performance, and how this rich-club structure corresponds to brain regions within the reading network. We examined a range of sub-skills related to reading in children with and without RD. Given a lack of evidence of categorical distinctions between typical readers and individuals with RD, and the advisability of imposing arbitrary cut-off scores for categorizing children into either group, here we have examined behavioral markers of reading as continuous variables. We then tested how individual differences in these sub-skills correlated with rich-club-wise connections which were categorized into specific types based on hub and non-hub nodes they linked. In addition, we investigated how this rich club structure relates to connectivity structure within the reading network.
A second set of exploratory analyses was also performed for the boys and girls group separately. This was intended to address prior findings that the rate of diagnosis of RD tends to be higher in boys (Flannery et al., 2000; Katusic et al., 2001; Rutter et al., 2004; Liederman et al., 2005). Moreover, some MRI studies in RD reported stronger associations between brain structure and reading in either male or female readers (Altarelli et al., 2013, 2014; Clark et al., 2014; Evans et al., 2014; Sandu et al., 2008; Su et al., 2018). Such findings highlighted the possibility that there could be different neuroanatomical bases of RD for boys than girls (Krafnick and Evans, 2019). Likewise, at least one study o functional connectivity in spoken language processing has raised the possibility of sex differences at the connectome level (Xu et al., 2020). To address these findings, this study performed an exploratory analysis that examined connectome-reading correlations separately for boys and girls.
2. Methods
2.1. Participants
An initial group of 73 school-age children (mean age: 11.15 years, standard deviation: 1.39, range from 8.83 to 14.68) were recruited into the study. Thirty-four of them were boys and 39 were girls. All participants in the study were native English speakers with normal hearing and uncorrected vision abilities, with no history of neurological disorders as assessed by parental report. All participants were combined from two datasets who completed the same reading tests and structural MRI scan sequences. One of the datasets included 44 children (10.51 ± 0.89 years old, range from 8.83 to 11.90 years) and the second dataset included 29 children (12.13 ± 1.45 years old, range from 10.03 to 14.68 years). Recruitment for the first dataset was targeted toward children with reading difficulties and as such, 18 participants had been previously identified as having reading difficulties by school professionals. The remaining participants in this dataset were children with a wide range of reading abilities, but any poor readers had not been formally identified with reading difficulties. The second dataset comprised children with a wide range of reading abilities, with some demonstrating a profile of abilities consistent with reading disability. As the first dataset included more children with reading difficulties and the mean age of children from the second dataset was significantly higher than the first, this study controlled for the confounding correlation between standard reading scores and age by removing 7 children from the second dataset whose ages were higher than 12 years old and who had standardized sight word reading and phonemic decoding scores higher than 100. Age and reading scores of the remaining 64 participants included in this study (mean age: 10.94 years, standard deviation: 1.26, range 8.83–14.68) were not correlated and were not confounded by recruitment group. This study was approved by the Office of Human Research Ethics of the Western Research Ethics Board. Informed consent was provided by a parent or guardian of each participant, and written and verbal assent was obtained from each participant.
2.2. Behavioral measures
The Sight Word Efficiency and Phonemic Decoding Efficiency subtests of the Test of Word Reading Efficiency 2 (TOWRE, Torgesen et al., 2012) were administered to examine sight word reading and decoding fluency. The Sight Word Efficiency subtest asked children to read familiar words as quickly and accurately as they could in 45 s. The Phonemic Decoding subtest asked children to read pseudowords as quickly and accurately as they could in 45 s. Reading comprehension ability was tested using the Woodcock Johnson III Test of Achievement Normative Update (McGrew et al., 2007; Woodcock et al., 2001) Passage Comprehension subtest by asking children to read a sentence or paragraph and provide a missing word. Rapid Automatized Naming (RAN) of letters was assessed by asking children to name each letter from a set of 4 (k, r, m, g) presented at random in a 5 × 10 grid of items as quickly as possible (Arnell et al., 2009). The score consisted of the number of correctly named items per second. All scores, except for RAN, were standardized based on age.
2.3. Imaging acquisition
Prior to MRI scanning, all children participated in a training session to familiarize them with the scanner environment, lying on a bed inside a simulated MRI scanner for around 5 min. An audiobook and recordings of MRI scanner sounds were played simultaneously, and feedback was provided to them about movement using an electromagnetic motion tracker. This training session also served as an informal assessment of whether each participant could remain sufficiently still during the MRI. No participants were excluded from the study based on excessive movement or reported discomfort during the training session. Children then underwent actual MRI scanning with a Siemens 3T Magnetom Prisma MRI scanner equipped with a 32-channel head coil at the Robarts Research Institute, Western University, London, Ontario, Canada. To control head movement, foam pads were used while participants lay awake on the scanner bed. Whole-brain 3D anatomical imaging was performed using a T1-weighted MPRAGE sequence (TR = 2.30 s, TE = 2.98 s, FOV = 256 × 256 mm, voxel size = 1 mm × 1 mm × 1 mm, 192 slices). The diffusion weighted (DW) images were acquired using a DTI scan sequence (TR = 3.0 s, TE =50.6 ms, FOV = 256 × 256 mm, voxel size = 2.04 mm × 2.04 mm x 2 mm, 64 slices, 56 directions with b = 1000s/mm2 and 8 directions with b = 0). A 6-minute resting-state fMRI scan was also acquired during the scanning session from children in the first dataset, and additional 3 fMRI tasks from children in the second dataset as part of another study, both part of a separate study not reported here. Total scanning time was approximately 30 min per child in the first dataset and around 60 min per children in the second dataset, including set-up time.
2.4. Image analysis
For each of the T1-weighted images, the robustfov command from FSL was applied to crop the images, ensuring better skull stripping results. The FSL Brain Extraction Tool (BET, Smith, 2002) was used to perform skull stripping. In terms of DW images, processing steps used p-code version of ExploreDTI (Leemans et al., 2009, http://www.exploredti.com) toolbox for MATLAB. First, NIFTI images were converted to MATLAB-compatible format. Head motion and eddy current correction were applied. To correct EPI distortions, DW images were also nonlinearly registered onto the cropped T1 images, resampled to 1 × 1 × 1 mm voxel size. Corrected DW images were then used to perform whole-brain tractography via the deterministic DTI algorithm. Fibers were reconstructed by starting from a seed voxel and followed the main diffusion direction until encountering a voxel with a fractional anisotropy (FA) lower than 0.1 or the turning angle exceeded 60 degrees. The length of the fibers ranged from 25 to 500 mm with a step size of 0.5 mm.
White matter connectome maps were constructed for each participant: nodes consisted of anatomical regions obtained by parcellating each brain image into 90 gray matter regions of interests (ROIs) based on the Automated Anatomical Labelling (AAL) template (Tzourio-Mazoyer et al., 2002). These were fit to individual brains using FMRIB’s Nonlinear Image Registration Tool (FNIRT, FSL; Jenkinson et al., 2012, http://www.fmrib.ox.ac.uk/fsl/). The FA map of each participant, which was extracted from corrected DW images using ExploreDTI, was registered to the cropped T1 images to acquire a transfer matrix. Next, the cropped T1 images were nonlinearly registered to the MNI (Montreal Neurological Institute) 152 template. The two transfer matrices were combined and inversed to generate a transformation matrix from the standard space to each participant’s native space. An AAL template in each native space was then acquired by applying the matrix to the atlas. Edges of the matrix were defined as the number of white matter streamlines connecting each pair of nodes, which quantifies the amount of reconstructed white matter streamlines starting from one node and terminating at the other. The present study used the number of streamlines to quantify edge weight, on the assumption that number of streamlines estimates fiber strength (Delettre et al., 2019; van den Heuvel et al., 2015), and therefore the efficiency of information transmission among brain regions. This captured both the volume and diffusion characteristics of white matter voxels linking pairs of nodes. This contrasts with DTI measures such as FA as edge weights, since these require averaging many FA values across all reconstructed streamlines while ignoring, for instance, the principal directions of diffusion that might provide additional information about connection efficiency. The flow chart of constructing the network shown in Fig. 1.
Fig. 1.
Workflow for building white matter network.
2.5. Network analysis
Before performing network analyses, each network was thresholded to remove any edge with fewer than 3 streamlines. This step aimed to minimize false positive tracts between brain regions and yielded a sparsely connected matrix.
We defined hub regions based on the degree of the nodes. The degree of any one node equals the number of other nodes directly connected to it:
where N is the set of all nodes in the network and is the binary value of the connection between node i and j. The distribution of degree within the connectome follows a truncated power law degree distribution, where most of the nodes have low degree and few of them have high degree. Nodes with a degree higher than a threshold of k could be defined as hubs. We considered a wide range of k-degree thresholds to avoid arbitrary selection. For each degree threshold, a set of hub regions was identified and a normalized rich-club coefficient was computed to examine the existence of rich-club structure. The rich-club coefficient (Opsahl et al., 2008) was defined as:
where refers all connections between any pair of nodes whose degree is higher than k, is the total weighted value of connections between each pair of those nodes, and is the ranked connections according to the weight of edges across whole network. The normalized rich-club coefficient was then computed by comparing the rich-club coefficient of the original network to that of 10,000 random networks with identical size and degree sequence:
where is the rich-club coefficient of the random network. When is higher than , one-sample t-tests were performed to compare the normalized rich-club coefficient and 1 (when is higher than ). The network was considered as being equipped with a rich-club structure if the normalized rich-club coefficient was significantly higher than 1.
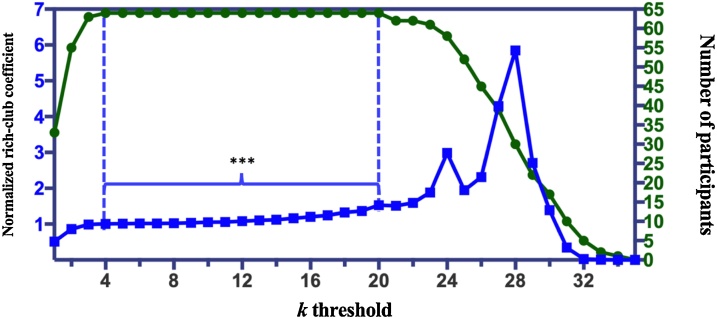
Across all thresholds where all participants exhibited rich-club structure, the threshold Km with the highest normalized rich-club coefficient was viewed as the most representative threshold and the subsequent analyses were performed under that threshold. As visualized in Fig. 2, all individuals’ networks contained rich-club connections under a wide range of k-degree thresholds ranging from 4 to 20 (green line). Within this range, the normalized rich-club coefficients () were all significantly higher than 1 (ts(63) > 4.49, ps < 2e−5), however we selected the point of highest k = 20 ( = 1.53, t(63) = 7.30, p = 3e-10) (Fig. 2, blue line). It ensures that the strongest rich-club structure was identified under the corresponding threshold. Hence, the Km was set as 20 for all analyses.
Fig. 2.
Trade-off between number of participants demonstrating rich-club connections (green) and the normalized rich-club coefficient (blue), across a wide range of k-degree threshold. All participants showed rich-club connections under thresholds between the two dashed lines. Normalized rich-club coefficients were significantly greater than 1 under thresholds which were labelled with ***; for this study we chose a k-threshold of 20 the highest coefficient point at which all participants showed rich-club structure.
When the hub regions were identified for each network, edges across the whole network could be categorized into three types (Fig. 3). Edges between two hub nodes were classified as rich-club connections. Edges between a hub a and a non-hub node were classified as feeder connections. Edges between two non-hub nodes were classified as local connections. The present study used mean number of streamlines across every edge in each type as metrics for statistical analyses.
Fig. 3.
a) Hypothetical rich-club structure showing different node and connection types; b) Chord diagram describing connections between hubs (red), reading network nodes (green) and other non-hub nodes (grey). The connection line thickness corresponds to the number of streamlines between nodes. Red links refers to connections between reading network nodes and hubs. Blue links refers to rich-club connections. Grey links represents the rest feeder connections and all local connections.
Rich-club structure is related to topological properties of the connectome. Specifically, more feeder connections reduce the shortest path length between a pair of nodes, inducing lower characteristic path length, which is equal to the average shortest path length between any pair of nodes across the whole network (Watts and Strogatz, 1998). To investigate how feeder connections changed shortest path length of rich-club nodes, we calculated local efficiency (Latora and Marchiori, 2001), which is the inverse shortest path length, of each rich-club nodes:
where is the shortest path length between node j and h. Efficiency of rich-club structure was computed by averaging local efficiency of all rich-club nodes.
The selection of nodes forming the reading network was based on a meta-analysis study that summarized 20 fMRI studies across multiple task types in children with typical reading abilities (Martin et al., 2015), a meta-analysis of functional neuroimaging studies in dyslexia (Maisog et al., 2008), as well as a recent study of white matter connectome in dyslexic children which reported differences within one subnetwork (Lou et al., 2019). Regions of interest identified in these studies were then colocated with anatomical regions as labelled in the AAL template. This included the pars triangularis and pars opercularis of inferior frontal gyrus (IFG), insula, fusiform gyrus, inferior parietal lobe, supramarginal gyrus, angular gyrus, Heschl’s gyrus, superior temporal gyrus (STG), middle temporal gyrus (MTG), inferior temporal gyrus, inferior occipital gyrus, precentral gyrus, and Rolandic operculum (which included ventral areas of pre- and post-central gyrus) in the left hemisphere. In addition, we included the left thalamus, given recent studies reporting its role in reading disabilities/dyslexia (Díaz et al., 2012; Müller-Axt et al., 2017; Paz-Alonso et al., 2018; Tschentscher et al., 2019). The connections between rich-club structure and reading network nodes were extracted from each participant. When examining feeder connections between rich-club nodes and reading network nodes, overlapping nodes between the two sub-networks were labelled as rich-club nodes instead of reading network nodes. Otherwise, connections between rich-club nodes and overlapping nodes were labelled as rich-club connections instead of feeders.
2.6. Statistical analyses
To assess the relationship between rich-club structure and reading subskills, we computed the mean number of streamlines for each of the three types of connections (rich-club, feeder and local) for each individual, and calculated Pearson’s correlations with each of the four behavioural reading scores (sight word reading, phonemic decoding, passage comprehension, and RAN), including sex, average whole-brain FA, and handedness as covariates. As no age-based norms exist for our RAN measure, age was included as an additional covariate for this measure.
A similar approach was taken to analyzing the structure of the reading network. Connections between rich-club nodes and reading network nodes, as well as the efficiency of reading network nodes, were extracted to examine their correlations with reading scores using partial Pearson’s correlations. Sex, average whole-brain FA, handedness, and the proportion of overlapped rich-club nodes between rich-club and reading network (Prc, number of overlapped rich-club nodes/total number of rich-club nodes) were set as covariates.
We also performed an exploratory analysis that examined these correlations separately in the male and female participants by running the above partial Pearson’s correlations in boys and girls separately. For all analyses, family-wise error was corrected with 10,000-permutation Monte-Carlo simulations.
3. Results
3.1. Demographic and behavioural measures
Descriptive statistics for demographic and behavioural measures are shown in Table 1.
Table 1.
Demographic and behavioural measures.
| N | Mean | SD | Range (min/max) | |
|---|---|---|---|---|
| Age (years) | 64 | 10.94 | 1.26 | 8.83/14.68 |
| Sex (boys/girls) | 31/33 | / | / | |
| Handedness (left/right) | 2/62 | / | / | |
| Sight Word Efficiency (standard score) | 64 | 93.02 | 21.35 | 55/139 |
| Phonemic Decoding (standard score) | 64 | 92.72 | 19.83 | 55/129 |
| Passage Comprehension (standard score) | 64 | 90.03 | 13.30 | 46/114 |
| Rapid Automatized Naming (RAN) (#correct/second) | 64 | 1.87 | 0.48 | 0.56/2.84 |
3.2. Rich-club structure and reading scores
With the Km threshold set as 20, the location of the group-level hubs was identified. Those hubs generally overlapped with findings in prior rich-club studies (van den Heuvel et al., 2012; van den Heuvel and Sporns, 2011), including bilateral superior frontal lobe, precuneus, supplemental motor area, and thalamus.
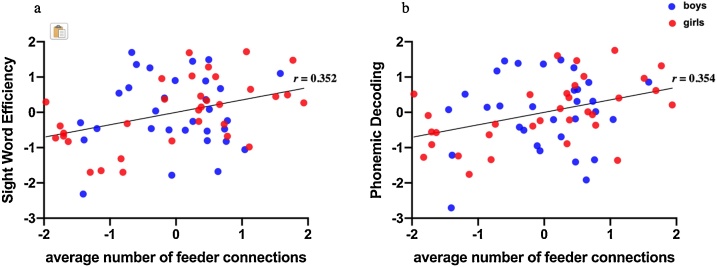
Partial Pearson’s correlation tests showed that the average number of streamlines of feeder connections was significantly correlated with standard scores on the Sight Word Efficiency (r(59) = 0.352, corrected p = 0.047) and Phonemic Decoding (r(59) = 0.354, corrected p = 0.045) tasks (Fig. 4). The average number of streamlines of feeder connections was also positively correlated with Passage Comprehension scores, however this did not survive correction for multiple comparisons (r(59) = 0.346, corrected p = 0.053). Rich-club and local connections were not correlated with any reading scores. RAN score was not correlated with any types of connections. The results are shown in Table 2.
Fig. 4.
Correlations between average number of streamlines of feeder connections and standard reading scores. (a) Sight Word Efficiency and (b) Phonemic Decoding. Values of feeder connections, standard Sight Word Efficiency, and standard Phonemic Decoding scores on the scatter plots are standard residuals after controlling for sex, mean whole-brain FA, and handedness.
Table 2.
Correlations and uncorrected p values between rich-club-wise connections and reading scores.
| Rich-club | Feeder | Local | |
|---|---|---|---|
| Sight word efficiency | 0.073 (0.578) | 0.352 (0.005)* | −0.007 (0.959) |
| Phoneme decoding | 0.094 (0.471) | 0.354 (0.005)* | −0.025 (0.850) |
| Passage comprehension | 0.026 (0.845) | 0.346 (0.006) | −0.016 (0.901) |
| Rapid Automatized Naming (RAN) | 0.180 (0.168) | 0.128 (0.329) | −0.015 (0.908) |
Correlations survived correction by 10,000-permutation Monte-Carlo simulation.
Before examining sex differences in those correlations, a multivariate analysis of variance (MANOVA) test was conducted to test if there were any sex differences in both reading scores and connectomic metrics. As indicated in Table S1 boys only differed from girls on standardized phonemic decoding, where they showed higher scores on average (F(1,62) = 8.087, p = 0.006); no other behavioral differences were observed As shown in Table 3, after splitting participants by sex, partial correlation results showed that the average number of streamlines of feeder connections was significantly correlated with standard Sight Word Efficiency (r(29) = 0.543, p = 0.002), Phonemic Decoding (r(29) = 0.533, p = 0.002) and Passage Comprehension (r(29) = 0.565, p < 0.001) in girls. No significant correlations were found in the boys group.
Table 3.
Correlations between average number of the feeder streamlines in boys and girls group.
| Boys |
Girls |
|||
|---|---|---|---|---|
| r | p | r | p | |
| Sight word efficiency | 0.066 | 0.726 | 0.556 | < 0.001* |
| Phoneme decoding | 0.024 | 0.897 | 0.533 | 0.0017* |
| Passage comprehension | 0.039 | 0.835 | 0.527 | 0.0019* |
| Rapid Automatized Naming (RAN) | −0.021 | 0.910 | 0.237 | 0.184 |
Correlations survived correction by 10,000-permutation Monte-Carlo simulation.
As expected, analyses in the full sample showed that the average number of streamlines of feeder connections was negatively correlated with characteristic path length (r(64) = -0.600, p < 0.001). Likewise, average local efficiency of rich-club nodes was also correlated with feeder connections (r(64) = 0.785, p < 0.001). However, similar correlations between other behavioural measures and local efficiency were not significant: Sight Word Efficiency (r(59) = 0.230, p = 0.072), Phonemic Decoding (r(59) = 0.230, p = 0.072) and Passage Comprehension (r(59) = 0.245, p = 0.055) scores.
3.3. Connections between rich-club and reading networks
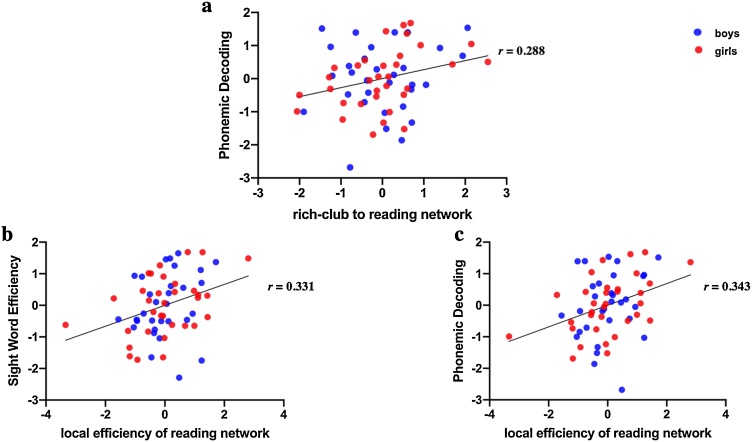
The connection strengths between the rich-club and reading network nodes were significantly correlated with TOWRE Phonemic Decoding score (r(58) = 0.288, corrected p = 0.026) (Fig. 5a), and marginally correlated with TOWRE Sight Word Efficiency and Passage Comprehension scores (Table 4).
Fig. 5.
Correlations between (a) standard Phonemic Decoding score and feeder connections from rich-club nodes to reading network, (b) mean local efficiency of the reading network and standard Sight Word Efficiency and (c) standard Phonemic Decoding scores. Values of standard Sight Word Efficiency and Phonemic Decoding scores, rich-club nodes to reading network, and mean local efficiency of reading network on the scatter plots are standard residuals after controlling for sex, mean whole-brain FA, handedness, and the proportion of overlapped rich-club nodes between rich-club and reading network.
Table 4.
Correlations between average number of streamlines in connections between hubs and reading network.
| All participants |
Boys |
Girls |
||||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| Sight word efficiency | 0.252 | 0.052 | 0.056 | 0.774 | 0.413 | 0.023* |
| Phoneme decoding | 0.288 | 0.026* | 0.151 | 0.434 | 0.414 | 0.023* |
| Passage comprehension | 0.175 | 0.182 | 0.051 | 0.792 | 0.274 | 0.143 |
| Rapid Automatized Naming (RAN) | 0.095 | 0.472 | −0.017 | 0.930 | 0.192 | 0.310 |
Correlations survived correction by 10,000-permutation Monte-Carlo simulation.
Follow-up analyses examined this pattern in girls vs. boys. As above, connections between rich-club and reading network were significantly correlated with standard Sight Word Efficiency (r(29) = 0.413, p = 0.023) and Phonemic Decoding (r(29) = 0.414, p = 0.023) scores only in girls. No significant correlations were found in the boys group (Table 4).
3.4. Efficiency of reading network
Mean local efficiency of the reading network was significantly correlated with both Sight Word Efficiency (r(58) = 0.331, corrected p = 0.016) and Phonemic Decoding score (r(58) = 0.343, corrected p = 0.013) (Fig. 5b and c). The correlation between mean local efficiency of reading network and Sight Word Efficiency score did not survive correction (r(58) = 0.273, corrected p = 0.054).
After splitting participants into boys and girls, partial correlation, including sex, average whole-brain FA, and handedness as covariates, results showed that mean local efficiency of reading network was significantly correlated with standard Sight Word Efficiency (r(29) = 0.439, p = 0.015) and Phonemic Decoding scores (r(29) = 0.491, p = 0.006) only in girls group. No significant correlations were found in the boys group.
3.5. Validation
To examine whether the tractography parameters influenced the topological properties of the connectome and subsequent correlations with behavioural measures, two additional whole-brain tractography calculations were performed, in which we either reduced the turning angle threshold to 30 degrees, or increased the minimal fiber length from 25 to 50 mm. This allowed us to better assess whether our results are fundamentally linked to tunable parameters in our tractography approach. Rich-club, feeder and local connections, and hub-reading-network connections were all positively correlated between the original and the two additional tractography calculations (rs > 0.59, ps < 2.9e-7). This indicated that the variability of number of streamlines between participants scaled in a predictable way when tractography parameters were modified. Similarly, significant connectome-reading correlations were not influenced by these changes. For the 30-degree turning angle threshold, these correlations were very similar to those observed in the original connectome feeder-SWE, feeder-PDE (r = 0.26, p = 0.043), hub-to-reading-network-PDE (r = 0.39, p = 0.002). A similar pattern was found for the 50 mm minimal length threshold: feeder-PDE (r = 0.30, p = 0.017), hub-to-reading-network-PDE (r = 0.30, p = 0.019), although the feeder-SWE correlation also increased (r = 0.30, p = 0.018) compared to our original analysis (r = 0.20, p = 0.106). In addition, the previously-noted significant correlation between local efficiency of reading network and PDE was not replicated with either the 30-degree turning angle or 50 mm streamline constraint. The failure of verification in nodal efficiency may be due to the modified parameters generating fewer streamlines, leading to limitation on the variance that could be captured by the statistical models.
In addition, the present study applied logarithm transforms to verify the usage of number of streamlines as edge weights (Sotiropoulos and Zalesky, 2019). Statistical analyses on networks which were weighed as logarithm-transformed numbers of streamlines revealed similar results to the number-of-streamline weighted network. Specifically, the number of feeder connections in the logarithm-transformed network was correlated with both SWE (r = 0.30, p = .019) and PDE scores (r = 0.27, p = .038). Local efficiency of the reading network was correlated with PDE score (r = 0.27, p = .040) as well. The correlation of number of streamlines joining hub and reading network nodes and PDE scores was in the same direction as in our initial analyses, but did not reach significance (r = 0.25, p = .057).
Finally, it is possible that our findings with regard to white matter connectivity within the reading network were susceptible to the choice of ROIs. To test this possibility, the ROI list was revised based on those listed in a different study (McNorgan et al., 2011), which proposed a more restricted subset of regions. This re-analysis found very similar results, which replicated all the significant correlations identified in Table 4, indicating that our findings were not affected by minor distinctions in the literature concerning the exact specification of reading network nodes.
4. Discussion
This study examined the relationship between connectome-wide rich-club structure and reading performance. Here we considered component processes of reading along a spectrum of good and poor readers, given evidence that reading disability is a graded disorder with no clear cutoff point, and that observed neurocognitive differences between good and poor readers lie along a continuum (Fletcher, 2012; Pugh et al., 2014; Shaywitz et al., 1992). Accordingly, across a number of measures we observed significant associations between network structure and performance on reading sub-tasks. In particular, the number of feeder streamlines was significantly correlated with sight word reading and phonemic decoding scores, measuring familiar and nonword reading, respectively. Phonemic decoding ability was also positively correlated with the number of streamlines between hubs and reading network nodes. In addition, sight word efficiency and phonemic decoding were positively correlated with the local efficiency of reading network nodes as well. Finally, exploratory measures indicated sex differences on the correlation between connectivity of rich-club structure and reading performance, marked by appreciably stronger effects in girls.
4.1. Feeder connections and reading
Feeder connections in the white matter connectome are pathways linking hub and non-hub regions, and serve as bridges between anatomical cores and peripheral stations (van den Heuvel et al., 2012). While not all types of connections exhibited associations with reading scores, significant correlations between number of streamlines per feeder connection and reading scores (sight word efficiency and phonemic decoding) indicate that rich-club structure is reflected in children’s reading skill. Rich-club structure has been viewed as the skeleton of the connectome for supporting high-level cognitive functions (Crossley et al., 2013; van den Heuvel et al., 2012). This structure enables the brain to process information in sparsely distributed modules and integrate the processed information across different modules via long-distance connections between hubs (van den Heuvel and Sporns, 2011). This structure represents an optimization among the competing needs of maintaining a sparsely connected matrix of brain regions, and minimizing the path length of connections among distal regions. Likewise, reading involves coordinating widespread brain regions which are responsible for its many component sub-processes (Paulesu et al., 2014). Hubs serve as a pivot to ensure optimized organization for high efficiency of transmitting information among distributed brain regions. As there was little overlap between the rich-club hubs and the reading network, connections between the two could be categorized as feeders. This suggests that processing reading materials requires feeder connections, beyond links within the reading network, for integrating reading components across various modules.
Prior work has found differences in connectivity of individual white matter tracts in RD, especially with regards to pathways within the reading network (e.g., Su et al., 2018; Zhao et al., 2016; for review see Vandermosten et al., 2012b). Likewise, Bathelt et al. (2018) demonstrated correlations between global measures of the white matter connectome and educational achievement, including children with a wide range of reading and math abilities. Lou et al. (2019) also reported global structure of the connectome correlates of word reading skill, as well as decreased number of white matter streamlines within the reading network. Accordingly, the present study is the first to more directly examine how reading is specifically related to rich-club structure within the brain’s entire white matter connectome, and to tie these results specifically to different component processes of reading ability.
Well-developed reading abilities could be sustained by this hierarchical structure while receiving and processing reading material within the reading network, involving not only anatomical connections within the reading network but also hubs that project links to reading network. Decreased feeder connections could be barriers for integrating specific reading components (e.g. phonological or orthographic information) from one module to another. Feeder connections affect the efficiency of information transmission across the whole brain by increasing or decreasing shortest path length between two nodes (Ball et al., 2014). Altered topological properties might be due to decreased feeder connections in poor readers. More feeder connections reduce the number of steps travelling from one peripheral node to another, especially when the shortest path contains hubs, yielding higher global efficiency of the network. Therefore, results from the present study are in line with previous white matter connectome studies, indicating that disruption of rich-club structure coexists with the alteration of topological properties of white matter network in children with RD. Additionally, among all four reading tests, sight word efficiency and phonemic decoding were observed to be positively correlated with number of feeder connections. Processing real words or pseudowords involves both ventral lateral extrastriate and left inferior occipito-temporal area, and dorsal inferior parietal lobule and superior temporal gyrus, where typical readers exhibited higher activation in typical readers than RD (Pugh et al., 2000). Stronger feeder connection ensures rapid communication among those areas which belong to diverse structural modules. The nominal correlation between number of feeder streamlines and passage comprehension may be due to the fact that passage comprehension requires a range of processes in addition to single-word recognition (Biancarosa & Snow, 2004; Swanson and Trahan, 1996), and the variation in comprehension scores cannot be solely explained by the feeder connection metric (Meyler et al., 2007).
Interestingly, the RAN task was not correlated with feeder connections in spite of prior work indicating it is a significant predictor of reading ability (Bowers and Wolf, 1993). This difference may reflect the automaticity of processes for letters with fewer interactions among high-level cognitive functions (Norton and Wolf, 2012). Overall, the result highlights that feeder connections play a specific role in word-level recognition rather than more broadly-construed reading subskills.
4.2. Connections between hub and reading network nodes
The second finding of this study is the positive correlation between the number of hub-reading-network streamlines and phonemic decoding. The location of the hubs identified in the present study mostly overlapped with previous studies of rich club structure in adults (van den Heuvel et al., 2012; van den Heuvel and Sporns, 2011), including bilateral superior frontal lobe, precuneus, supplemental motor area, and thalamus. Children, even newborn babies, exhibit a similar hub distribution (Ball et al., 2014; Grayson et al., 2014). Interestingly, the left thalamus was the exclusive hub that overlapped with the reading network. Therefore, white matter pathways between hubs and reading network nodes could be mostly classified as feeder connections, suggesting that feeder connections between hubs and the reading network contributed to individual variability in phonemic decoding ability. Phonemic decoding measures the translation of written words into spoken words without meaning cues, via association between letters and phonemic representations (Byrne, 1998). The present study illustrated that phonemic decoding involves not only direct connections within the reading network, but also detouring pathways outside the reading network. One explanation is that brain regions for processing letters and sounds pertain to different modules, and more feeder connections facilitate communication across these modules (van den Heuvel and Sporns, 2013). Another explanation comes from the navigation model, which depicts a communication strategy for passing information from one node to target within in a large-scale network (Kleinberg, 2000). Optimizing the route from a seed node to the destination requires knowledge of global topology of the network, which is impractical for the network itself. Instead, navigating in the network follows the rule that information is transferred to the next node geometrically closer to the destination (In a social network, for instance, one person has no idea about the entire picture of the relationship network. Hence, if one intends to find a pathway to contact a stranger, they would contact an acquaintance (local information) who might have the closest relationship to the target individual). For efficient communication within a large-scale brain network, both the topology and geometry of the network are also utilized for travelling from a seed node to the desired destination (Seguin et al., 2018). Direct bundles of fibers for reading, such as superior longitudinal fasciculus and arcuate fasciculus, minimize the number of edges between two brain nodes topologically. However, as reading involves multiple brain regions and connections, travelling only along this route is not sufficient for supporting the corresponding process and also ignores geometry distance of the entire network. It is therefore suggested that readers with better reading skills, especially with respect to phonological processes, can better exploit connections outside the reading network, achieving an optimal combination between topology path length and geometry distance in the connectome.
Unlike phonemic decoding, sight word efficiency was not significantly correlated with number of hub-reading-network streamlines. This differed from the pattern between reading scores and feeder connections, and may related to the procedure of word recognition. Word-level reading in RD or poor readers can be similar to novel-word reading, due to difficulty recognizing target words and increased dependence on the grapheme-phoneme correspondence rules. Good readers, on the other hand, have mastered grapheme-phoneme conversion and can accordingly apply a more direct orthographic pathway to recognize a familiar word (Frith, 1985). This is related to the reduced involvement of between-module communication during word-level reading in good readers, which leads to a decrease in the magnitude of the positive correlation between hub-reading-work connections and sight word reading.
4.3. Topological properties
Sight Word Efficiency and Phonemic decoding scores were also positively correlated with the mean local efficiency of all nodes in the reading network. This is generally concordant with prior studies examining the relationship between word reading performance and whole-brain topological properties. For example, Lou et al. (2019), also used deterministic tractography, applied a constrained spherical deconvolution (CSD) algorithm with turning angle threshold of 35 degrees and minimal fiber length of 25 mm (Lou et al., 2019). Although the turning angle threshold was lower than the present study, that earlier study also reported similar correlations between network efficiency and word-level reading scores. Decreased whole-brain local efficiency of other modalities of the connectome was also reported in Chinese native speakers with RD (Liu et al., 2015; Liu et al., 2016). Higher efficiency indicates better transfer of information between nodes (Bullmore and Sporns, 2009; Latora and Marchiori, 2001). Better physical organization across the whole brain accordingly supports higher ability of text reading, which is partly aligned with findings of the present study. Previous studies calculated the efficiency across the whole brain, which mixed information from irrelevant nodes and edges. The present study found contributions from topological properties of the connectome in more precise areas, which was aligned with prior study illustrating fewer number of streamlines in a subnetwork that largely overlapped with reading network in children with RD (Lou et al., 2019). To sum up, the efficiency of the reading network is associated with word-level reading abilities, including sight word reading and phonemic decoding skills, and more general reading activity relies on the efficiency of broader brain area which includes hubs.
4.4. Sex differences
Even though there was no priori hypothesis of sex differences in the relationship between the connectome and reading skills in this study, exploratory analyses suggested significant correlations persisted when limited to girls but were not significant when restricted to boys. We did find an effect of sex on one of our reading measures (the PDE subtest of TOWRE, which measures nonword decoding). However, this finding alone cannot explain our results since all subsequent analyses of sex differences were conducted within-group. We also assessed whether differences in variance could explain girl-specific significance in connectome-reading correlations. Levene’s test showed that there were no sex differences in variance with regard to age, reading scores or connectome measures. This indicated that sex differences on word-reading-connectome correlations did not originate from issues or range restriction in measures of behavioral, demographic or connectomic measures in either girls or boys. We caution however that examining such effects necessarily reduced our statistical power and thus these results call for further replication in larger samples.
The possibility of sex differences in reading disability is not a new one of course. To explain higher prevalence of RD in males, Geschwind (1981) proposed that it reflects either distinct etiology in male and female individuals with dyslexia, or that severe reading disability is over-represented in male individuals. Geschwind and Galaburda (1985) also proposed a hormonal account, arguing that female sex-linked hormones increase the resilience to disruptions of brain development. Therefore, female readers with RD may present more severe neural disruptions compared with male readers who show an equal degree of reading disabilities (Ramus, 2006), which may suggest that neuroanatomical differences between RD and controls, as well as brain-reading correlations, could be more easily observed in female readers. This is supported by evidence from previous studies, which reported deviations in brain morphology in female brains, but not male brains (Sandu et al., 2008; Altarelli et al., 2013; Su et al., 2018). The present results are consistent with this: observed connectome-reading correlations were only significant in girls. However, several studies reported sex effects in the opposite direction, where neural differences in RD were more severe in males (Altarelli et al., 2014; Clark et al., 2014; Evans et al., 2014). The inconsistent results on sex effects may due to the large amount of variability across those studies, such as age, sample size, and brain measures (Krafnick and Evans, 2019). Hence, our observed sex differences in connectome-reading correlations should be approached with caution.
At the macro-scale network level, one recent study of the functional connectome under a semantic fMRI task reported that functional brain networks for language exhibited differences in functional connection and topological properties between male and female participants (Xu et al., 2020). This effect may be rooted in the type of structural basis noted here. However, the available connectome model has limited power to explain such a structure-function relationship (Suárez et al., 2020). Further studies integrating the two modalities could be helpful to better grasp whether effects of sex on the relationship between the structural connectome and RD are robust, and if so, to more fully articulate their neurobiological origin.
Although the participants in this study exhibited a wide range of reading abilities, they were not divided into RD and typical readers for group analyses as previous white matter and connectome studies in RD. However, findings in the present study were in line with that from prior studies, which reported significant correlations between white matter connectivity and reading score when RD and typical readers were pooled into single group (Deutsch et al., 2005; Klingberg et al., 2000; Niogi and McCandliss, 2006; Rimrodt et al., 2010; Steinbrink et al., 2008; Vandermosten et al., 2012a). In addition, correlations between connectome-wise measures and reading scores were also reported in participants showing high variance in reading abilities (Lou et al., 2019; Bathelt et al., 2018). Overall the results suggest that such findings are not indicative of a qualitative difference between high- and low-achieving readers, and instead reflect a continuum in the neural correlates of reading across all skill levels.
One limitation of the present study is that we were not able to assess socioeconomic status (SES) of participants. It has been shown that SES is related to variance of reading achievements (Peterson and Pennington, 2015). This effect may also be reflected in neural differences, especially as it pertains to low versus middle-SES (Noble et al., 2005). The present study did not directly assess SES in children, and therefore it is not possible to evaluate its effect in this sample. With that in mind, other studies of white matter and reading have demonstrated that the inferior longitudinal fasciculus, a ventral reading-related white matter pathway, was related to SES in children with low-SES (Ozernov‐Palchik et al., 2019). Although a similar correlation was not observed in other reading-related white matter pathways, it might induce alterations at whole-brain connectome level. For example, Romeo et al. (2018) reported that the cortical thickness of bilateral perisylvian and supramarginal regions were related to SES. As the cortical thickness is associated with white matter pathways under corresponding cortex, SES may also contribute to white matter connectome development. Future connectome-wide studies of reading and SES are therefore warranted.
5. Conclusions
This study investigated the relationship between rich-club structure of the white matter network and reading skills in children with RD. Feeder connections were positively correlated with both the sight word efficiency and phonemic decoding scores. Among all feeder connections, the number of streamlines in those linking hubs and reading network nodes was also positively correlated with phonemic decoding score. Sight word efficiency and phonemic decoding was also related to mean local efficiency of the reading network. In summary, the hierarchical rich-club structure exhibited associations with reading skills, suggesting that white matter fibers outside the reading network also contributed to reading performance and broadening the view of how whole-brain connectivity contributes to reading success.
Data statement
Our ethics approval does not permit sharing raw data in a public repository. However, our data are available to researchers upon reasonable request, subject to approval from our local research ethics authority.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The work was supported by a Western Graduate Research Scholarship to CL, an NSERC Discovery Grant to MFJ, a Canada First Research Excellence Fund to BrainsCAN, funding from the Klaus J. Jabobs Foundation to DA. LP was supported by a Brain and Mind Institute Post-doctoral fellowship as well as a Children's Health Research Institute (CHRI) Trainee Award.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.dcn.2021.100957.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Altarelli I., Monzalvo K., Iannuzzi S., Fluss J., Billard C., Ramus F. A functionally guided approach to the morphometry of occipitotemporal regions in developmental dyslexia: evidence for differential effects in boys and girls. J. Neurosci. 2013;33:11296–11301. doi: 10.1523/JNEUROSCI.5854-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarelli I., Leroy F., Monzalvo K., Fluss J., Billard C., Dehaene-Lambertz G. Planum temporale asymmetry in developmental dyslexia: revisiting an old question. Hum. Brain Mapp. 2014;35:5717–5735. doi: 10.1002/hbm.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnell K.M., Joanisse M.F., Klein R.M., Busseri M.A., Tannock R. Decomposing the relation between Rapid Automatized Naming (RAN) and reading ability. Can. J. Exp. Psychol. 2009;63(3):173. doi: 10.1037/a0015721. [DOI] [PubMed] [Google Scholar]
- Bailey S.K., Aboud K.S., Nguyen T.Q., Cutting L.E. Applying a network framework to the neurobiology of reading and dyslexia. J. Neurodev. Disord. 2018;10(1):37. doi: 10.1186/s11689-018-9251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G., Aljabar P., Zebari S., Tusor N., Arichi T., Merchant N. Rich-club organization of the newborn human brain. Proc. Natl. Acad. Sci. U. S. A. 2014;111(20):7456. doi: 10.1073/pnas.1324118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P.J. Inferring microstructural features and the physiological state of tissues from diffusion-weighted images. NMR Biomed. 1995;8:333–344. doi: 10.1002/nbm.1940080707. [DOI] [PubMed] [Google Scholar]
- Basser P.J., Mattiello J., Lebihan D. MR diffusion tensor spectroscopy and imaging. Biol. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathelt J., Gathercole S.E., Butterfield S., team, t. C, Astle D.E. Children’s academic attainment is linked to the global organization of the white matter connectome. Dev. Sci. 2018;21(5):e12662. doi: 10.1111/desc.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancarosa G., Snow C.E. Alliance for Excellent Education; New York: 2004. Reading Next: a Vision for Action and Research in Middle and High School Literacy: a Report from Carnegie Corporation of. [Google Scholar]
- Boets B., Vandermosten M., Poelmans H., Luts H., Wouters J., Ghesquière P. Preschool impairments in auditory processing and speech perception uniquely predict future reading problems. Res. Dev. Disabil. 2011;32(2):560. doi: 10.1016/j.ridd.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Boets B., Beeck H.Od., Vandermosten M., Scott S.K., Gillebert C.R., Mantini D. Intact but less accessible phonetic representations in adults with dyslexia. Science. 2013;342(6163):1251–1254. doi: 10.1126/science.1244333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers P.G., Wolf M. Theoretical links among naming speed, precise timing mechanisms and orthographic skill in dyslexia. Read. Writ. 1993;5:69–85. doi: 10.1007/BF01026919. [DOI] [Google Scholar]
- Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structrual and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Byrne B.J. Psychology Press; 1998. The Foundation of Literacy: the Child’s Acquisition of the Alphabetic Principle. [Google Scholar]
- Carter J.C., Lanham D.C., Cutting L.E., Clements-Stephens A.M., Chen X.J., Hadzipasic M., Kim J., Denckla M.B., Kaufmann W.E. A dual DTI approach to analyzing white matter in children with dyslexia. Psychiatr. Res.-Neuroimaging. 2009;172:215–219. doi: 10.1016/j.pscychresns.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K., Helland T., Specht K., Narr K., Manis F., Toga A., Hugdahl K. Neuroanatomical precursors of dyslexia identified from pre-reading through to age 11. Brain. 2014;137(12):3136–3141. doi: 10.1093/brain/awu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Vértes P.E., Winton-Brown T.T., Patel A.X., Ginestet C.E. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc. Natl. Acad. Sci. U. S. A. 2013;110(28):11583–11588. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Scott J., Carletti F., Fox P.T., McGuire P., Bullmore E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137(8):2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C., Messé A., Dell L., Foubet O., Heuer K., Larrat B., Meriaux S., Mangin J., Reillo I., de Juan Romero C., Borrell V., Toro V., Hilgetag C.C. Comparison between diffusion MRI tractography and histological tract-tracing of cortico-cortical structural connectivity in the ferret brain. Netw. Neurosci. 2019;3(4):1038–1050. doi: 10.1162/netn_a_00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch G.K., Dougherty R.F., Bammer R., Siok W.T., Gabrieli J.D.E., Wandell B. Children’s reading performance is correlated with white matter structure measured by diffusion tensor imaging. Cortex. 2005;41:354–363. doi: 10.1016/s0010-9452(08)70272-7. [DOI] [PubMed] [Google Scholar]
- Díaz B., Hintz F., Kiebel S.J., von Kriegstein K. Dysfunction of the auditory thalamus in developmental dyslexia. Proc. Natl. Acad. Sci. U. S. A. 2012;109(34):13841–13846. doi: 10.1073/pnas.1119828109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T.M., Flowers D.L., Napoliello E.M., Eden G.F. Sex-specific gray matter volume differences in females with developmental dyslexia. Brain Struct. Funct. 2014;219:1041–1054. doi: 10.1007/s00429-013-0552-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn E.S., Shen X., Holahan J.M., Scheinost D., Lacadie C., Papademetris X. Disruption of functional networks in dyslexia: a whole-brain, data-driven analysis of connectivity. Biol. Psychiatry. 2014;76(5):397–404. doi: 10.1016/j.biopsych.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery K.A., Liederman J., Daly L., Schultz J. Male prevalence for reading disability is found in a large sample of black and white children free from ascertainment bias. J. Int. Neuropsychol. Soc. 2000;6:433–442. doi: 10.1017/S1355617700644016. [DOI] [PubMed] [Google Scholar]
- Fletcher J.M. vol. 4. 2012. Classification and identification of learning disabilities. (Learning about Learning Disabilities). [Google Scholar]
- Frith U. Beneath the surface of developmental dyslexia. In: Patterson K.E., Marschall J.C., Coltheart M., editors. Surface Dyslexia: Cognitive and Neuropsychological Studies of Phonological Reading. Erlbaum; Hillsdale, NJ: 1985. pp. 301–330. [Google Scholar]
- Geschwind N. A reaction of conference. In: Ansara A., Geschwind N., Galaburda A., Albert M., Gartrell N., editors. Sex Differences in Dyslexia. Orton Dyslexia Society; Baltimore: 1981. pp. 13–18. [Google Scholar]
- Geschwind N., Galaburda A.M. Cerebral lateralization. Biological mechanismsassociations, and pathology: I. A hypothesis and a program for research. Arch. Neurol. 1985;42:428–459. doi: 10.1001/archneur.1985.04060050026008. [DOI] [PubMed] [Google Scholar]
- Gong G., He Y., Concha L., Lebel C., Gross D.W., Evans A.C., Beaulieu C. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex. 2008;19(3):524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D.S., Ray S., Carpenter S., Iyer S., Dias T.G.C., Stevens C. Structural and functional rich club organization of the brain in children and adults. PLoS One. 2014;9(2):e88297. doi: 10.1371/journal.pone.0088297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffa A., Baumann P.S., Ferrari C., Do K.Q., Conus P., Thiran J.P., Hagmann P. Characterizing the connectome in schizophrenia with diffusion spectrum imaging. Hum. Brain Mapp. 2015;36(1):354–366. doi: 10.1002/hbm.22633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P., Cammoun L., Gigandet X., Meuli R., Honey C.J., Wedeen V.J., Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.J., De Wael R.V., Bethlehem R.A., Lariviere S., Paquola C., Valk S.L. Atypical functional connectome hierarchy in autism. Nat. Commun. 2019;10(1):1–13. doi: 10.1038/s41467-019-08944-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E., Woolrich M.W., Smith S.M. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jobard G., Crivello F., Tzourio-Mazoyer N. Evaluation of the dual route theory of reading: a metanalysis of 35 neuroimaging studies. Neuroimage. 2003;20(2):693–712. doi: 10.1016/S1053-8119(03)00343-4. [DOI] [PubMed] [Google Scholar]
- Katusic S.K., Colligan R.C., Barbaresi W.J., Schaid D.J., Jacobsen S.J. Incidence of reading disability in a population-based birth cohort, 1976-1982, Rochester. Minn. Mayo Clin. Proc. Mayo Clin. 2001;76:1081–1092. doi: 10.4065/76.11.1081. [DOI] [PubMed] [Google Scholar]
- Kleinberg J.M. Navigation in a small world. Nature. 2000;406(6798):845. doi: 10.1038/35022643. [DOI] [PubMed] [Google Scholar]
- Klingberg T., Hedehus M., Temple E., Salz T., Gabriell J.D.E., Moseley M.E., Poldrack R.A. Microstructure of temporo-parietal white matter as a basis for reading ability: Evidence from diffusion tensor magnetic resonance imaging. Neuron. 2000;25:493–500. doi: 10.1016/s0896-6273(00)80911-3. [DOI] [PubMed] [Google Scholar]
- Krafnick A.J., Evans T.M. Neurobiological sex differences in developmental dyslexia. Front. Psychol. 2019;9:2669. doi: 10.3389/fpsyg.2018.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V., Marchiori M. Efficient behavior of small-world networks. Phys. Rev. Lett. 2001;87:1–4. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Leemans A., Jeurissen B., Sijbers J., Jones D.K. ExploreDTI: a graphical toolbox for processing, analyzing, and visualizing diffusion MR data. Proceedings of the 17th Scientific Meeting, International Society for Magnetic Resonance in Medicine; Honolulu, USA; 2009. 3537. [Google Scholar]
- Liederman J., Kantrowitz L., Flannery K. Male vulnerability to reading disability is not likely to be a myth a call for new data. J. Learn. Disabil. 2005;38:109–129. doi: 10.1177/00222194050380020201. [DOI] [PubMed] [Google Scholar]
- Liu K., Shi L., Chen F., Waye M.M.Y., Lim C.K.P., Cheng P.-w. Altered topological organization of brain structural network in Chinese children with developmental dyslexia. Neurosci. Lett. 2015;589:169–175. doi: 10.1016/j.neulet.2015.01.037. [DOI] [PubMed] [Google Scholar]
- Liu L., Li H., Zhang M., Wang Z., Wei N., Liu L. Aberrant topologies and reconfiguration pattern of functional brain network in children with second language reading impairment. Dev. Sci. 2016;19(4):657–672. doi: 10.1111/desc.12440. [DOI] [PubMed] [Google Scholar]
- Lou C., Duan X., Altarelli I., Sweeney J.A., Ramus F., Zhao J. White matter network connectivity deficits in developmental dyslexia. Hum. Brain Mapp. 2019;40(2):505–516. doi: 10.1002/hbm.24390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon G.R., Shaywitz S.E., Shaywitz B.A. A definition of dyslexia. Ann. Dyslexia. 2003;53:1–14. [Google Scholar]
- Maisog José M. A meta‐analysis of functional neuroimaging studies of dyslexia. Ann. N. Y. Acad. Sci. 2008;1145(1):237–259. doi: 10.1196/annals.1416.024. (2008) [DOI] [PubMed] [Google Scholar]
- Martin A., Schurz M., Kronbichler M., Richlan F. Reading in the brain of children and adults: a meta‐analysis of 40 functional magnetic resonance imaging studies. Hum. Brain Mapp. 2015;36(5):1963–1981. doi: 10.1002/hbm.22749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew K.S., Woodcock R.W., Schrank K.A. Riverside Pub.; 2007. Woodcock-Johnson III Normative Update Technical Manual. [Google Scholar]
- McNorgan C., Alvarez A., Bhullar A., Gayda J., Booth J. Prediction of reading skill several years later depends on age and brain region: implications for developmental models of reading. J. Neurosci. 2011;31(26):9641–9648. doi: 10.1523/JNEUROSCI.0334-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaglia J.D., Lynall M.-E., Bassett D.S. Cognitive network neuroscience. J. Cogn. Neurosci. 2015;27(8):1471–1491. doi: 10.1162/jocn_a_00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyler A., Keller T.A., Cherkassky V.L., Lee D., Hoeft F., Whitfield-Gabrieli S. Brain activation during sentence comprehension among good and poor readers. Cereb. Cortex. 2007;17(12):2780–2787. doi: 10.1093/cercor/bhm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Axt C., Anwander A., von Kriegstein K. Altered structural connectivity of the left visual thalamus in developmental dyslexia. Curr. Biol. 2017;27(23):3692–3698. doi: 10.1016/j.cub.2017.10.034. e3694. [DOI] [PubMed] [Google Scholar]
- Niogi S.N., Mccandliss B.D. Left lateralized white matter microstructure accounts for individual differences in reading ability and disability. Neuropsychologia. 2006;44(11):2178–2188. doi: 10.1016/j.neuropsychologia.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Noble K.G., Norman M.F., Farah M.J. Neurocognitive correlates of socioeconomic status in kindergarten children. Dev. Sci. 2005;8(1):74–87. doi: 10.1111/j.1467-7687.2005.00394.x. [DOI] [PubMed] [Google Scholar]
- Norton E.S., Wolf M. Rapid automatized naming (RAN) and reading fluency: implications for understanding and treatment of reading disabilities. Annu. Rev. Psychol. 2012;63(1):427–452. doi: 10.1146/annurev-psych-120710-100431. [DOI] [PubMed] [Google Scholar]
- Odegard T.N., Farris E.A., Ring J., McColl R., Black J. Brain connectivity in nonreading impaired children and children diagnosed with developmental dyslexia. Neuropsychologia. 2009;47:1972–1977. doi: 10.1016/j.neuropsychologia.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Opsahl T., Colizza V., Panzarasa P., Ramasco J.J. Prominence and control: the weighted rich-club effect. Phys. Rev. Lett. 2008;101(16):168702. doi: 10.1103/PhysRevLett.101.168702. [DOI] [PubMed] [Google Scholar]
- Ozernov‐Palchik O., Norton E.S., Wang Y., Beach S.D., Zuk J., Wolf M., Gabrieli J.D., Gaab N. The relationship between socioeconomic status and white matter microstructure in pre‐reading children: a longitudinal investigation. Hum. Brain Mapp. 2019;40(3):741–754. doi: 10.1002/hbm.24407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulesu E., Danelli L., Berlingeri M. Reading the dyslexic brain: multiple dysfunctional routes revealed by a new meta-analysis of PET and fMRI activation studies. Front. Hum. Neurosci. 2014;8:1–20. doi: 10.3389/fnhum.2014.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Alonso P.M., Oliver M., Lerma-Usabiaga G., Caballero-Gaudes C., Quiñones I., Suárez-Coalla P. Neural correlates of phonological, orthographic and semantic reading processing in dyslexia. Neuroimage Clin. 2018;20:433–447. doi: 10.1016/j.nicl.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen Steven E., Sporns O. Brain networks and cognitive architectures. Neuron. 2015;88(1):207–219. doi: 10.1016/j.neuron.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R., Pennington B. Developmental dyslexia. Annu. Rev. Clin. Psychol. 2015;11:283–307. doi: 10.1146/annurev-clinpsy-032814-112842. [DOI] [PubMed] [Google Scholar]
- Price C.J., Devlin J.T. The interactive account of ventral occipitotemporal contributions to reading. Trends Cogn. Sci. 2011;15(6):246–253. doi: 10.1016/j.tics.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh K.R., Mencl W.E., Jenner A.R., Katz L., Frost S.J., Lee J.R. 2000. Functional Neuroimaging Studies of Reading and Reading Disabilities (Developmental Dyslexia) [DOI] [PubMed] [Google Scholar]
- Pugh K.R., Frost S.J., Rothman D.L., Hoeft F., Del Tufo S.N., Mason G.F. Glutamate and choline levels predict individual differences in reading ability in emergent readers. J. Neurosci. 2014;34(11):4082–4089. doi: 10.1523/jneurosci.3907-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Gu B., Ding G., Gong G., Lu C., Peng D. More bilateral, more anterior: alterations of brain organization in the large-scale structural network in Chinese dyslexia. Neuroimage. 2016;124(Pt A):63. doi: 10.1016/j.neuroimage.2015.09.011. [DOI] [PubMed] [Google Scholar]
- Ramus F. Developmental dyslexia: specific phonological deficit or general sensorimotor dysfunction? Curr. Opin. Neurobiol. 2003;13(2):212–218. doi: 10.1016/s0959-4388(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Ramus F. Genes, brain, and cognition: a roadmap for the cognitive scientist. Cognition. 2006;101(2):247–269. doi: 10.1016/j.cognition.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ramus F., Altarelli I., Jednorog K., Zhao J., Scotto di Covella L. Neuroanatomy of developmental dyslexia: pitfalls and promise. Neurosci. Biobehav. Rev. 2018;84:434–452. doi: 10.1016/j.neubiorev.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Ray S., Miller M., Karalunas S., Robertson C., Grayson D.S., Cary R.P., Hawkey E., Painter J.G., Kriz D., Fombonne E., Nigg J.T., Fair D.A. Structural and functional connectivity of the human brain in autism spectrum disorders and attention‐deficit/hyperactivity disorder: a rich club‐organization study. Hum. Brain Mapp. 2014;35:6032–6048. doi: 10.1002/hbm.22603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T., Stevenson J., Crouch J., Johnson L., C, Maravilla K., Stock P. Tract-based spatial statistics of diffusion tensor imaging in adults with dyslexia. Am. J. Neuroradiol. 2008;29(6):1134–1139. doi: 10.3174/ajnr.A1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimrodt S.L., Peterson D.J., Denckla M.B., Kaufmann W.E., Cutting L.E. White matter microstructural differences linked to left perisylvian language network in children with dyslexia. Cortex. 2010;46(6):739–749. doi: 10.1016/j.cortex.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G., Perry A., Lord A., Frankland A., Leung V., Holmes-Preston E. Structural dysconnectivity of key cognitive and emotional hubs in young people at high genetic risk for bipolar disorder. Mol. Psychiatry. 2018;23(2):413–421. doi: 10.1038/mp.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo R.R., Christodoulou J.A., Halverson K.K., Murtagh J., Cyr A.B., Schimmel C. Socioeconomic status and reading disability: neuroanatomy and plasticity in response to intervention. Cereb. Cortex. 2018;28(7):2297–2312. doi: 10.1093/cercor/bhx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M., Caspi A., Fergusson D., Horwood L.J., Goodman R., Maughan B. Sex differences in developmental reading disability: new findings from 4 epidemiological studies. JAMA. 2004;291:2007–2012. doi: 10.1001/jama.291.16.2007. [DOI] [PubMed] [Google Scholar]
- Sandu A.-L., Specht K., Beneventi H., Lundervold A., Hugdahl K. Sex-differences in grey-white matter structure in normal-reading and dyslexic adolescents. Neurosci. Lett. 2008;438:80–84. doi: 10.1016/j.neulet.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Schlaggar B.L., Mccandliss B.D. Development of neural systems for reading. Annu. Rev. Neurosci. 2007;30(1):475. doi: 10.1146/annurev.neuro.28.061604.135645. [DOI] [PubMed] [Google Scholar]
- Seguin C., van den Heuvel M.P., Zalesky A. Navigation of brain networks. Proc. Natl. Acad. Sci. U. S. A. 2018;115(24):6297–6302. doi: 10.1073/pnas.1801351115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz B.A., Fletcher J.M., Holahan J.M., Shaywitz S.E. Discrepancy compared to low achievement definitions of reading disability:results from the connecticut longitudinal study. J. Learn. Disabil. 1992;25(10):639–648. doi: 10.1177/002221949202501003. [DOI] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowling M.J. 2nd ed. 2000. Dyslexia. [Google Scholar]
- Sotiropoulos S.N., Zalesky A. Building connectomes using diffusion MRI: why, how and but. NMR Biomed. 2019;32:e3752. doi: 10.1002/nbm.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O., Tononi G., Kötter R. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 2005;1(4):e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O., Honey C.J., Kötter R. Identification and classification of hubs in brain networks. PLoS One. 2007;2(10):e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrink C., Vogt K., Kastrup A., Muller H.-P., Juengling F.D., Kassubek J., Riecker A. The contribution of white and gray matter differences to developmental dyslexia: insight from DTI and VBM at 3.0 T. Neuropsychologia. 2008;46:3170–3178. doi: 10.1016/j.neuropsychologia.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Su M., Zhao J., Thiebaut de Schotten M., Zhou W., Gong G., Ramus F., Shu H. Alterations in white matter pathways underlying phonological and morphological processing in Chinese developmental dyslexia. Dev. Cogn. Neurosci. 2018;31:11–19. doi: 10.1016/j.dcn.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez L.E., Markello R.D., Betzel R.F., Misic B. Linking structure and function in macroscale brain networks. Trends Cogn. Sci. 2020;24(4):302–315. doi: 10.1016/j.tics.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Swanson H.L., Trahan M. Learning disabled and average readers’ working memory and comprehension: does metacognition play a role? Br. J. Educ. Psychol. 1996;66(3):333–355. doi: 10.1111/j.2044-8279.1996.tb01201.x. [DOI] [PubMed] [Google Scholar]
- Torgesen J.K., Wagner R., Rashotte C. 2012. Test of Word Reading Efficiency: (TOWRE-2): Pearson Clinical Assessment. [Google Scholar]
- Tschentscher N., Ruisinger A., Blank H., Díaz B., von Kriegstein K. Reduced structural connectivity between left auditory thalamus and the motion-sensitive planum temporale in developmental dyslexia. J. Neurosci. 2019;39(9):1720–1732. doi: 10.1523/jneurosci.1435-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Etard F.C.O., Delcroix N., Mazoyer B., Joliot M. Antomated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O. Rich-club organization of the human connectome. J. Neurosci. 2011;31(44):15775–15786. doi: 10.1523/jneurosci.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O. Network hubs in the human brain. Trends Cogn. Sci. 2013;17(12):683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O. A cross-disorder connectome landscape of brain dysconnectivity. Nat. Rev. Neurosci. 2019;20(7):435–446. doi: 10.1038/s41583-019-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M.P., Kahn R.S., Goñi J., Sporns O. High-cost, high-capacity backbone for global brain communication. Proc. Natl. Acad. Sci. U. S. A. 2012;109(28):11372–11377. doi: 10.1073/pnas.1203593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M.P., Sporns O., Collin G., Scheewe T., Mandl R.C., Cahn W. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry. 2013;70(8):783–792. doi: 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., de Reus M.A., Feldman Barrett L., Scholtens L.H., Coopmans F.M., Schmidt R., Preuss T.M., Rilling J.K., Li L. Comparison of diffusion tractography and tract‐tracing measures of connectivity strength in rhesus macaque connectome. Hum. Brain Mapp. 2015;36:3064–3075. doi: 10.1002/hbm.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermosten M., Boets B., Poelmans H., Sunaert S., Wouters J., Ghesquière P. A tractography study in dyslexia: neuroanatomic correlates of orthographic, phonological and speech processing. Brain. 2012;135:935–948. doi: 10.1093/brain/awr363. [DOI] [PubMed] [Google Scholar]
- Vandermosten M., Boets B., Wouters J., Ghesquière P. A qualitative and quantitative review of diffusion tensor imaging studies in reading and dyslexia. Neurosci. Biobehav. Rev. 2012;36:1532–1552. doi: 10.1016/j.neubiorev.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Watts D.J., Strogatz S.H. Collective dynamics of’ small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Woodcock R.W., McGrew K.S., Mather N. 2001. Woodcock-Johnson III Tests of Achievement. [Google Scholar]
- Xu M., Liang X., Ou J., Li H., Luo Y.-j., Tan L.H. Sex differences in functional brain networks for language. Cereb. Cortex. 2020;30(3):1528–1537. doi: 10.1093/cercor/bhz184. [DOI] [PubMed] [Google Scholar]
- Zhao J., Schotten M.Td., Altarelli I., Dubois J., Ramus F. Altered hemisphperic lateralization of white matter pathways in developmental dyslexia: evidence from spherical deconvolution tractography. Cortex. 2016;76:51–62. doi: 10.1016/j.cortex.2015.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.